The majority of characterized B. animalis strains have been isolated from human fecal samples. In order to explore genome variability within this species, we isolated 15 novel strains from the gastrointestinal tracts of different animals, including mammals and birds. The present study allowed us to reconstruct the pangenome of this taxon, including the genome contents of 56 B. animalis strains. Through careful assessment of subspecies-specific core genes of the B. animalis subsp. animalis/lactis taxon, we identified genes encoding enzymes involved in carbohydrate transport and metabolism, while unveiling specific gene acquisition and loss events that caused the evolutionary emergence of these two subspecies.
KEYWORDS: Bifidobacterium, bifidobacteria, pangenome, phylogeny, probiotic
ABSTRACT
Bifidobacteria are members of the gut microbiota of animals, including mammals, birds, and social insects. In this study, we analyzed and determined the pangenome of Bifidobacterium animalis species, encompassing B. animalis subsp. animalis and the B. animalis subsp. lactis taxon, which is one of the most intensely exploited probiotic bifidobacterial species. In order to reveal differences within the B. animalis species, detailed comparative genomics and phylogenomics analyses were performed, indicating that these two subspecies recently arose through divergent evolutionary events. A subspecies-specific core genome was identified for both B. animalis subspecies, revealing the existence of subspecies-defining genes involved in carbohydrate metabolism. Notably, these in silico analyses coupled with carbohydrate profiling assays suggest genetic adaptations toward a distinct glycan milieu for each member of the B. animalis subspecies, resulting in a divergent evolutionary development of the two subspecies.
IMPORTANCE The majority of characterized B. animalis strains have been isolated from human fecal samples. In order to explore genome variability within this species, we isolated 15 novel strains from the gastrointestinal tracts of different animals, including mammals and birds. The present study allowed us to reconstruct the pangenome of this taxon, including the genome contents of 56 B. animalis strains. Through careful assessment of subspecies-specific core genes of the B. animalis subsp. animalis/lactis taxon, we identified genes encoding enzymes involved in carbohydrate transport and metabolism, while unveiling specific gene acquisition and loss events that caused the evolutionary emergence of these two subspecies.
INTRODUCTION
Bifidobacteria are Gram-positive, anaerobic, nonmotile, and non-spore-forming bacteria, which are commonly found in the gastrointestinal tracts (GITs) of various animals, the human oral cavity, and sewage (1). Bifidobacterial species residing in the human GIT are believed to support host health in providing energy and nutrients, modulating the immune system and adjusting the gut physiology of the host (2–5). Currently, 72 different species of bifidobacteria have been identified and, depending on the species, more or less characterized (6). Among this large number of bifidobacterial taxa, just a few species, including Bifidobacterium animalis (7, 8), Bifidobacterium bifidum (9, 10), Bifidobacterium breve (11), and Bifidobacterium longum (12, 13), have been exploited as health-promoting bacteria. In particular, B. animalis strains have been extensively used as active ingredients in a variety of functional foods (14, 15). The B. animalis species consists of two subspecies, B. animalis subsp. animalis and B. animalis subsp. lactis (16). Of these two taxa, only members of B. animalis subsp. lactis have been utilized for their health-promoting purposes (17). To date, a number of scientific publications have investigated the purported probiotic features of a number of B. animalis subsp. lactis strains, such as their protective behavior against periodontitis (18), their ability to improve GIT health in abdominal discomfort and obesity disorder states (19, 20), and the inhibition of pathogenic bacteria (21).
Before the advent of next-generation sequencing (NGS) methods, the classification criteria to discriminate (what were then called) B. lactis and B. animalis were based on phenotypic characteristics, such as morphology and carbohydrate fermentation abilities. However, 16S rRNA gene sequence comparison, combined with DNA-DNA hybridization of the type strains of these two taxa, led to the proposal to consider B. lactis as a junior, synonymous taxon of the B. animalis species (22). Subsequently, using a polyphasic approach, B. animalis and B. lactis were reclassified as B. animalis subsp. animalis and B. animalis subsp. lactis, respectively (16). Various genomic studies have revealed the existence of a high level of genome synteny between the two B. animalis subspecies (23, 24), as well as similar levels of acid, heat, and oxygen tolerance (25, 26).
To date, comprehensive comparative genomic analyses of bifidobacterial taxa have been performed (23, 27–29). In this context, members of the B. bifidum, B. breve, and B. longum species have been shown to exhibit a closed pangenome structure, revealing the presence of specific genetic strategies to establish and persist in the human gut, such as through the production of various types of pili (30, 31) or metabolic capabilities toward particular host glycans (32, 33). In the same fashion, members of the B. animalis subsp. lactis taxon have been investigated through genomic decoding. Notably, such analyses involved B. animalis subsp. lactis strains, which were isolated from the human GIT and dairy products. Overall, their genetic characterization highlighted the presence of a very modest number of genomic differences (23). Conversely, genotypic and phenotypic analyses of B. animalis subsp. lactis strains from commercial products and animals revealed some distinct differences in fermentation profiles and peptide mass fingerprints (34). In contrast to these investigations involving B. animalis subsp. lactis, very little investigative work has been done on members of the B. animalis subsp. animalis taxon.
The aim of this study was to investigate the genetic biodiversity of the B. animalis species by decoding genome sequences of isolates collected from the GITs of various animals, including mammals and birds. The identification of the genomic makeup of members belonging to either of the two subspecies is considered crucial in order to provide information regarding the subspecies-specific repertoire of genes that may have caused their evolutionary differentiation. Furthermore, such genomic analyses, combined with carbohydrate profiling experiments, support the hypothesis that the two B. animalis subspecies have been subject to genetic adaptations to environments that had a distinct glycan content.
RESULTS AND DISCUSSION
Isolation and genetic characterization of the B. animalis species.
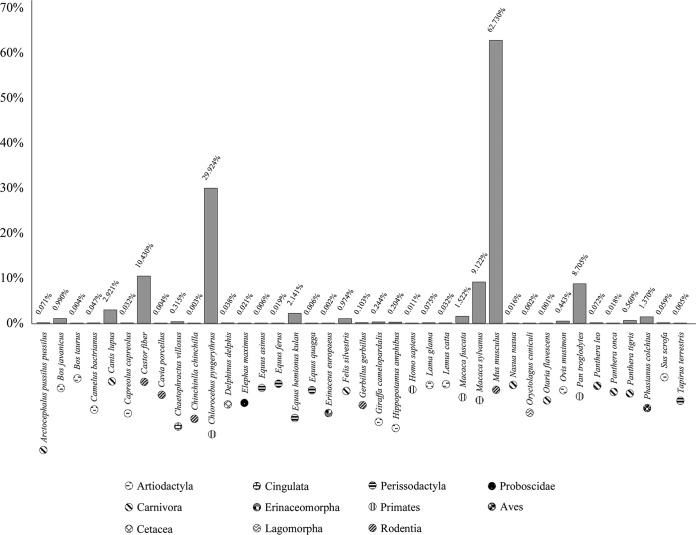
To investigate the occurrence of B. animalis in the gut of animals, we screened the internally transcribed spacer (ITS) sequence profiling data derived from fecal samples of four mammalian and bird species, together with the bifidobacterial community data previously determined by Milani et al. (35) (Fig. 1). In this context, B. animalis was detected in 55% of such fecal samples, with a higher occurrence in the fecal samples of dogs (Canis lupus), onagers (Equus hemionus kulan), monkeys (Chlorocebus pygerytrhus, Macaca fuscata, Macaca sylvanus, and Pan troglodytes), and mice (Mus musculus) (Fig. 1). These data revealed a cosmopolitan lifestyle of this taxon, underlining the potential high genetic adaptation of B. animalis strains to different (host) environments.
FIG 1.
B. animalis profiling data obtained from fecal samples of different animals. In this bar plot, the x axis represents the animals tested for the presence of B. animalis, while the y axis represents the percentage of B. animalis compared to other Bifidobacterium species present in the samples. Each pattern represents an animal order, as indicated in the key.
In order to investigate the genetic contents of the B. animalis species, including representatives of both B. animalis subsp. animalis and B. animalis subsp. lactis taxa, we applied a bifidobacterial isolation protocol on fecal samples of animal species displaying a high abundance of these taxa. The above-mentioned analyses (see Materials and Methods) (36, 37) allowed the isolation of 15 novel B. animalis strains from birds (Phasianus colchicus) and various Mammalia, such as canine breeds, i.e., German shepherd, Pomeranian, Alaskan malamute, and flat-coated retriever, and three different nonhuman primates, i.e., Pan troglodytes, Chlorocebus pygerythrus, and Macaca sylvanus. Moreover, B. animalis subsp. lactis/animalis strains were isolated from fecal samples of rabbits (Oryctolagus cuniculus), beavers (Castor fiber), and pigs (Sus scrofa domesticus) (Table 1 and Fig. 1). Interestingly, we were also able to isolate different B. animalis strains from stool samples of animals in which the ITS bifidobacterial profiling analysis indicated a low relative abundance of this species, i.e., Sus scrofa (0.06%) and Oryctolagus cuniculis (0.002%). This may be due to the better growth performance (e.g., high tolerance to environmental stresses) of members of the B. animalis species compared to other bifidobacteria (38–40). A comparative genomic analysis between newly isolated strains was complemented with the inclusion of publicly available genomic repertoire of 41 B. animalis strains, thereby exemplifying a broad ecological representation, including the GITs of human and other animals (e.g., rats and chickens) (41), as well as different food matrices (e.g., milk and yogurt) and human vaginal swabs (23, 42) (Table 1). This information further validates the notion that B. animalis seems to be genetically adapted to a large number of habitats. Notably, the ORFeome of B. animalis subsp. animalis strains, defined as the complete set of open reading frames (ORFs) in genomes of the same species, was shown to be substantially larger compared to that of B. animalis subsp. lactis strains, suggesting that members of the B. animalis subsp. animalis taxon exhibit a more extensive level of genetic diversity.
TABLE 1.
B. animalis strains/genomes used in this studya
| Species and strain | Ecological origin | Genome size (Mb) | No. of ORFs | GC content (%) | No. of tRNAs | rRNA locus | Coverage depth (fold) | No. of contigs | Source or reference |
|---|---|---|---|---|---|---|---|---|---|
| B. animalis subsp. animalis | |||||||||
| 2022B | Castor fiber feces | 2.4 | 1,935 | 61.08 | 65 | 2 | 142 | 17 | RSDC00000000 |
| 2006B | Canis lupus familiaris (German shepherd) feces | 2.16 | 1,747 | 61.24 | 56 | 3 | 195 | 47 | RSDB00000000 |
| ATCC 25527 | Human feces | 1.93 | 1,622 | 61.35 | 52 | 2 | 24 | ||
| ATCC 27672 | Rat feces | 1.99 | 1,611 | 60.97 | 52 | 1 | NCBI database | ||
| IM386 | Human feces | 1.93 | 1,623 | 61.35 | 52 | 1 | NCBI database | ||
| LMG10508 | Rat feces | 1.92 | 1,619 | 60.53 | 52 | 2 | NCBI database | ||
| MCC0483 | Rat feces | 2.18 | 1,922 | 60.97 | 53 | 1 | NCBI database | ||
| MCC0499 | Rat feces | 2.13 | 1,870 | 61.05 | 62 | 1 | NCBI database | ||
| MCC1489 | Guinea pig feces | 1.91 | 1,619 | 61.35 | 52 | 2 | NCBI database | ||
| YL2 | Rat feces | 2.02 | 1,705 | 61.1 | 52 | 3 | 75 | ||
| B. animalis subsp. lactis | |||||||||
| 646 | Human feces | 1.92 | 1,673 | 61.4 | 52 | 4 | NCBI database | ||
| 1316B | Phasianus colchicus feces | 1.92 | 1,556 | 60.47 | 52 | 3 | 195 | 14 | RSDA00000000 |
| 1395B | Oryctolagus cuniculus feces | 1.92 | 1,557 | 60.47 | 52 | 2 | 99 | 12 | RSCZ00000000 |
| 1528B | Sus scrofa domesticus feces | 1.95 | 1,600 | 61.47 | 55 | 2 | 97 | 12 | RSCY00000000 |
| 1802B | Macaca sylvanus feces | 1.92 | 1,557 | 61.36 | 52 | 2 | 132 | 15 | RSCX00000000 |
| 1808B | Chlorocebus pygerytrhus feces | 1.92 | 1,556 | 61.35 | 52 | 2 | 87 | 15 | RSCW00000000 |
| 1811B | Chlorocebus pygerytrhus feces | 1.68 | 1,560 | 61.37 | 52 | 2 | 156 | 16 | RSCV00000000 |
| 1813B | Pan troglodytes feces | 1.68 | 1,557 | 61.36 | 52 | 2 | 247 | 12 | RSCU00000000 |
| 1821B | Pan troglodytes feces | 1.75 | 1,636 | 60.71 | 53 | 2 | 278 | 40 | RSCT00000000 |
| 1843B | Pan troglodytes feces | 1.68 | 1,557 | 61.36 | 52 | 2 | 85 | 14 | RSCS00000000 |
| 1869B | Pan troglodytes feces | 1.92 | 1,557 | 61.36 | 52 | 2 | 229 | 14 | RSCR00000000 |
| 2007B | Canis lupus familiaris (Pomeranian) feces | 1.97 | 1,599 | 61.28 | 52 | 1 | 194 | 25 | RSCQ00000000 |
| 2010B | Canis lupus familiaris (Alaskan malamute) feces | 1.98 | 1,607 | 61.21 | 52 | 2 | 121 | 29 | RSCP00000000 |
| 2011B | Canis lupus familiaris (Flat coated retriever) feces | 2.08 | 1,700 | 61.32 | 54 | 1 | 181 | 44 | RSCO00000000 |
| A6 | Human feces | 1.96 | 1,651 | 61.38 | 52 | 5 | NCBI database | ||
| AD011 | Infant feces | 1.93 | 1,642 | 61.38 | 52 | 2 | 76 | ||
| ATCC 27536 | Chicken feces | 1.91 | 1,632 | 61.35 | 52 | 1 | NCBI database | ||
| ATCC 27673 | Fermented milk sample | 1.95 | 1,685 | 61.52 | 52 | 3 | 77 | ||
| ATCC 27674 | Rabbit feces | 1.91 | 1,629 | 61.35 | 52 | 1 | NCBI database | ||
| B420 | Human feces | 1.94 | 1,633 | 61.37 | 52 | 3 | 78 | ||
| BB-12 | Food matrices | 1.97 | 1,639 | 61.38 | 52 | 3 | 79 | ||
| BF052 | Feces of breast-fed infant | 1.94 | 1,632 | 61.38 | 52 | 3 | NCBI database | ||
| Bi-07 | Human feces | 1.94 | 1,831 | 61.38 | 52 | 3 | 78 | ||
| Bifido_08 | Human feces | 1.95 | 1,757 | 61.32 | 52 | 4 | NCBI database | ||
| Bifido_11 | Human feces | 1.94 | 1,702 | 61.32 | 52 | 4 | NCBI database | ||
| Bl_04 | Human feces | 1.94 | 1,633 | 61.38 | 52 | 3 | 5 | ||
| Bl12 | Human colonoscopic sample | 1.94 | 1,633 | 61.37 | 52 | 3 | NCBI database | ||
| BL3 | Human feces | 1.94 | 1,639 | 61.38 | 52 | 3 | 80 | ||
| BLC1 | Human feces | 1.94 | 1,630 | 61.37 | 52 | 3 | 81 | ||
| BS01 | Human feces | 1.93 | 1,632 | 61.37 | 52 | 1 | NCBI database | ||
| CECT8145 | Infant feces | 1.96 | 1,766 | 61.38 | 52 | 1 | NCBI database | ||
| CNCMI-2494 | Human feces | 1.94 | 1,635 | 61.38 | 52 | 3 | 82 | ||
| DS1_2 | Human feces | 1.92 | 1,636 | 61.36 | 52 | 2 | NCBI database | ||
| DS11_2 | Human feces | 1.92 | 1,637 | 61.36 | 52 | 2 | NCBI database | ||
| DS15_2 | Human feces | 1.92 | 1,635 | 61.37 | 52 | 3 | NCBI database | ||
| DS2_2 | Human feces | 1.92 | 1,634 | 61.35 | 52 | 2 | NCBI database | ||
| DS24_2 | Human feces | 1.92 | 1,670 | 61.35 | 52 | 1 | NCBI database | ||
| DS27_2 | Human feces | 1.92 | 1,642 | 61.35 | 52 | 1 | NCBI database | ||
| DS28_2 | Human feces | 1.92 | 1,633 | 61.35 | 52 | 2 | NCBI database | ||
| DSM10140 | Human feces | 1.94 | 1,635 | 61.37 | 51 | 3 | 42 | ||
| HN019 | Human feces | 1.92 | 1,645 | 61.35 | 52 | 1 | NCBI database | ||
| KLDS2.0603 | Human feces | 1.95 | 1,646 | 61.37 | 52 | 2 | NCBI database | ||
| LMG P-17502_1 | Food sample | 1.92 | 1,628 | 61.36 | 52 | 2 | NCBI database | ||
| LMG P-17502_2 | Food sample | 1.92 | 1,628 | 61.36 | 52 | 1 | NCBI database | ||
| RH | Human feces | 1.93 | 1,629 | 61.37 | 52 | 2 | NCBI database | ||
| V9 | Human feces | 1.94 | 1,633 | 61.38 | 52 | 3 | NCBI database |
The references are based on the decoding genomes project according to the NCBI database.
Pangenome and core genome analyses of B. animalis species.
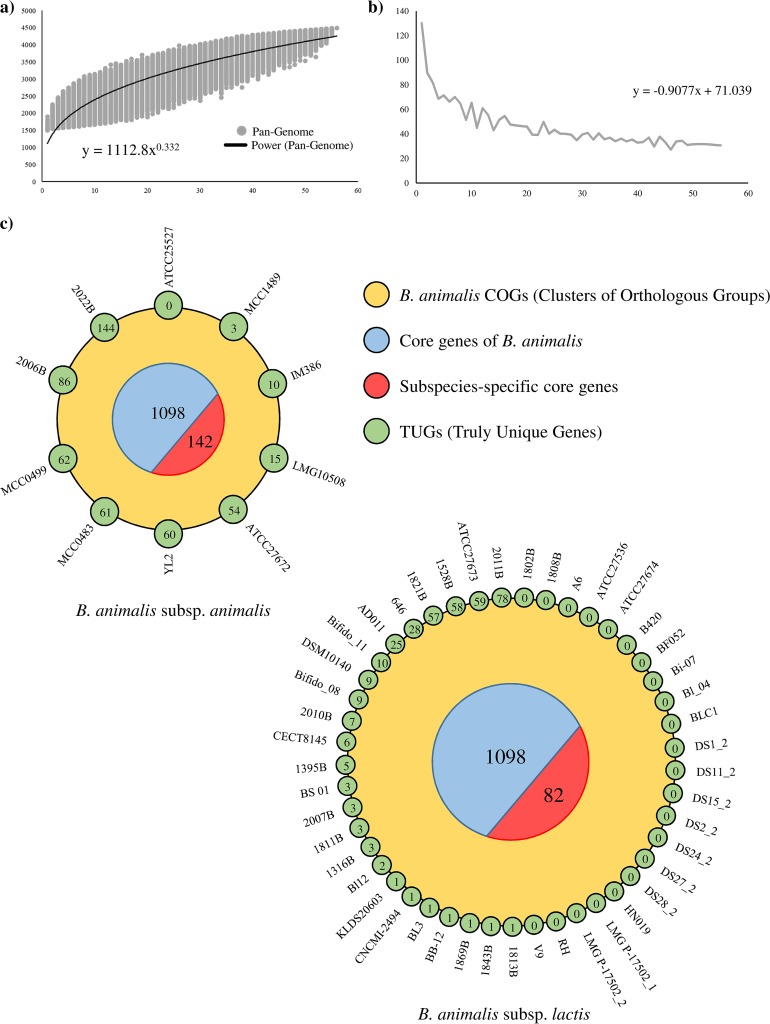
The reconstructed genomic data sets of the B. animalis species, encompassing a total of 56 chromosomal sequences, represents the genetic catalogue for this bifidobacterial species. The genetic makeup of the whole taxon was employed to predict the pangenome of the B. animalis species, i.e., the available collection of genes from strains of a given species (43). Moreover, these data were used to predict also the core genome, i.e., the collection of gene families shared between organisms of a given species, i.e., the B. animalis taxon, as based on the clusters of orthologous groups (COGs) (44). The pangenome size, consisting of 4,486 COGs, when plotted on a log-log scale as a function of the number of analyzed genomes, suggests that the power trend line has almost reached a plateau (Fig. 2). The average number of new genes discovered by sequential addition of genome sequences decreased from 130 COGs upon the addition of another genome, to 30 COGs in the final addition (Fig. 2). Thus, these findings indicate that genome sequencing of additional (novel) B. animalis strains are expected to increase the pangenome size by <0.7% (Fig. 2). Furthermore, the 56 B. animalis genomes were screened to identify shared orthologous genes, as well as unique genes. In silico analyses reveals that 1,098 ORFs were shared between the assessed strains, representing the core genome of this species. The functional examination of the core genome, based on the eggNOG database (45), reveals that 26.1% of the identified core genes are predicted to encode housekeeping functions and enzymatic activities related to amino acid and carbohydrate metabolism and their corresponding transport.
FIG 2.
Pangenome and core genome of the B. animalis species. (a) Pangenome of the B. animalis species. (b) Average of new genes upon sequential addition of the B. animalis genomes. (c) Two Venn diagrams representing shared orthologous, as well as unique, genes among the 56 B. animalis genomes. Numbers in blue circular segments represent the core genes of the B. animalis taxon, while numbers in red circular segments symbolize the subspecies-specific core genes. Moreover, the numbers of unique genes are highlighted in small green circles.
When we separately analyzed the core-genome of strains belonging to a specific B. animalis subspecies, subspecies-specific core genes could be identified (Fig. 2). In this context, 142 subspecies-specific genes were retrieved in the genomes of the B. animalis subsp. animalis subspecies, while just 82 were detected in the chromosome sequences of B. animalis subsp. lactis members. The existence of specific conserved genes among the two subspecies is suggestive of an evolutionary separation between these bifidobacterial taxa. Specifically, genes that have driven this differentiation are expected to be among the subspecies-specific core and include genes that are predicted to encode transporters and carbohydrate active proteins, i.e., 51 in the B. animalis subsp. animalis-specific and 31 in the B. animalis subsp. lactis-specific core genomes, respectively (see Table S2 in the supplemental material). Interestingly, the higher number of the above-mentioned genes in the B. animalis subsp. animalis-specific core genome compared to the corresponding number in the B. animalis subsp. lactis-specific core genome suggests that B. animalis subsp. animalis strains are able to metabolize a larger number of glycan substrates compared to B. animalis subsp. lactis strains (Table S2). Furthermore, the subspecies-specific core genomes include various DNA binding proteins, with a distinctly higher abundance in B. animalis subsp. animalis (13 genes) compared to B. animalis subsp. lactis (three genes), five of which belong to the MarR family of transcriptional regulators (Table S2). Altogether, the observed differences in the number of subspecies-specific core genes between the B. animalis subsp. animalis and B. animalis subsp. lactis were shown to be statistically significant (P < 0.05).
An in silico approach was used to calculate the average nucleotide identity (ANI) values, defined as a measure of nucleotide-level genomic similarity between the coding regions of two genomes, between B. animalis genomes (46), showing a highly syntenic genome structure among members of this species, with associated ANI values ranging from 95.81 to 99.99%. Moreover, different ranges of ANI values were identified between strains belonging to B. animalis subsp. animalis and B. animalis subsp. lactis. Interestingly, the lowest ANI value between B. animalis subsp. lactis genomes was 98.7%, while for B. animalis subsp. animalis genomes this number was 96.1%. These data reflect the differences between these two subspecies, highlighting a highly syntenic genome structure among members of the B. animalis subsp. lactis subspecies. This statement was further validated by the pangenome analysis mentioned above that allowed us to highlight truly unique genes (TUGs) of each B. animalis strain (Fig. 2). In this context, a variable number of TUGs, ranging from 0 genes for 23 B. animalis subsp. lactis strains to 144 genes for B. animalis subsp. animalis 2022B, were detected (Fig. 2). Thus, the absence of TUGs within the majority of B. animalis subsp. lactis strains supports the previously noted high isogenic nature of members of this taxon (23). Furthermore, the ANI analysis highlights that genomes of two B. animalis subsp. lactis strains, i.e., ATCC 27674 and CNCM I-2494, displayed a genetic identity of 99.9% compared to that of the prototypical probiotic bifidobacterial strain, i.e., B. animalis subsp. lactis BB-12 (17). Thus, we can speculate that the latter strains exhibit similar probiotic characteristics (47). Nevertheless, additional functional genomics analyses coupled with in vivo studies should be performed in order to confirm this notion.
Phylogenetic analyses of the B. animalis species.
Recently, a phylogenomic assessment of members of the genus Bifidobacterium allowed the identification of nine phylogenetic groups (6). Notably, B. animalis subsp. lactis and B. animalis subsp. animalis taxa are members of the Bifidobacterium pseudolongum group, which also includes Bifidobacterium choerinum, Bifidobacterium cuniculi, Bifidobacterium gallicum, Bifidobacterium magnum, Bifidobacterium pseudolongum subsp. globosum, and Bifidobacterium pseudolongum subsp. pseudolongum (48). Accordingly, we reevaluated the evolutionary development of the 56 B. animalis strains analyzed here using a phylogenomic approach, which also included the genome sequences of the bifidobacterial type strains belonging to the B. pseudolongum phylogenetic group.
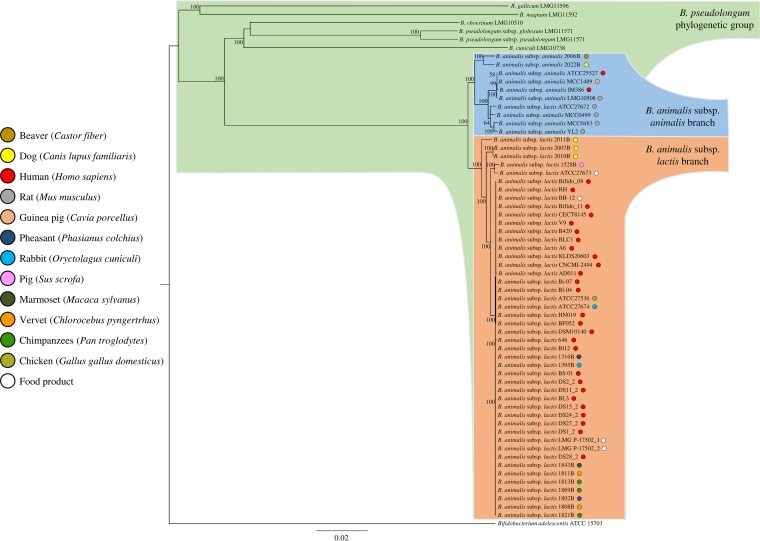
In silico analyses identified 667 orthologous genes, which were shared among sequenced genomes of the B. pseudolongum group, which were then used to build a so-called supertree (Fig. 3). This supertree showed that all 15 B. animalis strains isolated in this study cocluster with other publicly available B. animalis genomes. Furthermore, a clear division was identified between genomes belonging to the B. animalis subsp. animalis subspecies and those encompassing the B. animalis subsp. lactis subspecies (Fig. 3). As previously observed through molecular typing approaches, B. animalis subsp. lactis ATCC 27672 clusters together with members of the B. animalis subsp. animalis group, suggesting a misclassification of this strain (39). Interestingly, B. animalis subsp. lactis 2011B clusters on a separate branch with respect to other B. animalis subsp. lactis strains, suggesting that this isolate may have followed a different evolutionary pathway compared to the other members of B. animalis subsp. lactis taxon.
FIG 3.
Phylogenomic tree of the B. animalis taxa. A proteomic tree was developed based on the concatenation of 667 B. animalis core genes identified in the B. pseudolongum group phylogenomic analysis. This tree was constructed by the neighbor-joining method, and the genome sequence of Bifidobacterium adolescentis ATCC 15703 was used as outgroup. Bootstrap percentages of >50 are shown at node points, based on 1,000 replicates. Colored small circles indicate the ecological origins of each bacteria.
In order to assess the level of genetic differences between each B. animalis subspecies, we analyzed single nucleotide polymorphisms (SNPs) among genomes of this taxon, using the software Mauve (49). The number of identified SNPs was higher in B. animalis subsp. animalis genomes (123,338 SNPs) compared to those detected in the B. animalis subsp. lactis chromosomes (52,162 SNPs). In this context, 59.5% of the B. animalis subsp. animalis SNPs were identified only in two strains, i.e., B. animalis subsp. animalis 2006B and B. animalis subsp. animalis 2022B, while 54.8% of the B. animalis subsp. lactis SNPs were detected in only three strains, i.e., B. animalis subsp. lactis 2010B, B. animalis subsp. lactis 2011B, and B. animalis subsp. lactis 2007B. It should be noted that some of these differences may be correlated with the quality of the deposited genome sequences, which may have been affected by a low sequencing fold coverage. Nonetheless, strains that display the highest number of SNPs in their genomes also reflect their apparent phylogenetic distinctiveness in the supertree of the B. pseudolongum group (Fig. 3), perhaps reflecting divergent evolution compared to other members of their subspecies. Furthermore, the performed phylogenetic analysis may assist in the selection of novel probiotic strains. In this context, 18 B. animalis subsp. lactis strains cluster in the BB-12 branch (Fig. 3). Their genomic relatedness was also highlighted in the pangenome analysis, where half of the B. animalis subsp. lactis strains does not show any TUGs (Fig. 2).
Glycobiome of the B. animalis species.
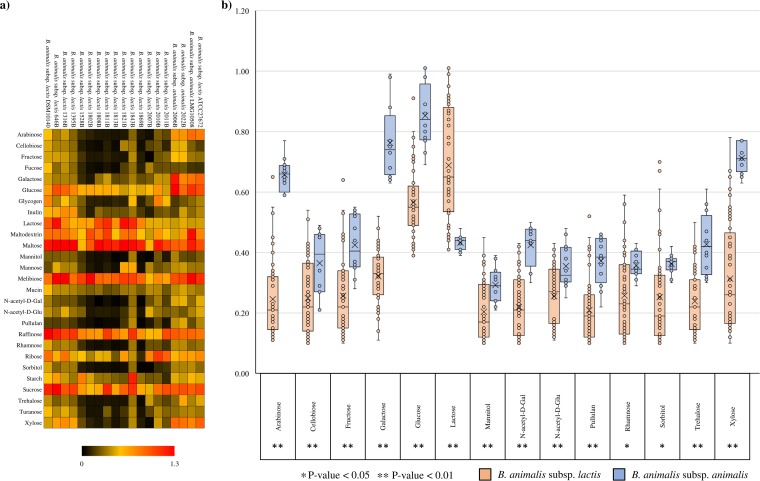
Bifidobacteria are known to metabolize a wide range of carbohydrates as a carbon and energy source, ranging from dietary to host-derived glycans (50–53). In order to assess carbohydrate fermentation capabilities of the two B. animalis subspecies, we performed growth experiments involving 19 B. animalis species cultivated on semisynthetic medium with different carbohydrates as the sole carbon source. In order to obtain a complete overview of such carbohydrate metabolic abilities, we included both plant- and host-derived glycans (Fig. 4). As displayed in Fig. 4, all B. animalis subsp. lactis strains were able to grow on a common set of sugars, such as lactose, maltose, raffinose, and sucrose. In contrast, B. animalis subsp. animalis strains was shown to metabolize a broader array of sugars, with a high growth performance in medium containing arabinose, galactose, glucose, maltose, melibiose, sucrose, or xylose (54). Furthermore, B. animalis subsp. lactis 646, B. animalis subsp. lactis 1316B, and B. animalis subsp. lactis 1395B, in contrast to other members of this subspecies, exhibited appreciable growth on xylose (Fig. 4).
FIG 4.
Evaluation of carbohydrate utilization by B. animalis strains. (a) Heat map representing the growth performances of B. animalis strains on different sugars. Cultures were grown in biologically independent triplicates. Different shadings represent the optical densities reached by the assessed cultures. (b) Whiskers plot based on optical density values of sugars with a P value of <0.05 between subspecies (Student t test). The x axis represents the sole carbon source used for the growth experiments, while the y axis shows the optical density values obtained for B. animalis subsp. animalis strains (blue) and B. animalis subs. lactis strains (orange). Points reflect the distribution of a data set, while the boxes represent 50% of the data set, distributed between the first and third quartiles. The median divides the boxes into the interquartile range, while the “X” represents the mean. The lines extending vertically outside the boxes show the outlier range.
Statistical analyses were performed to corroborate the observed growth differences between B. animalis subsp. lactis and B. animalis subsp. animalis strains on different sugars. As shown in Fig. 4, a significant growth difference (P < 0.05) for 14 carbohydrates was observed, with the highest growth performances of B. animalis subsp. animalis strains (compared to B. animalis subsp. lactis strains) in medium containing arabinose, fructose, galactose, glucose, pullulan, trehalose, or xylose (Table S1). On the other hand, B. animalis subsp. lactis strains were shown to grow significantly better (compared to B. animalis subsp. animalis strains) in MRS medium supplemented with lactose (Fig. 4). Moreover, in five cases, the obtained growth performances were shown to be highly significantly different, with P values of <0.001 (Fig. 4). Notably, none of B. animalis subsp. lactis strains was able to utilize mucin, N-acetyl-d-galactosamine, and N-acetyl-d-glucosamine, which indicates that the tested strains possess limited metabolic capabilities with regard to host-derived glycans (Fig. 4).
In order to validate the observed metabolic differences of the B. animalis subspecies, we predicted the glycosyl hydrolase (GH) enzymes involved in carbohydrate breakdown and belonging to the subspecies-specific core genes, as mentioned above. The in silico analyses were performed using the carbohydrate-active enzymes (CAZy) database (55) involving the 56 B. animalis genomes mentioned above. Interestingly, 13 subspecies-specific core genes of B. animalis subsp. animalis genomes are predicted to be involved in sugar metabolism, while 5 genes indicated as carbohydrate-active enzymes are present in the subspecies-specific core genes of B. animalis subsp. lactis genomes. Among these subspecies-specific carbohydrate-active enzymes, we retrieved seven GH-encoding genes in B. animalis subsp. animalis genomes and four within B. animalis subsp. lactis strains. Interestingly, one of the seven B. animalis subsp. animalis-specific GH belongs to the GH2 family, which typically represent β-galactosidase (56) and exo-β-glucosaminidase (57) activities, confirming the observed high metabolic capabilities of this taxon toward galactose- and glucose-containing sugars (Fig. 4). Furthermore, two B. animalis subsp. animalis GH-specific genes belong to the GH3 family, representing β-glucosidases and xylosidases (58), and the GH43 family, representing xylosidases (58) and arabinosidases (59), which are involved in the metabolism of xylose- and arabinose-containing glycans. Therefore, in silico analyses showed a larger number of GH-encoding genes among the B. animalis subsp. animalis subspecies-specific core genes compared to the B. animalis subsp. lactis subspecies-specific core genes, confirming the observed broader carbohydrate-dependent growth performances displayed by this taxon.
Evolutionary gain gene and loss gene analysis.
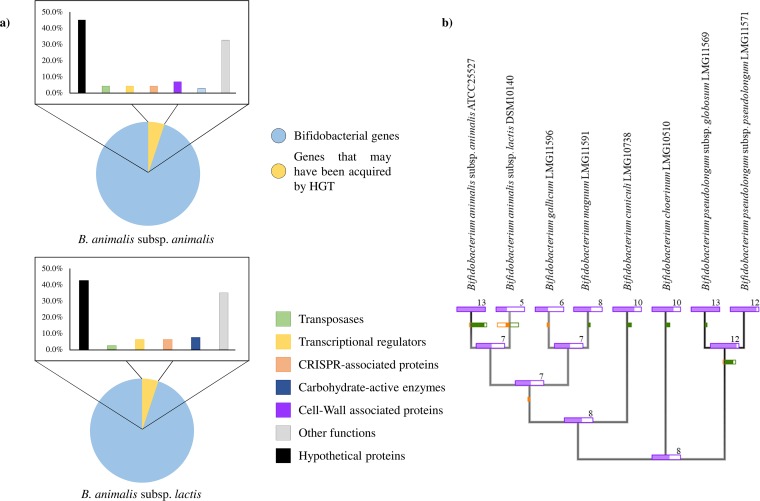
In order to identify genes that may have been acquired by horizontal genes transfer (HGT), the genomes of the type strains of B. animalis subsp. animalis/lactis were analyzed with the software suite COLOMBO v4.0 (60). Interestingly, 80 genes, representing 5.1% of the B. animalis subsp. lactis genes, seem to have alien origins of which 42.5% encode hypothetical proteins. Moreover, 7.5% of the genes that may have been acquired by HGT are predicted to be enzymes involved in carbohydrate metabolism, while 12.6% represent genes encoding transcriptional regulators, and genes involved in CRISPR-Cas systems (Fig. 5). In the case of B. animalis subsp. animalis, 4.6% of the genes seem to have been acquired by HGT, of which 45.1% represent hypothetical proteins. Moreover, 4.2% of these genes encode transposase and 8.4% are predicted to be involved in CRISPR-Cas and in transcriptional regulation. These data suggest that HGT events represent a minor force in the evolution of genomes of B. animalis species.
FIG 5.
Evolutionary gene gain and gene loss analysis within the B. pseudolongum phylogenetic group, as based on predicted subspecies-specific GHs. (a) Genes predicted to have been acquired by HGT events in type strains of the B. animalis species. Bar plots represent in different colors the functional annotations of the predicted genes. (b) Tree based on the core genome of the B. pseudolongum phylogenetic group. The different sticks represent the predicted subspecies-specific GHs within B. pseudolongum phylogenetic group. Each node reports the number of predicted GHs identified in the type strains tested. Green and orange bars on the edge leading to each node indicate gains and losses.
To evaluate the acquisition and loss of the subspecies-specific GH genes through the B. pseudolongum phylogenetic group, we analyzed the predicted subspecies-specific carbohydrate-active enzymes using Count software (61). This evolutionary development analysis is based on the core-gene sequences retrieved from the type strains of the B. pseudolongum phylogenetic group. As indicated in Fig. 5, the B. animalis subsp. animalis taxon seems to have acquired five carbohydrate-active enzymes during evolution compared to the common ancestor of the phylogenetic group. Furthermore, the B. animalis subsp. lactis taxon was shown to be the subspecies with the higher prevalence of subspecies-specific GH gene loss, encompassing five specific GHs (Fig. 5). These findings suggest that the B. animalis subspecies has followed a different evolutionary path, confirming our observed differences between these two taxa identified in the phylogenomic analyses.
Conclusions.
Isolation of 15 B. animalis strains from the GITs of different animals and representing the B. animalis subsp. animalis and B. animalis subsp. lactis taxa revealed the cosmopolitan lifestyle of this species. Genome sequencing of the collected strains allowed us to reconstruct the genomic data set of the B. animalis species, including 41 publicly available B. animalis genomes, unveiling that further genome sequencing of novel B. animalis strains will only slightly contribute to increase the pan-genome size. Nonetheless, phylogenetic analysis based on core genome sequences, among the 56 bifidobacterial genomes, showed a clear differentiation between the B. animalis subsp. animalis and B. animalis subsp. lactis branch. In fact, genome comparison of each strain showed the presence of a subspecies-specific core genome, representing the genetic differences between these two subspecies. Furthermore, the performed phylogenetic analysis highlights a cluster composed of 18 B. animalis subsp. lactis isolates that represent potential novel probiotic strains. Interestingly, a large proportion of the subspecies-specific genes of either B. animalis subspecies seems to be involved in sugar transport and metabolism. In this context, a larger number of such subspecies-specific transporter and GH activities was found in B. animalis subsp. animalis genomes. Growth performances on various sugars as their sole carbon source confirmed the ability of B. animalis subsp. animalis taxon to metabolize a broader set of sugars, e.g., arabinose, galactose, glucose, maltose, melibiose, sucrose, and xylose, whereas B. animalis subsp. lactis strains seems to be more specialized using a smaller number of sugars, such as lactose, maltose, raffinose, and sucrose. Altogether, these results seem to highlight a better ecological fitness of B. animalis subsp. animalis taxon compared to B. animalis subsp. lactis taxon. Moreover, a gene acquisition and loss analysis based on subspecies-specific glycosyl hydrolase genes revealed that B. animalis subsp. animalis taxon seems to have acquired several GHs through HGT, whereas B. animalis subsp. lactis species appears to have suffered loss of GH-encoding genes. Thus, these findings confirm the evolutionary differentiation between these two subspecies as highlighted in both phylogenetic and genomic analyses.
MATERIALS AND METHODS
Bifidobacterial selection.
In order to explore genome variability of the B. animalis species, 15 novel strains were isolated from fecal samples collected from different animals. Samples were composed of 10 g of fresh fecal material, which is a sufficient quantity to represent the overall biodiversity of the fecal microbiota as reported in a previously published study (62). One gram of fecal sample from each collected animal was mixed with 9 ml of phosphate-buffered saline (pH 6.5). Serial dilution and subsequent plating were performed using de Man–Rogosa–Sharpe (MRS) agar, supplemented with 50 μg/ml mupirocin (Delchimica, Italy) and 0.05% (wt/vol) l-cysteine hydrochloride. Agar plates were incubated for 48 h at 37°C in a chamber (Concept 400; Ruskin) with an anaerobic atmosphere (2.99% H2, 17.01% CO2, and 80% N2). Morphologically different colonies that developed on MRS plates were randomly picked and restreaked in order to isolate purified bacterial strains. All isolates were subjected to DNA isolation and characterized as previously described by Turroni et al. (63). The B. animalis strains isolated in this study are listed in Table 1, together with other strains used for in silico analyses.
Bifidobacterial ITS profiling.
Partial ITS sequences were amplified from extracted DNA using the primer pair Probio-bif_Uni/Probio-bif_Rev (64). Resulting reads were analyzed by means of an updated bifidobacterial ITS database encompassing all publicly available bifidobacterial genomes and a custom bioinformatics script as previously described (64). ITS bifidobacterial profiling of mammalian species and birds were coupled with data of mammalian bifidobacterial communities as previously determined by Milani et al. (35).
Genome sequencing and assemblies.
DNA extracted from bifidobacterial isolates was subjected to whole-genome sequencing using MiSeq (Illumina, UK) at GenProbio srl (Parma, Italy) according to the supplier’s protocol (Illumina, UK). Fastq files of the paired-end reads obtained from targeted genome sequencing of isolated strains were utilized as input for genome assemblies through the MEGAnnotator pipeline (65). SPAdes software was used for de novo assembly of each bifidobacterial genome sequence (66, 67), while protein-encoding ORFs were predicted using Prodigal (68). The coverage depth of these newly isolated 15 B. animalis chromosomes ranged from 85- to 278-fold, which upon assembly generated 47 to 12 contigs (Table 1). The number of predicted ORFs ranged from 1,556 of B. animalis subsp. lactis 1808B to 1,935 of B. animalis subsp. animalis 2022B (Table 1). In order to ensure data consistency, B. animalis chromosomes retrieved from public databases were reannotated using the same bioinformatics pipeline applied for the 15 B. animalis strains isolated in the present study.
Comparative genomics.
A pangenome calculation was performed using the pan-genome analysis pipeline PGAP (69), including each B. animalis genome collected from this study (Table 1). Each predicted proteome of a given B. animalis strain was screened for orthologues against the proteome of every collected B. animalis strain by means of BLAST analysis (70) (cutoff, E value of <1 × 10−4 and 50% identity over at least 80% of both protein sequences). The resulting output was then clustered into protein families by means of MCL (graph theory-based Markov clustering algorithm) (71), using the gene family method. A pangenome profile was built using all possible BLAST combinations for each genome being sequentially added. Using this approach, unique protein families encoded by the analyzed B. animalis genomes were also identified. Protein families shared between analyzed genomes allowed us to identify the core genome of the B. animalis species. Each set of orthologous proteins, belonging to the core genome, was aligned using Mafft software (72), and phylogenetic trees were constructed using ClustalW (73). Based on these comparative analyses, a B. animalis supertree was constructed and visualized using FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
Carbohydrate growth assays.
Bifidobacterial strains were cultivated on semisynthetic MRS medium supplemented with a 1% (wt/vol) concentration of a particular sugar, and the optical densities (measured at a wavelength of 600 nm) were recorded using a plate reader (BioTek, Winooski, VT). The plate reader was read in intermittent mode, with absorbance readings performed at 3-min intervals for three times after 48 h of growth, where each reading was ahead of 30 s of shaking at medium speed. Cultures were grown in biologically independent triplicates, and the resulting growth data were expressed as the means of these replicates. Carbohydrates were purchased from Sigma and Carbosynth (Berkshire, UK). Carbohydrate-active enzymes were identified based on similarity to the Carbohydrate-Active enZYmes (CAZy) database entries.
SNP identification.
Multiple alignment of conserved genomic sequence with rearrangements (Mauve) software (74) was employed to perform whole-genome sequence alignments between bifidobacterial genome sequences. SNPs reported by Mauve were manually evaluated to identify polymorphisms between subspecies.
Gene gain or loss through evolutionary reconstruction.
Identification of genes that are predicted to be acquired by an HGT event was performed using COLOMBO v4.0 (60). Evolution-driven acquisition and loss of GH-encoding genes among members of the B. pseudolongum phylogenetic group was performed with Count (61) software using Wagner’s parsimony.
Statistical analyses.
SPSS software (IBM, Italy) was used to perform statistical analysis between B. animalis subsp. animalis strains group and B. animalis subsp. lactis group by Student t test. Furthermore, t test assumption was verified using the unequal variances Welch t test analysis to validate samples that exhibit unequal variance in the sample size (Table S1).
Data availability.
Newly isolated B. animalis genomes were sequenced and deposited at DDBJ/ENA/GenBank under the accession numbers reported in Table 1 (BioProject PRJNA506409).
Supplementary Material
ACKNOWLEDGMENTS
We thank GenProbio SRL for financial support of the Laboratory of Probiogenomics. Part of this research was conducted using the High Performance Computing (HPC) facility of the University of Parma. This study was funded by the EU Joint Programming Initiative—A Healthy Diet for a Healthy Life (JPI HDHL, http://www.healthydietforhealthylife.eu/) support to D.V.S. (in conjunction with Science Fondation Ireland [SFI], grant 15/JP-HDHL/3280) and to M.V. (in conjunction with MIUR, Italy). D.V.S. is a member of The APC Microbiome Institute funded by Science Foundation Ireland (SFI), through the Irish Government’s National Development Plan (grant SFI/12/RC/2273). The authors declare that they have no competing interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02806-18.
REFERENCES
- 1.Milani C, Lugli GA, Duranti S, Turroni F, Bottacini F, Mangifesta M, Sanchez B, Viappiani A, Mancabelli L, Taminiau B, Delcenserie V, Barrangou R, Margolles A, van Sinderen D, Ventura M. 2014. Genomic encyclopedia of type strains of the genus Bifidobacterium. Appl Environ Microbiol 80:6290–6302. doi: 10.1128/AEM.02308-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferrario C, Statello R, Carnevali L, Mancabelli L, Milani C, Mangifesta M, Duranti S, Lugli GA, Jimenez B, Lodge S, Viappiani A, Alessandri G, Dall’Asta M, Del Rio D, Sgoifo A, van Sinderen D, Ventura M, Turroni F. 2017. How to feed the mammalian gut microbiota: bacterial and metabolic modulation by dietary fibers. Front Microbiol 8:1749. doi: 10.3389/fmicb.2017.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, Belzer C, Delgado Palacio S, Arboleya Montes S, Mancabelli L, Lugli GA, Rodriguez JM, Bode L, de Vos W, Gueimonde M, Margolles A, van Sinderen D, Ventura M. 2017. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev 81:e00036-17. doi: 10.1128/MMBR.00036-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.LeBlanc JG, Milani C, de Giori GS, Sesma F, van Sinderen D, Ventura M. 2013. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol 24:160–168. doi: 10.1016/j.copbio.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Ventura M, O’Toole PW, de Vos WM, van Sinderen D. 2018. Selected aspects of the human gut microbiota. Cell Mol Life Sci 75:81–82. doi: 10.1007/s00018-017-2669-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lugli GA, Milani C, Duranti S, Mancabelli L, Mangifesta M, Turroni F, Viappiani A, van Sinderen D, Ventura M. 2018. Tracking the taxonomy of the genus bifidobacterium based on a phylogenomic approach. Appl Environ Microbiol 84:e02249-17. doi: 10.1128/AEM.02249-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Invernici MM, Salvador SL, Silva PHF, Soares MSM, Casarin R, Palioto DB, Souza SLS, Taba M Jr, Novaes AB Jr, Furlaneto FAC, Messora MR. 2018. Effects of Bifidobacterium probiotic on the treatment of chronic periodontitis: a randomized clinical trial. J Clin Periodontol 45:1198–1210. doi: 10.1111/jcpe.12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Solano-Aguilar G, Shea-Donohue T, Madden KB, Quinones A, Beshah E, Lakshman S, Xie Y, Dawson H, Urban JF. 2018. Bifidobacterium animalis subspecies lactis modulates the local immune response and glucose uptake in the small intestine of juvenile pigs infected with the parasitic nematode Ascaris suum. Gut Microbes 9:422–436. doi: 10.1080/19490976.2018.1460014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turroni F, Duranti S, Bottacini F, Guglielmetti S, Van Sinderen D, Ventura M. 2014. Bifidobacterium bifidum as an example of a specialized human gut commensal. Front Microbiol 5:437. doi: 10.3389/fmicb.2014.00437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Andrés J, Manzano S, García C, Rodríguez JM, Espinosa-Martos I, Jiménez E. 2018. Modulatory effect of three probiotic strains on infants’ gut microbial composition and immunological parameters on a placebo-controlled, double-blind, randomised study. Benef Microbes 9:573–584. doi: 10.3920/BM2017.0132. [DOI] [PubMed] [Google Scholar]
- 11.Bottacini F, Zomer A, Milani C, Ferrario C, Lugli GA, Egan M, Ventura M, van Sinderen D. 2017. Global transcriptional landscape and promoter mapping of the gut commensal Bifidobacterium breve UCC2003. BMC Genomics 18:991. doi: 10.1186/s12864-017-4387-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inturri R, Ventura M, Ruas-Madiedo P, Lugli GA, Blandino G. 2017. Complete genome sequence of Bifidobacterium longum W11 (LMG P-21586), used as a probiotic strain. Genome Announc 5:e01659-16. doi: 10.1128/genomeA.01659-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carbuhn AF, Reynolds SM, Campbell CW, Bradford LA, Deckert JA, Kreutzer A, Fry AC. 2018. Effects of probiotic (Bifidobacterium longum 35624) supplementation on exercise performance, immune modulation, and cognitive outlook in Division I female swimmers. Sports (Basel) 6:E116. doi: 10.3390/sports6040116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waitzberg DL, Quilici FA, Michzputen S, Friche Passos MDC. 2015. The effect of probiotic fermented milk that includes Bifidobacterium lactis Cncm I-2494 on the reduction of gastrointestinal discomfort and symptoms in adults: a narrative review. Nutr Hosp 32:501–509. doi: 10.3305/nh.2015.32.2.9232. [DOI] [PubMed] [Google Scholar]
- 15.Lee A, Lee YJ, Yoo HJ, Kim M, Chang Y, Lee DS, Lee JH. 2017. Consumption of dairy yogurt containing Lactobacillus paracasei ssp. paracasei, Bifidobacterium animalis ssp. lactis and heat-treated Lactobacillus plantarum improves immune function including natural killer cell activity. Nutrients 9:E558. doi: 10.3390/nu9060558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masco L, Ventura M, Zink R, Huys G, Swings J. 2004. Polyphasic taxonomic analysis of Bifidobacterium animalis and Bifidobacterium lactis reveals relatedness at the subspecies level: reclassification of Bifidobacterium animalis as Bifidobacterium animalis subsp. animalis subsp. nov. and Bifidobacterium lactis as Bifidobacterium animalis subsp. lactis subsp. nov. Int J Syst Evol Microbiol 54:1137–1143. doi: 10.1099/ijs.0.03011-0. [DOI] [PubMed] [Google Scholar]
- 17.Jungersen M, Wind A, Johansen E, Christensen JE, Stuer-Lauridsen B, Eskesen D. 2014. The science behind the probiotic strain Bifidobacterium animalis subsp. lactis BB-12(R). Microorganisms 2:92–110. doi: 10.3390/microorganisms2020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliveira LF, Salvador SL, Silva PH, Furlaneto FA, Figueiredo L, Casarin R, Ervolino E, Palioto DB, Souza SL, Taba M Jr, Novaes AB Jr, Messora MR. 2017. Benefits of Bifidobacterium animalis subsp. lactis probiotic in experimental periodontitis. J Periodontol 88:197–208. doi: 10.1902/jop.2016.160217. [DOI] [PubMed] [Google Scholar]
- 19.Eskesen D, Jespersen L, Michelsen B, Whorwell PJ, Muller-Lissner S, Morberg CM. 2015. Effect of the probiotic strain Bifidobacterium animalis subsp. lactis, BB-12(R), on defecation frequency in healthy subjects with low defecation frequency and abdominal discomfort: a randomised, double-blind, placebo-controlled, parallel-group trial. Br J Nutr 114:1638–1646. doi: 10.1017/S0007114515003347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martorell P, Llopis S, González N, Chenoll E, López-Carreras N, Aleixandre A, Chen Y, Karoly ED, Ramón D, Genovés S. 2016. Probiotic strain Bifidobacterium animalis subsp. lactis CECT 8145 reduces fat content and modulates lipid metabolism and antioxidant response in Caenorhabditis elegans. J Agric Food Chem 64:3462–3472. doi: 10.1021/acs.jafc.5b05934. [DOI] [PubMed] [Google Scholar]
- 21.O’Mahony D, Murphy S, Boileau T, Park J, O’Brien F, Groeger D, Konieczna P, Ziegler M, Scully P, Shanahan F, Kiely B, O’Mahony L. 2010. Bifidobacterium animalis AHC7 protects against pathogen-induced NF-κB activation in vivo. BMC Immunol 11:63. doi: 10.1186/1471-2172-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai Y, Matsumoto M, Benno Y. 2000. Bifidobacterium lactis Meile et al. 1997 is a subjective synonym of Bifidobacterium animalis (Mitsuoka 1969) Scardovi and Trovatelli 1974. Microbiol Immunol 44:815–820. doi: 10.1111/j.1348-0421.2000.tb02568.x. [DOI] [PubMed] [Google Scholar]
- 23.Milani C, Duranti S, Lugli GA, Bottacini F, Strati F, Arioli S, Foroni E, Turroni F, van Sinderen D, Ventura M. 2013. Comparative genomics of Bifidobacterium animalis subsp. lactis reveals a strict monophyletic bifidobacterial taxon. Appl Environ Microbiol 79:4304–4315. doi: 10.1128/AEM.00984-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loquasto JR, Barrangou R, Dudley EG, Roberts RF. 2011. Short communication: the complete genome sequence of Bifidobacterium animalis subspecies animalis ATCC 25527T and comparative analysis of growth in milk with B. animalis subspecies lactis DSM 10140T. J Dairy Sci 94:5864–5870. doi: 10.3168/jds.2011-4499. [DOI] [PubMed] [Google Scholar]
- 25.Matsumoto M, Ohishi H, Benno Y. 2004. H+-ATPase activity in Bifidobacterium with special reference to acid tolerance. Int J Food Microbiol 93:109–113. doi: 10.1016/j.ijfoodmicro.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 26.Simpson PJ, Stanton C, Fitzgerald GF, Ross RP. 2005. Intrinsic tolerance of Bifidobacterium species to heat and oxygen and survival following spray drying and storage. J Appl Microbiol 99:493–501. doi: 10.1111/j.1365-2672.2005.02648.x. [DOI] [PubMed] [Google Scholar]
- 27.Duranti S, Milani C, Lugli GA, Turroni F, Mancabelli L, Sanchez B, Ferrario C, Viappiani A, Mangifesta M, Mancino W, Gueimonde M, Margolles A, van Sinderen D, Ventura M. 2015. Insights from genomes of representatives of the human gut commensal Bifidobacterium bifidum. Environ Microbiol 17:2515–2531. doi: 10.1111/1462-2920.12743. [DOI] [PubMed] [Google Scholar]
- 28.Bottacini F, O’Connell Motherway M, Kuczynski J, O’Connell K, Serafini F, Duranti S, Milani C, Turroni F, Lugli G, Zomer A, Zhurina D, Riedel C, Ventura M, Sinderen D. 2014. Comparative genomics of the Bifidobacterium breve taxon. BMC Genomics 15:170. doi: 10.1186/1471-2164-15-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Callaghan A, Bottacini F, O’Connell Motherway M, van Sinderen D. 2015. Pangenome analysis of Bifidobacterium longum and site-directed mutagenesis through by-pass of restriction-modification systems. BMC Genomics 16:832. doi: 10.1186/s12864-015-1968-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turroni F, Serafini F, Foroni E, Duranti S, O’Connell Motherway M, Taverniti V, Mangifesta M, Milani C, Viappiani A, Roversi T, Sanchez B, Santoni A, Gioiosa L, Ferrarini A, Delledonne M, Margolles A, Piazza L, Palanza P, Bolchi A, Guglielmetti S, van Sinderen D, Ventura M. 2013. Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium-host interactions. Proc Natl Acad Sci U S A 110:11151–11156. doi: 10.1073/pnas.1303897110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, O’Brien F, Flynn K, Casey PG, Munoz JA, Kearney B, Houston AM, O’Mahony C, Higgins DG, Shanahan F, Palva A, de Vos WM, Fitzgerald GF, Ventura M, O’Toole PW, van Sinderen D. 2011. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci U S A 108:11217–11222. doi: 10.1073/pnas.1105380108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turroni F, Bottacini F, Foroni E, Mulder I, Kim JH, Zomer A, Sanchez B, Bidossi A, Ferrarini A, Giubellini V, Delledonne M, Henrissat B, Coutinho P, Oggioni M, Fitzgerald GF, Mills D, Margolles A, Kelly D, van Sinderen D, Ventura M. 2010. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc Natl Acad Sci U S A 107:19514–19519. doi: 10.1073/pnas.1011100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turroni F, Milani C, van Sinderen D, Ventura M. 2011. Genetic strategies for mucin metabolism in Bifidobacterium bifidum PRL2010: an example of possible human-microbe co-evolution. Gut Microbes 2:183–189. doi: 10.4161/gmic.2.3.16105. [DOI] [PubMed] [Google Scholar]
- 34.Bunesova V, Killer J, Javurkova B, Vlkova E, Tejnecky V, Musilova S, Rada V. 2017. Diversity of the subspecies Bifidobacterium animalis subsp. lactis. Anaerobe 44:40–47. doi: 10.1016/j.anaerobe.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 35.Milani C, Mangifesta M, Mancabelli L, Lugli GA, James K, Duranti S, Turroni F, Ferrario C, Ossiprandi MC, van Sinderen D, Ventura M. 2017. Unveiling bifidobacterial biogeography across the mammalian branch of the tree of life. ISME J 11:2834–2847. doi: 10.1038/ismej.2017.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turroni F, Milani C, Duranti S, Mancabelli L, Mangifesta M, Viappiani A, Lugli GA, Ferrario C, Gioiosa L, Ferrarini A, Li J, Palanza P, Delledonne M, van Sinderen D, Ventura M. 2016. Deciphering bifidobacterial-mediated metabolic interactions and their impact on gut microbiota by a multi-omics approach. ISME J 10:1656–1668. doi: 10.1038/ismej.2015.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferrario C, Milani C, Mancabelli L, Lugli GA, Turroni F, Duranti S, Mangifesta M, Viappiani A, Sinderen D, Ventura M. 2015. A genome-based identification approach for members of the genus Bifidobacterium. FEMS Microbiol Ecol 91:fiv009. doi: 10.1093/femsec/fiv009. [DOI] [PubMed] [Google Scholar]
- 38.Ventura M, Reniero R, Zink R. 2001. Specific identification and targeted characterization of Bifidobacterium lactis from different environmental isolates by a combined multiplex-PCR approach. Appl Environ Microbiol 67:2760–2765. doi: 10.1128/AEM.67.6.2760-2765.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ventura M, Zink R. 2003. Comparative sequence analysis of the tuf and recA genes and restriction fragment length polymorphism of the internal transcribed spacer region sequences supply additional tools for discriminating Bifidobacterium lactis from Bifidobacterium animalis. Appl Environ Microbiol 69:7517–7522. doi: 10.1128/AEM.69.12.7517-7522.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Briczinski EP, Loquasto JR, Barrangou R, Dudley EG, Roberts AM, Roberts RF. 2009. Strain-specific genotyping of Bifidobacterium animalis subsp. lactis by using single-nucleotide polymorphisms, insertions, and deletions. Appl Environ Microbiol 75:7501–7508. doi: 10.1128/AEM.01430-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Odamaki T, Horigome A, Sugahara H, Hashikura N, Minami J, Xiao JZ, Abe F. 2015. Comparative genomics revealed genetic diversity and species/strain-level differences in carbohydrate metabolism of three probiotic bifidobacterial species. Int J Genomics 2015:567809. doi: 10.1155/2015/567809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barrangou R, Briczinski EP, Traeger LL, Loquasto JR, Richards M, Horvath P, Coute-Monvoisin AC, Leyer G, Rendulic S, Steele JL, Broadbent JR, Oberg T, Dudley EG, Schuster S, Romero DA, Roberts RF. 2009. Comparison of the complete genome sequences of Bifidobacterium animalis subsp. lactis DSM 10140 and Bl-04. J Bacteriol 191:4144–4151. doi: 10.1128/JB.00155-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tettelin H, Riley D, Cattuto C, Medini D. 2008. Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol 11:472–477. doi: 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 44.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M, Scarselli M, Margarit y Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, O’Connor KJ, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc Natl Acad Sci U S A 102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Powell S, Forslund K, Szklarczyk D, Trachana K, Roth A, Huerta-Cepas J, Gabaldon T, Rattei T, Creevey C, Kuhn M, Jensen LJ, von Mering C, Bork P. 2014. eggNOG v4.0: nested orthology inference across 3686 organisms. Nucleic Acids Res 42:D231–D239. doi: 10.1093/nar/gkt1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richter M, Rossello MR. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eales J, Gibson P, Whorwell P, Kellow J, Yellowlees A, Perry RH, Edwards M, King S, Wood H, Glanville J. 2017. Systematic review and meta-analysis: the effects of fermented milk with Bifidobacterium lactis CNCM I-2494 and lactic acid bacteria on gastrointestinal discomfort in the general adult population. Ther Adv Gastroenterol 10:74–88. doi: 10.1177/1756283x16670075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lugli GA, Milani C, Turroni F, Duranti S, Mancabelli L, Mangifesta M, Ferrario C, Modesto M, Mattarelli P, Jiří K, van Sinderen D, Ventura M. 2017. Comparative genomic and phylogenomic analyses of the Bifidobacteriaceae family. BMC Genomics 18:568. doi: 10.1186/s12864-017-3955-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duranti S, Turroni F, Lugli GA, Milani C, Viappiani A, Mangifesta M, Gioiosa L, Palanza P, van Sinderen D, Ventura M. 2014. Genomic characterization and transcriptional studies of the starch-utilizing strain Bifidobacterium adolescentis 22L. Appl Environ Microbiol 80:6080–6090. doi: 10.1128/AEM.01993-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Milani C, Lugli GA, Duranti S, Turroni F, Mancabelli L, Ferrario C, Mangifesta M, Hevia A, Viappiani A, Scholz M, Arioli S, Sanchez B, Lane J, Ward DV, Hickey R, Mora D, Segata N, Margolles A, van Sinderen D, Ventura M. 2015. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci Rep 5:15782. doi: 10.1038/srep15782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turroni F, Strati F, Foroni E, Serafini F, Duranti S, van Sinderen D, Ventura M. 2012. Analysis of predicted carbohydrate transport systems encoded by Bifidobacterium bifidum PRL2010. Appl Environ Microbiol 78:5002–5012. doi: 10.1128/AEM.00629-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pokusaeva K, Fitzgerald GF, van Sinderen D. 2011. Carbohydrate metabolism in bifidobacteria. Genes Nutr 6:285–306. doi: 10.1007/s12263-010-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Egan M, Bottacini F, O’Connell Motherway M, Casey PG, Morrissey R, Melgar S, Faurie JM, Chervaux C, Smokvina T, van Sinderen D. 2018. Staying alive: growth and survival of Bifidobacterium animalis subsp. animalis under in vitro and in vivo conditions. Appl Microbiol Biotechnol 102:10645–10663. doi: 10.1007/s00253-018-9413-7. [DOI] [PubMed] [Google Scholar]
- 55.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Landman OE. 1957. Properties and induction of β-galactosidase in Bacillus megaterium. Biochim Biophys Acta 23:558–569. doi: 10.1016/0006-3002(57)90377-3. [DOI] [PubMed] [Google Scholar]
- 57.Nanjo F, Katsumi R, Sakai K. 1990. Purification and characterization of an exo-β-d-glucosaminidase, a novel type of enzyme, from Nocardia orientalis. J Biol Chem 265:10088–10094. [PubMed] [Google Scholar]
- 58.Chinchetru MA, Cabezas JA, Calvo P. 1989. Purification and characterization of a broad specificity beta-glucosidase from sheep liver. Int J Biochem 21:469–476. doi: 10.1016/0020-711X(89)90126-2. [DOI] [PubMed] [Google Scholar]
- 59.Weinstein L, Albersheim P. 1979. Structure of plant cell walls. IX. Purification and partial characterization of a wall-degrading endo-arabanase and an arabinosidase from Bacillus subtilis. Plant Physiol 63:425–432. doi: 10.1104/pp.63.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Waack S, Keller O, Asper R, Brodag T, Damm C, Fricke WF, Surovcik K, Meinicke P, Merkl R. 2006. Score-based prediction of genomic islands in prokaryotic genomes using hidden Markov models. BMC Bioinformatics 7:142. doi: 10.1186/1471-2105-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Csuros M. 2010. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics 26:1910–1912. doi: 10.1093/bioinformatics/btq315. [DOI] [PubMed] [Google Scholar]
- 62.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI. 2008. Evolution of mammals and their gut microbes. Science 320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Turroni F, Marchesi JR, Foroni E, Gueimonde M, Shanahan F, Margolles A, van Sinderen D, Ventura M. 2009. Microbiomic analysis of the bifidobacterial population in the human distal gut. ISME J 3:745–751. doi: 10.1038/ismej.2009.19. [DOI] [PubMed] [Google Scholar]
- 64.Milani C, Lugli GA, Turroni F, Mancabelli L, Duranti S, Viappiani A, Mangifesta M, Segata N, van Sinderen D, Ventura M. 2014. Evaluation of bifidobacterial community composition in the human gut by means of a targeted amplicon sequencing (ITS) protocol. FEMS Microbiol Ecol 90:493–503. doi: 10.1111/1574-6941.12410. [DOI] [PubMed] [Google Scholar]
- 65.Lugli GA, Milani C, Mancabelli L, van Sinderen D, Ventura M. 2016. MEGAnnotator: a user-friendly pipeline for microbial genomes assembly and annotation. FEMS Microbiol Lett 363:fnw049. doi: 10.1093/femsle/fnw049. [DOI] [PubMed] [Google Scholar]
- 66.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, Stepanauskas R, Clingenpeel SR, Woyke T, McLean JS, Lasken R, Tesler G, Alekseyev MA, Pevzner PA. 2013. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol 20:714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao Y, Wu J, Yang J, Sun S, Xiao J, Yu J. 2012. PGAP: pan-genomes analysis pipeline. Bioinformatics 28:416–418. doi: 10.1093/bioinformatics/btr655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 71.Vlietstra WJ, Zielman R, van Dongen RM, Schultes EA, Wiesman F, Vos R, van Mulligen EM, Kors JA. 2017. Automated extraction of potential migraine biomarkers using a semantic graph. J Biomed Inform 71:178–189. doi: 10.1016/j.jbi.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 72.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss, and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Uchimura Y, Wyss M, Brugiroux S, Limenitakis JP, Stecher B, McCoy KD, Macpherson AJ. 2016. Complete genome sequences of 12 species of stable defined moderately diverse mouse microbiota 2. Genome Announc 4:e00951-16. doi: 10.1128/genomeA.00951-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim JF, Jeong H, Yu DS, Choi SH, Hur CG, Park MS, Yoon SH, Kim DW, Ji GE, Park HS, Oh TK. 2009. Genome sequence of the probiotic bacterium Bifidobacterium animalis subsp. lactis AD011. J Bacteriol 191:678–679. doi: 10.1128/JB.01515-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Loquasto JR, Barrangou R, Dudley EG, Stahl B, Chen C, Roberts RF. 2013. Bifidobacterium animalis subsp. lactis ATCC 27673 is a genomically unique strain within its conserved subspecies. Appl Environ Microbiol 79:6903–6910. doi: 10.1128/AEM.01777-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stahl B, Barrangou R. 2012. Complete genome sequences of probiotic strains Bifidobacterium animalis subsp. lactis B420 and Bi-07. J Bacteriol 194:4131–4132. doi: 10.1128/JB.00766-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Garrigues C, Johansen E, Pedersen MB. 2010. Complete genome sequence of Bifidobacterium animalis subsp. lactis BB-12, a widely consumed probiotic strain. J Bacteriol 192:2467–2468. doi: 10.1128/JB.00109-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kang J, Chung WH, Lim TJ, Lim S, Nam YD. 2017. Complete genome sequence of the Bifidobacterium animalis subspecies lactis BL3, preventive probiotics for acute colitis and colon cancer. New Microbes New Infect 19:34–37. doi: 10.1016/j.nmni.2017.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bottacini F, Dal Bello F, Turroni F, Milani C, Duranti S, Foroni E, Viappiani A, Strati F, Mora D, van Sinderen D, Ventura M. 2011. Complete genome sequence of Bifidobacterium animalis subsp. lactis BLC1. J Bacteriol 193:6387–6388. doi: 10.1128/JB.06079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chervaux C, Grimaldi C, Bolotin A, Quinquis B, Legrain-Raspaud S, van Hylckama Vlieg JE, Denariaz G, Smokvina T. 2011. Genome sequence of the probiotic strain Bifidobacterium animalis subsp. lactis CNCM I-2494. J Bacteriol 193:5560–5561. doi: 10.1128/JB.05716-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Newly isolated B. animalis genomes were sequenced and deposited at DDBJ/ENA/GenBank under the accession numbers reported in Table 1 (BioProject PRJNA506409).