Abstract
Common IRF5 variants associated with multiple immune-mediated diseases are a major determinant of inter-individual variability in pattern-recognition receptor (PRR)-induced cytokines in macrophages. PRR-initiated pathways also contribute to bacterial clearance, and dysregulation of bacterial clearance can contribute to immune-mediated diseases. However, the role of IRF5 in macrophage-mediated bacterial clearance is not well-defined. Furthermore, it is unclear if macrophages from individuals who are carriers of low-IRF5 expressing genetic variants associated with protection for immune-mediated diseases might be at a disadvantage in bacterial clearance. We found that IRF5 was required for optimal bacterial clearance in PRR-stimulated, M1-differentiated human macrophages. Mechanisms regulated by IRF5 included inducing reactive oxygen species (ROS) through p40phox, p47phox and p67phox, NOS2, and autophagy through ATG5. Complementing these pathways in IRF5-deficient M1 macrophages restored bacterial clearance. Further, these antimicrobial pathways required the activation of IRF5-dependent MAPK, NFκB and Akt2 pathways. Importantly, relative to high IRF5-expressing rs2004640/rs2280714 TT/TT immune-mediated disease risk-carrier human macrophages, M1-differentiated GG/CC carrier macrophages demonstrated less ROS, NOS2, and autophagy pathway induction, and consequently, reduced bacterial clearance. Increasing IRF5 expression to the rs2004640/rs2280714 TT/TT levels restored these antimicrobial pathways. We define mechanisms wherein common IRF5 genetic variants modulate bacterial clearance, thereby highlighting that immune-mediated disease risk IRF5 carriers might be relatively protected from microbial-associated diseases.
Introduction
Bacterial products stimulate pattern-recognition receptors (PRRs) to secrete pro-inflammatory cytokines. These PRR responses must be tightly regulated during infection and on mucosal surfaces as excessive cytokine production can result in ongoing inflammation and tissue damage. As such, many gene variants in pathways modulating PRR-initiated responses are associated with immune-mediated diseases (1). Given the importance of balancing inflammation with effective bacterial clearance, variants regulating PRR-initiated outcomes resulting in decreased inflammation may be protective to immune-mediated diseases. Simultaneously, these gene-variant-regulated reduced PRR outcomes may increase susceptibility to microbial-mediated diseases (2) as PRR-initiated pathways contribute to bacterial clearance. For example, SOCS1/LITAF/RMI2 region variants that confer decreased inflammatory bowel disease (IBD) risk (2) are associated with increased leprosy risk (3). Mouse models can demonstrate a similar dichotomy. For example, TNF-deficient mice are protected from LPS-induced lethality, but are more susceptible to infection with Listeria monocytogenesis or Mycobacterium tuberculosis (4,5). Conversely, IRAKM−/− mice show increased experimental systemic lupus erythematosus (SLE) (6), but more effectively clear Pseudomonas aeruginosa relative to wild type mice (7).
IFN regulatory factor (IRF5) is critical for multiple PRR-induced functions, including cytokine secretion (8,9), glycolysis (10) and myeloid cell-mediated T cell activation in a co-culture system (9). IRF5 can regulate these outcomes both as a proximal signaling molecule (8,10–12) and as a transcription factor (1,8,9,13). Consistent with its critical role in PRR-initiated outcomes, non-coding IRF5 genetic variants resulting in increased IRF5 mRNA (14) and protein (10) expression are associated with multiple immune-mediated diseases characterized by increased inflammation, including SLE, IBD, rheumatoid arthritis, Sjogren’s syndrome, primary biliary cirrhosis, systemic sclerosis and multiple sclerosis (15). These IRF5 variants modulate PRR-induced cytokines in human myeloid-derived cells (16,17). In part due to the common distribution of both the risk- and non-risk rs2004640 IRF5 variants and that IRF5 variants show one of the strongest expression quantitative trait loci (eQTLs) in the genome (18), the rs2004640 polymorphism is a major determinant of the variance in inter-individual PRR-induced cytokine secretion (16). PRR-initiated pathways can also regulate bacterial clearance in myeloid cells. However, limited studies have examined IRF5 in bacterial clearance. In mice, IRF5 upregulates NOS2 and Th1 responses following Leishmania donovani infection (19). Moreover, IRF5−/− mice show decreased type I IFN induction upon New Castle disease virus infection (20). Consistently, in vitro, IRF5 in mouse myeloid cells is required for optimal M. tuberculosis or S. pyogenes-mediated type I IFN induction (21–23). However, these studies did not examine the macrophage-intrinsic role and mechanisms for IRF5 in bacterial clearance. Furthermore, it is not known if IRF5 plays a role in bacterial clearance in human macrophages and if macrophages from disease-associated IRF5 genetic carriers show altered bacterial clearance. Mouse and human cell inflammatory outcomes can dramatically differ (24), such that it is critical to examine these questions in human cells. As such, many questions remain unanswered. Is there a human macrophage-intrinsic role for IRF5 in bacterial clearance? What are the mechanisms through which IRF5 mediates putative bacterial clearance in macrophages? How might this clearance be regulated by IRF5 in distinct macrophage subsets? Do immune-mediated disease-protective IRF5 genetic variants associated with decreased IRF5 expression and decreased PRR-induced cytokine secretion also show decreased bacterial clearance and mechanisms mediating this clearance? Can IRF5 expression modulation in IRF5 polymorphism carrier cells regulate these outcomes?
In this study, we found that IRF5 was required for optimal bacterial clearance in primary human monocyte-derived macrophages (MDMs) and particularly in PRR-stimulated M1-differentiated macrophages. Mechanisms mediating IRF5-dependent bacterial clearance in M1-differentiated macrophages included induction of members of the NADPH oxidase complex, NOS2, and autophagy pathways. Importantly, M1-polarized macrophages from rs2004640/rs2280714 GG/CC carriers protective for immune-mediated diseases and expressing lower IRF5 levels showed decreased bacterial clearance, and decreased induction of ROS, RNS and autophagy pathways relative to TT/TT immune-mediated disease risk carriers. Therefore, we define how IRF5 and the immune disease-associated IRF5 variants regulate macrophage-intrinsic bacterial clearance and identify mechanisms leading to these outcomes. These findings highlight that the immune-disease risk variant expressing higher IRF5 shows a benefit with respect to macrophage-mediated clearance of bacteria, and conversely, that IRF5 variant carriers protected from immune-mediated diseases might be at increased risk for microbial diseases.
Materials and Methods
Patient recruitment and genotyping
Informed consent was obtained per protocol approved by the institutional review board at Yale University. Healthy individuals were genotyped by Taqman (Life Technologies, Grand Island, NY) or Sequenom Platform (Sequenom, San Diego, CA).
Primary myeloid cell culture
Human peripheral blood mononuclear cells (PBMCs) were isolated from peripheral blood using Ficoll-Paque (GE Dharmacon, Lafayette, CO). Monocytes were purified from PBMCs by adhesion and tested for purity (>98% by CD11c expression). Monocytes were differentiated in M-CSF (10ng/ml) (Shenandoah Biotechnology, Warwick, PA) and culture conditions as in (10) for 7 days to generate MDMs. MDMs were treated for 24h with 100ng/ml Escherichia coli LPS (Sigma-Aldrich, St. Louis, MO) and 20ng/ml IFNγ (R&D Systems, Minneapolis, MI) for M1 polarization.
Transfection of vector constructs and small interfering RNAs (siRNAs)
100nM (unless otherwise indicated) scrambled or siGENOME SMARTpool siRNA against IRF5, ERK, p38, JNK, NEMO, Akt2, p40phox, p47phox, p67phox, NOS2 or ATG5 (Dharmacon, Lafayette, CO) (4 pooled siRNAs for each gene) or 2μg vectors expressing ATG5 ((Addgene plasmid #24922; kindly deposited by Toren Finkel) (25)), NOS2 (generous gift of Tony Eissa, (26)), p47phox, p67phox ((generous gifts of Celine DerMardirossian, (27)), IRF5 (Genecopoeia, Rockville, MD), or empty vector were transfected into macrophages using Amaxa nucleofector technology (Amaxa, San Diego, CA). In the experiments that examined IRF5-dependent outcomes in M1 polarized macrophages, MDMs were first polarized into M1-differentiated macrophages for 24h, then transfected with 100nM IRF5 siRNA for 48h, and then treated with LPS for the indicated times (Fig. 1A), unless otherwise stated.
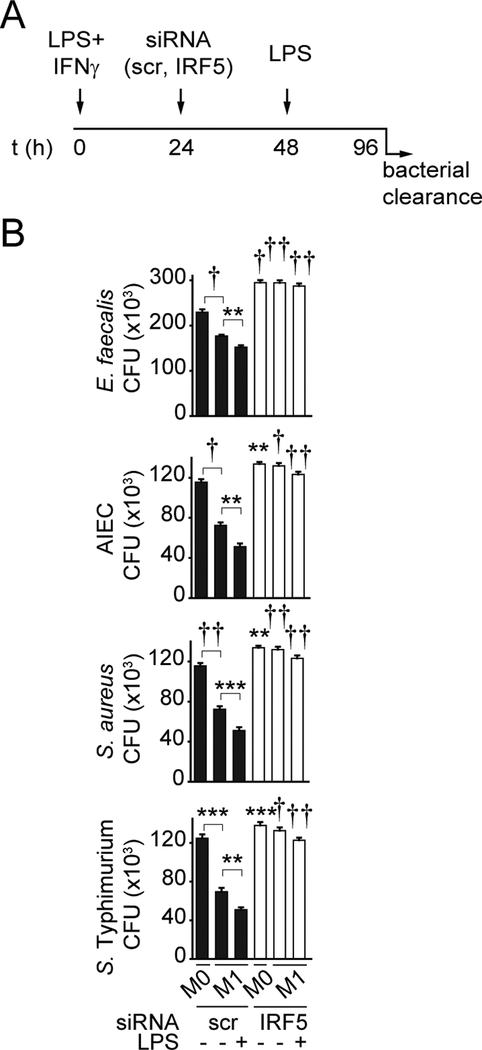
Figure 1. IRF5 is required for optimal intracellular bacterial clearance.
(A) Timeline schematic for M1 polarization, IRF5 knockdown and LPS treatment. (B) MDMs (n=6 donors, similar results in n=12–16) were left untreated (M0) or polarized into M1 macrophages and then transfected with scrambled or IRF5 siRNA. Cells were then left untreated or treated with 0.1μg/ml LPS for 48h and co-cultured with E. faecalis, AIEC, S. aureus or S. Typhimurium. Colony forming units (CFU)+SEM. Significance is compared to scrambled siRNA-transfected cells for each respective condition (i.e. M0, M1 and M1+LPS) or as indicated. Scr, scrambled; **, p<0.01; ***, p<0.001; †, p<1×10−4; ††, p<1×10−5.
Protein detection
Proteins were detected in permeabilized cells by flow cytometry (gated on live cells) with Alexa Fluor 647, phycoerythrin-, Alexa Fluor 488 or biotin-labeled antibodies to phospho-ERK (E10), phospho-p38 (3D7), phospho-JNK (G9), phospho-IκBα (14D4) (Cell Signaling Technology, Danvers, MA), or antibodies to IRF5 (1H6) (Novus, Littleton, CO), p40phox (B-8), p47phox (A-7), p67phox (D-6) (Santa Cruz Biotechnology, Santa Cruz, CA), NOS2, LC3II (Cell Signaling), phospho-Akt2 or ATG5 (EPR1755) (Abcam, Cambridge, MA). Western blot was performed as per (10). Blots were incubated with IRF5 (E1N9G), GAPDH (6C5, EMD Millipore) or the antibodies above.
mRNA expression analysis
Total RNA was isolated using Trizol reagent (Thermo Fisher Scientific). Quantitative PCR was performed using All-In-One qPCR Mix (Genecopoeia) on the ABI Prism 7000 (Applied Biosystems). Samples were normalized to GAPDH. Primer sequences are available upon request.
Intracellular ROS measurement
ROS was measured by flow cytometry using 10μM 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) (Invitrogen) following manufacturer’s instructions or by nitrozolium blue (Sigma Aldrich) as in (28).
Intracellular bacterial clearance
Macrophages were cultured in triplicate with Enterococcus faecalis, adherent invasive Escherichia coli (AIEC) strain LF82 (a generous gift from Dr. Emiko Mizoguchi), or Salmonella enterica serovar Typhimurium at 10:1 MOI, or Staphylococcus aureus at 1:1 MOI for 1h, washed with PBS, and incubated in HBSS with 50μg/ml gentamicin for an additional hour. Cells were washed, lysed with 1% Triton X-100 (Sigma) and plated on MacConkey or LB agar.
Statistical analysis
Significance was assessed using two-tailed t-test. p < 0.05 was considered significant.
Results
IRF5 mediates intracellular bacterial clearance in human macrophages.
To examine the contribution of IRF5 in intracellular bacterial clearance in human macrophages, we first corroborated the role of IRF5 in the differentiation of select macrophage subsets and then examined how it regulates the enhanced bacterial clearance under select conditions. Macrophages consist of multiple subtypes depending on environmental conditions. In particular, pro-inflammatory M1 macrophages more effectively clear bacteria compared to non-polarized macrophages (29). We confirmed that upon culturing MDMs under M1 differentiation conditions (generated by IFNγ and LPS), M1 markers were increased relative to M0 macrophages (MDMs) (Supplementary Fig. 1A). M1 macrophages secrete more TLR4-induced pro-inflammatory and less anti-inflammatory cytokines relative to undifferentiated MDMs, and we confirmed this was the case (Supplementary Fig. 1B). We (10) and others (9,30) have found that IRF5 is required for optimal M1 differentiation and that IRF5 expression is increased upon M1 differentiation. We confirmed this increased IRF5 expression by flow cytometry (Supplementary Fig. 1C) and used Western blot as an independent approach (Supplementary Fig. 1D). We also confirmed our previous finding (10) that IRF5 expression is lower in M2-differentiated macrophages relative to undifferentiated MDMs (Supplementary Fig. 1C). Consistent with the IRF5 expression regulation, we observed that LPS-induced IRF5 phosphorylation, which contributes to IRF5 downstream outcomes (13), was higher in M1-differentiated macrophages compared to MDMs and M2-differentiated macrophages (Supplementary Fig. 1E). To confirm that IRF5 regulates M1 differentiation, we used siRNA to reduce IRF5 expression and established effective IRF5 knockdown through both Western blot and flow cytometry in undifferentiated (M0) macrophages (Supplementary Fig. 1F&G) and in macrophages in M1-polarizing conditions (Supplementary Fig. 1H&I). We will examine IRF5 protein expression by flow cytometry in the studies that follow given the accurate per cell normalization and quantitation advantages (31), and the consistent results between flow cytometry and Western blot in the preceding studies. We also confirmed that cell viability was unaffected following IRF5 knockdown (Supplementary Fig. 1J). As expected, IRF5 was required for optimal induction of M1 polarization markers (Supplementary Fig. 1A) and TLR4-induced M1-associated pro-inflammatory cytokines (Supplementary Fig. 1B). As a comparison, we examined M2 macrophages as IRF5 has not generally been associated with M2 polarization (9,30). IRF5 was required for the upregulation of M2-associated chemokine (e.g. CCL18, CCL22, CCL23) and cytokine (e.g. IL10 and TGFB) transcripts, and TLR4-induced IL10, IL1Ra and TGFβ protein secretion (Supplementary Fig. 1A&B), but not for M2-associated CD36, MAF, MRC, SLC38A6 or TGM2 transcripts (Supplementary Fig. 1A). This indicates that IRF5 deficiency does not universally impair macrophage outcomes. To further ensure that IRF5 knockdown does not result in global macrophage dysfunction, we measured LPS-induced Akt1 phosphorylation, an IRF5-independent event (10), and confirmed that it remained intact in IRF5-deficient MDMs (Supplementary Fig. 1K).
We (32) and others (33) have found that chronic PRR stimulation of macrophages increases antimicrobial mechanisms and bacterial clearance efficacy. Chronic PRR stimulation can be observed with ongoing infection or in environments with persistent bacterial exposure such as at mucosal sites, including in intestinal tissues. Therefore, we examined clearance of the resident intestinal luminal bacteria E. faecalis in MDMs (M0), M1 macrophages, and M1 macrophages chronically treated with LPS (Fig. 1A). We confirmed that compared to M0 macrophages, M1 macrophages exhibited enhanced bacterial clearance, which was further enhanced by chronic LPS treatment (Fig. 1B). We then examined the role of IRF5 in bacterial clearance under these conditions utilizing IRF5 knockdown. Given that IRF5 is required for optimal M1 polarization, we asked if IRF5 contributes to bacterial clearance independent of its role in M1 polarization. We therefore knocked down IRF5 following M1 polarization in the subsequent studies, unless otherwise indicated. IRF5 contributed to bacterial clearance in M0 macrophages, even more so in M1-differentiated macrophages, and particularly following chronic LPS treatment of M1-differentiated macrophages (Fig. 1B). We observed similar requirements with additional bacteria, including adherent-invasive E. coli (AIEC), which is enhanced in the ilea of Crohn’s disease patients (34), the resident bacteria S. aureus, and the intestinal pathogen S. Typhimurium (Fig. 1B). We asked if the reduced LPS-enhanced bacterial clearance observed following IRF5 knockdown was due to decreased TLR4 (receptor recognizing LPS) surface expression and found that this was not the case (Supplementary Fig. 1L). Taken together, these results indicate that IRF5 is required for optimal intracellular bacterial clearance in unpolarized MDMs and in M1-differentiated macrophages and its contribution is more pronounced in PRR-stimulated M1 macrophages.
IRF5 is required for TLR4-induced MAPK, NFκB and Akt2 pathway activation, and these pathways contribute to bacterial clearance in M1 macrophages.
We next assessed mechanisms through which IRF5 contributes to bacterial clearance in PRR-stimulated M1 macrophages. IRF5 can modulate myeloid cell outcomes both as a transcription factor (1,8,9,13) and by directly regulating proximal signaling pathways (8,10–12). We previously found that IRF5 is required for PRR-induced MAPK, NFκB and Akt2 signaling in non-polarized macrophages, as well as in M1-polarized macrophages when IRF5 is knocked down prior to M1 polarization (10), and that IRF5-dependent Akt2 activation is required for M1 polarization (10). To ensure that these signaling pathways were also IRF5-dependent in already differentiated M1 macrophages, we examined their activation when IRF5 is knocked down after M1 polarization. LPS-induced MAPK, NFκB and Akt2 activation was reduced under these conditions (Fig. 2A–C). Similar results were observed utilizing an independent approach by Western blot (Fig. 2D).
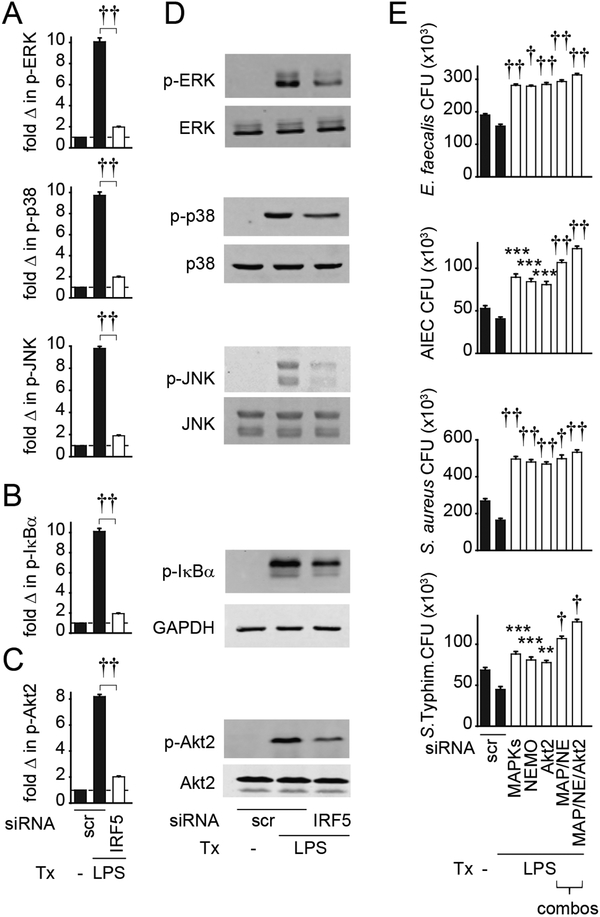
Figure 2. IRF5 is required for optimal LPS-induced MAPK, NFκB and Akt2 pathway activation in M1 macrophages, and these pathways are required for enhanced bacterial clearance in LPS-treated M1 macrophages.
(A-C) MDMs (n=6 donors, similar results seen in an independent n=6) were polarized into M1 macrophages, transfected with scrambled or IRF5 siRNA, and then treated with 0.1μg/ml LPS for 15 min. Summarized data are represented as the fold (A) phospho-ERK, phospho-p38, phospho-JNK, (B) phospho-IκBα, or (C) phospho-Akt2 change normalized to untreated cells (represented by the dotted line at 1) + SEM. (D) MDMs were treated as in (A-C) and the indicated proteins assessed by Western blot. (E) MDMs (n=6, similar results in n=12 for MAPK/NEMO and full combination) were polarized into M1 macrophages, transfected with scrambled or combined ERK, p38, JNK (MAPK), NEMO (NE), or Akt2 siRNA alone or in various combinations as indicated, then left untreated or treated with 0.1μg/ml LPS for 48h and co-cultured with E. faecalis, AIEC, S. aureus or S. Typhimurium. CFU+SEM. Significance is compared to scrambled siRNA-transfected, LPS-treated cells or as indicated. Scr, scrambled; p-, phospho-; tx, treatment. **, p<0.01; ***, p<0.001; †, p<1×10−4; ††, p<1×10−5.
We next asked if MAPK, NFκB and Akt2 pathways were required for the enhanced bacterial clearance observed upon LPS treatment of M1 polarized macrophages. We therefore first polarized MDMs to M1 macrophages, and then knocked down expression of each of these signaling pathways. Knockdown of the MAPK, NFκB or Akt2 pathways following M1 polarization decreased LPS-enhanced bacterial clearance (Fig. 2E). The decrease was more pronounced when the pathways were knocked down in combination, indicating that they cooperate for this outcome (Fig. 2E). Taken together, these results indicate that IRF5 is required for LPS-induced activation of MAPK, NFκB and Akt2 pathways in M1 polarized macrophages, and these signaling pathways, in turn, are required for optimal bacterial clearance in LPS-treated, M1-differentiated macrophages.
IRF5-mediated bacterial clearance is dependent on the NADPH oxidase and NOS2 pathways.
We next investigated the mechanisms through which IRF5 mediates bacterial clearance in PRR-treated M1 macrophages. Reactive oxygen species (ROS) and reactive nitrogen species (RNS) can contribute to bacterial clearance and they cooperate to maintain homeostasis in environments heavily populated by microbes, such as the intestine (35). Furthermore, ROS is induced with chronic PRR ligand exposure (36). To our knowledge IRF5 regulation of ROS production has not been reported. IRF5 knockdown in LPS-treated M1 macrophages resulted in reduced ROS as assessed by two independent approaches (Fig. 3A&B). One mechanism for ROS production is through regulation of the NADPH oxidase complex. Chronic LPS treatment of M1 macrophages induced p40phox, p47phox, and p67phox transcripts and this induction was reduced upon IRF5 knockdown (Fig. 3C). In contrast, p22phox was not regulated by IRF5 and LPS did not induce gp91phox (Fig. 3C). IRF5 regulates NOS2, an RNS-producing enzyme, in the liver of Leishmania donovani-infected mice, and IRF5 enhances NOS2 promoter activity in HEK293 cells (19). We found that LPS treatment of M1 human macrophages upregulated NOS2, but that IRF5-deficient M1 macrophages failed to induce NOS2 (Fig. 3D).
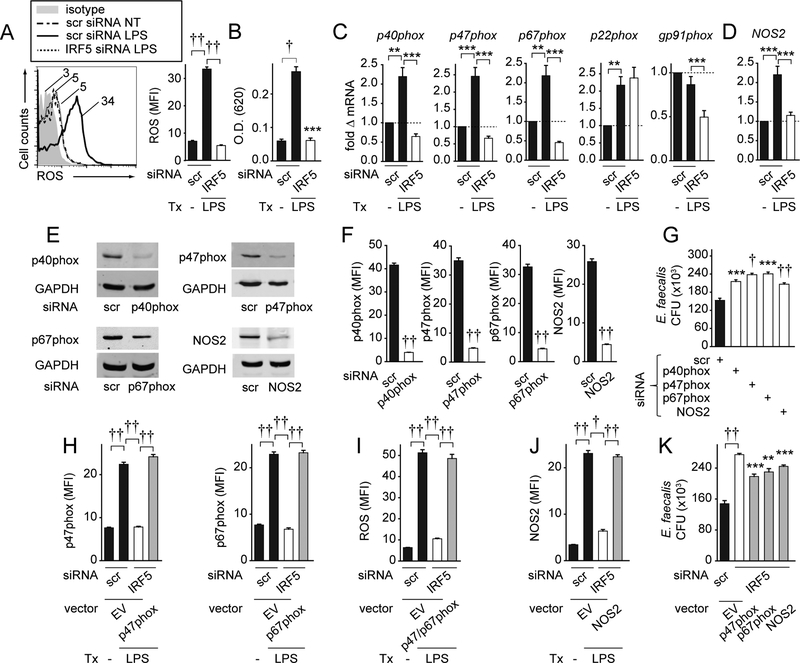
Figure 3. IRF5 is required for optimal induction of NADPH oxidase subunits and NOS2 which contribute to bacterial clearance in PRR-stimulated M1 macrophages.
(A-D) MDMs were polarized into M1 macrophages, transfected with scrambled or IRF5 siRNA, and then left untreated or treated with 0.1μg/ml LPS for 48h. (A) Representative flow cytometry for the ROS-detecting dye H2DCFDA with MFI values and summary graph with MFI+SEM (n=6 donors). (B) Summary graph for nitroblue tetrazolium+SEM (n=6). (C) p40phox, p47phox, p67phox, gp91phox, p22phox and (D) NOS2 mRNA expression. Fold mRNA change compared to scrambled siRNA-transfected, untreated cells+SEM (n=6). (E-G) MDMs were polarized into M1 macrophages, transfected with scrambled or the indicated siRNA, and then treated with 0.1μg/ml LPS for 48h. p40phox, p47phox, p67phox, and NOS2 protein expression was assessed by: (E) Western blot or (F) flow cytometry with MFI+SEM (n=6). (G) Cells were cultured with E. faecalis. CFU+SEM (n=8). (H-K) MDMs were polarized into M1 macrophages, transfected with scrambled or IRF5 siRNA±p47phox-, p67phox- or NOS2-expressing vectors or empty vector (EV) then left untreated or treated with 0.1μg/ml LPS for 48h. (H) p47phox and p67phox protein (n=6), (I) ROS (n=6), and (J) NOS2 protein (n=6) expression was assessed by flow cytometry. MFI+SEM. (K) Cells (n=8, similar results in an independent n=4) were cultured with E. faecalis. CFU+SEM. Significance is compared to IRF5 siRNA, EV-transfected cells or as indicated. Tx, treatment; scr, scrambled. **, p<0.01; ***, p<0.001; †, p<1×10−4; ††, p<1×10−5.
We next sought to confirm that the protein pathways encoded by the antimicrobial transcripts regulated by IRF5 (Fig 3C–D) were required for optimal bacterial clearance in LPS-treated M1 macrophages. We confirmed silencing of p40phox, p47phox, p67phox and NOS2 protein by two independent approaches: Western blot (Fig. 3E) and flow cytometry (Fig. 3F). We will use flow cytometry to examine these proteins in the experiments that follow. Each protein was required for optimal clearance of E. faecalis (Fig. 3G), AIEC, S. aureus and S. Typhimurium (data not shown). To clearly establish that IRF5 contributes to bacterial clearance through upregulation of the implicated NADPH oxidase subunits and NOS2, we transfected p47phox and p67phox, the IRF5-dependent NADPH oxidase subunits that contributed most to bacterial clearance in Fig. 3G, into IRF5-deficient LPS-treated M1 macrophages to restore expression of these proteins to physiological levels (Fig. 3H). We confirmed that the complementation of p47phox and p67phox expression restored ROS production in LPS-treated M1 macrophages (Fig. 3I). We also complemented NOS2 expression to physiological levels in IRF5-deficient LPS-treated M1 macrophages (Fig. 3J). Restoration of each protein partially rescued bacterial clearance in IRF5-deficient LPS-treated M1 macrophages (Fig. 3K). Taken together, in LPS-treated M1 macrophages, IRF5 is required for optimal ROS production through the upregulation of NADPH oxidase subunits and for optimal induction of the RNS-producing enzyme NOS2, thereby resulting in more efficient bacterial clearance.
IRF5-mediated bacterial clearance in PRR-stimulated M1 macrophages is dependent on autophagy proteins.
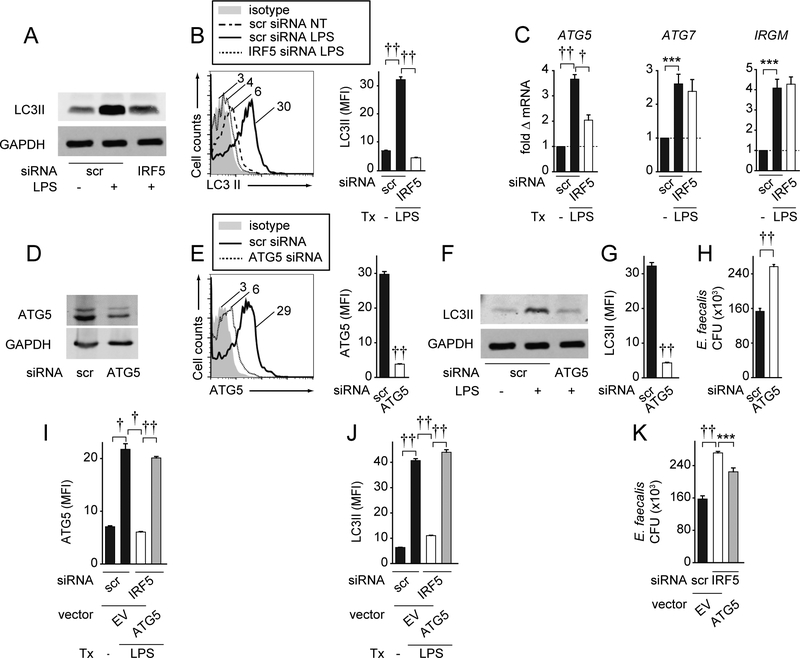
Autophagy is another key bacterial clearance mechanism regulated through PRR stimulation (1). One report showed that IRF5 impairs Hepatitis C virus-induced autophagy in MH-14 and C-5B transfected cell lines (37); impaired autophagy could reduce effective bacterial clearance. However, how IRF5 regulates autophagy following PRR stimulation in human macrophages has not been reported. We found that IRF5 was, in fact, required for optimal induction of the autophagy marker LC3II upon LPS treatment of M1 macrophages per both Western blot (Fig. 4A) and intracellular flow cytometry (Fig. 4B). To define mechanisms wherein IRF5 enhances autophagy, we examined the expression of the autophagy-related genes ATG5, ATG7, and IRGM in M1-polarized macrophages followed by IRF5 knockdown. ATG5 expression increased upon LPS treatment of M1 macrophages, and optimal ATG5 expression induction required IRF5 (Fig. 4C). In contrast, LPS-induced ATG7 and IRGM did not require IRF5 (Fig. 4C). To determine if ATG5 contributes to intracellular bacterial clearance in M1 macrophages, we silenced ATG5, and confirmed reduced ATG5 protein expression through two independent approaches, Western blot and flow cytometry (Fig. 4D&E). We ensured that ATG5 knockdown decreased the expression of the autophagy marker LC3II in LPS-treated M1 macrophages as assessed by both Western blot and flow cytometry (Fig. 4F&G). We next ensured that ATG5 was required for optimal E. faecalis clearance in human LPS-treated M1 macrophages (Fig. 4H). Finally, complementing ATG5 expression in IRF5-deficient LPS-treated M1 macrophages to physiological levels (Fig. 4I), restored LC3II expression (Fig. 4J) and partially rescued E. faecalis clearance (Fig. 4K). Taken together, IRF5 is required for optimal upregulation of ATG5 expression, which in turn, regulates autophagy pathways and bacterial clearance in human M1 macrophages.
Figure 4. IRF5 is required for optimal induction of the autophagy protein ATG5, thereby contributing to bacterial clearance in PRR-stimulated M1 macrophages.
(A-C) MDMs were polarized into M1 macrophages, transfected with scrambled or IRF5 siRNA, and then treated with 0.1μg/ml LPS for 48h. (A-B) LC3II protein expression was assessed by: (A) Western blot or (B) flow cytometry with representative plot and summary graph of MFI+SEM (n=6 donors). (C) ATG5, ATG7, and IRGM fold mRNA induction compared to scrambled siRNA-transfected, untreated cells+SEM (n=6). (D-H) MDMs were polarized into M1 macrophages, transfected with scrambled or ATG5 siRNA, then treated with 0.1μg/ml LPS for 48h. ATG5 and LC3II protein expression was assessed by: (D,F) Western blot, or (E,G) flow cytometry with MFI+SEM (n=6). (H) Cells were cultured with E. faecalis. CFU+SEM (n=8). (I-K) MDMs were polarized into M1 macrophages, transfected with scrambled or IRF5 siRNA± ATG5-expressing vector or empty vector (EV), then treated with 0.1μg/ml LPS for 48h. (I) ATG5 (n=6) and (J) LC3II (n=6) protein expression was assessed by flow cytometry with MFI+SEM. (K) Cells (n=8, similar results in an independent n=4) were cultured with E. faecalis. CFU+SEM. Scr, scrambled; tx, treatment. ***, p<0.001; †, p<1×10−4; ††, p<1×10−5.
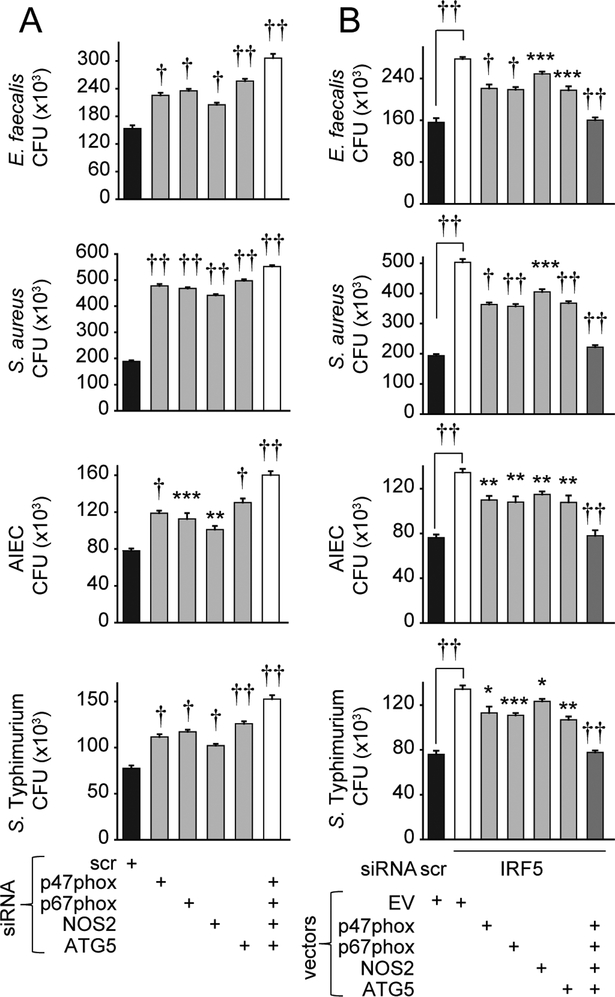
Complementing each of the IRF5-dependent antimicrobial pathways alone only partially rescued bacterial clearance in IRF5-deficient macrophages (Figs. 3K & 4K). We therefore asked if IRF5-dependent p47phox, p67phox, NOS2 and ATG5 upregulation cooperate to mediate optimal bacterial clearance in LPS-treated M1 macrophages. Clearance of each E. faecalis, S. aureus, AIEC and S. Typhimurium was most impaired when all the pathways were disrupted in combination (Fig. 5A). Consistently, restoring expression of the IRF5-dependent proteins in combination in LPS-treated M1 macrophages deficient in IRF5 more fully rescued the impaired bacterial clearance relative to that observed with restoration of each protein alone (Fig. 5B). Furthermore, and consistent with the requirement for the IRF5-dependent MAPK, NFκB and Akt2 pathways in optimal bacterial clearance in LPS-treated M1 macrophages, MAPK and NFκB pathway knockdown or Akt2 knockdown in M1-differentiated macrophages reduced the LPS-induced ROS and p47phox, p67phox pathways, NOS2 expression, and LC3II and ATG5 pathways observed in M1 macrophages (Supplementary Fig. 2A–E). Therefore, IRF5 regulates intracellular bacterial clearance in human macrophages through at least three mechanisms: ROS, RNS and autophagy pathway induction, which in turn, are regulated by IRF5-dependent MAPK, NFκB and Akt2 signaling. Importantly, these IRF5-dependent antimicrobial mechanisms cooperate to mediate bacterial clearance in LPS-treated M1 macrophages.
Figure 5. ROS, RNS and autophagy pathways cooperate for optimal IRF5-dependent bacterial clearance.
(A) MDMs (n=4 donors) were transfected with scrambled or the indicated siRNA, polarized into M1 macrophages, then treated with 0.1μg/ml LPS for 48h. (B) MDMs (n=8) were polarized into M1 macrophages, transfected with scrambled or IRF5 siRNA±p47phox-, p67phox-, NOS2-, or ATG5-expressing vectors alone or in combination, or empty vector (EV), then treated with 0.1μg/ml LPS for 48h. (A-B) Cells were cultured with E. faecalis, AIEC, S. aureus and S. Typhimurium. CFU+SEM. Significance is compared in ‘A’ to scrambled siRNA-transfected cells, or in ‘B’ to IRF5 siRNA, EV-transfected cells or as indicated. Scr, scrambled. *, p<0.05; **, p<0.01; ***, p<0.001; †, p<1×10−4; ††, p<1×10−5.
IRF5 is required for bacterial clearance pathways in IFNγ-treated macrophages
To ensure that IRF5 was required for PRR-induced outcomes in macrophages differentiated to an M1-like phenotype through another approach, we polarized MDMs with IFNγ (38), and then knocked down IRF5 and treated cells with LPS. IRF5 was required for optimal levels of bacterial clearance (Supplementary Fig. 2F), signaling (Supplementary Fig. 2G–I), and bacterial clearance mechanisms (Supplementary Fig. 2J) under these conditions. We also verified that IRF5-dependent MAPK, NFκB and Akt2 signaling pathways were required for bacterial clearance in these IFNγ-polarized macrophages (Supplementary Fig. 2K). Taken together, IRF5 deficiency in IFNγ-polarized macrophages results in decreased PRR-induced bacterial clearance-related outcomes.
IRF5 deficiency prior to M1 polarization results in decreased bacterial clearance pathways
Under physiological conditions cells from immune-mediated disease protective IRF5 genetic variant carriers exhibit reduced IRF5 expression throughout the immune response, including before macrophage polarization. We therefore wanted to confirm that the most important outcomes we had defined also demonstrated dependency on IRF5 when IRF5 was knocked down prior to M1 polarization. We first assessed if the IRF5-dependent signaling pathways we had identified were required for M1 differentiation. We effectively knocked down intermediates essential for MAPK, NFκB and Akt2 pathway activation as evidenced by lack of activation of the respective pathways (Supplementary Fig. 3A–C) and ensured that cell viability was intact (Supplementary Fig. 3D). MAPK and NFκB pathways were essential for human M1 macrophage polarization (Supplementary Fig. 3E), and in fact, contributed to M1 polarization to a similar degree as did the Akt2 pathway (Supplementary Fig. 3E), which we had previously identified to be required for optimal M1 polarization (10). We then examined bacterial clearance pathways and observed that PRR-induced bacterial clearance (Supplementary Fig. 3F), activation of signaling pathways (Supplementary Fig. 3G–I), and bacterial clearance mechanisms (Supplementary Fig. 3J–O) were dependent on IRF5 under these conditions. Furthermore, restoration of ROS, RNS and autophagy pathways in cells where IRF5 was knocked down prior to M1 polarization restored bacterial clearance (Supplementary Fig. 3P). Therefore, IRF5-dependent signaling pathways are required for M1 differentiation, and IRF5 deficiency prior to M1 differentiation results in defects in both M1 polarization and the induction of antibacterial clearance mechanisms observed in M1 macrophages.
PRR-stimulated MDMs and M1 macrophages from immune-mediated disease-protective IRF5 rs2004640/rs2280714 GG/CC carriers show decreased bacterial clearance and ROS, RNS and autophagy pathway induction relative to TT/TT carriers.
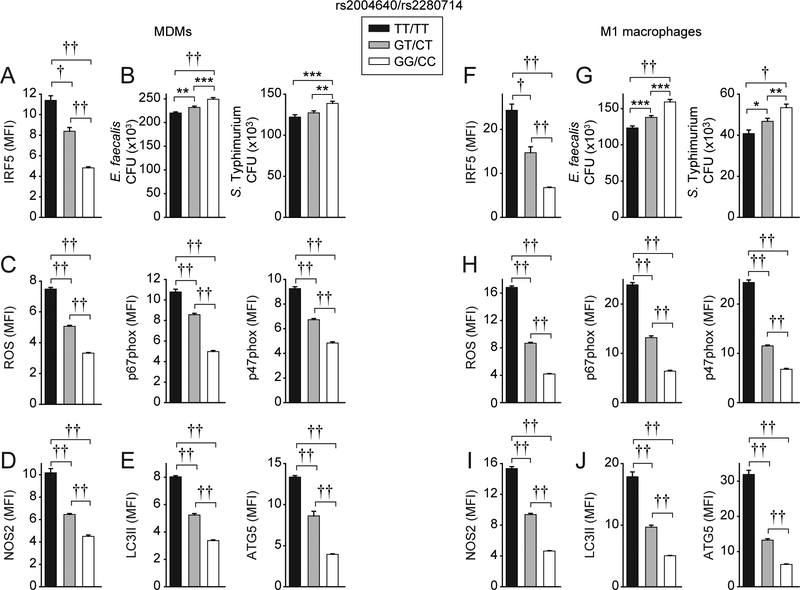
Given our results that IRF5 is required for bacterial clearance and relevant mechanisms in human macrophages, we hypothesized that immune-mediated disease protective rs2004640/rs2280714 GG/CC carrier macrophages that show reduced IRF5 expression (10, 14), which we confirmed in both undifferentiated MDMs (Fig. 6A) and in macrophages following M1 polarization and LPS stimulation (Fig. 6F), would show decreased bacterial clearance. LPS-treated undifferentiated MDMs and M1-differentiated macrophages from these carriers showed decreased E. faecalis and S. Typhimurium clearance relative to TT/TT carriers, with GT/CT heterozygotes showing an intermediate phenotype (Fig. 6B&G). Consistently, GG/CC carrier LPS-treated undifferentiated MDMs and M1 macrophages showed decreased induction of each of the IRF5-dependent mechanisms we had identified, including ROS and p47phox and p67phox (Fig. 6C&H), NOS2 (Fig. 6D&I), and LC3II and ATG5 (Fig. 6E&J) pathways. Taken together, these data indicate that rs2004640/rs2280714 GG/CC low IRF5-expressing undifferentiated MDMs and M1 macrophages show decreased bacterial clearance and bacterial clearance mechanisms relative to TT/TT immune-mediated disease risk carrier cells.
Figure 6. MDMs and M1 macrophages from immune-mediated disease-protective rs2004640/rs2280714 GG/CC carriers show reduced intracellular bacterial clearance and ROS, RNS and autophagy pathway induction.
MDMs from rs2004640/rs2280714 TT/TT, GT/CT and GG/CC carriers (n=15/genotype) were left undifferentiated or were differentiated under M1 polarizing conditions and then treated with 0.1 μg/ml LPS for 48h. (A,F) IRF5 expression was examined by flow cytometry. MFI+SEM. (B,G) Cells were cultured with E. faecalis or S. Typhimurium. CFU+SEM. (C-E, H-J) Summarized MFI of ROS or the indicated proteins as assessed by flow cytometry+SEM. *, p<0.05; **, p<0.01; ***, p<0.001; †, p<1×10−4; ††, p<1×10−5.
Modulation in IRF5 expression by the rs2004640/rs2280714 variant accounts for the genotype-dependent differences in bacterial clearance mechanisms.
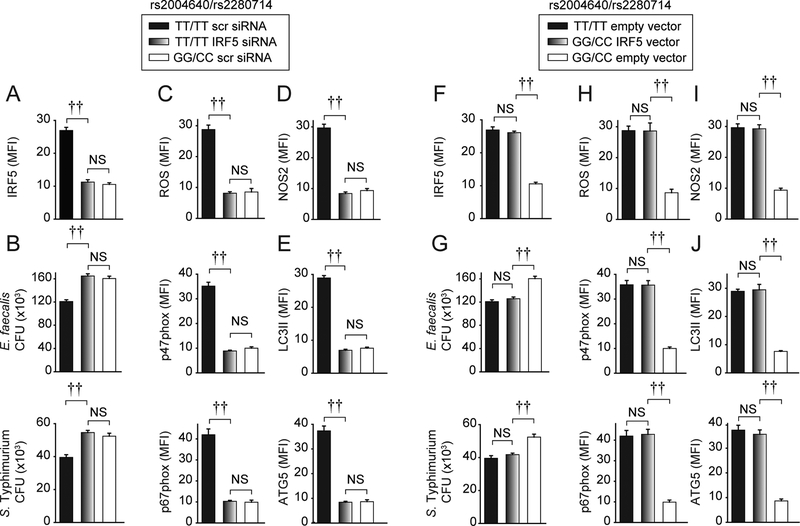
We next assessed if modulation in IRF5 expression levels by rs2004640/rs2280714 accounts for the genotype-dependent regulation of bacterial clearance. We used a knockdown approach to decrease IRF5 levels in macrophages from high IRF5-expressing rs2004640/rs2280714 TT/TT carriers to the levels observed in GG/CC carriers (Fig. 7A). This reduced bacterial clearance (Fig. 7B) and bacterial clearance mechanisms (Fig. 7C–E) to levels comparable to GG/CC carrier LPS-treated M1 macrophages. We also utilized a complementary approach and transfected an IRF5-expressing vector in low IRF5-expressing GG/CC carrier macrophages to increase IRF5 expression to levels observed in TT/TT carriers (Fig. 7F). This increased bacterial clearance (Fig. 7G) and relevant mechanisms (Fig. 7H–J) to levels comparable to those observed in TT/TT carrier LPS-treated M1 macrophages. Taken together, the modulation in IRF5 expression levels by the rs2004640/rs2280714 variant accounts for the differences in IRF5-dependent bacterial clearance mechanisms across the rs2004640/rs2280714 genotypes in LPS-treated M1 macrophages.
Figure 7. Rs2004640/rs2280714 genotype-dependent regulation of bacterial clearance, ROS, RNS and autophagy proteins is due to IRF5 expression modulation.
MDMs from rs2004640/rs2280714 TT/TT or GG/CC carriers (n=15/genotype) were transfected with 25 nM scrambled or IRF5 siRNA (A-E), or empty vector or IRF5 vector (F-J), and then polarized to M1 macrophages and treated with 0.1μg/ml LPS for 48h. (A,F) Summarized MFI of IRF5 as assessed by flow cytometry+SEM. (B,G) Cells were cultured with E. faecalis or S. Typhimurium. CFU+SEM. (C-E, H-J) Summarized MFI of ROS or the indicated proteins as assessed by flow cytometry+SEM. NS, not significant; scr, scrambled. ††, p<1×10−5.
Discussion
The immune system must effectively balance handling microbial infections while simultaneously controlling the resulting pro-inflammatory responses. Consistently, polymorphisms resulting in less microbial-induced inflammation can reduce risk of immune-mediated diseases, but simultaneously be detrimental to optimal clearance of infectious challenges. In this study, we find that IRF5, a protein required for PRR-induced cytokines (8,9) and associated with multiple immune-mediated diseases (15), is also required for optimal bacterial clearance in primary human M1-differentiated macrophages (Supplementary Fig. 4). We identify mechanisms through which IRF5 contributes to bacterial clearance, including ROS production mediated by p40phox, p47phox, p67phox upregulation, induction of NOS2, and induction of autophagy pathways mediated through ATG5 upregulation. We find that these pathways cooperate for optimal bacterial clearance. Furthermore, the induction of antimicrobial pathways and bacterial clearance in LPS-treated M1 macrophages required IRF5-dependent MAPK, NFκB, and Akt2 pathways. Importantly, bacterial clearance and the identified IRF5-dependent clearance mechanisms were modulated by IRF5 genotype, which regulates IRF5 expression, thereby highlighting that carriers of these low-expressing IRF5 polymorphisms might be at increased risk for bacterial infection.
Whereas IRF5 regulates M1 macrophage polarization (9,10,30), and has not generally been associated with M2 polarization (9,30), we identify that IRF5 is required for certain M2-associated chemokines and cytokines in human macrophages, while other M2 markers are IRF5-independent (Supplementary Fig. 1A&B). A few studies suggest that IRF5 might be required for aspects of M2 polarization. They include a report that IRF5 is upregulated in M2 human macrophages (39) and another describing a requirement for IRF5 in secretion of IL10, an M2-associated cytokine, in PRR-stimulated mouse BMDMs (40), which we also observe in human MDMs (16). A reverse regulation of IL10 by IRF5 was observed in another study of human macrophages (9). These discrepancies might be due to differences in conditions used for macrophage polarization, differences in concentrations, origins and the duration of the PRR stimuli used, or other differences in experimental conditions. The differential IRF5 dependency in select M2 macrophage outcomes may reflect that M2 polarization consists of a spectrum of subtypes, arising as an outcome of different stimulation conditions activating distinct signaling pathways (41). For example, endotoxin tolerance upregulates some (e.g. CCL22), but not all (e.g. CD206) M2 markers in human myeloid cells (42). Importantly, we found that IRF5 was required for both LPS-induced M1- and M2-associated cytokines (Supplementary Fig. 1B), highlighting that IRF5 has a central role in PRR-induced cytokines regardless of macrophage polarization. With respect to the mechanisms through which IRF5 regulates M1 polarization, we had previously identified that IRF5-dependent Akt2 activation was required for the enhanced glycolysis in M1 macrophages (10). We now find that IRF5-dependent MAPK and NFκB signaling is also required for optimal M1 polarization (Supplementary Fig. 3E). Consistent with its contribution to proximal signaling pathways in human macrophages, IRF5 is phosphorylated within 15min of LPS treatment (Supplementary Fig. 1E). The NFκB pathway is required for M1 polarization of mouse macrophages (43), but to our knowledge, its requirement for human M1 polarization has not yet been reported. The requirement for IRF5 in MAPK and NFκB signaling has demonstrated variable results; IRF5 did not contribute to activation of these pathways in mouse B cells (8), yet it was required for these pathways in human macrophages (10) and interacted with intermediates activating these pathways in 293T cell lines, and mouse and human macrophages (10–12). These differences might be partly dependent on cell type, species and stimuli.
We identify that IRF5 and IRF5 polymorphisms regulate autophagy (Fig. 4–7). As autophagy mediates not only microbial clearance but numerous other pathways important for inflammatory and infectious diseases (e.g. cellular stress, apoptosis and inflammasome regulation (44)), IRF5 may play a role in these pathways as well. Moreover, we observe that autophagy, ROS, and RNS pathways cooperate to mediate the IRF5-dependent role in bacterial clearance. These findings are consistent with the observation that mice with combined deficiency in NADPH oxidase and NOS2 show worse bacterial clearance in vivo compared to single knockout mice (35). Importantly, we found that complementing each pathway alone in IRF5-deficient M1 macrophages only partially restores bacterial clearance, whereas combined complementation optimizes clearance in IRF5-deficient cells (Fig. 5). Such regulation of broad processes by IRF5 may be in part due to the ability of IRF5 to affect multiple proximal signaling pathways, including MAPKs, NFκB and Akt2 pathways, and multiple transcriptional programs (8,10–12). Importantly, IRF5 knockdown did not globally affect macrophage function as Akt1 signaling (Supplementary Fig. 1K) and select M2-associated markers (Supplementary Fig. 1A) were intact. While macrophages can demonstrate a spectrum of differentiation conditions, we utilized common IFNγ- and LPS-conditioned macrophages, and then exposed the cells to additional LPS treatment to simulate conditions of ongoing microbial exposure. As such conditions may not be universally encountered, we also examined and observed the dependency on IRF5 for microbial clearance in undifferentiated macrophages and in macrophages differentiated with IFNγ alone (Supplementary Fig. 2F–K). Given its ability to regulate multiple bacterial clearance pathways simultaneously, IRF5 might represent an effective therapeutic target when treating infectious diseases.
We previously found that IRF5 is a major genetic determinant of the inter-individual variance in PRR-induced cytokine secretion (16). This is in part due to the similar frequencies of the IRF5 rs2004640 risk and non-risk alleles, such that the three genotypes are commonly distributed across the population. In this study we find that IRF5 immune-mediated disease risk variants are associated with increased bacterial-induced ROS, RNS and autophagy-associated proteins, and increased macrophage-mediated clearance of multiple bacteria. Conversely, this would imply that the IRF5 variants protective for immune-mediated diseases may be disadvantageous in select infectious diseases. Whether common genetic variants confer a slight increase in susceptibility to transient bacterial infections can be difficult to assess and track (45) and is therefore not as well studied as how these variants affect chronic immune-mediated diseases or chronic infectious diseases (e.g. M. tuberculosis). However, that PRR-associated signaling intermediates affect acute bacterial clearance is evidenced by studies involving individuals harboring dramatic loss-of-function coding mutations resulting in increased frequency of infections, as is observed in MYD88 and IRAK4 mutation carriers who show increased susceptibility to pyogenic infections (46). Furthermore, uncommon mutations significantly reducing ROS production and autophagy, pathways that we identify to be regulated by IRF5, are associated with increased risk for infectious diseases (2,47). The dichotomy in regulation of inflammatory versus infectious disease susceptibility has been observed for SOCS1/LITAF/RMI2 and IL12B region polymorphisms associated with IBD (2), but protective for leprosy (3,48). IL12B polymorphisms are associated with risk for M. leprae, M. tuberculosis and S. enteridis infections (49). IRF5 regulates multiple other cytokines in addition to IL12 (Supplementary Fig. 1B), suggesting that variants in IRF5 might affect the balance between inflammatory and infectious diseases through additional pathways. Our findings identify critical functions for IRF5 and IRF5 immune-mediated disease variants in immunological processes critical for bacterial clearance. These findings advance our understanding of the emerging roles of IRF5 variants in the balance of immune- and microbial-mediated diseases and indicate that when considering targeting this pathway for immune-mediated diseases, caution might be needed due to potential risk for microbial infections.
Supplementary Material
Acknowledgements:
We thank Tony Eissa, Celine DerMardirossian and Emiko Mizoguchi for reagents, and the blood donors for their participation.
This work was supported by R01AI120369, R01DK099097, DK062422, and DKP30–34989.
Abbreviations:
- IRF
interferon-regulatory factor
- NOD
nucleotide-oligomerization-domain
- PRR
pattern-recognition receptor
- MDMs
monocyte-derived macrophages
References
- 1.Abraham C, and Medzhitov R. 2011. Interactions between the host innate immune system and microbes in inflammatory bowel disease. Gastroenterology 140: 1729–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. 2012. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491: 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu H, Irwanto A, Fu X, Yu G, Yu Y, Sun Y, Wang C, Wang Z, Okada Y, Low H, et al. 2015. Discovery of six new susceptibility loci and analysis of pleiotropic effects in leprosy. Nat Genet 47: 267–271. [DOI] [PubMed] [Google Scholar]
- 4.Wajant H, Pfizenmaier K, and Scheurich P. 2003. Tumor necrosis factor signaling. Cell Death Differ 10: 45–65. [DOI] [PubMed] [Google Scholar]
- 5.Pasparakis M, Alexopoulou L, Episkopou V, and Kollias G. 1996. Immune and inflammatory responses in TNF alpha-deficient mice: a critical requirement for TNF alpha in the formation of primary B cell follicles, follicular dendritic cell networks and germinal centers, and in the maturation of the humoral immune response. J Exp Med 184: 1397–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lech M, Kantner C, Kulkarni OP, Ryu M, Vlasova E, Heesemann J, Anz D, Endres S, Kobayashi KS, Flavell RA, et al. 2011. Interleukin-1 receptor-associated kinase-M suppresses systemic lupus erythematosus. Ann Rheum Dis 70: 2207–2217. [DOI] [PubMed] [Google Scholar]
- 7.Deng JC, Cheng G, Newstead MW, Zeng X, Kobayashi K, Flavell RA, and Standiford TJ. 2006. Sepsis-induced suppression of lung innate immunity is mediated by IRAK-M. J Clin Invest 116: 2532–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takaoka A, Yanai H, Kondo S, Duncan G, Negishi H, Mizutani T, Kano S, Honda K, Ohba Y, Mak TW, et al. 2005. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature 434: 243–249. [DOI] [PubMed] [Google Scholar]
- 9.Krausgruber T, Blazek K, Smallie T, Alzabin S, Lockstone H, Sahgal N, Hussell T, Feldmann M, and Udalova IA. 2011. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat Immunol 12: 231–238. [DOI] [PubMed] [Google Scholar]
- 10.Hedl M, Yan J, and Abraham C. 2016. IRF5 and IRF5 Disease-Risk Variants Increase Glycolysis and Human M1 Macrophage Polarization by Regulating Proximal Signaling and Akt2 Activation. Cell Rep 16: 2442–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue M, Arikawa T, Chen YH, Moriwaki Y, Price M, Brown M, Perfect JR, and Shinohara ML. 2014. T cells down-regulate macrophage TNF production by IRAK1-mediated IL-10 expression and control innate hyperinflammation. Proc Natl Acad Sci U S A 111: 5295–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balkhi MY, Fitzgerald KA, and Pitha PM. 2008. Functional regulation of MyD88-activated interferon regulatory factor 5 by K63-linked polyubiquitination. Mol Cell Biol 28: 7296–7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barnes BJ, Kellum MJ, Field AE, and Pitha PM. 2002. Multiple regulatory domains of IRF-5 control activation, cellular localization, and induction of chemokines that mediate recruitment of T lymphocytes. Mol Cell Biol 22: 5721–5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham RR, Kozyrev SV, Baechler EC, Reddy MV, Plenge RM, Bauer JW, Ortmann WA, Koeuth T, Gonzalez Escribano MF, Pons-Estel B, et al. 2006. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat Genet 38: 550–555. [DOI] [PubMed] [Google Scholar]
- 15.Eames HL, Corbin AL, and Udalova IA. 2016. Interferon regulatory factor 5 in human autoimmunity and murine models of autoimmune disease. Transl Res 167: 167–182. [DOI] [PubMed] [Google Scholar]
- 16.Hedl M, and Abraham C. 2012. IRF5 risk polymorphisms contribute to interindividual variance in pattern recognition receptor-mediated cytokine secretion in human monocyte-derived cells. J Immunol 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niewold TB, Kelly JA, Kariuki SN, Franek BS, Kumar AA, Kaufman KM, Thomas K, Walker D, Kamp S, Frost JM, et al. 2012. IRF5 haplotypes demonstrate diverse serological associations which predict serum interferon alpha activity and explain the majority of the genetic association with systemic lupus erythematosus. Ann Rheum Dis 71: 463–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, and Cheung VG. 2004. Genetic analysis of genome-wide variation in human gene expression. Nature 430: 743–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paun A, Bankoti R, Joshi T, Pitha PM, and Stager S. 2011. Critical role of IRF-5 in the development of T helper 1 responses to Leishmania donovani infection. PLoS Pathog 7: e1001246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paun A, Reinert JT, Jiang Z, Medin C, Balkhi MY, Fitzgerald KA, and Pitha PM. 2008. Functional characterization of murine interferon regulatory factor 5 (IRF-5) and its role in the innate antiviral response. J Biol Chem 283: 14295–14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pandey AK, Yang Y, Jiang Z, Fortune SM, Coulombe F, Behr MA, Fitzgerald KA, Sassetti CM, and Kelliher MA. 2009. NOD2, RIP2 and IRF5 play a critical role in the type I interferon response to Mycobacterium tuberculosis. PLoS Pathog 5: e1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gratz N, Hartweger H, Matt U, Kratochvill F, Janos M, Sigel S, Drobits B, Li XD, Knapp S, and Kovarik P. 2011. Type I interferon production induced by Streptococcus pyogenes-derived nucleic acids is required for host protection. PLoS Pathog 7: e1001345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castiglia V, Piersigilli A, Ebner F, Janos M, Goldmann O, Dambock U, Kroger A, Weiss S, Knapp S, Jamieson AM, et al. 2016. Type I Interferon Signaling Prevents IL-1beta-Driven Lethal Systemic Hyperinflammation during Invasive Bacterial Infection of Soft Tissue. Cell Host Microbe 19: 375–387. [DOI] [PubMed] [Google Scholar]
- 24.Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, et al. 2013. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A 110: 3507–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, and Finkel T. 2008. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A 105: 3374–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Musial A, and Eissa NT. 2001. Inducible nitric-oxide synthase is regulated by the proteasome degradation pathway. J Biol Chem 276: 24268–24273. [DOI] [PubMed] [Google Scholar]
- 27.Gianni D, DerMardirossian C, and Bokoch GM. 2011. Direct interaction between Tks proteins and the N-terminal proline-rich region (PRR) of NoxA1 mediates Nox1-dependent ROS generation. Eur J Cell Biol 90: 164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munoz M, Cedeno R, Rodriguez J, van der Knaap WPW, Mialhe E, and Bachere E. 2000. Measurement of reactive oxygen intermediate production in haemocytes of the penaeid shrimp, Penaeus vannamei. Aquaculture 191: 89–107. [Google Scholar]
- 29.Benoit M, Desnues B, and Mege JL. 2008. Macrophage polarization in bacterial infections. J Immunol 181: 3733–3739. [DOI] [PubMed] [Google Scholar]
- 30.Dalmas E, Toubal A, Alzaid F, Blazek K, Eames HL, Lebozec K, Pini M, Hainault I, Montastier E, Denis RG, et al. 2015. Irf5 deficiency in macrophages promotes beneficial adipose tissue expansion and insulin sensitivity during obesity. Nat Med. [DOI] [PubMed] [Google Scholar]
- 31.Saeys Y, Gassen SV, and Lambrecht BN. 2016. Computational flow cytometry: helping to make sense of high-dimensional immunology data. Nat Rev Immunol 16: 449–462. [DOI] [PubMed] [Google Scholar]
- 32.Lahiri A, and Abraham C. 2014. Activation of pattern recognition receptors up-regulates metallothioneins, thereby increasing intracellular accumulation of zinc, autophagy, and bacterial clearance by macrophages. Gastroenterology 147: 835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foster SL, Hargreaves DC, and Medzhitov R. 2007. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature 447: 972–978. [DOI] [PubMed] [Google Scholar]
- 34.Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, Bringer MA, Swidsinski A, Beaugerie L, and Colombel JF. 2004. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology 127: 412–421. [DOI] [PubMed] [Google Scholar]
- 35.Shiloh MU, MacMicking JD, Nicholson S, Brause JE, Potter S, Marino M, Fang F, Dinauer M, and Nathan C. 1999. Phenotype of mice and macrophages deficient in both phagocyte oxidase and inducible nitric oxide synthase. Immunity 10: 29–38. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Zhang P, Wang C, Han C, Meng J, Liu X, Xu S, Li N, Wang Q, Shi X, et al. 2013. Immune responsive gene 1 (IRG1) promotes endotoxin tolerance by increasing A20 expression in macrophages through reactive oxygen species. J Biol Chem 288: 16225–16234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cevik O, Li D, Baljinnyam E, Manvar D, Pimenta EM, Waris G, Barnes BJ, and Kaushik-Basu N. 2017. Interferon regulatory factor 5 (IRF5) suppresses hepatitis C virus (HCV) replication and HCV-associated hepatocellular carcinoma. J Biol Chem 292: 21676–21689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T, et al. 2014. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41: 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lacey DC, Achuthan A, Fleetwood AJ, Dinh H, Roiniotis J, Scholz GM, Chang MW, Beckman SK, Cook AD, and Hamilton JA. 2012. Defining GM-CSF- and macrophage-CSF-dependent macrophage responses by in vitro models. J Immunol 188: 5752–5765. [DOI] [PubMed] [Google Scholar]
- 40.Watkins AA, Yasuda K, Wilson GE, Aprahamian T, Xie Y, Maganto-Garcia E, Shukla P, Oberlander L, Laskow B, Menn-Josephy H, et al. 2015. IRF5 deficiency ameliorates lupus but promotes atherosclerosis and metabolic dysfunction in a mouse model of lupus-associated atherosclerosis. J Immunol 194: 1467–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, and Locati M. 2004. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol 25: 677–686. [DOI] [PubMed] [Google Scholar]
- 42.Pena OM, Pistolic J, Raj D, Fjell CD, and Hancock RE. 2011. Endotoxin tolerance represents a distinctive state of alternative polarization (M2) in human mononuclear cells. J Immunol 186: 7243–7254. [DOI] [PubMed] [Google Scholar]
- 43.Ricote M, Li AC, Willson TM, Kelly CJ, and Glass CK. 1998. The peroxisome proliferator-activated receptor-gamma is a negative regulator of macrophage activation. Nature 391: 79–82. [DOI] [PubMed] [Google Scholar]
- 44.Rubinsztein DC, Bento CF, and Deretic V. 2015. Therapeutic targeting of autophagy in neurodegenerative and infectious diseases. J Exp Med 212: 979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lipoldova M, and Demant P. 2006. Genetic susceptibility to infectious disease: lessons from mouse models of leishmaniasis. Nat Rev Genet 7: 294–305. [DOI] [PubMed] [Google Scholar]
- 46.Casanova JL, Abel L, and Quintana-Murci L. 2011. Human TLRs and IL-1Rs in host defense: natural insights from evolutionary, epidemiological, and clinical genetics. Annu Rev Immunol 29: 447–491. [DOI] [PubMed] [Google Scholar]
- 47.Newburger PE, Skalnik DG, Hopkins PJ, Eklund EA, and Curnutte JT. 1994. Mutations in the promoter region of the gene for gp91-phox in X-linked chronic granulomatous disease with decreased expression of cytochrome b558. J Clin Invest 94: 1205–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu H, Irwanto A, Tian H, Fu X, Yu Y, Yu G, Low H, Chu T, Li Y, Shi B, et al. 2012. Identification of IL18RAP/IL18R1 and IL12B as leprosy risk genes demonstrates shared pathogenesis between inflammation and infectious diseases. Am J Hum Genet 91: 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Picard C, Fieschi C, Altare F, Al-Jumaah S, Al-Hajjar S, Feinberg J, Dupuis S, Soudais C, Al-Mohsen IZ, Genin E, et al. 2002. Inherited interleukin-12 deficiency: IL12B genotype and clinical phenotype of 13 patients from six kindreds. Am J Hum Genet 70: 336–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.