Abstract
Kinase enzymes phosphorylate protein substrates in a highly ordered manner to control cell signaling. Unregulated kinase activity is associated with a variety of disease states, most notably cancer, making the characterization of kinase activity in cellulo critical to understand disease formation. However, the paucity of available tools has prevented a full mapping of the substrates and interacting proteins of kinases involved in cellular function. Recently we developed kinase-catalyzed crosslinking to covalently connect substrate and kinase in a phosphorylation-dependent manner. Here, we report a new method combining kinase-catalyzed crosslinking and immunoprecipitation (K-CLIP) to identify kinase-substrate pairs and kinase-associated proteins. K-CLIP was applied to the substrate p53, which is robustly phosphorylated. Both known and unknown kinases of p53 were isolated from cell lysates using K-CLIP. In follow-up validation studies, MRCKbeta was identified as a new p53 kinase. Beyond kinases, a variety of p53 and kinase-associated proteins were also identified using K-CLIP, which provided a snapshot of cellular interactions. The K-CLIP method represents an immediately useful chemical tool to identify kinase-substrate pairs and multi-proteins complexes in cells, which will embolden cell signaling research and enhance our understanding of kinase activity in normal and disease states.
Introduction
Phosphorylation plays a key role in regulating protein activity by altering enzymatic function, cellular localization, molecular interactions, among many others. In fact, the complicated network of protein-protein interactions mediating cell signaling are often controlled by phosphorylation events. Given the essential role of phosphorylation in cellular events, the kinase enzymes that control protein phosphorylation are highly regulated to assure proper cell function.1 Unregulated kinase activity is associated with a variety of disease states, making kinases a significant drug target.2 Characterizing kinases and protein phosphorylation generally represents a critical aspect of disease-related cell biology research.
Despite the governing role of kinases and protein phosphorylation in cell function, identifying the upstream kinase responsible for phosphorylating a specific protein substrate is a challenge. Signaling networks can have multiple complementary pathways, which can result in multiple kinases acting on a specific substrate depending on the cellular environment.3 In addition, the interaction between kinase and substrate is transient,4 which can complicate their characterization using traditional methods, such as immunoprecipitation or two-hybrid methods. Beyond kinase-substrate characterization, the multi-protein cellular complexes associated with kinase or phosphorylated substrate during phosphorylation represent essential players in cell biological events.5 Yet, identifying dynamic multi-protein complexes in the context of a specific phosphorylation event is also challenging. Methods to monitor both kinase-substrate pairs and the proteins in proximity to a kinase and substrate during dynamic phosphorylation have the potential to untangle the complicated web of protein-protein interactions governing signaling and other cellular events.
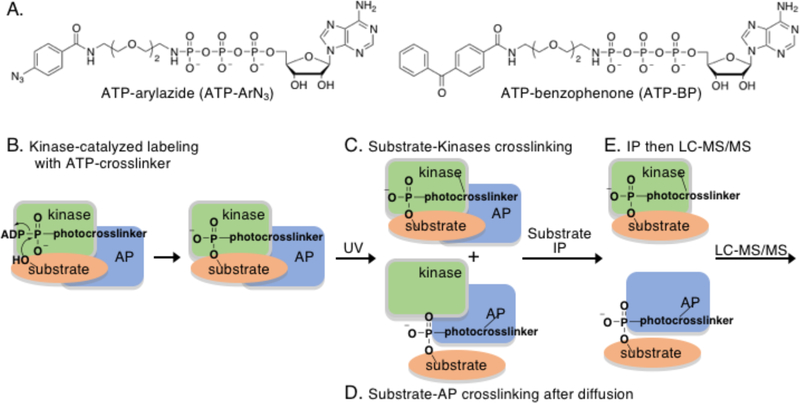
To address the critical need for kinase-substrate identification methods, several chemical approaches have been developed, including allele-specific kinase mutants and mechanism-based crosslinkers.6–7 As an alternative, we recently reported kinase-catalyzed crosslinking where kinase and substrate are covalently conjugated to stabilize the typically transient interaction and promote purification and identification.8–10 Kinase-catalyzed crosslinking utilizes a modified version of the ATP (adenosine 5’-triphosphate) cosubstrate where the γ-phosphate contains a photocrosslinking group (ATP-arylazide (ATP-ArN3) or ATP-benzophenone (ATP-BP), Figure 1A) separated by a linker.11 The ATP analog acts as a kinase cosubstrate resulting in the attachment of the photocrosslinking group onto the protein substrate (Figure 1B). Simultaneous UV irradiation during the kinase reaction activates the photoreactive crosslinking group for covalent conjugation of kinase to substrate (Figure 1C). The requirements of both photocrosslinker transfer to the substrate (Figure 1B) and photoactivated conjugation to the kinase (Figure 1C) makes kinase-catalyzed crosslinking a distinctive method for kinase-substrate stabilization and subsequent identification.
Figure 1.
A) The chemical structures of the two ATP-crosslinking analogs used in this work. B) Kinase-catalyzed labeling of a substrate with the ATP-crosslinking analog forms a substrate product with the crosslinker group covalently attached. C) Simultaneous UV irradiation results in covalent conjugation of the kinase and substrate through the photocrosslinking group. D) Because the kinase-substrate interaction is transient, a large fraction of the substrate is expected to dissociate and diffuse away from the kinase after catalysis. UV irradiation after diffusion will result in covalent attachment of the substrate to any substrate- and/or kinase-associated protein (AP). E) After kinase-catalyzed crosslinking, immunoprecipitation (IP) of the substrate protein will isolate both the attached kinase and associate proteins, which can be characterized by LC-MS/MS.
In addition to kinase-substrate conjugation, kinase-catalyzed crosslinking offers a second unique and powerful feature, which is the identification of associated proteins of the kinase or substrate during the phosphorylation event.10 Because kinase-substrate interactions are unstable and transient, the substrate is likely to dissociate from the kinase quickly after catalysis (Figure 1D). When the photocrosslinking group is UV-activated after kinase dissociation, the substrate will conjugate to proteins in close proximity to the phosphorylated site. Therefore, kinase-catalyzed crosslinking has the unique ability to covalently connect substrate to both its phosphorylating kinases and proteins associated with that kinase at the time of phosphorylation. Given the need for methods to characterize protein-protein interactions involved in dynamic kinase functions, kinase-catalyzed crosslinking offers a powerful platform to discover phosphorylation-mediated cellular complexes.
A requirement for productive use of kinase-catalysed crosslinking is a means to selectively isolate only conjugated proteins of interest among the sea of crosslinked complexes in lysates. In prior work, we used a biotinylated peptide substrates as a handle to isolate kinases specific to a single phosphosite.10 Here we combined crosslinking with immunoprecipitation in a method called K-CLIP (Kinase-catalyzed CrossLinking and ImmunoPrecipitation) to isolate and characterize select full-length substrate-conjugated complexes from a complex cellular mixture. Immunoprecipitation (IP) with an antibody selective to a desired substrate or kinase of interest will isolate only substrate- or kinase-bound complexes (Figure 1E). After K-CLIP, liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis will identify kinases and associated proteins covalently linked to the substrate or kinase.
To establish K-CLIP for identification of kinase-substrate complexes, p53 was tested here as a substrate model system. The p53 protein acts as a key tumor suppressor by inducing senescence or apoptosis through its transcriptional activation function.12 In fact, mutation of p53 is frequently observed in cancers.13 Beyond apoptosis, p53 has also been implicated in a variety of additional biological events, including DNA damage response, aging, and fertility. Due to its pivotal role in cell biology, p53 is tightly regulated, particularly through binding of MDM2 (murine double minute 2), which promotes ubiquitin-mediated proteomosomal degration.14–16 In addition, p53 is regulated through phosphorylation, with many characterized phosphorylation sites and kinases.17–18 With a variety of kinases documented, p53 represents an ideal model substrate to establish K-CLIP for kinase identification. Likewise, the interactome network of p53 has been extensively characterized, making p53 an ideal substrate to test the ability of K-CLIP to monitor associated proteins.
After K-CLIP with p53, two known (DNAPK and PKR) and one previously unknown (MRCKβ) kinases of p53 were identified. Several secondary confirmation studies were performed to validate MRCKβ as a p53 kinase. A variety of associated proteins were also observed. An interactome analysis documented that 81% of the K-CLIP protein hits are known to physically associate either directly or indirectly with p53. These results demonstrated the utility of K-CLIP to observe both active kinases and associated proteins of p53-kinase complexes. In addition, kinase-catalyzed crosslinking was also established to validate a suspected kinase-substrate pair in lysates. Importantly, the discovery of MRCKβ and new p53 interacting proteins lead to several novel biological hypothesis, including the revelation that MRCKβ governs p53 protein levels. Taken together, these studies substantiate kinase-catalyzed crosslinking and K-CLIP as powerful tools to validate suspected kinase substrates, identify unknown kinase-substrate pairs, monitor multi-protein complexes in cellular mixtures, and discover unanticipated mechanisms in cell biology.
Experimental Section
Cell culture procedures
RKO cells obtained from ATCC (ATCC CRL-2577) were grown in EMEM media supplemented with 10% FBS and 1% antibiotic-antimycotic. Cells were cultured in a 37°C incubator under a 5% CO2 environment with 95% relative humidity. For (±)-Nutlin-3 treatment, RKO cells (1.5 × 106) were seeded in a 75 cm2 tissue culture flasks in growth medium (10 mL) for 24 hours prior to treatment. (±)-Nutlin-3 (Cayman Chemicals, 10 mM stock solution in DMSO) was added to the media to produce final concentrations of either 10 μM or 20 μM and incubated for 24 hours. Control cells were treated with an equivalent amount of DMSO (Figure S1). After 24 hours, the growth media was removed, and the flask was rinsed with PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) before trypsinization and harvesting by centrifugation. Collected cells were washed with cold PBS buffer and then lysed immediately, as described below, or stored at −80 °C until needed. The nutlin-treated RKO cells were lysed in lysis buffer (200 μL; 50 mM HEPES, pH 8.0, 150 mM NaCl, 10% glycerol, 0.5% Triton X-100) containing freshly added protease inhibitor cocktail V (1X, Calbiochem) with rocking at 4 °C for 10 minutes. The soluble fraction was collected by centrifugation and the concentration of total protein was determined using a Bradford assay.19 Lysates were used immediately or stored at −80°C in single use aliquots.
Kinase-catalyzed crosslinking
Kinase-catalyzed crosslinking reactions contained untreated (4 mg total protein) or nutlin-treated RKO cell lysates (20 μg total protein for gel-based analysis or 500 μg for LC-MS/MS studies) in K-CLIP buffer (50 mM HEPES, pH 8.0, 150 mM NaCl, 50 mM KCl, 10 mM MgCl2 10 % glycerol, 0.5 % Triton X-100). The kinase reaction was initiated by addition of ATP (10 mM), ATP-ArN3 (10 mM), or ATP-BP (10 mM) on ice. Crosslinking was then initiated within 3–5 minutes of ATP or ATP analog addition by incubating the reaction mixtures for 2 hrs at 30 °C under a handheld UV lamp (365 nm). The final reaction volume was 25 μL. Heat denatured lysates were obtained by incubation of nutlin-treated lysates at 95°C for 5 minutes. The reaction products were separated using 10% SDS-PAGE, electrotransferred onto PVDF membrane (Immobilon-PSQ), and visualized using p53 (Santa Cruz, either rabbit polyclonal SC-6243 or mouse monoclonal SC-55476), DNA-PK (Cell Signaling, 4602P), or PLK3 (Genetex, GTX111495) antibodies.
Immunoprecipitation and Coimmunoprecipitation
For immunoprecipitation as part of a K-CLIP study, antibody-loaded agarose beads were first generated by initially washing protein A/G plus agarose beads (50 μL, SantaCruz, Cat No. SC-2003) twice with coupling buffer (500 μL; 100 mM sodium phosphate, 150 mM NaCl, pH 7.2). The p53 antibody (25 μL, Santa Cruz, SC-6243 or SC-55476) in coupling buffer (1 mL) was incubated with the washed beads for 2 hrs at 4°C with rocking. After washing twice with coupling buffer (1 mL), the antibody-loaded agarose beads were resuspended in coupling buffer (500 μL), added to the kinase-catalyzed crosslinking reaction mixture generated above, and incubated overnight at 4°C with rocking. For coimmunoprecipitation studies, washed protein A/G plus agarose beads (40 μL, SantaCruz, Cat No. SC-2003) were incubated with p53 (20 μL, Santa Cruz, SC-6243 or SC-55476) or MRCKβ (20 μL, SignalChem, C28–11G-05) antibody in coupling buffer (1 mL) for 2 hr at 4°C with rocking. After washing twice with coupling buffer (1 mL), the antibody-loaded agarose beads were resuspended in coupling buffer and incubated with untreated RKO lysates (4 mg, 1 mL total volume) overnight at 4°C with rocking. After washing the immunoprecipitated beads twice with coupling buffer (1 mL), the bound beads were resuspended in K-CLIP buffer (20 μL) containing 2% (w/v) SDS (sodium dodecyl sulfate; electrophoresis grade), 10% (v/v) glycerol, and DTT or beta-mercaptoethanol (50 mM) and incubated at 95°C for 2 minutes to denature and release the bound proteins from the beads. Proteins were separated by 4–12% gradient (K-CLIP for LC-MS/MS) or 10% (coimmunoprecipitation) SDS-PAGE and visualized with SyproRuby stain. For western blotting, proteins were transferred to the PVDF membrane (Immobilon PSQ, EMD Millipore) with visualized using p53 (Santa Cruz, SC-6243 or SC-55476) or MRCKβ (SignalChem, C28–11G-05) antibodies.
In-Gel Protein Digestion
For LC-MS/MS analysis, the in-gel protein digestion method was adapted from Shevchenko et al.20 The gel was stored until processing (up to one week) in destaining solution (7% acetic acid (Mallinckrodt Chemicals) and 10% methanol (EMD) in ultrapure H2O). Prior to band excision, the gel was washed with 10% methanol in ultrapure water twice for a minimum of 2 hours. The proteins in the gel were visualized on an FBTIV-88 ultraviolet trans-illuminator and the gel lane was excised from the 55 kDa molecular weight marker to the top of gel. Gel slices were washed with hydration buffer (100 μL; 1:1 (v/v) solution of 50 mM ammonium bicarbonate and acetonitrile) for 10 minutes. Gel slices were then dehydrated with acetonitrile (100 μL) with agitation. The acetonitrile was removed and the hydration/dehydration sequence was repeated, followed by dying in a speedvac concentrator (SDP131DDA, Thermo Scientific Savant) for 10 minutes. Dried gel pieces were rehydrated in hydration buffer (100 μL) before cysteines were reduced in reducing buffer (100 μL; 50 mM TCEP and 25 mM ammonium bicarbonate) at 37 °C for 20 minutes. After cooling for 10 minutes, alkylation was performed by adding iodoacetamide (100 μL; 100 mM in 25 mM ammonium bicarbonate buffer) and incubating at room temperature in the dark for 30 minutes. Gel particles were then rehydrated, dehydrated, and dried, as described. Activated proteomics grade trypsin (20 μg/mL; Sigma) in digestion buffer (75 μL; 40 mM ammonium bicarbonate, 9% acetonitrile) was added and incubated on ice for 1 hour. Additional digestion buffer (50 μL) was added and then tubes were incubated at 37 °C overnight. The digested peptide solution was collected and the gel fragments were further extracted with formic acid (75 μL; 0.1% (v/v) formic acid in (65:35) water:acetonitrile) by sonication for 5 minutes. The combined peptide solution was dried in vaccuo and then resuspended in a solution of 5% acetonitrile, 0.1% formic acid and 0.05% trifluoroacetic acid (20 μL) for LC-MS/MS analysis.
LC-MS/MS Analysis
Peptide mixtures after protein digestion were desalted in-line and separated by reverse phase chromatography (Acclaim PepMap100 C18 pre-column, Acclaim PepMapRSLC C18 analytical column, Thermo Scientific) on a nanoflow HPLC instrument (Easy-nLC 1000, Thermo Scientific), followed by ionization with a Nanospray Flex Ion Source (Proxeon Biosystems A/S) and introduction into a Q Exactive mass spectrometer (Thermo Scientific). A linear gradient of 95% buffer A (water, 0.1% formic acid) with 5% buffer B (acetonitrile) to 90% buffer A:10% buffer B over 2 minutes, followed by 68% buffer A: 32% buffer B over 30 minutes at a flow rate of 300 nL/min, was used for separation. Pumps and instrument methods were controlled with Thermo Xcalibur 2.2 SP1.48 software (Thermo). The instrument was operated in positive mode and data was acquired using a data-dependent top 12 method. The 12 most abundant ions from the full MS scan (375–1600 m/z) were selected for fragmentation by high-energy collisional dissocation (HCD). Dynamic exclusion was enabled with 12.0 s duration and precursor isolation width set to 3.0 m/z. Resolution of full MS and HCD scans was 70,000 at 375 m/z and 17,500 at 200 m/z, respectively. Normalized collision energy for HCD spectra was 30 eV and data were acquired in profile mode. Isotope exclusion, singly charged and unrecognized charged ion exclusion was enabled.
Analysis of raw data was performed with Proteome Discoverer 1.4.0.288 (Thermo) using the Mascot search algorithm (Matrix Science). The SwissProt_2013_04 database was used against human protein sequences and the reverse decoy protein database used to calculate false discovery rates (FDR). Secondary analysis was performed using Scaffold 4.0.5 (Proteome Software) with X! Tandem software (The Global Proteome Machine Organization) for subset database searching. Minimum protein identification probability was set to ≥99% with 2 unique peptides per protein and ≥80% minimum peptide identification probability. Mascot and X! Tandem algorithms were searched with a precursor mass tolerance of 10 ppm and fragment mass tolerance of 0.02 Da. Target FDR was set to 0.01 (strict) and 0.05 (relaxed). Carbamidomethylation of cysteine (+57) was set as a fixed modification. Variable modifications included oxidation of methionine (+16), N-terminal acetylation (+42), and phosphorylation on serine, threonine, and tyrosine (+80). Enzyme specificity was defined for trypsin as C-terminal to lysine and arginine, with 1 missed cleavage site allowed. The parameters for the data analysis of each set of proteins obtained were mentioned in the tables.
Recombinant p53 purification
A hexahistidine tagged p53 fusion protein was expressed by transforming the pET15b-p53(1–393) plasmid (Addgene)21 into the BL21(DE3)pLysS strain. Expression was performed in Luria Broth (LB, 2 g Bacto-tryptone, 1 g yeast extract, 2 g NaCl in 200 mL water) at 37°C for 4 hours at an OD600 of 0.5 using 1.0 mM IPTG. Cells were lysed by sonication in p53 lysis buffer (50 mM Tris, pH 8, 300 mM NaCl, 1 mM EDTA, 0.5% Triton-X 100, 20 ng/mL lysozyme, and 1X protease inhibitor (Xpert)). The insoluble fraction containing p53 was collected by centrifugation and p53 was solubilized by incubation in guanidine buffer (6 M guanidine HCl, 25 mM Tris, pH 8, 300 mM NaCl, 5 mM imidazole) for 1 hour at room temperature. The p53 protein was purified via the hexa histidine tag using Ni-NTA resin (1 mL packed beads) pre-equilibrated in guanidine buffer. Unbound proteins were removed by washing with guanidine buffer (2 mL) three times, followed urea wash buffer (2 mL; 8 M urea, 25 mM Tris, pH 8, 300 mM NaCl, 20 mM imidazole) four times. The bound p53 was refolded on resin by successive washings with each of the following buffers: four times with a 1:1 mixture (2 mL) of urea wash buffer:native folding buffer (25 mM Tris, pH 8, 300 mM NaCl, 20 mM imidazole, and 1% triton-X 100), five times with 1:7 mixture (2 mL) of urea buffer:native folding buffer, and five times with native folding buffer (2 mL). The folded p53 was eluted using elution buffer (10 mL; 25 mM Tris, pH 8, 500 mM NaCl, 300 mM imidazole, 1% triton-X 100). The purified p53 solution was desalted and concentrated with a centrifugal device (Amicon Ultra 10,000 molecular weight cut off) using storage buffer (25 mM Tris, pH 7.5, 300 mM NaCl, 50 mM glycine, 0.5 % triton-X 100, and 5 % glycerol). Recombinant p53 was stored in aliquots at −20°C until use in in vitro kinase assays.
In vitro kinase assays
MRCKβ (0.025 μg, SignalChem) was incubated with p53 (0.5 μg, Santacruz) and ATP (4 mM) at 31°C for 4 hours with shaking at 300 rpm in kinase assay buffer (25 mM MOPS, pH 7.2, 12.5 mM β-glycerol-phosphate, 25 mM MgCl2, 5 mM EGTA, 2 mM EDTA, 0.25 mM DTT). For comparison, DNAPK (0.025 μg, Life Technologies) was incubated with p53 (0.5 μg, Santacruz) and MRCKβ (0.025 μg, SignalChem) was incubated its known substrate MYPT1 (0.5 μg, EMD Millipore) under the same reaction conditions. The total reaction volume was 30 μL. Larger scale reactions (Figure S12B) were also performed by incubating MRCKβ (1.5 μg, SignalChem) with bacterially expressed p53 (5.5 μg) and ATP (4 mM) at 31°C for 4 hours with shaking at 300 rpm in kinase assay buffer. Samples were then separated with 10% SDS-PAGE. Phosphoproteins were visualized by Pro-Q® Diamond Phosphoprotein gel stain (ThermoFisher Scientific). Total proteins were visualized by Sypro® Ruby gel stain (ThermoFisher Scientific).
MRCKβ knock down
RKO cells at 70% confluency in a 75 cm2 flask were treated with a pool of MRCKβ-targeting siRNA (25 nM, Dharmacon, M-004075–02–0005) or a control non-targeting pool of siRNA (25 nM, Dharmacon, D-001206–14–05), along with a transfection reagent (10, Dharmacon, T-2001–02). As another control, cells were treated with the transfection reagent alone. After 72 hours at 37 °C, cells were harvested and lysed, as described. Proteins were separated by 10% SDS-PAGE, transferred to the PVDF membrane (Immobilon P, EMD Millipore), and visualized using p53 (Santa Cruz, SC-6243 or SC-55476) or MRCKβ (SignalChem, C28–11G-05) antibodies.
Results
Kinase-catalyzed crosslinking with RKO lysates and p53
As a first step towards studying p53 phosphorylation, we generated cells expressing wild type p53. Colon carcinoma RKO cells were selected for these experiments due to their wide use in prior p53 studies.22 Colon carcinoma RKO cells were treated with the MDM2 inhibitor (±)-Nutlin-3 (Figure S1A) to block the interaction of MDM2 and p53.15–16 Because MDM2 promotes p53 degradation under normal cell conditions (Figure S1C, lane 1), the disruption of MDM2/p53 interactions stabilized p53 for cellular accumulation (Figure S1C, lanes 2 and 3).23
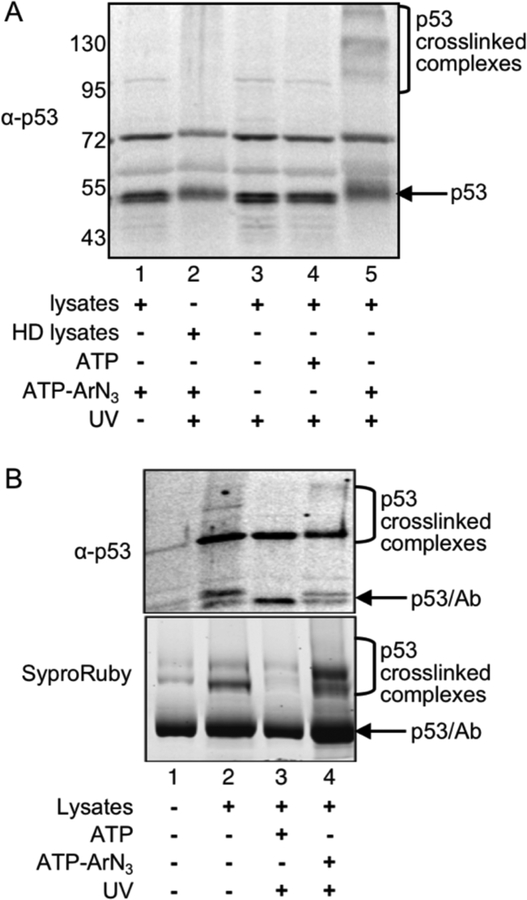
For kinase-catalyzed crosslinking, ATP-ArN3 was incubated with nutlin-treated RKO lysates under UV irradiation. Proteins were separated by SDS-PAGE before western blotting to identify p53-specific crosslinked bands. In the presence of ATP-ArN3 and UV, higher molecular weight crosslinked bands specific to p53 were observed (Figure 2A, lane 5). As controls, crosslinked bands were absent without UV light (Figure 2A, lane 1) or ATP-ArN3 (Figure 2A, lanes 3 and 4). Heat denatured lysates were unable to produce p53-crosslinked bands (Figure 2A, lane 2), which indicated that the photocrosslinking was kinase enzyme dependent. The gel analysis established that kinase-catalyzed crosslinking in lysates produced higher molecular weight complexes that can be recognized by antibody detection.
Figure 2:
A) Photocrosslinking reactions were performed using the indicated reaction components, followed by SDS-PAGE separation and visualization with a p53 antibody. HD lysates indicates heat-denatured lysates (lane 2). Repetitive trials are shown in Figure S2. B) K-CLIP was performed by immunoprecipitating p53 after kinase-catalyzed crosslinking of nutlin-treated RKO lysates. Immunoprecipitates were separated by SDS-PAGE and visualized using a p53 antibody and SyproRuby total protein stain. All lanes contain anti-p53 and Protein A/G agarose beads (lane 1), with the other components indicated under each lane (lanes 2–4). Results using a rabbit polyclonal antibody for immunoprecipitation are shown here, whereas results from use of a mouse monoclonal antibody are displayed in Figure S3. Protein bands from SyproRuby stained gels were excised, trypsin digested, and analyzed by LC-MS/MS to identify crosslinked proteins. For both gel images, p53, contaminating heavy chain of the antibody (Ab) after IP, and the high molecular weight photocrosslinked complexes containing p53 are indicated.
K-CLIP identified p53 kinases
Next, kinase-catalyzed crosslinking was coupled with p53 antibody immunoprecipitation and gel analysis to identify p53 crosslinked proteins using K-CLIP. Immunoprecipitation was performed using both mouse monoclonal (Figure 2B) and rabbit polyclonal antibodies (Figure S3) because each recognized different epitopes on the p53 antigen. Similar to photocrosslinking alone, higher molecular weight bands were observed after crosslinking and immunoprecipitation (Figure 2B, lane 4). As critical controls, fewer higher molecular weight bands were observed without ATP-ArN3 (Figure 2B, lanes 2 and 3). The K-CLIP method reproduced the data obtained with photocrosslinking alone, while also allowing for isolation and MS analysis of the crosslinked proteins.
To identify the proteins crosslinked to p53 in the K-CLIP reactions using LC-MS/MS, protein bands were excised from the ATP-ArN3 and ATP lanes of the SyproRuby stained gels (Figure 2B and S2, lanes 3 and 4). The ATP-containing reactions were used as an uncrosslinked negative control to reveal only phosphorylation-dependent crosslinked proteins. After in-gel digestion, the peptide mixtures were analyzed by LC-MS/MS. Proteins observed in the ATP-ArN3 crosslinked reactions (columns B and D in Tables 1 and S1), but not in the ATP uncrosslinked reactions (columns A and C in Tables 1 and S1) and seen reproducibly in two trials were considered hits. Among the list of protein hits were three kinases and 125 non-kinase proteins (Table 1 and S1), including p53 (Figures S4 and S5). The three kinases identified were DNA-dependent protein kinase (PRKDC or DNA-PK, Figures S6 and S7), Interferon-induced double-stranded RNA-activated protein kinase (EIF2AK2 or PKR, Figures S8 and S9), and myotonic dystrophy-related Cdc42-binding kinases beta (CDC42BPB or MRCKβ, Figures S10 and S11). Of the three identified kinases, DNA-PK and PKR are known to phosphorylate p53,24–28 whereas MRCKβ is a potential new p53 kinase.
Table 1.
List of kinases identified using p53 K-CLIP a
| Protein | Accession | Known? | A | B | C | D |
|---|---|---|---|---|---|---|
| DNAPK | P78527 | Yes | 0 | 7 | 0 | 47 |
| PKR | P19525 | Yes | 0 | 1 | 0 | 3 |
| MRCKβ | Q9Y5S2 | No | 0 | 3 | 0 | 1 |
Lettered columns display the number of peptides observed for the indicated proteins in uncrosslinked (A and C) or crosslinked (B and D) K-CLIP reactions with a rabbit (A and B) or mouse (C and D) p53 antibody immunoprecipitation. All three kinases were observed in only crosslinked reactions reproducibly in both trials. MS data are shown in Figures S4–S11.
MRCKβ is a p53 kinases
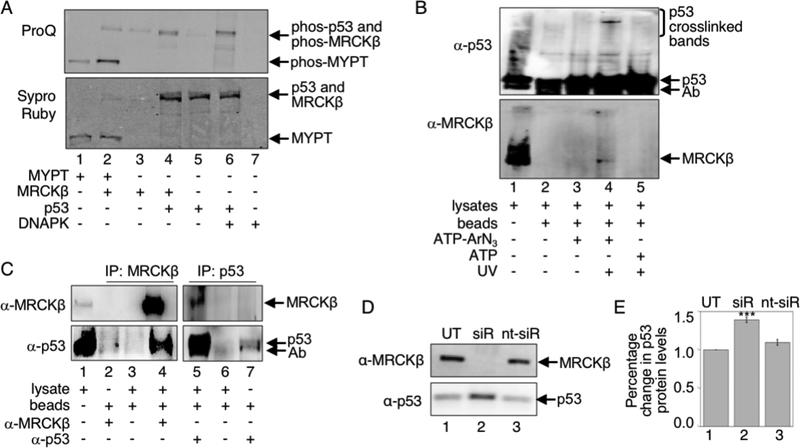
The K-CLIP study identified MRCKβ as a possible kinase of p53. To test if MRCKβ phosphorylates p53, in vitro kinase assays were performed. Recombinant MRCKβ and ATP were incubated with recombinant p53 and phosphorylation was monitored using the ProQ phosphoprotein stain (Figure 3A). Phosphorylation of p53 was observed in the presence but not in the absence of MRCKβ (Figure 3A, compare lanes 4 and 5). To compare levels of phosphorylation of p53 by MRCKβ to levels of phosphorylation of p53 by a known p53 kinase, recombinant DNAPK and ATP were incubated with recombinant p53 and phosphorylation was monitored. The level of phosphorylation of p53 by DNAPK was comparable to the level of phosphorylation of p53 by MRCKβ (Figure 3A, compare lanes 4 and 6). To also compare the levels of MRCKβ phosphorylation of p53 to a known MRCKβ substrate, recombinant MYPT1 was tested. Phosphorylation of MYPT1 by MRCKβ was comparable to p53 phosphorylation by MRCKβ (Figure 3A, compared lanes 2 and 4). To confirm these studies, another larger scale assay using bacterially expressed and purified p5321 was performed. As expected, phosphorylation of p53 was observed in the presence but not in the absence of MRCKβ (Figure S12B, lane 2 compared to lane 1). The in vitro kinase assays document that p53 is a substrate of MRCKβ, as predicted by the K-CLIP study.
Figure 3:
A) Recombinant MYPT1 (lanes 1–2) or p53 (lanes 4–6) was incubated with recombinant MRCKβ (lanes 2–4) or DNAPK (lanes 6–7), along with ATP. After separation by SDS-PAGE, phosphoproteins were visualized with ProQ Diamond stain (top) and total proteins were observed with SyproRuby stain (bottom). Full gel image and repetitive trials are shown in Figure S12A. B) K-CLIP was performed with untreated RKO cell lysates (4 mg) after crosslinking using ATP-ArN3 (lanes 3–4) or ATP (lane 5) either in the absence (lanes 1–3) or presence (lanes 4–5) of UV. After SDS-PAGE separation, both p53 (α-p53) and MRCKβ (α-MRCK) were visualized using Western blot analysis. Repetitive trials are shown in Figures S13. C) Coimmunoprecipitation was performed with untreated RKO cell lysates (4 mg) and either MRCKβ (lanes2 and 4) or p53 (lanes 5 and 7) antibodies, along with protein A/G agarose beads (lanes 2–7). After SDS-PAGE separation, Western blot analysis was performed with the MRCKβ (α-MRCK) or p53 (α-p53) antibodies. Repetitive trials are shown in Figures S14. D) RKO cells were untreated (UT), treated with a pool of MRCKβ-targeting siRNA (siR), or treated with a control non-targeting pool of siRNA, along with a transfection reagent. After 72 hours at 37 °C, cells were harvested and lysed, before cellular proteins were separated by SDS-PAGE and visualized with both MRCKβ (α-MRCK) and p53 (α-p53) antibodies. E) Three independent trials from part D (Figure S15A) were quantified and the percentage change in p53 protein levels were plotted as mean and standard error. Data were analyzed by Prism software for statistical significance (***p<0.001). For all gel images, protein bands are indicated with arrows, including contaminating antibody heavy chain (Ab), which co-migrated with p53.
As secondary confirmation that p53 is an MRCKβ substrate, K-CLIP was used as a lysate-based kinase-substrate validation tool. After crosslinking, immunoprecipitation with the p53 antibody, and SDS-PAGE separation, western blot analysis identified higher molecular weight p53-reactive bands (Figure 3B, lane 4), which disappeared in the absence of UV light (Figure 3B, lanes 3) or ATPArN3 (Figure 3B, lanes 5). Importantly, MRCKβ was also observed in immunoprecipitates after K-CLIP (Figure 3B, lane 4), which disappeared in the absence of UV light (Figure 3B, lanes 3) or ATPArN3 (Figure 3B, lanes 5). The K-CLIP study further confirmed that MRCKβ and p53 are a kinase-substrate pair under cellular conditions.
Finally, to test whether p53 physical interacts with MRCKβ in a cellular complex, a coimmunoprecipitation experiment was performed. RKO lysates were incubated with protein A/G agarose beads with and without either p53 antibody or MRCKβ antibody, before SDS-PAGE separation and Western blot analysis to visualized both p53 and MRCKβ. Consistent with the presence of p53 and MRCKβ in a complex in cells, p53 was observed when MRCKβ was immunoprecipitated (Figure 3C, lane 4). Likewise, MRCKβ was coimmunoprecipitated when a p53 antibody was used (Figure 3C, lane 5). These coimmunoprecipitation experiments indicate that p53 physically interacts with MRCKβ, which are consistent with possibility that MRCKβ is a p53 kinase. Taken together, the in vitro kinase assays, K-CLIP confirmation studies, and coimmunoprecipitation experiments suggest that MRCKβ is a p53 kinase.
MRCKβ affects p53 proteins levels
Previous reports established that MRCKβ is a downstream effector of Cdc42, which regulate cell adhesion, invasion, and motility. Similar to MRCKβ, prior studies document that p53 activity lies downstream of Cdc42,29 and implicate p53 as a player in cancer cell invasion and motility.30 Mechanistically, p53 levels were augmented after Cdc42 knockdown,31–32 suggesting that the Cdc42 pathway targets p53 for degradation. Given the results here indicating that MRCKβ is a p53 kinase, we speculated that one effect of the Cdc42 pathway involves MRCKβ-mediated phosphorylation of p53, which targets p53 for degradation. Therefore, we sought to test this hypothesis by monitoring the p53 protein levels as a function of MRCKβ.
To test whether phosphorylation by MRCKβ influences p53 protein levels, RKO cells were treated with MRCKβ siRNA for 60 hours to knockdown MRCKβ protein levels, followed by lysis, separation of cellular proteins by SDS-PAGE separation, and western blot visualization of both MRCKβ and p53. Cells treated with MRCKβ siRNA showed reduced levels of MRCKβ expression compared to cells treated with non-targeting control siRNA or transfection reagent only (Figures 3D and S15A, compare lane 2 to lanes 1 and 3), confirming successful knockdown (Figure S15B). By monitoring p53 protein levels in those same cells, MRCKβ knockdown cells resulted in elevated p53 protein levels compared to the p53 levels in control cells (Figure 3D, lane 2 compared to lanes 1 and 3). Quantification of three independent trials (Figure S15A) showed an increase in p53 protein levels of 41 ± 4 % with MRCKβ knockdown (Figure 3E, lane 2). The data are consistent with a model where MRCKβ-mediated phosphorylation targets p53 for degradation.
Kinase-catalyzed crosslinking as a kinase validation tool
In addition to kinase-substrate discovery, kinase-catalyzed crosslinking has application as a lysate-based kinase substrate validation tool. In this case, after crosslinking and SDS-PAGE separation, western blot analysis of the high molecular weight complexes will identify the presence of suspected kinase and substrate. To test kinase-catalyzed crosslinking as a validation tool, two known p53 kinases were tested. DNA-PK was chosen as a positive test case because it was seen in the earlier K-CLIP study (Table 1). The p53 kinase PLK3 (Polo-like kinase 3) was selected as a negative test case due to its absence in the earlier K-CLIP study. These two kinases will establish if kinase-catalyzed crosslinking is able to observe active kinase-substrate pairs present under specific cellular conditions.
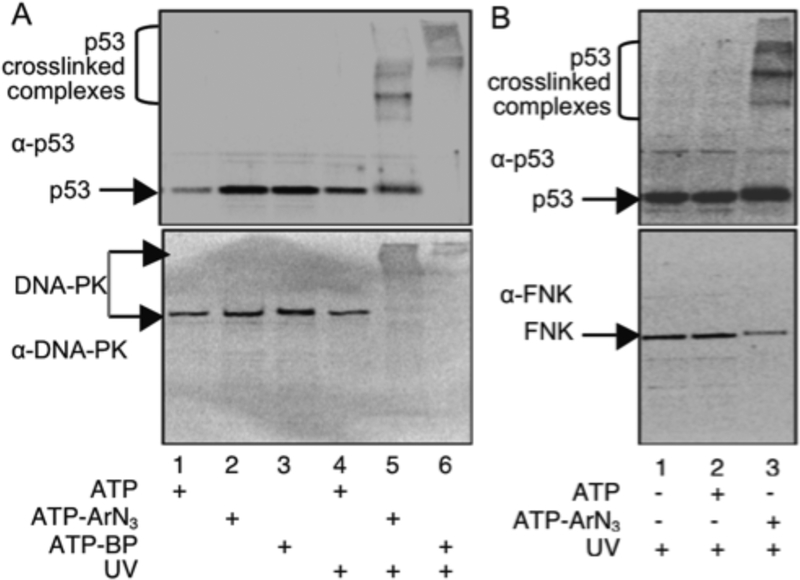
After kinase-catalyzed crosslinking and protein separation by SDS-PAGE, higher molecular weight crosslinked bands recognized by the p53 antibody were observed only in the presence of UV and ATP-ArN3 (Figure 4A, lane 5, top), consistent with earlier studies (Figure 2). Western blot analysis of the same reactions with an antibody to DNA-PK identified a higher molecular weight DNA-PK reactive band (Figure 4A, lane 5, bottom), which was absent without UV (Figure 4A, lane 2, bottom) or ATP-ArN3 (Figure 4, lanes 1 and 4, bottom). For further confirmation, the same experiment was performed with an alternative photocrosslinking ATP-analog, ATP-BP (Figure 1A). Again, DNA-PK was observed in the crosslinked complex, but no other controls (Figure 5A, lane 6 versus lane 3). Due to the presence of higher order crosslinked complexes with ATP-BP, which were absent with ATP-ArN3 (Figure 4A, lane 6 compared to lane 5), as was previously reported,9 ATP-ArN3 was preferred for kinase-catalyzed crosslinking. These results confirm that kinase-catalyzed crosslinking with either ATP-ArN3 or ATP-BP analogs can be used to validate an active p53 kinase.
Figure 4:
Kinase-catalyzed crosslinking was performed using HeLa cell lysates (all lanes) and the indicated ATP analog with or without UV, followed by SDS-PAGE separation and visualization with a p53 antibody (A and B, top), and either a DNA-PK (A, bottom) or PLK3 (B, bottom) antibody. The high molecular weight crosslinked complexes, p53, DNAPK, and PLK3 are indicated with arrows. Repetive trials are shown in Figure S16.
Figure 5:
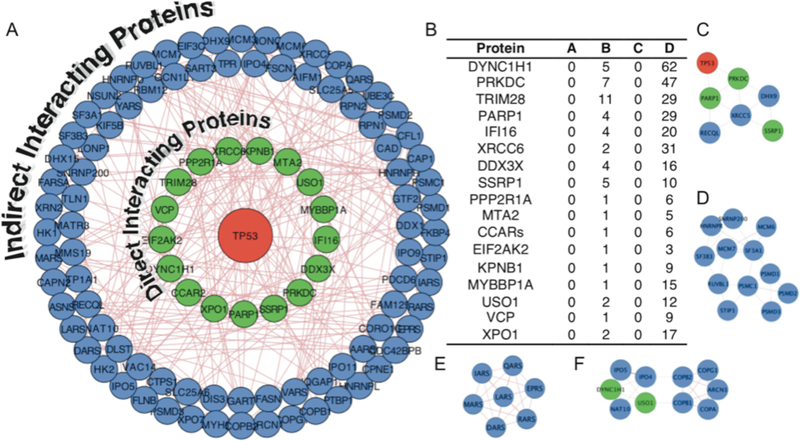
A) The 128 proteins enriched using K-CLIP (Table S1) were analyzed to identify known physical protein-protein interactions of p53 (red) using GeneMANIA in Cytoscape. The p53 interacting proteins (81% of hits) were classified into direct (green inner circle) or indirect (blue outer circles) interactors. Line thickness and length were adjusted arbitrary to improve clarity. The Cytoscape file is available as a supplementary document. D) Table of the seventeen direct physical interactors of p53. Lettered columns display the number of peptides observed for the indicated proteins in uncrosslinked (A and C) or crosslinked (B and D), with rabbit (A and B) or mouse (C and D) p53 antibody immunopreciptiation. All K-CLIP hits were observed in only crosslinked reactions reproducibly in both trials (Table S1). C-F) Clusters of protein-protein interactions identified using the MCODE application in Cytoscape, which are related to DNA damage and repair (C), the spliceosome and proteosome complexes (D), tRNA aminylacylation (E), and protein transport (F).
PLK3 (also known as FNK) kinase phosphorylates p53 under oxidative conditions.33–34 Due to the lack of oxidative stress during the K-CLIP study, PLK3 was not likely to be actively phosphorylating p53, which explains the absence of PLK3 as a K-CLIP hit. To demonstrate that kinase-catalyzed crosslinking validates only kinase-substrate pairs actively undergoing phosphorylation in the lysates, western blot analysis was performed with the PLK3 antibody after crosslinking. While higher molecular weight complexes containing p53 were observed (Figure 4B, lane 3, top), no PLK3 antibody reactive bands were detected at that same higher molecular weight region (Figure 4B, lane 3, bottom). However, a band at roughly 72 kDa corresponding to PLK3 was observed in all reactions (Figure 4B, bottom gel), confirming that PLK3 is present in the lysates. These data document the utility of kinase-catalyzed crosslinking to study the presence of active kinase-substrate partners under specific cellular conditions.
K-CLIP identified p53-interacting proteins
In addition to kinases, 125 non-kinase proteins were observed reproducibly in only crosslinked reactions (Table S1). The presence of non-kinase proteins is expected due to the fact that the kinase-substrate interaction is unstable and transient, allowing p53 to diffuse away from the kinase after catalysis (Figure 1D). Then, upon UV-mediated activation, p53 crosslinks to proteins in close proximity, such as interacting proteins. To assess if the enriched, non-kinase K-CLIP hits have known interactions with p53, an interactome analysis was performed using the GeneMania application in Cytoscape.35–37 All proteins with known physical interactions to p53 were mapped (green inner circles in Figure 5A). Among the 128 enriched proteins, seventeen (13%) interact with p53 directly (Figure 5B), including the two kinases DNA-PK (PRKDC) and PKR (EIF2CK2). In addition to documented direct physical interactors of p53, proteins that indirectly interact with p53 via multi-protein complexes were also observed (blue outer circles in Figure 5A). Considering both direct and indirect physical interactions, 81% of the K-CLIP hits are accounted for in the p53 interactome. The fact that a high percentage of hits are known physical interacting proteins of p53 confirms that protein-protein interactions beyond kinases can be monitored using K-CLIP.
The MCODE application in Cytoscape38 was next used to identify clusters of interacting proteins and predict cellular complexes among the K-CLIP hits. A cluster of DNA damage and repair proteins, including p53 and DNA-PK, was identified (Figure 5C), which is consistent with the widely accepted role of p53 in DNA damage.39 The presence of this DNA repair cluster suggests that p53-dependent DNA damage repair was active during the course of the experiment. In another cluster (Figure 5E), seven out of eight AIMP2-associated ARS proteins (IRS, LRS, EPRS, RRS, MRS, DRS, and QRS), along with four additional ARS proteins (ARS, VRS, YRS, and FRS), were observed by K-CLIP. Prior work documented that p53 interacts with a multi-protein aminoacyl tRNA synthetase (ARS) complex via the scaffolding protein AIMP2 (ARS-interacting, multifunctional proteins 2),40 consistent with the presence of this ARS protein cluster. Two additional clusters were also observed by MCODE with function in RNA splicing and proteosomal degradation (Figure 5D) and protein transport (Figure 5F). Although not in an MCODE cluster, DDX1 interacts with the known p53 binder HNRPK,41–42 suggesting the presence of a cellular complex comprising p53, DDX1, and HNRPK. In total, the K-CLIP data suggests that p53 was in proximity to DNA repair, ARS complex, spliceosome, proteosome, and protein transport proteins during the experiment, suggesting a role of p53 in these cellular activities.
As well as known direct and indirect binding partners and multi-protein clusters, many proteins observed by K-CLIP are structurally or functionally related to reported p53-interacting proteins. DDX3X, DDX1, and DDX41 were observed by K-CLIP and are in the same DEAD box helicase family as the p53-associated proteins DDX3, DDX5, and DDX17, which either stabilize or coactivate p53 transcriptional activity.43–44 Likewise, the known association of p53 with RNA helicase activity implicates other helicases identified by K-CLIP as p53 binders, such as DHX15, U520, DHX9, and RECQ1. Finally, UBE3B is an E3 ligase, which is similar to the p53-interacting UBE3A ligase.45 In addition to these functional connections, several proteins are transcription factors like p53, including TIF1B, GTF2I, SSRP1, and MBB1A.46–47 The presence of known interactors, along with proteins structurally or functionally-related to known p53 associated partners, establishes K-CLIP for studying protein-protein interactions and multi-protein cellular complexes, in addition to kinase-substrate pairs.
Kinase-associated complexes identified by K-CLIP
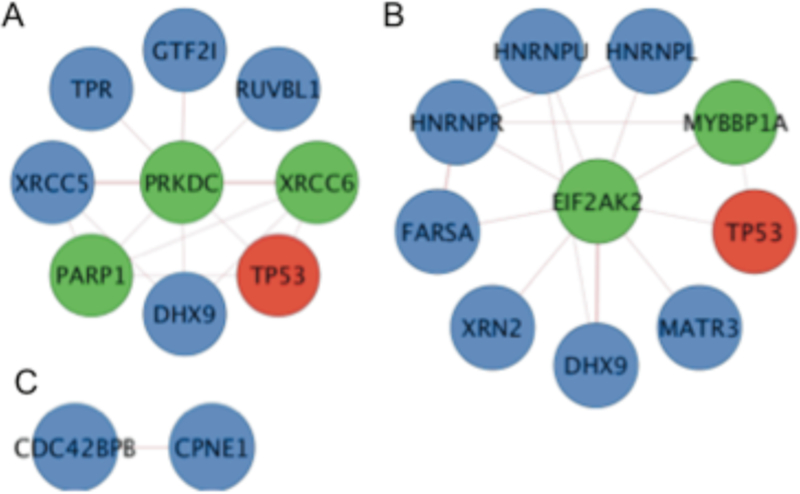
In addition to identifying the associated proteins of p53, K-CLIP can also identify interacting proteins of the actively phosphorylating p53 kinases, which has the potential to uncover the cellular complexes governing p53 phosphorylation. To reveal possible cellular complexes of each p53 kinase, the 125 non-kinase proteins observed by K-CLIP were analyzed by Cytoscape for known interaction with the three identified p53 kinases-DNA-P, PKR, and MRCKβ. As expected, the two proteins necessary to recruit DNA-PK to the ends of double stranded DNA and enhance its kinase activity, XRCC5 and XRCC6 (Ku 80 and Ku70),48 were observed (F igure 6A), indicating that DNA-PK was in an active state. Consistent with the earlier cluster analysis (Figure 6C), DNA-PK associates with PARP1, which is phosphorylated by DNA-PK to influence its poly(ADP-ribosylation) activity and regulate the initiation of DNA strand break repair.49 Several proteins linked to transcriptional activation were also observed in the DNA-PK cluster, including the general transcription factor TFII-I (GTF2I), the RNA/DNA helicase DHX9 (RHA),50 and the DNA helicase RuvBL1, which is found in the NuA4 histone acetyltransferase complex.51 The presence of DNA repair and transcriptional activator proteins in the vicinity of actively phosphorylating DNA-PK is consistent with the role of DNA-PK and p53 in DNA repair and transcription. On the other hand, the presence of TPR (translocated promoter region), which is a component of the nuclear pore complex involved in trafficking proteins and mRNA across the nuclear envelope,52 suggests that DNA-PK and p53 were actively transported to the nucleus during phosphorylation.
Figure 6: Direct interacting proteins of the kinases identified by p53 K-CLIP.
A) DNA-PK (PRKDC) has reported interaction with eight of the K-CLIP hits, including p53 (TP53, red). B) PKR (EIF2AK2) interacts directly with nine of the K-CLIP hit proteins, including p53 (TP53, red). C) MRCKβ (CDC42BPB) has only a single known interaction among the K-CLIP hit proteins, copine 1 (CPNE1). As in Figure 5, p53 is red and known direct and indirect interacting proteins of p53 are in green and blue, respectively.
Multiple direct interacting proteins of PKR (EIF2AK2) were also among the K-CLIP hits (Figure 6B), in addition to p53 (TP53). PKR interacted with HNRPL, HNRPR, and HNRPU, which are RNA-binding heterogeneous nucleoriboproteins (HNRP) found in the spliceosome. Consistent with spliceosome association, prior work documented direct interaction of PKR with spliceosome proteins.53 In addition, HNRPK modulates the transcriptional activation function of p53 and is in the same HNRP family as those identified K-CLIP.42 The K-CLIP data implicate that PKR and p53 were spliceosome associated at the time of phosphorylation.
Of the three kinases analyzed, the new p53 kinase MRCKβ had the fewest known direct interactors among the K-CLIP hits, with only a single protein, copine-1 (CPNE1). The few protein hits is not surprising considering that MRCKβ has only 21 known direct physical interactions according to the BioGRID database (http://thebiogrid.org).54 Copine-1 is a phospholipid-binding protein thought to be involved in cell signaling by localizing proteins to the membrane in a calcium-dependent manner.55–56 MRCKβ interacts with membrane-associated GTPase, CDC42, to promote actin-myosin contraction, cytokinesis, and cell motility.57–58 Given the membrane localization of both MRCKβ and copine-1, the K-CLIP study suggests that MRCKβ was membrane associated at the time of p53 phosphorylation.
Discussion
A variety of methods have been used to identify kinase-substrate pairs. Classical methods, including yeast two-hybrid and pull-down assays, have shown success.59 However, the transient kinase-substrate interaction can be lost or overlooked during purification or gene expression. Consensus sequences for many protein kinases have been determined using combinatorial peptide libraries, which allows sequence searching to identify kinase substrates.60–61 Unfortunately, the consensus sequences of all human protein kinases are not yet available. As alternatives, several chemical methods have been developed to convert the unstable kinase-substrate interaction into a stable complex using covalent crosslinking, which facilitates purification and identification. Most notably, analog-sensitive kinase mutants6 and a mechanism-based cross-linker7 have been used to identify kinase substrate pairs. While these chemical tools are powerful, they require mutant kinases or substrates. In contrast, K-CLIP is capable of identifying kinase-substrate pairs without overexpression or mutagenesis. In addition, K-CLIP can be used with any lysate or homogenate containing the kinase or substrate of interest. A key feature of K-CLIP is the requirement of kinase catalysis for crosslinking, making all proteins identified conjugated by an active kinase to a substrate. K-CLIP provides a general method for exploring phosphorylation-mediated cell biology.
Here, K-CLIP was used to identify kinases of p53. Two known p53 kinases, DNA-PK and PKR, were observed by K-CLIP, which validate the method for kinase-substrate identification. DNA-PK is activated in response to DNA damage, including double stranded breaks and single strand nicks and gaps.24–26 K-CLIP using ATP-ArN3 or ATP-BP requires UV light exposure to photoactivate the crosslinking groups, and UV light leads to DNA damage and p53 phosphorylation.62 The fact that UV light was used during crosslinking is consistent with activation of DNA-PK due to UV-mediated DNA damage and subsequent p53 phosphorylation. Likewise, eleven ARS proteins were observed (Figure 5E), and previous reports documented that p53 interacts in a UV-dependent manner with the scaffolding protein AIMP2 to bind the multi-protein ARS complex.40 The data suggest that the conditions of crosslinking will influence the interactions observed by K-CLIP. In this case, the UV irradiation used for crosslinking allowed DNA repair activities and AIMP2 interactions to be monitored.
In addition to known p53 kinases, K-CLIP identified MRCKβ as a potentially new p53 kinase. In vitro kinase assays and K-CLIP studies confirmed that MRCKβ phosphorylates p53 (Figure 3A, 3B, S12, and S13). Consistent with their cellular relationship, MRCKβ and p53 coimmunoprecipitated from RKO cells (Figure 3D and S14). MRCKβ plays a role in cytoskeleton organization during mitosis and cell migration.63 The MRCKβ homolog MRCKα (61% similarity) is overexpressed in a variety of invasive and metastatic cancers, which implicates a role for MRCK kinases in cancer cell invasion.63 MRCKβ binds to the membrane-associated GTPase Cdc42 in its active GTP-bond form to promote phosphorylation of the regulatory myosin light chain (MYL9), leading to actin-myosin contraction, cytokinesis, and cell motility.57–58 Consistent with the role of MRCK in actin-myosin organization, myosin-9 (MYH9) was identified by p53 K-CLIP (Table S1), suggesting that MRCKβ may form a complex with myosin-9 and p53.
Both MRCKβ and p53 are linked to cancer cell invasion and motility through the Cdc24 pathway.30,·63 For example, overexpression of p53 inhibited Cdc42-mediated filopodia formation, while p53 knockout resulted in constitutive filopodia formation.29 Further studies on the Cdc42 pathway indicated that p53 activity lies downstream of Cdc42, suggesting a mechanism independent of p53 transcriptional function.29 Given the results here indicating that MRCKβ is a p53 kinase, we speculated that cell migration mediated by the Cdc42 pathway involves phosphorylation of p53 by activated MRCKβ. Prior work also indicated that p53 levels were augmented after Cdc42 knockdown,31–32 suggesting that the Cdc42 pathway targets p53 for degradation. Taken together, we hypothesized that Cdc42-MRCKβ phosphorylates p53 targets p53 for degradation to enhance cell motility. Consistent with this hypothesis, knockdown of MRCKβ resulted in elevated p53 protein levels (Figure 3D). Additional experiments exploring p53 phosphorylation and degradation related to MRCKβ-dependent cell motility are needed to further test this proposed model. However, the K-CLIP data implicate p53 as a negative regulator of Cdc42-mediated cell motility and suggest that p53-deficient cancer cells display enhanced cell spreading and metastasis. These studies document the value of K-CLIP to facilitate discovery of novel hypothesis in cell biology related to phosphorylation-mediated signaling.
Beyond the three kinases identified by K-CLIP, 125 non-kinase proteins were also observed (Table S1). Most of these non-kinase hits bind directly or indirectly to p53 (81%, Figure 5A). In addition, the many proteins in interacting clusters, or structurally or functionally related to known p53 interacting proteins, further confirmed the interactome analysis. Among these proteins, most are involved in metabolic function according to gene ontology (GO) analysis (Figure S17),64 consistent with the known activities of p53 in cellular events. Of particular note, many proteins associated with DNA damage and repair were identified (Figure 5C). Deficiency and/or mutation of p53 are found is roughly 50 % of cancer cells,65 and has been linked to the loss of proper DNA damage events in the cell. The K-CLIP data are consistent with the role of phosphorylation-mediated regulation of p53 upon DNA damage.
In addition to DNA damage proteins, a cluster containing spliceosome and proteosome proteins was identified by K-CLIP (Figure 5D). Prior work reported that p53 activity is upregulated by disruption of splicing,66 and p53 interacts with the proteosomal regulatory protein PSME3, consistent with the K-CLIP study.67 Likewise, a complex of transport proteins was observed with K-CLIP (Figure 5F), including NAT10, which was recently shown to bind and acetylate p53.68 The p53 protein contains a nuclear localization signal to bind importins, and two importin family members were observed in the transport complex.69 The presence of transport proteins among the K-CLIP hits implicates p53 phosphorylation as a regulator of its nuclear transport. Beyond known interactions and functions of p53, new roles of p53 were implicated by the K-CLIP study. For example, a number of coatomer proteins were observed (Figure 5F; COPA, COPB1, COPB2, COPG1, and COPD/ARCN1), although the role of p53 in coatomer function is unexplored. The close proximity of p53 to coatomer proteins in the K-CLIP study suggests a role for p53 phosphorylation in coatomer function. In summary, the K-CLIP method identified both known and unanticipated p53 interacting proteins. The study reinforced the known functions, while also implicating new roles for p53 phosphorylation in cellular events.
A variety of methods are available to monitor or identify protein interaction networks. In addition to classic methods, such as immunoprecipitation and two-hybrid methods, modern approaches include proximity-dependent biotinylation (BioID technology).70–71 K-CLIP represents a unique alternative to these methods by requiring kinase-catalyzed phosphorylation for crosslinking. With K-CLIP, non-kinase proteins identified were likely located in close proximity to a phosphorylated residue. Consequently, K-CLIP offers the unprecedented ability to discover or monitor dynamic protein-protein interactions located adjacent to a phosphorylation event. Another important feature of K-CLIP is the use of affinity purification, which promotes identification of proteins regardless of cellular abundance. In fact, an abundance analysis of the 128 K-CLIP hits documents that even low abundance proteins were observed (Figure S18, 2 to 2029 ppm abundance values for K-CLIP proteins compared to the full 0.01 to 10,000 range of RKO cells),72 consistent with the abundance-independence of the technology. Given the paucity of tools to discover unanticipated protein-protein interactions, and particularly those of low abundance near a dynamically regulated phosphorylation site, K-CLIP provides a powerful discovery tool to assist in mapping cellular pathways.
In addition to use of K-CLIP as a tool to monitor and identify dynamic kinase-substrate and protein-protein interactions, kinase-catalyzed crosslinking was also used to validate the relationship of a speculated kinase-substrate pair. The critical requirement of kinase-catalyzed crosslinking is that the substrates must be actively phosphorylated by a kinase under the conditions of the experiment. In the work reported here, DNA-PK and p53 were observed in complexes after kinase-catalyzed crosslinking (Figure 4A) and K-CLIP (Table 1), whereas PLK3 was absent despite its presence in the lysates (Figure 4B and Table 1). Likewise, MRCKβ was confirmed as a p53 substrates using K-CLIP (Figure 3B). Therefore, unlike other in vitro phosphorylation assays, kinase-catalyzed crosslinking reflects the dynamic kinase activity in a cellular context, providing a novel tool for validating kinase-substrate pairs.
Conclusion
K-CLIP provides a versatile tool to explore phosphorylation-mediated cell biology. K-CLIP requires only a relevant cell lysate and an antibody to the protein of interest, without the need for overexpression, knockdown, or mutagenesis. By covalently linking substrate with kinases and associated proteins, the method is capable of revealing the multi-protein complexes near a phosphorylation site in a specific cellular context. In addition to identifying the cellular kinases of a substrate of interest, as described here for p53, K-CLIP can also be used to identify the variety of cellular substrates of a kinase of interest, which is an application ongoing in our laboratory. In total, K-CLIP represents a multifunctional and enabling tool for studying complicated phosphorylation-mediated signaling networks in cell biology.
Supplementary Material
Acknowledgements
We thank N. Archige, A. Fouda, A. Gamage, and V. Ramanayake-Mudiyanselage for comments on the manuscript.
Funding Sources
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R01GM079529. Wayne State University Proteomics Core for mass spectrometric analysis was also supported by NIH Center Grant P30 ES020957, the NIH Cancer Center Support Grant P30 CA022453, and the NIH Shared Instrumentation Grant S10 OD010700. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We also thank Wayne State University for funding,
ABBREVIATIONS
- K-CLIP
Kinase-catalyzed Crosslinking and Immunoprecipation
- ATP
Adenosine 5’-triphosphate
- ATP-ArN3
ATP-arylazide
- ATP-BP
ATP-benzophenone
- UV
ultraviolet
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- ATCC
American Type Culture Collection
- DNA-PK or PRKDC
DNA-dependent protein kinase
- EIF2AK2 or PKR
Interferon-induced double-stranded RNA-activated protein kinase
- CDC42BPB or MRCKβ
myotonic dystrophy-related Cdc42-binding kinases beta
- PLK3
Polo-like kinase 3
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- ARS
aminoacyl tRNA synthetase complex
- AIMP2
ARS-interacting, multifunctional proteins 2
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
-Additional experimental details and supporting data (PDF)
References
- 1.Hunter T, Protein kinases and phosphatases: The yin and yang of protein phosphorylation and signaling. Cell 1995, 80, 225–236. [DOI] [PubMed] [Google Scholar]
- 2.Cohen P; Alessi DR, Kinase drug discovery--what’s next in the field? ACS chemical biology 2013, 8 (1), 96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gowda C; Soliman M; Kapadia M; Ding Y; Payne K; Dovat S, Casein Kinase II (CK2), Glycogen Synthase Kinase-3 (GSK-3) and Ikaros mediated regulation of leukemia. Advances in biological regulation 2017, 65, 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaffer J; Adams JA, Detection of conformational changes along the kinetic pathway of protein kinase A using a catalytic trapping technique. Biochemistry 1999, 38 (37), 12072–9. [DOI] [PubMed] [Google Scholar]
- 5.Scott JD; Pawson T, Cell signaling in space and time: where proteins come together and when they’re apart. Science 2009, 326 (5957), 1220–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blethrow JD; Glavy JS; Morgan DO; Shokat KM, Covalent capture of kinase-specific phosphopeptides reveals Cdk1-cyclin B substrates. Proc Natl Acad Sci U S A 2008, 105 (5), 1442–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maly DJ; Allen JA; Shokat KM, A Mechanism-Based Cross-Linker for the Identification of Kinase-Substrate Pairs. Journal of the American Chemical Society 2004, 126 (30), 9160–9161. [DOI] [PubMed] [Google Scholar]
- 8.Suwal S; Pflum MH, Phosphorylation-dependent kinase-substrate cross-linking. Angew Chem Int Ed Engl 2010, 49 (9), 1627–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garre S; Senevirathne C; Pflum MK, A comparative study of ATP analogs for phosphorylation-dependent kinase-substrate crosslinking. Bioorg Med Chem 2014, 22 (5), 1620–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dedigama-Arachchige PM; Pflum MK, K-CLASP: A Tool to Identify Phosphosite Specific Kinases and Interacting Proteins. ACS chemical biology 2016, 11 (12), 3251–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suwal S; Senevirathne C; Garre S; Pflum MK, Structural Analysis of ATP Analogues Compatible with Kinase-Catalyzed Labeling. Bioconjug Chem 2012, 23 (12), 2386–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meek David W., Regulation of the p53 response and its relationship to cancer. Biochemical Journal 2015, 469 (3), 325–346. [DOI] [PubMed] [Google Scholar]
- 13.Petitjean A; Mathe E; Kato S; Ishioka C; Tavtigian SV; Hainaut P; Olivier M, Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat 2007, 28 (6), 622–9. [DOI] [PubMed] [Google Scholar]
- 14.Hu W; Feng Z; Levine AJ, The Regulation of Multiple p53 Stress Responses is Mediated through MDM2. Genes & cancer 2012, 3 (3–4), 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vassilev LT; Vu BT; Graves B; Carvajal D; Podlaski F; Filipovic Z; Kong N; Kammlott U; Lukacs C; Klein C; Fotouhi N; Liu EA, In Vivo Activation of the p53 Pathway by Small-Molecule Antagonists of MDM2. Science 2004, 303 (5659), 844–848. [DOI] [PubMed] [Google Scholar]
- 16.Thompson T; Tovar C; Yang H; Carvajal D; Vu BT; Xu Q; Wahl GM; Heimbrook DC; Vassilev LT, Phosphorylation of p53 on Key Serines Is Dispensable for Transcriptional Activation and Apoptosis. Journal of Biological Chemistry 2004, 279 (51), 53015–53022. [DOI] [PubMed] [Google Scholar]
- 17.Kruse J-P; Gu W, Modes of p53Regulation. Cell 2009, 137 (4), 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maclaine NJ; Hupp TR, The regulation of p53 by phosphorylation: a model for how distinct signals integrate into the p53 pathway. Aging 2009, 1 (5), 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradford MM, A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976, 72, 248–54. [DOI] [PubMed] [Google Scholar]
- 20.Shevchenko A; Wilm M; Vorm O; Mann M, Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Analytical chemistry 1996, 68 (5), 850–8. [DOI] [PubMed] [Google Scholar]
- 21.Ayed A; Mulder FA; Yi GS; Lu Y; Kay LE; Arrowsmith CH, Latent and active p53 are identical in conformation. Nature structural biology 2001, 8 (9), 756–60. [DOI] [PubMed] [Google Scholar]
- 22.Smith ML; Chen IT; Zhan Q; O Connor PM; Fornace AJ Jr., Involvement of the p53 tumor suppressor in repair of u.v.-type DNA damage. Oncogene 1995, 10 (6), 1053–9. [PubMed] [Google Scholar]
- 23.Michael D; Oren M, The p53-Mdm2 module and the ubiquitin system. Seminars in cancer biology 2003, 13 (1), 49–58. [DOI] [PubMed] [Google Scholar]
- 24.Woo RA; Jack MT; Xu Y; Burma S; Chen DJ; Lee PW, DNA damage-induced apoptosis requires the DNA-dependent protein kinase, and is mediated by the latent population of p53. The EMBO journal 2002, 21 (12), 3000–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woo RA; McLure KG; Lees-Miller SP; Rancourt DE; Lee PW, DNA-dependent protein kinase acts upstream of p53 in response to DNA damage. Nature 1998, 394 (6694), 700–4. [DOI] [PubMed] [Google Scholar]
- 26.Achanta G; Pelicano H; Feng L; Plunkett W; Huang P, Interaction of p53 and DNA-PK in response to nucleoside analogues: potential role as a sensor complex for DNA damage. Cancer research 2001, 61 (24), 8723–9. [PubMed] [Google Scholar]
- 27.Cuddihy AR; Wong AH; Tam NW; Li S; Koromilas AE, The double-stranded RNA activated protein kinase PKR physically associates with the tumor suppressor p53 protein and phosphorylates human p53 on serine 392 in vitro. Oncogene 1999, 18 (17), 2690–702. [DOI] [PubMed] [Google Scholar]
- 28.Lees-Miller SP; Sakaguchi K; Ullrich SJ; Appella E; Anderson CW, Human DNA-activated protein kinase phosphorylates serines 15 and 37 in the amino-terminal transactivation domain of human p53. Mol Cell Biol 1992, 12 (11), 5041–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gadea G; Lapasset L; Gauthier-Rouviere C; Roux P, Regulation of Cdc42-mediated morphological effects: a novel function for p53. EMBO J 2002, 21 (10), 2373–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gadea G; de Toledo M; Anguille C; Roux P, Loss of p53 promotes RhoA-ROCK-dependent cell migration and invasion in 3D matrices. The Journal of cell biology 2007, 178 (1), 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park SY; Lee JH; Ha M; Nam JW; Kim VN, miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol 2009, 16 (1), 23–9. [DOI] [PubMed] [Google Scholar]
- 32.Ma J; Xue Y; Liu W; Yue C; Bi F; Xu J; Zhang J; Li Y; Zhong C; Chen Y, Role of activated Rac1/Cdc42 in mediating endothelial cell proliferation and tumor angiogenesis in breast cancer. PloS one 2013, 8 (6), e66275. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Xie S; Wu H; Wang Q; Cogswell JP; Husain I; Conn C; Stambrook P; Jhanwar-Uniyal M; Dai W, Plk3 functionally links DNA damage to cell cycle arrest and apoptosis at least in part via the p53 pathway. The Journal of biological chemistry 2001, 276 (46), 43305–12. [DOI] [PubMed] [Google Scholar]
- 34.Xie S; Wang Q; Wu H; Cogswell J; Lu L; Jhanwar-Uniyal M; Dai W, Reactive oxygen species-induced phosphorylation of p53 on serine 20 is mediated in part by polo-like kinase-3. The Journal of biological chemistry 2001, 276 (39), 36194–9. [DOI] [PubMed] [Google Scholar]
- 35.Warde-Farley D; Donaldson SL; Comes O; Zuberi K; Badrawi R; Chao P; Franz M; Grouios C; Kazi F; Lopes CT; Maitland A; Mostafavi S; Montojo J; Shao Q; Wright G; Bader GD; Morris Q, The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010, 38 (Web Server issue), W214–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montojo J; Zuberi K; Rodriguez H; Kazi F; Wright G; Donaldson SL; Morris Q; Bader GD, GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics 2010, 26 (22), 2927–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shannon P; Markiel A; Ozier O; Baliga NS; Wang JT; Ramage D; Amin N; Schwikowski B; Ideker T, Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research 2003, 13 (11), 2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bader GD; Hogue CW, An automated method for finding molecular complexes in large protein interaction networks. BMC bioinformatics 2003, 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kastan MB; Onyekwere O; Sidransky D; Vogelstein B; Craig RW, Participation of p53 protein in the cellular response to DNA damage. Cancer Res 1991, 51 (23 Pt 1), 6304–11. [PubMed] [Google Scholar]
- 40.Han JM; Park BJ; Park SG; Oh YS; Choi SJ; Lee SW; Hwang SK; Chang SH; Cho MH; Kim S, AIMP2/p38, the scaffold for the multi-tRNA synthetase complex, responds to genotoxic stresses via p53. Proc Nati Acad Sci U S A 2008, 105 (32), 11206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen HC; Lin WC; Tsay YG; Lee SC; Chang CJ, An RNA helicase, DDX1, interacting with poly(A) RNA and heterogeneous nuclear ribonucleoprotein K. The Journal of biological chemistry 2002, 277 (43), 40403–9. [DOI] [PubMed] [Google Scholar]
- 42.Moumen A; Masterson P; O‘Connor MJ; Jackson SP, hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell 2005, 123 (6), 1065–78. [DOI] [PubMed] [Google Scholar]
- 43.Bates GJ; Nicol SM; Wilson BJ; Jacobs AM; Bourdon JC; Wardrop J; Gregory DJ; Lane DP; Perkins ND; Fuller-Pace FV, The DEAD box protein p68: a novel transcriptional coactivator of the p53 tumour suppressor. EMBO J 2005, 24 (3), 543–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun M; Zhou T; Jonasch E; Jope RS, DDX3 regulates DNA damage-induced apoptosis and p53 stabilization. Biochimica et biophysica acta 2013, 1833 (6), 1489–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cooper B; Schneider S; Bohl J; Jiang Y; Beaudet A; Vande Pol S, Requirement of E6AP and the features of human papillomavirus E6 necessary to support degradation of p53. Virology 2003, 306 (1), 87–99. [DOI] [PubMed] [Google Scholar]
- 46.Barel M; Balbo M; Gauffre A; Frade R, Binding sites of the Epstein-Barr virus and C3d receptor (CR2, CD21) for its three intracellular ligands, the p53 anti-oncoprotein, the p68 calcium binding protein and the nuclear p120 ribonucleoprotein. Molecular Immunology 1995, 32 (6), 389–397. [DOI] [PubMed] [Google Scholar]
- 47.Kumazawa T; Nishimura K; Kuroda T; Ono W; Yamaguchi C; Katagiri N; Tsuchiya M; Masumoto H; Nakajima Y; Murayama A; Kimura K; Yanagisawa J, Novel Nucleolar Pathway Connecting Intracellular Energy Status with p53 Activation. Journal of Biological Chemistry 2011, 286 (23), 20861–20869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gottlieb TM; Jackson SP, The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell 1993, 72 (1), 131–42. [DOI] [PubMed] [Google Scholar]
- 49.Ariumi Y; Masutani M; Copeland TD; Mimori T; Sugimura T; Shimotohno K; Ueda K; Hatanaka M; Noda M, Suppression of the poly(ADP-ribose) polymerase activity by DNA-dependent protein kinase in vitro. Oncogene 1999, 18 (32), 4616–25. [DOI] [PubMed] [Google Scholar]
- 50.Nakajima T; Uchida C; Anderson SF; Lee CG; Hurwitz J; Parvin JD; Montminy M, RNA helicase A mediates association of CBP with RNA polymerase II. Cell 1997, 90 (6), 1107–12. [DOI] [PubMed] [Google Scholar]
- 51.Jha S; Shibata E; Dutta A, Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol Cell Biol 2008, 28 (8), 2690–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bangs P; Burke B; Powers C; Craig R; Purohit A; Doxsey S, Functional analysis of Tpr: identification of nuclear pore complex association and nuclear localization domains and a role in mRNA export. The Journal of cell biology 1998, 143 (7), 1801–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blalock WL; Piazzi M; Bavelloni A; Raffini M; Faenza I; D‘Angelo A; Cocco L, Identification of the PKR nuclear interactome reveals roles in ribosome biogenesis, mRNA processing and cell division. Journal of cellular physiology 2014, 229 (8), 1047–60. [DOI] [PubMed] [Google Scholar]
- 54.Chatr-Aryamontri A; Oughtred R; Boucher L; Rust J; Chang C; Kolas NK; O‘Donnell L; Oster S; Theesfeld C; Sellam A; Stark C;Breitkreutz BJ; Dolinski K; Tyers M, The BioGRID interaction database: 2017 update. Nucleic Acids Res 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tomsig JL; Snyder SL; Creutz CE, Identification of targets for calcium signaling through the copine family of proteins. Characterization of a coiled-coil copine-binding motif. The Journal of biological chemistry 2003, 278 (12), 10048–54. [DOI] [PubMed] [Google Scholar]
- 56.Tomsig JL; Sohma H; Creutz CE, Calcium-dependent regulation of tumour necrosis factor-alpha receptor signalling by copine. Biochem J 2004, 378 (Pt 3), 1089–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leung T; Chen XQ; Tan I; Manser E; Lim L, Myotonic dystrophy kinase-related Cdc42-binding kinase acts as a Cdc42 effector in promoting cytoskeletal reorganization. Molecular and cellular biology 1998, 18 (1), 130–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilkinson S; Paterson HF; Marshall CJ, Cdc42-MRCK and Rho-ROCK signalling cooperate in myosin phosphorylation and cell invasion. Nature cell biology 2005, 7 (3), 255–61. [DOI] [PubMed] [Google Scholar]
- 59.Fields S; Song O, A novel genetic system to detect protein-protein interactions. Nature 1989, 340 (6230), 245–6. [DOI] [PubMed] [Google Scholar]
- 60.Amanchy R; Periaswamy B; Mathivanan S; Reddy R; Tattikota SG; Pandey A, A curated compendium of phosphorylation motifs. Nature biotechnology 2007, 25 (3), 285–6. [DOI] [PubMed] [Google Scholar]
- 61.Pflum MK; Tong JK; Lane WS; Schreiber SL, Histone deacetylase 1 phosphorylation promotes enzymatic activity and complex formation. The Journal of biological chemistry 2001, 276 (50), 47733–41. [DOI] [PubMed] [Google Scholar]
- 62.Shieh SY; Ikeda M; Taya Y; Prives C, DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 1997, 91 (3), 325–34. [DOI] [PubMed] [Google Scholar]
- 63.Unbekandt M; Olson MF, The actin-myosin regulatory MRCK kinases: regulation, biological functions and associations with human cancer. J Mol Med (Berl) 2014, 92 (3), 217–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mi H; Muruganujan A; Thomas PD, PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res 2013, 41 (Database issue), D377–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hollstein M; Rice K; Greenblatt MS; Soussi T; Fuchs R; Sorlie T; Hovig E; Smith-Sorensen B; Montesano R; Harris CC, Database of p53 gene somatic mutations in human tumors and cell lines. Nucleic Acids Res 1994, 22 (17), 3551–5. [PMC free article] [PubMed] [Google Scholar]
- 66.Allende-Vega N; Dayal S; Agarwala U; Sparks A; Bourdon JC; Saville MK, p53 is activated in response to disruption of the pre-mRNA splicing machinery. Oncogene 2013, 32 (1), 1–14. [DOI] [PubMed] [Google Scholar]
- 67.Zhang Z; Zhang R, Proteasome activator PA28 gamma regulates p53 by enhancing its MDM2-mediated degradation. EMBO J 2008, 27 (6), 852–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu X; Tan Y; Zhang C; Zhang Y; Zhang L; Ren P; Deng H; Luo J; Ke Y; Du X, NAT10 regulates p53 activation through acetylating p53 at K120 and ubiquitinating Mdm2. EMBO reports 2016, 17 (3), 349–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Komlodi-Pasztor E; Trostel S; Sackett D; Poruchynsky M; Fojo T, Impaired p53 binding to importin: a novel mechanism of cytoplasmic sequestration identified in oxaliplatin-resistant cells. Oncogene 2009, 28 (35), 3111–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Roux KJ; Kim DI; Raida M; Burke B, A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. The Journal of Cell Biology 2012, 196 (6), 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Varnaite R; MacNeill SA, Meet the neighbours: mapping local protein interactomes by proximity-dependent labelling with BioID. Proteomics 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Geiger T; Wehner A; Schaab C; Cox J; Mann M, Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol Cell Proteomics 2012,11 (3), M111 014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.