Abstract
Background
Recent studies have revealed sexually dimorphic associations between the carbamoyl‐phosphate synthase 1 locus, intermediates of the metabolic pathway leading from choline to urea, and risk of coronary artery disease (CAD) in women. Based on evidence from the literature, the atheroprotective association with carbamoyl‐phosphate synthase 1 could be mediated by the strong genetic effect of this locus on increased circulating glycine levels.
Methods and Results
We sought to identify additional genetic determinants of circulating glycine levels by carrying out a meta‐analysis of genome‐wide association study data in up to 30 118 subjects of European ancestry. Mendelian randomization and other analytical approaches were used to determine whether glycine‐associated variants were associated with CAD and traditional risk factors. Twelve loci were significantly associated with circulating glycine levels, 7 of which were not previously known to be involved in glycine metabolism (ACADM,PHGDH,COX18‐ADAMTS3,PSPH,TRIB1,PTPRD, and ABO). Glycine‐raising alleles at several loci individually exhibited directionally consistent associations with decreased risk of CAD. However, these effects could not be attributed directly to glycine because of associations with other CAD‐related traits. By comparison, genetic models that only included the 2 variants directly involved in glycine degradation and for which there were no other pleiotropic associations were not associated with risk of CAD or blood pressure, lipid levels, and obesity‐related traits.
Conclusions
These results provide additional insight into the genetic architecture of glycine metabolism, but do not yield conclusive evidence for a causal relationship between circulating levels of this amino acid and risk of CAD in humans.
Keywords: causality, coronary artery disease, genome‐wide association study, glycine, Mendelian randomization, meta‐analysis
Subject Categories: Genetic, Association Studies; Coronary Artery Disease; Biomarkers; Metabolism; Meta Analysis
Clinical Perspective
What Is New?
The study identifies 12 genetic determinants of circulating glycine levels, 7 of which are novel and not previously known to be involved in the metabolism of this amino acid.
Biological mechanisms for half of the loci associated with circulating glycine levels are not directly evident.
What Are the Clinical Implications?
Although findings from this study provide additional insight into the genetic architecture of glycine metabolism, they do not yield conclusive evidence for a causal relationship between circulating levels of this amino acid and coronary artery disease in humans.
Introduction
Metabolites derived from gut microbiome and hepatic‐mediated metabolism of dietary choline and l‐carnitine, such as trimethylamine N‐oxide and betaine, have recently been shown to be proatherogenic in mice and novel biomarkers of coronary artery disease (CAD) risk in humans.1, 2, 3 In searching for genetic determinants of these metabolites, we identified sexually dimorphic associations between the carbamoyl‐phosphate synthase 1 (CPS1) locus and not only plasma trimethylamine N‐oxide and betaine levels, but also other intermediates in the metabolic pathway leading from choline to urea.4 We and others further noted that, of the various other biomarkers/metabolites that had previously been linked to CPS1,5, 6, 7, 8, 9, 10, 11, 12, 13, 14 the strongest effect size and most significant association was with circulating glycine levels in women.4, 15, 16, 17 Most important, the lead CPS1 variant also exhibited a strikingly significant female‐specific association with decreased risk of CAD.4 However, the direction of the associations between CPS1 and the various biomarkers and metabolites was opposite to what would be expected for a variant that decreased risk of CAD.
One explanation for the protective association of CPS1 with CAD could be the strong genetic effect of this locus on increased circulating glycine levels.4 For example, previous in vitro and in vivo studies have shown that glycine reduces inflammation and oxidative stress in endothelial cells, activated macrophages, and other leukocytes.18, 19, 20, 21, 22 Furthermore, platelet aggregation of both human and rodent platelets can be prevented by glycine in a dose‐dependent manner through mechanisms involving the glycine receptor.23 Interestingly, the same glycine‐raising CPS1 variant has been associated with reduced platelet counts.24 Alternatively, glycine has been reported to have antihypertensive effects in mice and humans.25 A recent epidemiological study also demonstrated an inverse relationship between plasma glycine levels and risk of an acute myocardial infarction.26 Taken together, these observations support the concept that glycine could have atheroprotective properties, but direct evidence for a causal relationship between this amino acid and risk of CAD is lacking.
In the present study, we used a meta‐analysis approach with genome‐wide association study (GWAS) data to identify additional genetics determinants of circulating glycine levels. The identified loci were then used to investigate the possible causal association between circulating glycine levels and risk of CAD and traditional risk factors. In total, 12 loci were identified for circulating glycine levels, 7 of which were novel and not previously known to be involved in glycine metabolism. However, various analytical approaches with glycine‐raising alleles at these loci did not provide conclusive evidence for a causal relationship between circulating glycine and risk of CAD in humans.
Methods
The statistical methods used in this study will be made available to other researchers for purposes of reproducing the results or replicating the analyses. The summary statistics of the meta‐analysis for circulating glycine levels will be made available through the NHGRI‐EBI Catalog of published GWASs (https://www.ebi.ac.uk/gwas/downloads/summary-statistics/).
Study Populations
The present analyses included 30 118 subjects of European ancestry from the GeneBank (GB),4 FINRISK 1997 and 2007 (FR97 and FR07),27 YFS (Cardiovascular Risk in Young Finns Study),28 NFBC1966 and NFBC1986 (Northern Finland Birth Cohort),29 and METSIM (Metabolic Syndrome in Men)30 studies. Details of subject recruitment and genotyping methodology for each cohort are provided in Data S1. For each cohort, written informed consent was obtained from all participants before being enrolled, and the studies were approved by the institutional review boards of the participating institutions. The present analysis was approved by the institutional review board of USC Keck School of Medicine.
Measurement of Circulating Glycine Levels
Glycine levels were quantified using stable isotope dilution high‐performance liquid chromatography with online electrospray ionization tandem mass spectrometry in the GB study4 and by quantitative high‐throughput NMR in the FR97, FR07, YFS NFBC66, NFBC86, and METSIM cohorts.31, 32
Data Harmonization and GWAS Analyses
Circulating glycine levels were first regressed on study‐specific covariates chosen by the investigators of each cohort. These included age and sex in GeneBank; age, sex, and time from last meal in FR97, FR07, YFS NFBC66, and NFBC8631; and age, age,2 and body mass index in METSIM.32 Inverse rank‐based normal transformations were carried out on the residuals after adjustment for covariates and used as the outcome in GWAS analyses by linear regression in each study.
Meta‐Analysis for Circulating Glycine Levels
We performed a fixed‐effects meta‐analysis for circulating glycine levels with 7 487 927 SNPs that were imputed using 1000 Genomes Project data and that were common to all data sets. This analysis was carried out assuming an additive model and after controlling for population structure within each study, as implemented in GWAMA (Genome‐Wide Association Meta‐Analysis) software.33 In addition to a combined meta‐analysis with all subjects, we also carried out a sex‐stratified fixed‐effects meta‐analysis. The genome‐wide threshold for significant association was set at P=5.0×10−8. A locus was defined as novel if the lead single‐nucleotide polymorphism (SNP) was in weak or no linkage disequilibrium (r 2≤0.1) with variants at genome‐wide significant loci previously reported for circulating glycine levels. Manhattan and quantile‐quantile plots were constructed using the “qqman” package in R (R Foundation for Statistical Computing, Vienna, Austria).34 To examine whether all novel loci identified in our meta‐analysis were also significantly associated with other traits (phenome‐wide association studies), we used publicly available databases, such as PhenoScanner,35 the UCSC Genome Browser (https://genome.ucsc.edu/), and the GWAS Catalog (https://www.ebi.ac.uk/gwas/home). The significance threshold for phenome‐wide association studies analyses was set to P=5.0×10−8 with a linkage disequilibrium cut off of r 2≥0.8 for proxy SNPs.
Proportion of Phenotypic Variance Explained
The proportion of variation in glycine levels explained by the identified variants was estimated using SumHer software.36 SNP heritability was calculated using a weighted linkage disequilibrium adjusted kinships model with the 12 glycine‐associated SNPs. 1000 Genomes Project–based imputed genotypes in ≈4500 subjects of European ancestry from the GB cohort were used as a reference panel for linkage disequilibrium (r 2) for these estimates.
Analysis of Variants With Risk of CAD and Traditional Risk Factors
Publicly available summary results from large‐scale GWAS in subjects of European ancestry37, 38, 39 were used to determine whether glycine‐associated variants were associated with risk of CAD and various lipid‐, metabolic‐, and blood‐pressure–related risk factors. Specifically, we tested associations using 3 analytical strategies with 4 genetic models that were based on various nested combinations of the 12 identified variants. Genetic model 1 included all 12 loci identified for glycine; model 2 was designed to specifically test only the 7 novel loci (ACADM, PHGDH, COX18‐ADAMTS3, PSPH, TRIB1, PTPRD, and ABO); and model 3 included only the 4 loci known to be related to glycine metabolism (PSPH, PHGDH, GLDC, and GCSH). Model 4 was the most restrictive and included only the 2 glycine‐associated loci that are known to be directly involved in the catabolism of glycine through the glycine cleavage system (GLDC and GCSH) and that did not exhibit pleiotropic effects with other traits or metabolites. In the first analytical approach, the average/overall association of CAD and its risk factors with glycine‐raising alleles in the 4 genetic models were evaluated by meta‐analysis, as implemented in the “meta” R package (https://cran.r-project.org/web/packages/meta/index.html). In the second approach, we generated genetic risk scores (GRS) with the identified variants for the same 4 genetic models to evaluate the cumulative joint effects of glycine‐raising alleles. Additive multi‐SNP GRS associations were estimated using the grs.summary function of the “gtx: Genetic ToolboX” R package (https://cran.r-project.org/web/packages/gtx). This approach approximates the regression of an intermediate trait or biomarker onto a GRS, which is based on the weighted sum of the single SNP coefficients derived from the association summary statistics.40 For the third strategy, we carried out weighted median and inverse variance weighted Mendelian randomization (MR) analyses with the 4 genetic models, as implemented in the “TwoSampleMR” R package.41 Because the weighted median MR method requires 3 or more variants, only the inverse variance weighted MR test was used for determining association of the 2 SNPs in model 4 with CAD and traditional risk factors.
Results
GWAS for Circulating Glycine Levels
To identify novel loci for circulating glycine levels, we carried out a meta‐analysis of GWAS summary‐level data with 7 487 927 genotyped and imputed SNPs in 30 118 subjects of European ancestry. Table 1 shows the characteristics of the study cohorts and data sets used for these analyses. A GWAS was carried out for circulating glycine levels in each cohort, followed by a fixed‐effects meta‐analysis. The genomic control factor (lambda, λ) in GB I (0.995), GB II (0.989), and the combination of the FR97, FR07, YFS NFBC66, and NFBC86 cohorts (1.039), and METSIM (1.014) were small or modest, thus decreasing the likelihood of identifying spurious associations attributed to population stratification (Figure S1). To further account for this potential confounder, we also applied genomic control to each study before the meta‐analysis. In total, 4934 variants distributed across 12 loci were associated with circulating glycine levels at the genome‐wide significance threshold (P=5.0×10−8; Figure 1, Table 2, and Table S1). Seven of these loci (ACADM, PHGDH, COX18‐ADAMTS3, PSPH, TRIB1, PTPRD, and ABO) were novel and identified as being associated with circulating glycine levels for the first time herein (Figure 1, Table 2, and Figure S2). The other 5 loci (CPS1, ALDH1L1, PPP1R3B‐LOC157273, GLDC, and GCSH) have previously been reported for circulating glycine levels, but the association signals became more significant in our meta‐analysis because of increased sample size (Figure 1, Table 2, and Figure S2). Overall, the 12 identified loci explained ≈15% of the variation in circulating glycine levels.
Table 1.
Description of Cohorts Used in Meta‐Analysis for Circulating Glycine Levels
| Cohort | No. of SNPs | N (Male/Female) | Metabolomics Platform |
|---|---|---|---|
| GB I | 8 986 545 | 391 (195/196) | HPLC‐MS |
| GB II | 8 986 545 | 885 (602/283) | HILIC‐MS |
| FR97 | 11 512 433 | 6631 (3198/3433) | NMR |
| FR07 | 11 512 433 | 4124 (1860/2264) | NMR |
| YFS | 11 512 433 | 1947 (1052/895) | NMR |
| NFBC66 | 11 512 433 | 4483 (2152/2331) | NMR |
| NFBC86 | 11 512 433 | 3112 (1508/1604) | NMR |
| METSIM | 16 888 882 | 8545 (8545/0) | NMR |
FR97 and FR07 indicates FINRISK; GB, GeneBank; HPLC‐MS, high‐performance liquid chromatography with mass spectrometry; METSIM, METabolic Syndrome In Men Study; NFBC, Northern Finland Birth Cohort; NMR, nuclear magnetic resonance; SNP, single‐nucleotide polymorphism; YFS, Cardiovascular Risk in Young Finns.
Figure 1.
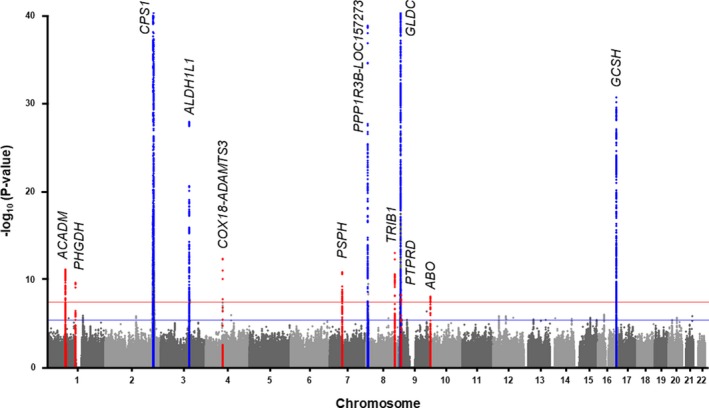
Results of GWAS meta‐analysis for circulating glycine levels. The Manhattan plot shows 7 novel significantly associated loci for circulating glycine levels (red dots) identified through meta‐analyses of GWAS data from 30 118 subjects in the GeneBank, FR97, FR07, YFS, NFBC66, NFBC86, and METSIM cohorts. The 5 previously known loci are indicated by blue dots and all increased in significance in the meta‐analysis. Genome‐wide thresholds for significant (P=5.0×10−8) and suggestive (P=5.0×10−6) association are indicated by the horizontal red and dark blue lines, respectively. P values are truncated at −log10 (P)=40. FR97 and FR07 indicates FINRISK; GWAS, genome‐wide association study; METSIM, METabolic Syndrome In Men Study; NFBC, Northern Finland Birth Cohort; YFS, Cardiovascular Risk in Young Finns.
Table 2.
Meta‐Analysis Identifies 12 Loci Significantly Associated With Circulating Glycine Levels
| Locus (Nearest Gene(s))a | Lead SNP | Position (bp)b | Effect/Other Allelec | EAF | β (SE) | P Value | Directiond |
|---|---|---|---|---|---|---|---|
| 1p31.1 (ACADM) | rs12126607 | 76 217 097 | A/G | 0.27 | 0.06 (0.01) | 1.1×10−11 | +−++ |
| 1p12 (PHGDH) | rs478093 | 120 255 126 | G/A | 0.67 | 0.06 (0.01) | 3.5×10−10 | ++++ |
| 2q34 (CPS1) | rs1047891 | 211 540 507 | A/C | 0.34 | 0.43 (0.01) | <1.0×10−300 | ++++ |
| 3q21.3 (ALDH1L1) | rs2364368 | 125 905 080 | T/A | 0.40 | 0.09 (0.01) | 2.2×10−28 | ++++ |
| 4q13.3 (COX18‐ADAMTS3) | rs143424675 | 73 749 419 | T/C | 0.03 | 0.19 (0.03) | 7.3×10−13 | +−++ |
| 7p11.2 (PSPH) | rs6955423 | 56 099 352 | A/G | 0.81 | 0.07 (0.01) | 2.3×10−11 | ++++ |
| 8p23.1 (PPP1R3B‐LOC157273) | rs2126263 | 9 181 611 | G/A | 0.15 | 0.16 (0.01) | 5.8×10−44 | ++++ |
| 8q24.13 (TRIB1) | rs28601761 | 126 500 031 | G/C | 0.41 | 0.06 (0.01) | 1.6×10−13 | +−++ |
| 9p24.1 (GLDC) | rs71503800 | 6 102 648 | T/C | 0.05 | 0.46 (0.02) | 8.5×10−121 | −++ |
| 9p24.1 (PTPRD) | rs12003835 | 8 424 378 | T/G | 0.03 | 0.15 (0.03) | 8.2×10−9 | −++ |
| 9q34.2 (ABO)e | rs492488 | 136 144 960 | G/A | 0.55 | 0.05 (0.01) | 1.2×10−8 | −+++ |
| 16q23.2 (GCSH) | rs11860711 | 81 132 493 | C/T | 0.80 | 0.12 (0.01) | 4.2×10−31 | ++++ |
EAF indicates effect allele frequency; SNP, single‐nucleotide polymorphism.
Novel loci identified in this study are highlighted in gray.
SNP base pair (bp) positions are given according to NCBI build 37 of the reference human genome sequence (hg19).
Effect allele refers to allele that increases glycine levels.
Direction of betas in the 4 data sets used for meta‐analysis are in the following order: GB I, GB II, Combination of FR97‐FR07‐YFS‐NFBC66‐NFBC86, and METSIM.
N=27 006 for chromosome 9q34.2 locus.
Based on previous observations that the CPS1 locus exhibited a pattern of sexually dimorphic associations with glycine, various other metabolites, and risk of CAD,4, 17 we also carried out meta‐analyses in men and women separately. Five and 9 regions were significantly associated with circulating glycine levels in females and males, respectively (Figures S3 and S4), all of which were also observed in the combined GWAS analysis with all subjects (Figure 1). With the exception of the previously observed stronger association signal for glycine levels at the CPS1 locus in women (β=0.572; P<1.0×10−300) compared with men (β=0.322; P=5.9×10−189), the effect sizes at the remaining 11 loci were similar in males and females with no significant evidence for heterogeneity (Table S2 and Figure S5). We next carried out a phenome‐wide association studies analysis based on publicly available data to determine whether any of the loci for glycine were associated with other traits. Six of the 12 loci (ACADM, CPS1, ALDH1L1, PPP1R3B‐LOC157273, TRIB1, and ABO) exhibited pleiotropic associations with blood cell counts or lipid levels, some of which were even more significant than the association signals for glycine (Table S3). Two other loci (PSPH and PHGDH) had also been associated with serine and homocysteine levels, which are metabolites related to glycine metabolism (Table S3). However, no genome‐wide significant associations have previously been reported for the 4 remaining loci (COX18‐ADAMTS3, GLDC, PTPRD, and GCSH).
Association of Loci for Circulating Glycine Levels With CAD and Traditional Risk Factors
We next sought to evaluate association of loci for glycine levels with risk of CAD and traditional risk factors. Of the 12 regions identified, glycine‐raising alleles of the lead variants at the CPS1, PSPH, TRIB1, and ABO loci individually yielded directionally consistent associations with decreased risk of CAD at the Bonferroni‐corrected threshold of P=4.2×10−3 for testing 12 loci (0.05/12; Table S4). We next tested 4 genetic models based on various nested combinations of the 12 glycine loci for association with risk of CAD using 3 analytical strategies (details provided in Methods). Consistent with the individual SNP results, meta‐analysis or GRS‐based joint SNP effects analysis of glycine‐raising alleles in all 4 genetic models yielded modest, but significant, associations (odds ratios, ≈0.98) with decreased risk of CAD (Figure 2). By comparison, weighted median and inverse variance weighted MR tests yielded much weaker or no evidence for a protective association of glycine‐raising alleles with CAD, including the most restrictive model constructed with only variants at the 2 glycine cleavage system loci (Figure 2). We next evaluated whether loci for glycine were associated with blood pressure, lipid levels, and obesity‐related traits using the same analytical strategies. Glycine‐raising alleles at several loci (CPS1, PPP1R3B‐LOC157273, TRIB1, and ABO) individually exhibited highly significant associations with decreased blood pressure and lipid levels (Table S5). The meta‐analysis and GRS‐based joint SNP effects analysis also provided evidence for similar associations with blood pressure and lipid levels, although these were only observed for the genetic models that included either all 12 glycine‐associated loci or the 7 novel loci. However, the 2 MR analyses provided no evidence that glycine‐raising alleles were causally associated with any of the selected traditional risk factors (Table S5).
Figure 2.
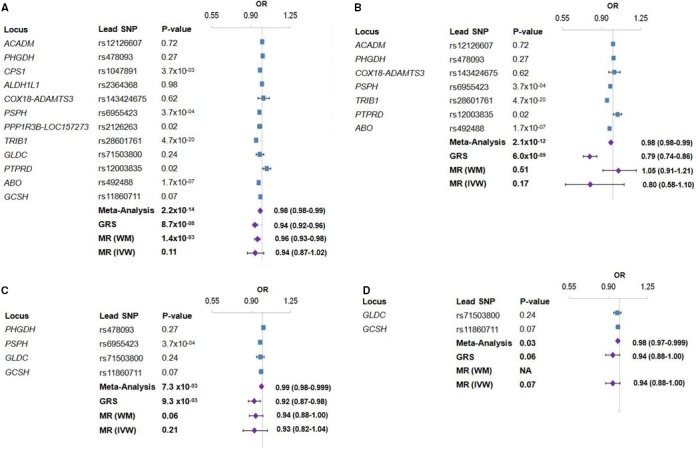
Association of loci identified for circulating glycine levels with risk of CAD. Individual associations between glycine‐raising alleles at each locus and risk of CAD are shown by blue squares in the forest plots. Purple diamonds indicate combined associations based on meta‐analysis, joint SNP effects with a genetic risk score (GRS), and weighted median (WM) or inverse variance weighted (IVW) Mendelian randomization (MR) test. Model 1 included all 12 glycine‐associated loci (A), model 2 included the 7 novel loci for glycine in this study (B), model 3 included the 4 loci known to be involved in glycine metabolism (C), and model 4 was constructed with only the 2 loci directly involved in the catabolism of glycine through the glycine cleavage complex (D). CAS indicates coronary artery disease; OR, odds ratio; SNP, single‐nucleotide polymorphism.
Discussion
In the present study, we used a meta‐analysis approach to identify 7 novel genomic regions associated with circulating glycine levels and strengthen the association signals at 5 previously known loci. Among all 12 loci, CPS1 and GLDC were the most strongly associated with glycine levels, with variants at the remaining 10 loci having anywhere between ≈60% and 90% lower effect sizes. Furthermore, sex‐stratified analyses confirmed the strong effect of CPS1 on glycine levels in women compared with men, but did not reveal sexually dimorphic associations with any of the remaining 11 loci. Follow‐up analyses with the identified loci also yielded evidence that glycine could be causally associated with risk of CAD, although the biological mechanism(s) through which this effect occurs remains to be determined.
Based on what is known about amino acid metabolism, plausible biological links could be inferred between several of the newly identified loci and glycine levels. For example, PHGDH and PSPH encode phosphoglycerate dehydrogenase and phosphoserine phosphatase, which catalyze the first and last reactions, respectively, in the 3‐step process leading to the synthesis of serine from 3‐phosphoglycerate.42 Although the PHGDH and PSPH loci have both been strongly associated with circulating serine or homocysteine levels,8, 15, 16, 43, 44, 45, 46, 47 they were not known to be associated with glycine levels before the results of our meta‐analysis. Interestingly, serine can serve as a substrate for the synthesis of glycine in a reversible reaction catalyzed by SHMT,48 and glycine levels have been reported to be lower in humans deficient for PHGDH or PSPH.49, 50, 51, 52 With respect to our results, the lead variant at PHDGH has yielded several highly significant (P values ranging from ≈1.0×10−10 to 1.0×10−34) cis expression quantitative trait loci where the glycine‐raising allele of rs478093 (G) increases PHGDH mRNA levels.35 This would presumably lead to increased production of serine and, by extension, glycine, thus providing a directionally consistent molecular mechanism for the observed association of the PHGDH locus with circulating glycine levels. However, even when taking into account previously identified associations at loci harboring enzymes involved in either glycine catabolism (GLDC, GCSH)53, 54 or downstream detoxification through the urea cycle (CPS1),55, 56 biological mechanisms for half of the loci associated with circulating glycine levels are not directly evident.
A primary goal of our study was to test whether glycine is a causal and protective biomarker of CAD risk. To address this question, we used the results of large GWAS meta‐analyses to determine whether loci identified for glycine levels were associated with CAD and traditional risk factors. Glycine‐raising alleles at 3 of the 7 novel loci (PSPH, TRIB1, and ABO) were individually associated with decreased risk of CAD at the Bonferroni‐corrected significance threshold (P=4.2×10−3), of which TRIB1 and ABO had been identified as CAD susceptibility loci in previous GWASs.37, 57, 58 Rather than glycine levels, it is likely that association of TRIB1 and ABO with CAD is attributed to their stronger effect sizes on lipid levels and hematological parameters12, 59, 60 and, in the case of ABO, numerous other CAD‐relevant traits.35 When all 12 loci or only the 7 novel loci were considered in combination, the meta‐analyses and joint SNP effects analyses also revealed association of glycine‐raising alleles with decreased risk of CAD. Because several of the loci included in these analyses (CPS1, PSPH, PPP1R3B‐LOC157273, TRIB1, and ABO) exhibited associations with other CAD‐related traits, either individually or in various combinations, it was not possible based on these results alone to conclude that glycine is the causal biomarker driving the association of these loci with CAD. Therefore, we assessed causality more directly with 2 different MR tests, which provided little to no evidence that glycine‐raising alleles were associated with risk of CAD or lipid levels, blood pressure, and obesity‐related traits. In this regard, the results of MR tests with the most restrictive genetic model that included only the 2 loci directly involved in glycine degradation (GLDC and GCSH) are particularly relevant. For example, no CAD‐related traits, aside from glycine levels, are known to be associated with the GLDC and GCSH loci, thus satisfying the lack of pleiotropy as 1 of the major assumptions in MR analysis. Moreover, the glycine‐raising alleles of rs71503800 at the GLDC locus and rs1047891 at the CPS1 locus have nearly equivalent effect sizes on circulating glycine levels. However, none of the analyses with rs71503800 at the GLDC locus yielded evidence for association of this variant with risk of CAD or traditional risk factors. Taken together, we conclude that evidence for a causal relationship between circulating glycine and risk of CAD is relatively weak and requires additional studies.
Whereas the present results have revealed novel genetic determinants of circulating glycine levels, our study should also be taken in the context of certain limitations. First, depending on the cohort, metabolomic analysis was carried out using different platforms and glycine was measured in either serum or plasma, some of which were not fasting samples. Although this may have led to identifying fewer significant associations for circulating glycine levels, our relatively large sample size in the meta‐analysis still provided sufficient power to detect robust associations at several previously known loci and 7 novel genomic regions. Second, the sex‐stratified analyses had approximately half the number of females than males, which likely decreased power to identify loci for circulating glycine levels that were either specific to, or more strongly associated in, 1 sex or the other. Third, all study subjects in our study were of European ancestry, and it is possible that the genetic association results for either circulating plasma glycine levels may not be generalizable to other populations. Last, our evaluation of the causal relationship between glycine and risk of CAD or traditional risk factors may have resulted in biased estimates because of pleiotropic effects, especially in models that included all 12 loci or the 7 newly identified SNPs, or because of weak instruments in nested models that included only the 4 or 2 loci directly involved in glycine metabolism.
In summary, the results of our study provide additional insight into the genetic architecture of glycine metabolism, but a more‐complete understanding of the mechanisms through which some of these loci influence circulating levels remains to be determined. Despite these genetic findings, we did not obtain conclusive evidence for a causal relationship between glycine and risk of CAD, raising the possibility that another unknown metabolite or biological pathway is driving the protective association of glycine‐raising alleles at the CPS1 locus with risk of CAD.
Sources of Funding
This work was supported, in part, by NIH grants R01HL133169, R01ES021801, R01ES025786, R01HL103866, P20HL113452, R01DK062370, and S10OD016346 and Transatlantic Networks of Excellence Awards from Foundation Leducq. The GeneBank study was supported, in part, by NIH grants P01HL098055, P01HL076491, and R01HL103931. Mass Spectrometry instrumentation used for the GeneBank study was housed in a facility supported, in part, through a Shimadzu Center of Excellence Award. Kettunen is supported through funds from the Academy of Finland (Grant Nos. 297338 and 307247) and Novo Nordisk Foundation (Grant No. NNF17OC0026062). Ala‐Korpela is supported by a Senior Research Fellowship from the National Health and Medical Research Council (NHMRC) of Australia (APP1158958) and also works in a unit that is supported by the University of Bristol and the UK Medical Research Council (MC_UU_12013/1). The Baker Institute is supported, in part, by the Victorian Government's Operational Infrastructure Support Program.
Disclosures
Z. Wang and Hazen are named as co‐inventors on pending and issued patents held by the Cleveland Clinic relating to cardiovascular diagnostics and therapeutics and have the right to receive royalty payment for inventions or discoveries related to cardiovascular diagnostics or therapeutics from Cleveland Heart Lab, Quest Diagnostics, and Procter & Gamble Company. Hazen also reports having been paid as a consultant from Procter & Gamble Company and having received research funds from Procter & Gamble Company and Roche. Kettunen reports owning a modest amount of stock options for Nightingale Health Ltd, a company offering metabolic profiling. The remaining authors have no disclosures to report.
Author's Affiliations
From the Departments of Preventive Medicine (Q.J., Y.H., P.H., N.C.W., J.G., J.A.H., H.A.) and Biochemistry & Molecular Medicine (Q.J., Y.H., P.H., N.C.W., J.G., J.A.H., H.A.), Keck School of Medicine, University of Southern California, Los Angeles, CA; Xiangya School of Medicine, Central South University, Hunan, China (P.H.); Computational Medicine, Faculty of Medicine, University of Oulu and Biocenter Oulu, Oulu, Finland (J.K., M.A.‐K., O.A., Q.W., M.‐R.J.); National Institute for Health and Welfare, Helsinki, Finland (J.K., M.P.); Systems Epidemiology, Baker Heart and Diabetes Institute, Melbourne, Victoria, Australia (M.A.‐K., Q.W.); NMR Metabolomics Laboratory, School of Pharmacy (M.A.‐K.) and School of Medicine (M.L.), University of Eastern Finland, Kuopio, Finland; Population Health Science, Bristol Medical School, University of Bristol, United Kingdom (M.A.‐K.); Medical Research Council Integrative Epidemiology Unit at the University of Bristol, United Kingdom (M.A.‐K.); Department of Epidemiology and Preventive Medicine, School of Public Health and Preventive Medicine, Faculty of Medicine, Nursing and Health Sciences, The Alfred Hospital, Monash University, Melbourne, Victoria, Australia (M.A.‐K.); Estonian Genome Center, University of Tartu, Estonia (M.P.); Institute for Molecular Medicine (FIMM), University of Helsinki, Finland (M.P.); Research Centre of Applied and Preventive Cardiovascular Medicine (O.R.) and Department of Medicine (J.V.), University of Turku, Finland; Department of Clinical Physiology (O.R.) and Division of Medicine (J.V.), Turku University Hospital, Turku, Finland; Department of Clinical Chemistry, Fimlab Laboratories and Faculty of Medicine and Health Technology, Finnish Cardiovascular Research Center–Tampere, Tampere University, Tampere, Finland (T.L.); Department of Epidemiology and Biostatistics, MRC‐PHE Centre for Environment and Health, School of Public Health, Imperial College London, London, United Kingdom (M.‐R.J.); Center for Life Course and Systems Epidemiology, University of Oulu, Finland (M.‐R.J.); Unit of Primary Care, Oulu University Hospital, Oulu, Finland (M.‐R.J.); Department of Biostatistics and Center for Statistical Genetics, University of Michigan, Ann Arbor, MI (M.B.); Department of Genetics, University of North Carolina, Chapel Hill, NC (K.L.M.); Genome Center, University of California, Davis, CA (O.F., S.L.H.); Departments of Cardiovascular Medicine (Z.W., W.H.W.T., S.L.H.) and Cellular & Molecular Medicine (W.H.W.T.), Cleveland Clinic, Cleveland, OH.
Supporting information
Data S1. Detailed description of cohorts.
Figure S1. Quantile‐quantile (Q‐Q) plot of GWAS meta‐analysis results for circulating glycine levels in 30 118 subjects. The observed vs the expected P values from the meta‐analyses for glycine levels are shown in the Q‐Q plot. These analyses yielded a genomic inflation factor (λ) of 1.035, indicating that the GWAS meta‐analyses were not confounded by underlying population stratification.
Figure S2. Twelve loci identified for circulating glycine associated levels. Regional plots for the ACADM, PHGDH, CPS1, ALDH1L1, COX18‐ADAMTS3, PSPH, PPP1R3B‐LOC157273, TRIB1, GLDC, PTPRD, ABO, and GCSH loci are shown in (A through L). Each region is centered on the lead SNP (purple diamond) and the genes in the interval are indicated in the bottom panel. The degree of linkage disequilibrium (LD) between the lead SNP and other variants is shown as r 2 values according to the color‐coded legend in the box.
Figure S3. Results of GWAS meta‐analysis for circulating glycine levels in women. A, The Manhattan plot shows 5 previously identified loci significantly associated with circulating glycine levels (blue dots) in a stratified GWAS analysis with 10 886 women. Red dots indicate association signals for the 7 novel identified in our meta‐analysis with all 30 118 subjects, all of which were only suggestively associated in women. Genome‐wide thresholds for significant (P=5.0×10−8) and suggestive (P=5.0×10−6) association are indicated by the horizontal red and dark blue lines, respectively. P values are truncated at −log10 (P)=40. B, The Q‐Q plot shows the observed vs the expected P values from the meta‐analyses for glycine levels in women. These analyses yielded a genomic inflation factor (λ) of 1.002, indicating that the GWAS meta‐analyses were not confounded by underlying population stratification.
Figure S4. Results of GWAS meta‐analysis for circulating glycine levels in men. A, The Manhattan plot shows 9 loci significantly associated with circulating glycine levels in a stratified GWAS analysis with 19 004 men. The 5 loci identified in previous studies are indicated by blue dots. The red dots indicate association signals at the 7 novel identified by our meta‐analysis with all 30 118 subjects, of which 4 were also significant in only men. Genome‐wide thresholds for significant (P=5.0×10−8) and suggestive (P=5.0×10−6) association are indicated by the horizontal red and dark blue lines, respectively. P values are truncated at −log10 (P)=40. B, The Q‐Q plot shows the observed vs the expected P values from the meta‐analyses for glycine levels in men. These analyses yielded a genomic inflation factor (λ) of 1.035, indicating that the GWAS meta‐analyses were not confounded by underlying population stratification.
Figure S5. Sex‐stratified results for 12 loci identified for circulating glycine levels. Effect sizes for the lead SNPs at the 12 loci identified for circulating glycine levels are shown in men (blue) and women (red) separately. With the exception of CPS1, which is associated with ≈2‐fold higher glycine levels in women compared with men, effect sizes at the 11 other loci were similar in males and females. EA indicates effect allele; OA, other allele.
Table S1. Results of 12 Loci Significantly Associated With Circulating Glycine Levels Stratified by Metabolomics Platform
Table S2. Results of 12 Loci Significantly Associated With Circulating Glycine Levels Stratified by Sex
Table S3. PheWAS Results for 12 Loci Significantly Associated With Circulating Glycine Levels
Table S4. Association of 12 Glycine‐Associated Loci With CAD in CARDIoGRAM+C4D and UK Biobank
Table S5. Individual and Joint SNP Effect Associations and Mendelian randomization analysis of Glycine‐Associated Loci With Traditionally CAD Risk Factors
Acknowledgments
We gratefully acknowledge the subjects in each of the cohorts for their participation in these studies
(J Am Heart Assoc. 2019;8:e011922. DOI: 10.1161/JAHA.119.011922.)
The authors' affiliations are proivded under “Authors' Affiliations” section before the References.
References
- 1. Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WH, DiDonato JA, Lusis AJ, Hazen SL. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, DiDonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WH, Bushman FD, Lusis AJ, Hazen SL. Intestinal microbiota metabolism of L‐carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19:576–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368:1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hartiala JA, Tang WH, Wang Z, Crow AL, Stewart AF, Roberts R, McPherson R, Erdmann J, Willenborg C, Hazen SL, Allayee H. Genome‐wide association study and targeted metabolomics identifies sex‐specific association of CPS1 with coronary artery disease. Nat Commun. 2016;7:10558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pare G, Chasman DI, Parker AN, Zee RR, Malarstig A, Seedorf U, Collins R, Watkins H, Hamsten A, Miletich JP, Ridker PM. Novel associations of CPS1, MUT, NOX4, and DPEP1 with plasma homocysteine in a healthy population: a genome‐wide evaluation of 13 974 participants in the Women's Genome Health Study. Circ Cardiovasc Genet. 2009;2:142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lange LA, Croteau‐Chonka DC, Marvelle AF, Qin L, Gaulton KJ, Kuzawa CW, McDade TW, Wang Y, Li Y, Levy S, Borja JB, Lange EM, Adair LS, Mohlke KL. Genome‐wide association study of homocysteine levels in Filipinos provides evidence for CPS1 in women and a stronger MTHFR effect in young adults. Hum Mol Genet. 2010;19:2050–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van Meurs JB, Pare G, Schwartz SM, Hazra A, Tanaka T, Vermeulen SH, Cotlarciuc I, Yuan X, Malarstig A, Bandinelli S, Bis JC, Blom H, Brown MJ, Chen C, Chen YD, Clarke RJ, Dehghan A, Erdmann J, Ferrucci L, Hamsten A, Hofman A, Hunter DJ, Goel A, Johnson AD, Kathiresan S, Kampman E, Kiel DP, Kiemeney LA, Chambers JC, Kraft P, Lindemans J, McKnight B, Nelson CP, O'Donnell CJ, Psaty BM, Ridker PM, Rivadeneira F, Rose LM, Seedorf U, Siscovick DS, Schunkert H, Selhub J, Ueland PM, Vollenweider P, Waeber G, Waterworth DM, Watkins H, Witteman JC, den Heijer M, Jacques P, Uitterlinden AG, Kooner JS, Rader DJ, Reilly MP, Mooser V, Chasman DI, Samani NJ, Ahmadi KR. Common genetic loci influencing plasma homocysteine concentrations and their effect on risk of coronary artery disease. Am J Clin Nutr. 2013;98:668–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Williams SR, Yang Q, Chen F, Liu X, Keene KL, Jacques P, Chen WM, Weinstein G, Hsu FC, Beiser A, Wang L, Bookman E, Doheny KF, Wolf PA, Zilka M, Selhub J, Nelson S, Gogarten SM, Worrall BB, Seshadri S, Sale MM; Genomics and Randomized Trials Network; Framingham Heart Study . Genome‐wide meta‐analysis of homocysteine and methionine metabolism identifies five‐one carbon metabolism loci and a novel association of ALDH1L1 with ischemic stroke. PLoS Genet. 2014;10:e1004214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kottgen A, Pattaro C, Boger CA, Fuchsberger C, Olden M, Glazer NL, Parsa A, Gao X, Yang Q, Smith AV, O'Connell JR, Li M, Schmidt H, Tanaka T, Isaacs A, Ketkar S, Hwang SJ, Johnson AD, Dehghan A, Teumer A, Pare G, Atkinson EJ, Zeller T, Lohman K, Cornelis MC, Probst‐Hensch NM, Kronenberg F, Tonjes A, Hayward C, Aspelund T, Eiriksdottir G, Launer LJ, Harris TB, Rampersaud E, Mitchell BD, Arking DE, Boerwinkle E, Struchalin M, Cavalieri M, Singleton A, Giallauria F, Metter J, de Boer IH, Haritunians T, Lumley T, Siscovick D, Psaty BM, Zillikens MC, Oostra BA, Feitosa M, Province M, de Andrade M, Turner ST, Schillert A, Ziegler A, Wild PS, Schnabel RB, Wilde S, Munzel TF, Leak TS, Illig T, Klopp N, Meisinger C, Wichmann HE, Koenig W, Zgaga L, Zemunik T, Kolcic I, Minelli C, Hu FB, Johansson A, Igl W, Zaboli G, Wild SH, Wright AF, Campbell H, Ellinghaus D, Schreiber S, Aulchenko YS, Felix JF, Rivadeneira F, Uitterlinden AG, Hofman A, Imboden M, Nitsch D, Brandstatter A, Kollerits B, Kedenko L, Magi R, Stumvoll M, Kovacs P, Boban M, Campbell S, Endlich K, Volzke H, Kroemer HK, Nauck M, Volker U, Polasek O, Vitart V, Badola S, Parker AN, Ridker PM, Kardia SL, Blankenberg S, Liu Y, Curhan GC, Franke A, Rochat T, Paulweber B, Prokopenko I, Wang W, Gudnason V, Shuldiner AR, Coresh J, Schmidt R, Ferrucci L, Shlipak MG, van Duijn CM, Borecki I, Kramer BK, Rudan I, Gyllensten U, Wilson JF, Witteman JC, Pramstaller PP, Rettig R, Hastie N, Chasman DI, Kao WH, Heid IM, Fox CS. New loci associated with kidney function and chronic kidney disease. Nat Genet. 2010;42:376–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Choe CU, Atzler D, Wild PS, Carter AM, Boger RH, Ojeda F, Simova O, Stockebrand M, Lackner K, Nabuurs C, Marescau B, Streichert T, Muller C, Luneburg N, De Deyn PP, Benndorf RA, Baldus S, Gerloff C, Blankenberg S, Heerschap A, Grant PJ, Magnus T, Zeller T, Isbrandt D, Schwedhelm E. Homoarginine levels are regulated by L‐arginine:glycine amidinotransferase and affect stroke outcome: results from human and murine studies. Circulation. 2013;128:1451–1461. [DOI] [PubMed] [Google Scholar]
- 11. Kleber ME, Seppala I, Pilz S, Hoffmann MM, Tomaschitz A, Oksala N, Raitoharju E, Lyytikainen LP, Makela KM, Laaksonen R, Kahonen M, Raitakari OT, Huang J, Kienreich K, Fahrleitner‐Pammer A, Drechsler C, Krane V, Boehm BO, Koenig W, Wanner C, Lehtimaki T, Marz W, Meinitzer A. Genome‐wide association study identifies 3 genomic loci significantly associated with serum levels of homoarginine: the AtheroRemo Consortium. Circ Cardiovasc Genet. 2013;6:505–513. [DOI] [PubMed] [Google Scholar]
- 12. Global Lipids Genetics Consortium , Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, Beckmann JS, Bragg‐Gresham JL, Chang HY, Demirkan A, Den Hertog HM, Do R, Donnelly LA, Ehret GB, Esko T, Feitosa MF, Ferreira T, Fischer K, Fontanillas P, Fraser RM, Freitag DF, Gurdasani D, Heikkila K, Hypponen E, Isaacs A, Jackson AU, Johansson A, Johnson T, Kaakinen M, Kettunen J, Kleber ME, Li X, Luan J, Lyytikainen LP, Magnusson PK, Mangino M, Mihailov E, Montasser ME, Muller‐Nurasyid M, Nolte IM, O'Connell JR, Palmer CD, Perola M, Petersen AK, Sanna S, Saxena R, Service SK, Shah S, Shungin D, Sidore C, Song C, Strawbridge RJ, Surakka I, Tanaka T, Teslovich TM, Thorleifsson G, Van den Herik EG, Voight BF, Volcik KA, Waite LL, Wong A, Wu Y, Zhang W, Absher D, Asiki G, Barroso I, Been LF, Bolton JL, Bonnycastle LL, Brambilla P, Burnett MS, Cesana G, Dimitriou M, Doney AS, Doring A, Elliott P, Epstein SE, Eyjolfsson GI, Gigante B, Goodarzi MO, Grallert H, Gravito ML, Groves CJ, Hallmans G, Hartikainen AL, Hayward C, Hernandez D, Hicks AA, Holm H, Hung YJ, Illig T, Jones MR, Kaleebu P, Kastelein JJ, Khaw KT, Kim E, Klopp N, Komulainen P, Kumari M, Langenberg C, Lehtimaki T, Lin SY, Lindstrom J, Loos RJ, Mach F, McArdle WL, Meisinger C, Mitchell BD, Muller G, Nagaraja R, Narisu N, Nieminen TV, Nsubuga RN, Olafsson I, Ong KK, Palotie A, Papamarkou T, Pomilla C, Pouta A, Rader DJ, Reilly MP, Ridker PM, Rivadeneira F, Rudan I, Ruokonen A, Samani N, Scharnagl H, Seeley J, Silander K, Stancakova A, Stirrups K, Swift AJ, Tiret L, Uitterlinden AG, van Pelt LJ, Vedantam S, Wainwright N, Wijmenga C, Wild SH, Willemsen G, Wilsgaard T, Wilson JF, Young EH, Zhao JH, Adair LS, Arveiler D, Assimes TL, Bandinelli S, Bennett F, Bochud M, Boehm BO, Boomsma DI, Borecki IB, Bornstein SR, Bovet P, Burnier M, Campbell H, Chakravarti A, Chambers JC, Chen YD, Collins FS, Cooper RS, Danesh J, Dedoussis G, de Faire U, Feranil AB, Ferrieres J, Ferrucci L, Freimer NB, Gieger C, Groop LC, Gudnason V, Gyllensten U, Hamsten A, Harris TB, Hingorani A, Hirschhorn JN, Hofman A, Hovingh GK, Hsiung CA, Humphries SE, Hunt SC, Hveem K, Iribarren C, Jarvelin MR, Jula A, Kahonen M, Kaprio J, Kesaniemi A, Kivimaki M, Kooner JS, Koudstaal PJ, Krauss RM, Kuh D, Kuusisto J, Kyvik KO, Laakso M, Lakka TA, Lind L, Lindgren CM, Martin NG, Marz W, McCarthy MI, McKenzie CA, Meneton P, Metspalu A, Moilanen L, Morris AD, Munroe PB, Njolstad I, Pedersen NL, Power C, Pramstaller PP, Price JF, Psaty BM, Quertermous T, Rauramaa R, Saleheen D, Salomaa V, Sanghera DK, Saramies J, Schwarz PE, Sheu WH, Shuldiner AR, Siegbahn A, Spector TD, Stefansson K, Strachan DP, Tayo BO, Tremoli E, Tuomilehto J, Uusitupa M, van Duijn CM, Vollenweider P, Wallentin L, Wareham NJ, Whitfield JB, Wolffenbuttel BH, Ordovas JM, Boerwinkle E, Palmer CN, Thorsteinsdottir U, Chasman DI, Rotter JI, Franks PW, Ripatti S, Cupples LA, Sandhu MS, Rich SS, Boehnke M, Deloukas P, Kathiresan S, Mohlke KL, Ingelsson E, Abecasis GR. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Danik JS, Pare G, Chasman DI, Zee RY, Kwiatkowski DJ, Parker A, Miletich JP, Ridker PM. Novel loci, including those related to Crohn disease, psoriasis, and inflammation, identified in a genome‐wide association study of fibrinogen in 17 686 women: the Women's Genome Health Study. Circ Cardiovasc Genet. 2009;2:134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sabater‐Lleal M, Huang J, Chasman D, Naitza S, Dehghan A, Johnson AD, Teumer A, Reiner AP, Folkersen L, Basu S, Rudnicka AR, Trompet S, Malarstig A, Baumert J, Bis JC, Guo X, Hottenga JJ, Shin SY, Lopez LM, Lahti J, Tanaka T, Yanek LR, Oudot‐Mellakh T, Wilson JF, Navarro P, Huffman JE, Zemunik T, Redline S, Mehra R, Pulanic D, Rudan I, Wright AF, Kolcic I, Polasek O, Wild SH, Campbell H, Curb JD, Wallace R, Liu S, Eaton CB, Becker DM, Becker LC, Bandinelli S, Raikkonen K, Widen E, Palotie A, Fornage M, Green D, Gross M, Davies G, Harris SE, Liewald DC, Starr JM, Williams FM, Grant PJ, Spector TD, Strawbridge RJ, Silveira A, Sennblad B, Rivadeneira F, Uitterlinden AG, Franco OH, Hofman A, van Dongen J, Willemsen G, Boomsma DI, Yao J, Swords Jenny N, Haritunians T, McKnight B, Lumley T, Taylor KD, Rotter JI, Psaty BM, Peters A, Gieger C, Illig T, Grotevendt A, Homuth G, Volzke H, Kocher T, Goel A, Franzosi MG, Seedorf U, Clarke R, Steri M, Tarasov KV, Sanna S, Schlessinger D, Stott DJ, Sattar N, Buckley BM, Rumley A, Lowe GD, McArdle WL, Chen MH, Tofler GH, Song J, Boerwinkle E, Folsom AR, Rose LM, Franco‐Cereceda A, Teichert M, Ikram MA, Mosley TH, Bevan S, Dichgans M, Rothwell PM, Sudlow CL, Hopewell JC, Chambers JC, Saleheen D, Kooner JS, Danesh J, Nelson CP, Erdmann J, Reilly MP, Kathiresan S, Schunkert H, Morange PE, Ferrucci L, Eriksson JG, Jacobs D, Deary IJ, Soranzo N, Witteman JC, de Geus EJ, Tracy RP, Hayward C, Koenig W, Cucca F, Jukema JW, Eriksson P, Seshadri S, Markus HS, Watkins H, Samani NJ; V.T.E. Consortium, Stroke Consortium, Wellcome Trust Case Control Consortium, C4D Consortium, CARDIoGRAM Consortium , Wallaschofski H, Smith NL, Tregouet D, Ridker PM, Tang W, Strachan DP, Hamsten A, O'Donnell CJ. Multiethnic meta‐analysis of genome‐wide association studies in >100 000 subjects identifies 23 fibrinogen‐associated loci but no strong evidence of a causal association between circulating fibrinogen and cardiovascular disease. Circulation. 2013;128:1310–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Illig T, Gieger C, Zhai G, Romisch‐Margl W, Wang‐Sattler R, Prehn C, Altmaier E, Kastenmuller G, Kato BS, Mewes HW, Meitinger T, de Angelis MH, Kronenberg F, Soranzo N, Wichmann HE, Spector TD, Adamski J, Suhre K. A genome‐wide perspective of genetic variation in human metabolism. Nat Genet. 2010;42:137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Suhre K, Shin SY, Petersen AK, Mohney RP, Meredith D, Wagele B, Altmaier E, Deloukas P, Erdmann J, Grundberg E, Hammond CJ, de Angelis MH, Kastenmuller G, Kottgen A, Kronenberg F, Mangino M, Meisinger C, Meitinger T, Mewes HW, Milburn MV, Prehn C, Raffler J, Ried JS, Romisch‐Margl W, Samani NJ, Small KS, Wichmann HE, Zhai G, Illig T, Spector TD, Adamski J, Soranzo N, Gieger C. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477:54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mittelstrass K, Ried JS, Yu Z, Krumsiek J, Gieger C, Prehn C, Roemisch‐Margl W, Polonikov A, Peters A, Theis FJ, Meitinger T, Kronenberg F, Weidinger S, Wichmann HE, Suhre K, Wang‐Sattler R, Adamski J, Illig T. Discovery of sexual dimorphisms in metabolic and genetic biomarkers. PLoS Genet. 2011;7:e1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Spittler A, Reissner CM, Oehler R, Gornikiewicz A, Gruenberger T, Manhart N, Brodowicz T, Mittlboeck M, Boltz‐Nitulescu G, Roth E. Immunomodulatory effects of glycine on LPS‐treated monocytes: reduced TNF‐alpha production and accelerated IL‐10 expression. FASEB J. 1999;13:563–571. [DOI] [PubMed] [Google Scholar]
- 19. Wheeler M, Stachlewitz RF, Yamashina S, Ikejima K, Morrow AL, Thurman RG. Glycine‐gated chloride channels in neutrophils attenuate calcium influx and superoxide production. FASEB J. 2000;14:476–484. [DOI] [PubMed] [Google Scholar]
- 20. Bruck R, Wardi J, Aeed H, Avni Y, Shirin H, Avinoach I, Shahmurov M, Hershkoviz R. Glycine modulates cytokine secretion, inhibits hepatic damage and improves survival in a model of endotoxemia in mice. Liver Int. 2003;23:276–282. [DOI] [PubMed] [Google Scholar]
- 21. Hasegawa S, Ichiyama T, Sonaka I, Ohsaki A, Okada S, Wakiguchi H, Kudo K, Kittaka S, Hara M, Furukawa S. Cysteine, histidine and glycine exhibit anti‐inflammatory effects in human coronary arterial endothelial cells. Clin Exp Immunol. 2012;167:269–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ruiz‐Ramirez A, Ortiz‐Balderas E, Cardozo‐Saldana G, Diaz‐Diaz E, El‐Hafidi M. Glycine restores glutathione and protects against oxidative stress in vascular tissue from sucrose‐fed rats. Clin Sci (Lond). 2014;126:19–29. [DOI] [PubMed] [Google Scholar]
- 23. Schemmer P, Zhong Z, Galli U, Wheeler MD, Xiangli L, Bradford BU, Conzelmann LO, Forman D, Boyer J, Thurman RG. Glycine reduces platelet aggregation. Amino Acids. 2013;44:925–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Polfus LM, Khajuria RK, Schick UM, Pankratz N, Pazoki R, Brody JA, Chen MH, Auer PL, Floyd JS, Huang J, Lange L, van Rooij FJ, Gibbs RA, Metcalf G, Muzny D, Veeraraghavan N, Walter K, Chen L, Yanek L, Becker LC, Peloso GM, Wakabayashi A, Kals M, Metspalu A, Esko T, Fox K, Wallace R, Franceshini N, Matijevic N, Rice KM, Bartz TM, Lyytikainen LP, Kahonen M, Lehtimaki T, Raitakari OT, Li‐Gao R, Mook‐Kanamori DO, Lettre G, van Duijn CM, Franco OH, Rich SS, Rivadeneira F, Hofman A, Uitterlinden AG, Wilson JG, Psaty BM, Soranzo N, Dehghan A, Boerwinkle E, Zhang X, Johnson AD, O'Donnell CJ, Johnsen JM, Reiner AP, Ganesh SK, Sankaran VG. Whole‐exome sequencing identifies loci associated with blood cell traits and reveals a role for alternative GFI1B splice variants in human hematopoiesis. Am J Hum Genet. 2016;99:481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. El Hafidi M, Perez I, Banos G. Is glycine effective against elevated blood pressure? Curr Opin Clin Nutr Metab Care. 2006;9:26–31. [DOI] [PubMed] [Google Scholar]
- 26. Ding Y, Svingen GF, Pedersen ER, Gregory JF, Ueland PM, Tell GS, Nygard OK. Plasma glycine and risk of acute myocardial infarction in patients with suspected stable angina pectoris. J Am Heart Assoc. 2016;5:e002621 DOI: 10.1161/JAHA.115.002621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vartiainen E, Laatikainen T, Peltonen M, Juolevi A, Mannisto S, Sundvall J, Jousilahti P, Salomaa V, Valsta L, Puska P. Thirty‐five‐year trends in cardiovascular risk factors in Finland. Int J Epidemiol. 2010;39:504–518. [DOI] [PubMed] [Google Scholar]
- 28. Raitakari OT, Juonala M, Ronnemaa T, Keltikangas‐Jarvinen L, Rasanen L, Pietikainen M, Hutri‐Kahonen N, Taittonen L, Jokinen E, Marniemi J, Jula A, Telama R, Kahonen M, Lehtimaki T, Akerblom HK, Viikari JS. Cohort profile: the cardiovascular risk in Young Finns Study. Int J Epidemiol. 2008;37:1220–1226. [DOI] [PubMed] [Google Scholar]
- 29. Rantakallio P. The longitudinal study of the northern Finland birth cohort of 1966. Paediatr Perinat Epidemiol. 1988;2:59–88. [DOI] [PubMed] [Google Scholar]
- 30. Laakso M, Kuusisto J, Stancakova A, Kuulasmaa T, Pajukanta P, Lusis AJ, Collins FS, Mohlke KL, Boehnke M. The Metabolic Syndrome in Men study: a resource for studies of metabolic and cardiovascular diseases. J Lipid Res. 2017;58:481–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kettunen J, Demirkan A, Wurtz P, Draisma HH, Haller T, Rawal R, Vaarhorst A, Kangas AJ, Lyytikainen LP, Pirinen M, Pool R, Sarin AP, Soininen P, Tukiainen T, Wang Q, Tiainen M, Tynkkynen T, Amin N, Zeller T, Beekman M, Deelen J, van Dijk KW, Esko T, Hottenga JJ, van Leeuwen EM, Lehtimaki T, Mihailov E, Rose RJ, de Craen AJ, Gieger C, Kahonen M, Perola M, Blankenberg S, Savolainen MJ, Verhoeven A, Viikari J, Willemsen G, Boomsma DI, van Duijn CM, Eriksson J, Jula A, Jarvelin MR, Kaprio J, Metspalu A, Raitakari O, Salomaa V, Slagboom PE, Waldenberger M, Ripatti S, Ala‐Korpela M. Genome‐wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat Commun. 2016;7:11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Teslovich TM, Kim DS, Yin X, Stancakova A, Jackson AU, Wielscher M, Naj A, Perry JRB, Huyghe JR, Stringham HM, Davis JP, Raulerson CK, Welch RP, Fuchsberger C, Locke AE, Sim X, Chines PS, Narisu N, Kangas AJ, Soininen P; Genetics of Obesity‐Related Liver Disease Consortium, Alzheimer's Disease Genetics Consortium (ADGC), DIAbetes Genetics Replication And Meta‐analysis (DIAGRAM) , Ala‐Korpela M, Gudnason V, Musani SK, Jarvelin MR, Schellenberg GD, Speliotes EK, Kuusisto J, Collins FS, Boehnke M, Laakso M, Mohlke KL. Identification of seven novel loci associated with amino acid levels using single‐variant and gene‐based tests in 8545 Finnish men from the METSIM study. Hum Mol Genet. 2018;27:1664–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Magi R, Morris AP. GWAMA: software for genome‐wide association meta‐analysis. BMC Bioinformatics. 2010;11:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Turner SD. qqman: an R package for visualizing GWAS results using Q‐Q and manhattan plots. J Open Source Software. 2018;3:731 Available at: 10.21105/joss.00731. [DOI] [Google Scholar]
- 35. Staley JR, Blackshaw J, Kamat MA, Ellis S, Surendran P, Sun BB, Paul DS, Freitag D, Burgess S, Danesh J, Young R, Butterworth AS. PhenoScanner: a database of human genotype‐phenotype associations. Bioinformatics. 2016;32:3207–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Speed D, Balding DJ. SumHer better estimates the SNP heritability of complex traits from summary statistics. Nat Genet. 2019;51:277–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. van der Harst P, Verweij N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ Res. 2018;122:433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Surakka I, Horikoshi M, Magi R, Sarin AP, Mahajan A, Lagou V, Marullo L, Ferreira T, Miraglio B, Timonen S, Kettunen J, Pirinen M, Karjalainen J, Thorleifsson G, Hagg S, Hottenga JJ, Isaacs A, Ladenvall C, Beekman M, Esko T, Ried JS, Nelson CP, Willenborg C, Gustafsson S, Westra HJ, Blades M, de Craen AJ, de Geus EJ, Deelen J, Grallert H, Hamsten A, Havulinna AS, Hengstenberg C, Houwing‐Duistermaat JJ, Hypponen E, Karssen LC, Lehtimaki T, Lyssenko V, Magnusson PK, Mihailov E, Muller‐Nurasyid M, Mpindi JP, Pedersen NL, Penninx BW, Perola M, Pers TH, Peters A, Rung J, Smit JH, Steinthorsdottir V, Tobin MD, Tsernikova N, van Leeuwen EM, Viikari JS, Willems SM, Willemsen G, Schunkert H, Erdmann J, Samani NJ, Kaprio J, Lind L, Gieger C, Metspalu A, Slagboom PE, Groop L, van Duijn CM, Eriksson JG, Jula A, Salomaa V, Boomsma DI, Power C, Raitakari OT, Ingelsson E, Jarvelin MR, Thorsteinsdottir U, Franke L, Ikonen E, Kallioniemi O, Pietiainen V, Lindgren CM, Stefansson K, Palotie A, McCarthy MI, Morris AP, Prokopenko I, Ripatti S; ENGAGE Consortium . The impact of low‐frequency and rare variants on lipid levels. Nat Genet. 2015;47:589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Loh PR, Kichaev G, Gazal S, Schoech AP, Price AL. Mixed‐model association for biobank‐scale datasets. Nat Genet. 2018;50:906–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dastani Z, Hivert MF, Timpson N, Perry JR, Yuan X, Scott RA, Henneman P, Heid IM, Kizer JR, Lyytikainen LP, Fuchsberger C, Tanaka T, Morris AP, Small K, Isaacs A, Beekman M, Coassin S, Lohman K, Qi L, Kanoni S, Pankow JS, Uh HW, Wu Y, Bidulescu A, Rasmussen‐Torvik LJ, Greenwood CM, Ladouceur M, Grimsby J, Manning AK, Liu CT, Kooner J, Mooser VE, Vollenweider P, Kapur KA, Chambers J, Wareham NJ, Langenberg C, Frants R, Willems‐Vandijk K, Oostra BA, Willems SM, Lamina C, Winkler TW, Psaty BM, Tracy RP, Brody J, Chen I, Viikari J, Kahonen M, Pramstaller PP, Evans DM, St Pourcain B, Sattar N, Wood AR, Bandinelli S, Carlson OD, Egan JM, Bohringer S, van Heemst D, Kedenko L, Kristiansson K, Nuotio ML, Loo BM, Harris T, Garcia M, Kanaya A, Haun M, Klopp N, Wichmann HE, Deloukas P, Katsareli E, Couper DJ, Duncan BB, Kloppenburg M, Adair LS, Borja JB; DIAGRAM Consortium, MAGIC Consortium, GLGC Consortium, MuTHER Consortium , Wilson JG, Musani S, Guo X, Johnson T, Semple R, Teslovich TM, Allison MA, Redline S, Buxbaum SG, Mohlke KL, Meulenbelt I, Ballantyne CM, Dedoussis GV, Hu FB, Liu Y, Paulweber B, Spector TD, Slagboom PE, Ferrucci L, Jula A, Perola M, Raitakari O, Florez JC, Salomaa V, Eriksson JG, Frayling TM, Hicks AA, Lehtimaki T, Smith GD, Siscovick DS, Kronenberg F, van Duijn C, Loos RJ, Waterworth DM, Meigs JB, Dupuis J, Richards JB, Voight BF, Scott LJ, Steinthorsdottir V, Dina C, Welch RP, Zeggini E, Huth C, Aulchenko YS, Thorleifsson G, McCulloch LJ, Ferreira T, Grallert H, Amin N, Wu G, Willer CJ, Raychaudhuri S, McCarroll SA, Hofmann OM, Segre AV, van Hoek M, Navarro P, Ardlie K, Balkau B, Benediktsson R, Bennett AJ, Blagieva R, Boerwinkle E, Bonnycastle LL, Bostrom KB, Bravenboer B, Bumpstead S, Burtt NP, Charpentier G, Chines PS, Cornelis M, Crawford G, Doney AS, Elliott KS, Elliott AL, Erdos MR, Fox CS, Franklin CS, Ganser M, Gieger C, Grarup N, Green T, Griffin S, Groves CJ, Guiducci C, Hadjadj S, Hassanali N, Herder C, Isomaa B, Jackson AU, Johnson PR, Jorgensen T, Kao WH, Kong A, Kraft P, Kuusisto J, Lauritzen T, Li M, Lieverse A, Lindgren CM, Lyssenko V, Marre M, Meitinger T, Midthjell K, Morken MA, Narisu N, Nilsson P, Owen KR, Payne F, Petersen AK, Platou C, Proenca C, Prokopenko I, Rathmann W, Rayner NW, Robertson NR, Rocheleau G, Roden M, Sampson MJ, Saxena R, Shields BM, Shrader P, Sigurdsson G, Sparso T, Strassburger K, Stringham HM, Sun Q, Swift AJ, Thorand B, Tichet J, Tuomi T, van Dam RM, van Haeften TW, van Herpt T, van Vliet‐Ostaptchouk JV, Walters GB, Weedon MN, Wijmenga C, Witteman J, Bergman RN, Cauchi S, Collins FS, Gloyn AL, Gyllensten U, Hansen T, Hide WA, Hitman GA, Hofman A, Hunter DJ, Hveem K, Laakso M, Morris AD, Palmer CN, Rudan I, Sijbrands E, Stein LD, Tuomilehto J, Uitterlinden A, Walker M, Watanabe RM, Abecasis GR, Boehm BO, Campbell H, Daly MJ, Hattersley AT, Pedersen O, Barroso I, Groop L, Sladek R, Thorsteinsdottir U, Wilson JF, Illig T, Froguel P, van Duijn CM, Stefansson K, Altshuler D, Boehnke M, McCarthy MI, Soranzo N, Wheeler E, Glazer NL, Bouatia‐Naji N, Magi R, Randall J, Elliott P, Rybin D, Dehghan A, Hottenga JJ, Song K, Goel A, Lajunen T, Doney A, Cavalcanti‐Proenca C, Kumari M, Timpson NJ, Zabena C, Ingelsson E, An P, O'Connell J, Luan J, Elliott A, McCarroll SA, Roccasecca RM, Pattou F, Sethupathy P, Ariyurek Y, Barter P, Beilby JP, Ben‐Shlomo Y, Bergmann S, Bochud M, Bonnefond A, Borch‐Johnsen K, Bottcher Y, Brunner E, Bumpstead SJ, Chen YD, Chines P, Clarke R, Coin LJ, Cooper MN, Crisponi L, Day IN, de Geus EJ, Delplanque J, Fedson AC, Fischer‐Rosinsky A, Forouhi NG, Franzosi MG, Galan P, Goodarzi MO, Graessler J, Grundy S, Gwilliam R, Hallmans G, Hammond N, Han X, Hartikainen AL, Hayward C, Heath SC, Hercberg S, Hillman DR, Hingorani AD, Hui J, Hung J, Kaakinen M, Kaprio J, Kesaniemi YA, Kivimaki M, Knight B, Koskinen S, Kovacs P, Kyvik KO, Lathrop GM, Lawlor DA, Le Bacquer O, Lecoeur C, Li Y, Mahley R, Mangino M, Martinez‐Larrad MT, McAteer JB, McPherson R, Meisinger C, Melzer D, Meyre D, Mitchell BD, Mukherjee S, Naitza S, Neville MJ, Orru M, Pakyz R, Paolisso G, Pattaro C, Pearson D, Peden JF, Pedersen NL, Pfeiffer AF, Pichler I, Polasek O, Posthuma D, Potter SC, Pouta A, Province MA, Rayner NW, Rice K, Ripatti S, Rivadeneira F, Rolandsson O, Sandbaek A, Sandhu M, Sanna S, Sayer AA, Scheet P, Seedorf U, Sharp SJ, Shields B, Sigurethsson G, Sijbrands EJ, Silveira A, Simpson L, Singleton A, Smith NL, Sovio U, Swift A, Syddall H, Syvanen AC, Tonjes A, Uitterlinden AG, van Dijk KW, Varma D, Visvikis‐Siest S, Vitart V, Vogelzangs N, Waeber G, Wagner PJ, Walley A, Ward KL, Watkins H, Wild SH, Willemsen G, Witteman JC, Yarnell JW, Zelenika D, Zethelius B, Zhai G, Zhao JH, Zillikens MC; DIAGRAM Consortium; GIANT Consortium; Global BPGen Consortium , Borecki IB, Meneton P, Magnusson PK, Nathan DM, Williams GH, Silander K, Bornstein SR, Schwarz P, Spranger J, Karpe F, Shuldiner AR, Cooper C, Serrano‐Rios M, Lind L, Palmer LJ, Hu FBs, Franks PW, Ebrahim S, Marmot M, Kao WH, Pramstaller PP, Wright AF, Stumvoll M, Hamsten A; Procardis C , Buchanan TA, Valle TT, Rotter JI, Penninx BW, Boomsma DI, Cao A, Scuteri A, Schlessinger D, Uda M, Ruokonen A, Jarvelin MR, Peltonen L, Mooser V, Sladek R; MAGIC investigators, GLGC Consortium , Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Chasman DI, Johansen CT, Fouchier SW, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Feitosa MF, Orho‐Melander M, Melander O, Li X, Li M, Cho YS, Go MJ, Kim YJ, Lee JY, Park T, Kim K, Sim X, Ong RT, Croteau‐Chonka DC, Lange LA, Smith JD, Ziegler A, Zhang W, Zee RY, Whitfield JB, Thompson JR, Surakka I, Spector TD, Smit JH, Sinisalo J, Scott J, Saharinen J, Sabatti C, Rose LM, Roberts R, Rieder M, Parker AN, Pare G, O'Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, McArdle W, Masson D, Martin NG, Marroni F, Lucas G, Luben R, Lokki ML, Lettre G, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Konig IR, Khaw KT, Kaplan LM, Johansson A, Janssens AC, Igl W, Hovingh GK, Hengstenberg C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Groop LC, Gonzalez E, Freimer NB, Erdmann J, Ejebe KG, Doring A, Dominiczak AF, Demissie S, Deloukas P, de Faire U, Crawford G, Chen YD, Caulfield MJ, Boekholdt SM, Assimes TL, Quertermous T, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Taylor HA Jr, Gabriel SB, Holm H, Gudnason V, Krauss RM, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Strachan DP, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, Kathiresan S. Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi‐ethnic meta‐analysis of 45,891 individuals. PLoS Genet. 2012;8:e1002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R, Tan VY, Yarmolinsky J, Shihab HA, Timpson NJ, Evans DM, Relton C, Martin RM, Davey Smith G, Gaunt TR, Haycock PC. The MR‐Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. de Koning TJ. Amino acid synthesis deficiencies. J Inherit Metab Dis. 2017;40:609–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rhee EP, Ho JE, Chen MH, Shen D, Cheng S, Larson MG, Ghorbani A, Shi X, Helenius IT, O'Donnell CJ, Souza AL, Deik A, Pierce KA, Bullock K, Walford GA, Vasan RS, Florez JC, Clish C, Yeh JR, Wang TJ, Gerszten RE. A genome‐wide association study of the human metabolome in a community‐based cohort. Cell Metab. 2013;18:130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xie W, Wood AR, Lyssenko V, Weedon MN, Knowles JW, Alkayyali S, Assimes TL, Quertermous T, Abbasi F, Paananen J, Haring H, Hansen T, Pedersen O, Smith U, Laakso M, Dekker JM, Nolan JJ, Groop L, Ferrannini E, Adam KP, Gall WE, Frayling TM, Walker M. Genetic variants associated with glycine metabolism and their role in insulin sensitivity and type 2 diabetes. Diabetes. 2013;62:2141–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J, Arnold M, Erte I, Forgetta V, Yang TP, Walter K, Menni C, Chen L, Vasquez L, Valdes AM, Hyde CL, Wang V, Ziemek D, Roberts P, Xi L, Grundberg E; Multiple Tissue Human Expression Resource (MuTHER) Consortium , Waldenberger M, Richards JB, Mohney RP, Milburn MV, John SL, Trimmer J, Theis FJ, Overington JP, Suhre K, Brosnan MJ, Gieger C, Kastenmuller G, Spector TD, Soranzo N. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46:543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Draisma HHM, Pool R, Kobl M, Jansen R, Petersen AK, Vaarhorst AAM, Yet I, Haller T, Demirkan A, Esko T, Zhu G, Bohringer S, Beekman M, van Klinken JB, Romisch‐Margl W, Prehn C, Adamski J, de Craen AJM, van Leeuwen EM, Amin N, Dharuri H, Westra HJ, Franke L, de Geus EJC, Hottenga JJ, Willemsen G, Henders AK, Montgomery GW, Nyholt DR, Whitfield JB, Penninx BW, Spector TD, Metspalu A, Slagboom PE, van Dijk KW, ‘t Hoen PAC, Strauch K, Martin NG, van Ommen GB, Illig T, Bell JT, Mangino M, Suhre K, McCarthy MI, Gieger C, Isaacs A, van Duijn CM, Boomsma DI. Genome‐wide association study identifies novel genetic variants contributing to variation in blood metabolite levels. Nat Commun. 2015;6:7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Long T, Hicks M, Yu HC, Biggs WH, Kirkness EF, Menni C, Zierer J, Small KS, Mangino M, Messier H, Brewerton S, Turpaz Y, Perkins BA, Evans AM, Miller LA, Guo L, Caskey CT, Schork NJ, Garner C, Spector TD, Venter JC, Telenti A. Whole‐genome sequencing identifies common‐to‐rare variants associated with human blood metabolites. Nat Genet. 2017;49:568–578. [DOI] [PubMed] [Google Scholar]
- 48. Wang W, Wu Z, Dai Z, Yang Y, Wang J, Wu G. Glycine metabolism in animals and humans: implications for nutrition and health. Amino Acids. 2013;45:463–477. [DOI] [PubMed] [Google Scholar]
- 49. Jaeken J, Detheux M, Van Maldergem L, Foulon M, Carchon H, Van Schaftingen E. 3‐Phosphoglycerate dehydrogenase deficiency: an inborn error of serine biosynthesis. Arch Dis Child. 1996;74:542–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pineda M, Vilaseca MA, Artuch R, Santos S, Garcia Gonzalez MM, Aracil A, Van Schaftingen E, Jaeken J. 3‐Phosphoglycerate dehydrogenase deficiency in a patient with West syndrome. Dev Med Child Neurol. 2000;42:629–633. [DOI] [PubMed] [Google Scholar]
- 51. van der Crabben SN, Verhoeven‐Duif NM, Brilstra EH, Van Maldergem L, Coskun T, Rubio‐Gozalbo E, Berger R, de Koning TJ. An update on serine deficiency disorders. J Inherit Metab Dis. 2013;36:613–619. [DOI] [PubMed] [Google Scholar]
- 52. Byers HM, Bennett RL, Malouf EA, Weiss MD, Feng J, Scott CR, Jayadev S. Novel report of phosphoserine phosphatase deficiency in an adult with myeloneuropathy and limb contractures. JIMD Rep. 2016;30:103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kikuchi G, Motokawa Y, Yoshida T, Hiraga K. Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc Jpn Acad Ser B Phys Biol Sci. 2008;84:246–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lamers Y, Williamson J, Gilbert LR, Stacpoole PW, Gregory JF III. Glycine turnover and decarboxylation rate quantified in healthy men and women using primed, constant infusions of [1,2‐(13)C2]glycine and [(2)H3]leucine. J Nutr. 2007;137:2647–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Freeman JM, Nicholson JF, Schimke RT, Rowland LP, Carter S. Congenital hyperammonemia. Association with hyperglycinemia and decreased levels of carbamyl phosphate synthetase. Arch Neurol. 1970;23:430–437. [DOI] [PubMed] [Google Scholar]
- 56. Colombo JP, Bachmann C, Schrammli A. Inborn defects of the mitochondrial portion of the urea cycle. Ann N Y Acad Sci. 1986;488:109–117. [DOI] [PubMed] [Google Scholar]
- 57. Reilly MP, Li M, He J, Ferguson JF, Stylianou IM, Mehta NN, Burnett MS, Devaney JM, Knouff CW, Thompson JR, Horne BD, Stewart AF, Assimes TL, Wild PS, Allayee H, Nitschke PL, Patel RS, Martinelli N, Girelli D, Quyyumi AA, Anderson JL, Erdmann J, Hall AS, Schunkert H, Quertermous T, Blankenberg S, Hazen SL, Roberts R, Kathiresan S, Samani NJ, Epstein SE, Rader DJ, Qasim AN, DerOhannessian SL, Qu L, Cappola TP, Chen Z, Matthai W, Hakonarson HH, Wilensky R, Kent KM, Lindsay JM, Pichard AD, Satler L, Waksman R. Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: two genome‐wide association studies. Lancet. 2011;377:383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, Saleheen D, Kyriakou T, Nelson CP, Hopewell JC, Webb TR, Zeng L, Dehghan A, Alver M, Armasu SM, Auro K, Bjonnes A, Chasman DI, Chen S, Ford I, Franceschini N, Gieger C, Grace C, Gustafsson S, Huang J, Hwang SJ, Kim YK, Kleber ME, Lau KW, Lu X, Lu Y, Lyytikainen LP, Mihailov E, Morrison AC, Pervjakova N, Qu L, Rose LM, Salfati E, Saxena R, Scholz M, Smith AV, Tikkanen E, Uitterlinden A, Yang X, Zhang W, Zhao W, de Andrade M, de Vries PS, van Zuydam NR, Anand SS, Bertram L, Beutner F, Dedoussis G, Frossard P, Gauguier D, Goodall AH, Gottesman O, Haber M, Han BG, Huang J, Jalilzadeh S, Kessler T, Konig IR, Lannfelt L, Lieb W, Lind L, Lindgren CM, Lokki ML, Magnusson PK, Mallick NH, Mehra N, Meitinger T, Memon FU, Morris AP, Nieminen MS, Pedersen NL, Peters A, Rallidis LS, Rasheed A, Samuel M, Shah SH, Sinisalo J, Stirrups KE, Trompet S, Wang L, Zaman KS, Ardissino D, Boerwinkle E, Borecki IB, Bottinger EP, Buring JE, Chambers JC, Collins R, Cupples LA, Danesh J, Demuth I, Elosua R, Epstein SE, Esko T, Feitosa MF, Franco OH, Franzosi MG, Granger CB, Gu D, Gudnason V, Hall AS, Hamsten A, Harris TB, Hazen SL, Hengstenberg C, Hofman A, Ingelsson E, Iribarren C, Jukema JW, Karhunen PJ, Kim BJ, Kooner JS, Kullo IJ, Lehtimaki T, Loos RJ, Melander O, Metspalu A, Marz W, Palmer CN, Perola M, Quertermous T, Rader DJ, Ridker PM, Ripatti S, Roberts R, Salomaa V, Sanghera DK, Schwartz SM, Seedorf U, Stewart AF, Stott DJ, Thiery J, Zalloua PA, O'Donnell CJ, Reilly MP, Assimes TL, Thompson JR, Erdmann J, Clarke R, Watkins H, Kathiresan S, McPherson R, Deloukas P, Schunkert H, Samani NJ, Farrall M; CARDIoGRAM+C4D Consortium . A comprehensive 1,000 Genomes‐based genome‐wide association meta‐analysis of coronary artery disease. Nat Genet. 2015;47:1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho‐Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee‐Hee Ong R, Croteau‐Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O'Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PK, Lucas G, Luben R, Loos RJ, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, Konig IR, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Cecile JWJA, Ingelsson E, Igl W, Kees Hovingh G, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Doring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJ, de Faire U, Crawford G, Collins FS, Chen YD, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA Jr, Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Astle WJ, Elding H, Jiang T, Allen D, Ruklisa D, Mann AL, Mead D, Bouman H, Riveros‐Mckay F, Kostadima MA, Lambourne JJ, Sivapalaratnam S, Downes K, Kundu K, Bomba L, Berentsen K, Bradley JR, Daugherty LC, Delaneau O, Freson K, Garner SF, Grassi L, Guerrero J, Haimel M, Janssen‐Megens EM, Kaan A, Kamat M, Kim B, Mandoli A, Marchini J, Martens JH, Meacham S, Megy K, O'Connell J, Petersen R, Sharifi N, Sheard SM, Staley JR, Tuna S, van der Ent M, Walter K, Wang SY, Wheeler E, Wilder SP, Iotchkova V, Moore C, Sambrook J, Stunnenberg HG, Di Angelantonio E, Kaptoge S, Kuijpers TW, Carrillo‐de‐Santa‐Pau E, Juan D, Rico D, Valencia A, Chen L, Ge B, Vasquez L, Kwan T, Garrido‐Martin D, Watt S, Yang Y, Guigo R, Beck S, Paul DS, Pastinen T, Bujold D, Bourque G, Frontini M, Danesh J, Roberts DJ, Ouwehand WH, Butterworth AS, Soranzo N. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell. 2016;167:1415–1429.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Detailed description of cohorts.
Figure S1. Quantile‐quantile (Q‐Q) plot of GWAS meta‐analysis results for circulating glycine levels in 30 118 subjects. The observed vs the expected P values from the meta‐analyses for glycine levels are shown in the Q‐Q plot. These analyses yielded a genomic inflation factor (λ) of 1.035, indicating that the GWAS meta‐analyses were not confounded by underlying population stratification.
Figure S2. Twelve loci identified for circulating glycine associated levels. Regional plots for the ACADM, PHGDH, CPS1, ALDH1L1, COX18‐ADAMTS3, PSPH, PPP1R3B‐LOC157273, TRIB1, GLDC, PTPRD, ABO, and GCSH loci are shown in (A through L). Each region is centered on the lead SNP (purple diamond) and the genes in the interval are indicated in the bottom panel. The degree of linkage disequilibrium (LD) between the lead SNP and other variants is shown as r 2 values according to the color‐coded legend in the box.
Figure S3. Results of GWAS meta‐analysis for circulating glycine levels in women. A, The Manhattan plot shows 5 previously identified loci significantly associated with circulating glycine levels (blue dots) in a stratified GWAS analysis with 10 886 women. Red dots indicate association signals for the 7 novel identified in our meta‐analysis with all 30 118 subjects, all of which were only suggestively associated in women. Genome‐wide thresholds for significant (P=5.0×10−8) and suggestive (P=5.0×10−6) association are indicated by the horizontal red and dark blue lines, respectively. P values are truncated at −log10 (P)=40. B, The Q‐Q plot shows the observed vs the expected P values from the meta‐analyses for glycine levels in women. These analyses yielded a genomic inflation factor (λ) of 1.002, indicating that the GWAS meta‐analyses were not confounded by underlying population stratification.
Figure S4. Results of GWAS meta‐analysis for circulating glycine levels in men. A, The Manhattan plot shows 9 loci significantly associated with circulating glycine levels in a stratified GWAS analysis with 19 004 men. The 5 loci identified in previous studies are indicated by blue dots. The red dots indicate association signals at the 7 novel identified by our meta‐analysis with all 30 118 subjects, of which 4 were also significant in only men. Genome‐wide thresholds for significant (P=5.0×10−8) and suggestive (P=5.0×10−6) association are indicated by the horizontal red and dark blue lines, respectively. P values are truncated at −log10 (P)=40. B, The Q‐Q plot shows the observed vs the expected P values from the meta‐analyses for glycine levels in men. These analyses yielded a genomic inflation factor (λ) of 1.035, indicating that the GWAS meta‐analyses were not confounded by underlying population stratification.
Figure S5. Sex‐stratified results for 12 loci identified for circulating glycine levels. Effect sizes for the lead SNPs at the 12 loci identified for circulating glycine levels are shown in men (blue) and women (red) separately. With the exception of CPS1, which is associated with ≈2‐fold higher glycine levels in women compared with men, effect sizes at the 11 other loci were similar in males and females. EA indicates effect allele; OA, other allele.
Table S1. Results of 12 Loci Significantly Associated With Circulating Glycine Levels Stratified by Metabolomics Platform
Table S2. Results of 12 Loci Significantly Associated With Circulating Glycine Levels Stratified by Sex
Table S3. PheWAS Results for 12 Loci Significantly Associated With Circulating Glycine Levels
Table S4. Association of 12 Glycine‐Associated Loci With CAD in CARDIoGRAM+C4D and UK Biobank
Table S5. Individual and Joint SNP Effect Associations and Mendelian randomization analysis of Glycine‐Associated Loci With Traditionally CAD Risk Factors


