The sulfate-dependent anaerobic methane oxidation (S-AOM) process catalyzed by anaerobic methanotrophic (ANME) archaea and sulfate-reducing bacteria (SRB) is a vital link connecting the global carbon and sulfur cycles. We conducted a research into the spatial-temporal pattern of S-AOM process and the distribution of ANME archaea in coastal sediments collected from the intertidal zone. The results implied that S-AOM process was a methane sink that cannot be overlooked in the intertidal ecosystem. We also found that ANME archaea were sensitive to oxygen and preferred anaerobic environmental conditions. This study will help us gain a better understanding of the global carbon-sulfur cycle and greenhouse gas emission reduction and introduce a new perspective into the enrichment of ANME archaea.
KEYWORDS: anaerobic methanotrophic archaea, intertidal zone, simulated column experiments, sulfate-dependent anaerobic methane oxidation
ABSTRACT
Methane is a primary greenhouse gas which is responsible for global warming. The sulfate-dependent anaerobic methane oxidation (S-AOM) process catalyzed by anaerobic methanotrophic (ANME) archaea and sulfate-reducing bacteria (SRB) is a vital link connecting the global carbon and sulfur cycles, and it is considered to be the overriding methane sink in marine ecosystem. However, there have been few studies regarding the role of S-AOM process and the distribution of ANME archaea in intertidal ecosystem. The intertidal zone is a buffer zone between sea and land and plays an important role in global geochemical cycle. In the present study, the abundance, potential methane oxidation rate, and community structure of ANME archaea in the intertidal zone were studied by quantitative PCR, stable isotope tracing method and high-throughput sequencing. The results showed that the potential S-AOM activity ranged from 0 to 0.77 nmol 13CO2 g−1 (dry sediment) day−1. The copy number of 16S rRNA gene of ANME archaea reached 106 ∼ 107 copies g−1 (dry sediment). The average contribution of S-AOM to total anaerobic methane oxidation was up to 34.5%, while denitrifying anaerobic methane oxidation accounted for the rest, which implied that S-AOM process was an essential methane sink that cannot be overlooked in intertidal ecosystem. The simulated column experiments also indicated that ANME archaea were sensitive to oxygen and preferred anaerobic environmental conditions. This study will help us gain a better understanding of the global carbon-sulfur cycle and greenhouse gas emission reduction and introduce a new perspective into the enrichment of ANME archaea.
IMPORTANCE The sulfate-dependent anaerobic methane oxidation (S-AOM) process catalyzed by anaerobic methanotrophic (ANME) archaea and sulfate-reducing bacteria (SRB) is a vital link connecting the global carbon and sulfur cycles. We conducted a research into the spatial-temporal pattern of S-AOM process and the distribution of ANME archaea in coastal sediments collected from the intertidal zone. The results implied that S-AOM process was a methane sink that cannot be overlooked in the intertidal ecosystem. We also found that ANME archaea were sensitive to oxygen and preferred anaerobic environmental conditions. This study will help us gain a better understanding of the global carbon-sulfur cycle and greenhouse gas emission reduction and introduce a new perspective into the enrichment of ANME archaea.
INTRODUCTION
Climate change has been a global environmental problem in recent decades, which is mainly caused by the increasing greenhouse gas concentration in the atmosphere. Among the three major greenhouse gases (CO2, CH4, and N2O), the contribution rate of methane to global warming is up to 16% (1), which ranks second, closely after carbon dioxide. Moreover, the emission of methane is still increasing at a rate of 0.5 to 1% yearly (2). It is estimated that about 600 Tg CH4 is discharged into the atmosphere annually (3), approximately two-thirds of which is from human activities, and the remainder is from natural habitats (4). Greenhouse gases play a crucial role in sustaining life on the earth. However, excessive greenhouse gases in the atmosphere may lead to a series of environmental problems, such as glacier melting, sea level rise, and global warming.
In the natural environment, methane can be removed by chemical reactions or biochemical reactions (5), and the biochemical reactions are catalyzed by microorganisms. Microbial methane oxidation includes aerobic methane oxidation and anaerobic methane oxidation (anaerobic oxidation of methane [AOM]). AOM consists primarily of denitrifying anaerobic methane oxidation (D-AOM), sulfate-dependent anaerobic methane oxidation (S-AOM) and iron-dependent anaerobic methane oxidation (iron-AOM). The D-AOM process, including nitrite-dependent anaerobic methane oxidation (nitrite-AOM) and nitrate-dependent anaerobic methane oxidation (nitrate-AOM), can remove methane and nitrite/nitrate simultaneously under anaerobic conditions and therefore plays a critical role in the carbon and nitrogen cycles (6–8). The S-AOM process is an important link connecting the global carbon and sulfur cycles. A total of 90% of the methane produced in the marine ecosystem is eventually oxidized by S-AOM process (9). Moreover, the iron-AOM process is an important interaction between the biogeochemical cycles of iron and carbon (10, 11).
In the 1970s, it was found that in marine sediments, the concentration of methane decreased gradually from sediment to water, while the sulfate concentration showed an opposite tendency, indicating that sulfate might be the electron acceptor of AOM (12). Since the anaerobic methane oxidation capability of anaerobic methanotrophic (ANME) archaea was discovered in the late 1990s (13), many studies on S-AOM process have been carried out. ANME archaea and sulfate-reducing bacteria (SRB) are primarily responsible for S-AOM process and show a close synergy in dual-species consortia (14–16). The ANME archaea are proved to oxidize methane in anaerobic conditions through the reverse methanogenic pathway (17–19), while SRB accept electrons and therefore reduce sulfate (20, 21). To date, four different ANME archaea clusters with S-AOM function have been discovered: ANME-1, ANME-2a/b, ANME-2c, and ANME-3.
More than 1,800 16S rRNA gene sequences were available from more than 50 different marine methane seeps, vents, and sulfate-methane transition zones, which differed from each other in environmental factors, such as temperature, methane flux, salinity, and pH (9). The results revealed that ANME-1 and ANME-2 archaea displayed a wide distribution spectrum. Nevertheless, ANME-3 archaea were mainly found in seabed mud volcano and occasionally in seep sediments (9). In most habitats, various ANME archaea could be detected at the same time (22–25). However, some studies also pointed out that only one kind of ANME archaea was dominant in the same habitat (22–24). Specifically, the abundance of ANME archaea was typically more than 1010 cell ⋅ cm−3 in cold seep ecosystems (24). Thus far, researches on S-AOM have been principally focused on cold seep ecosystems, sulfate methane transition zones (SMTZs), hydrothermal vents, the deep biosphere, marine water columns, and some terrestrial ecosystems (16, 22–32). It has been demonstrated that methane was mainly removed by S-AOM process in the marine ecosystem, which consumed up to 90% of the methane produced before entering the atmosphere. Therefore, S-AOM process is an important methane sink in marine habitats.
The intertidal ecosystem is rich in sulfate, and methane is available by the anaerobic decomposition of organic matter in sediments. This makes S-AOM process possible in intertidal zones. However, there have been few studies about the role of S-AOM process and the distribution of ANME archaea in the intertidal ecosystem. Here, we conducted a research into the microbial quantity, community structure, and the potential contribution of methane oxidation in the intertidal zone on the Zhoushan archipelago. In addition, considering impacts of tides on the dissolved oxygen concentration in the sediments, we designed three simulated columns (aerobic, microaerobic, anoxic) to test the influence of oxygen on niche differentiation of ANME archaea.
RESULTS
Potential S-AOM activities in the intertidal zone.
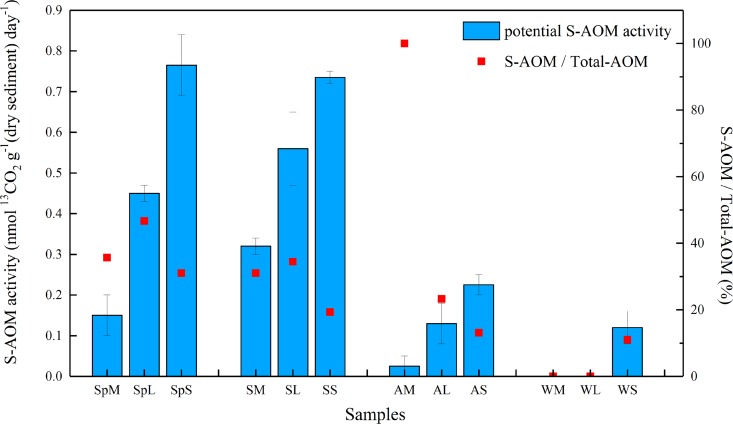
Aerobic methane oxidation, D-AOM and S-AOM activities were successfully measured while no obvious iron- and manganese-dependent AOM activities were detected by the stable isolate tracer experiment (see Fig. S1 in the supplemental material). The potential aerobic methane oxidation rates ranged from 2.12 to 52.27 nmol 13CO2 g−1 (dry sediment) day−1, and the D-AOM rates ranged from 0.00 to 1.70 nmol 13CO2 g−1 (dry sediment) day−1, which were higher than the potential S-AOM rates in most samples (Table 1). The potential S-AOM activities of sediments collected in four seasons showed significant differences (Fig. 1). In all four seasons, the S-AOM activities of the samples collected in the subtidal zone were apparently higher than those in the mid-tide zone and the low-tide zone, which indicated that the environmental factors formed by different intertidal locations had remarkable effects on the S-AOM process. Specifically, the activity of Sample SpS was the highest, which was 0.77 ± 0.08 nmol 13CO2 g−1 (dry sediment) day−1, followed by that of sample SS (Table 1). Overall, the average potential S-AOM activities in summer intertidal sediments ranked first, followed by those in spring, autumn, and winter. Notably, no obvious S-AOM activities were detected in the WM and WL samples (the various sample designations are defined in Materials and Methods), which might be caused by the low temperature, around 9°C, of sediments in the intertidal zone during wintertime.
TABLE 1.
Potential methane oxidation activity and the ratio of potential S-AOM activity to total-AOM activity in sediments collected from the Zhoushan archipelago intertidal zonea
| Parameter | nmol 13CO2 cm−3 (dry sediment) day−1 |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Spring |
Summer |
Autumn |
Winter |
|||||||||
| SpM | SpL | SpS | SM | SL | SS | AM | AL | As | WM | WL | WS | |
| S-AOM | 0.15 ± 0.05 | 0.45 ± 0.02 | 0.77 ± 0.08 | 0.32 ± 0.02 | 0.56 ± 0.09 | 0.74 ± 0.02 | 0.03 ± 0.02 | 0.13 ± 0.05 | 0.23 ± 0.03 | 0.00 | 0.00 | 0.12 ± 0.04 |
| Mean | 0.46 | 0.54 | 0.17 | 0.04 | ||||||||
| D-AOM | 0.27 ± 0.05 | 0.51 ± 0.18 | 1.70 ± 0.30 | 0.71 ± 0.07 | 1.07 ± 0.11 | 3.07 ± 0.55 | 0.00 | 0.43 ± 0.08 | 1.48 ± 0.12 | 0.00 | 0.35 ± 0.04 | 0.97 ± 0.15 |
| Aerobic methane oxidation | 27.06 ± 3.59 | 24.13 ± 5.85 | 20.90 ± 1.87 | 52.27 ± 3.00 | 31.29 ± 5.82 | 9.34 ± 0.83 | 17.56 ± 2.32 | 5.71 ± 1.12 | 4.55 ± 0.32 | 14.07 ± 1.71 | 3.82 ± 0.57 | 2.12 ± 0.15 |
| Fe-AOM | 0 | 0 | 0 | 0 | ||||||||
| Mn-AOM | 0 | 0 | 0 | 0 | ||||||||
| Total methane oxidation | 27.48 | 25.09 | 23.37 | 53.30 | 32.92 | 13.15 | 17.59 | 6.27 | 6.26 | 14.07 | 4.17 | 3.21 |
| Total-AOM | 0.42 | 0.96 | 2.47 | 1.03 | 1.63 | 3.81 | 0.03 | 0.56 | 1.71 | 0.00 | 0.35 | 1.09 |
| S-AOM/total AOM | 0.36 | 0.47 | 0.31 | 0.31 | 0.34 | 0.19 | 1.00 | 0.23 | 0.13 | 0.00 | 0.00 | 0.11 |
Values are expressed as means ± the standard deviations where applicable.
FIG 1.
Potential S-AOM rates and ratio of potential S-AOM activity to total AOM activity in sediments collected from the Zhoushan archipelago intertidal zone. The rates were calculated as the maximum slope of 13CO2 production during the slurry incubations. SpM (spring mid-tide zone), SpL (spring low-tide zone), and SpS (spring subtidal zone) represent the samples collected in spring; SM, SL, and SS represent the samples collected in summer; AM, AL, and AS represent the samples collected in autumn; and WM, WL, and WS represent the samples collected in winter. Error bars represent the standard deviations.
The ratios of potential S-AOM activities to total potential anaerobic methane oxidation activities (total-AOM) were calculated by using the following formula: the ratio = S-AOM/(S-AOM + D-AOM). The maximal ratio appeared at point AM, indicating that S-AOM process was the most important way of anaerobic methane removal at this point. In addition, the ratios at points SpL and SL were the highest in the spring and summer samples, respectively, with values of 0.47 and 0.34 (Table 1), implying that S-AOM process might be a significant methane sink in the intertidal zone. The ratios of S-AOM to total-AOM in both spring and summer samples hit the highest in the low-tide zone, but there was no similar pattern in autumn and winter samples.
Abundance of ANME archaea in the intertidal zone.
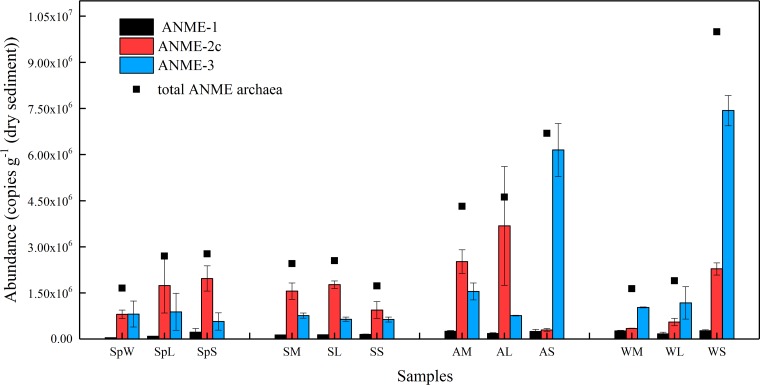
The abundance of total ANME archaea varied significantly in four seasons, ranging from 1.64 × 106 to 9.99 × 106 copies g−1 (dry sediment) (Fig. 2). The abundance of ANME archaea showed an increasing trend in order of the mid-tide zone, the low-tide zone, and the subtidal zone except in summer. ANME-2c archaea were the dominant ANME archaea in most spring, summer, and autumn samples, whereas ANME-3 archaea were the major ANME type in winter. However, considering the high similarity between the 16S rRNA genes of ANME-3 archaea and some methanogens (9), there might be an overestimate of the abundance of ANME-3 clade. The abundance of ANME-1 archaea was consistently the lowest in all sampling sites, ranging from 4.12 × 104 to 2.73 × 105 copies g−1 (dry sediment). It is worth noting that ANME archaea accounted for a large proportion of the total archaea, with the highest ratio of 47.77% in SpS and the lowest ratio of 1.92% in WM revealed by quantitative PCR (qPCR) results (Fig. S2), which indicated that ANME archaea played an important role in intertidal sediments. However, the relative abundance of ANME archaea detected by high-throughput sequencing was relatively low because of the different coverages of the primers used in this work (Fig. S3).
FIG 2.
16S rRNA gene copy numbers of ANME-1, ANME-2c, and ANME-3 in sediments collected from the Zhoushan archipelago intertidal zone. Black squares represent the total ANME archaea abundance. Error bars represent the standard deviations.
Pearson correlation analysis revealed both temperature and total organic matter (TOM) content had an extremely significant positive correlation with the abundance of ANME-1 and ANME-2c archaea (P < 0.01) (Table S1). In addition, ANME-1 archaeal abundance was positively correlated with SO42–, NO3–, and total phosphorus (TP) (P < 0.05). Moreover, temperature was significantly positively correlated with potential S-AOM activity, as well as potential D-AOM activity (P < 0.05), and TOM content showed an extremely significant positive correlation with potential S-AOM activity (P < 0.01).
Diversity and community structure of ANME archaea in the intertidal zone.
A total of 886 to 1,670 ANME archaeal 16S rRNA gene sequences were recovered from all 12 samples through Illumina high-throughput sequencing (Table S2). The numbers of operational taxonomic units (OTU) of each sample ranged from 39 to 70 with a cutoff 3% sequence differences. The OTU number was highest at points AL, WL, and AS and lowest at points SpS, SM, and AM. It is noteworthy that among all sampling points, the OTU numbers for sampling points SpL, SL, AL, and WL from the low-tide zone were the highest in each season. Moreover, the higher Chao1 indexes in low-tide zone (P < 0.05) also indicated that ANME archaea from this zone showed higher richness than those from the other two zones. The Shannon indexes in the low-tide zone were higher than those in the mid-tide zone and the subtidal zone (P < 0.05). We speculated that the frequent tide changes in the low-tide zone resulted in the high diversity of ANME archaea in this region.
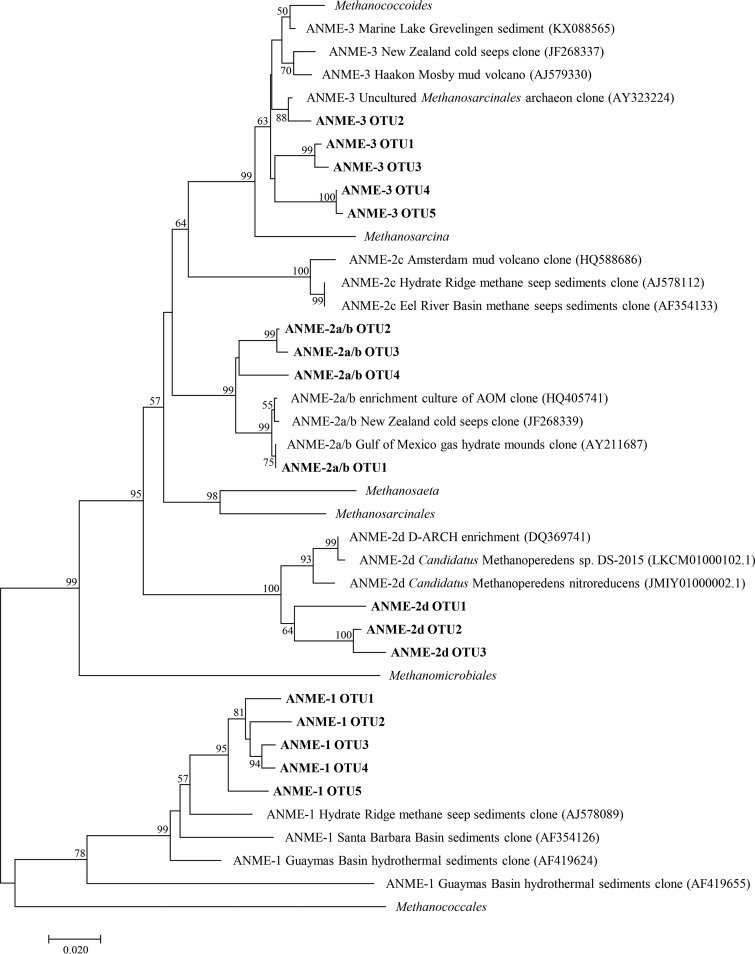
The 16S rRNA gene sequences of ANME-1, ANME-2a/b, ANME-2d, and ANME-3 clades were successfully retrieved by the primers Arc519F and Arc915R. Five OTU belonging to ANME-1 clade, 4 OTU belonging to ANME-2a/b clade, 3 OTU belonging to ANME-2d clade, and 5 OTU belonging to ANME-3 clade were selected for phylogenetic analysis (Fig. 3). The 5 OTU related to ANME-1 cluster exhibited high similarity to the sequence recovered from Hydrate Ridge methane seep sediments (AJ578089) (22), while the 5 OTU related to ANME-3 cluster were also closely related to Methanococcoides. The ANME-2a/b and ANME-2d clades were related to ANME-3 clade and showed a relatively distant phylogenetic relationship with ANME-1 clade.
FIG 3.
Neighbor-joining phylogenetic tree showing the phylogenetic affiliations of ANME archaeal 16S rRNA gene sequences recovered from the Zhoushan archipelago intertidal zone. The bootstrap values included 1,000 replicates, and the scale bar represents 2% sequence divergence.
Pearson correlation analysis showed that the SO42– content in sediments was positively correlated with the ANME archaea OTU number (P < 0.01) and the Shannon index (P < 0.05) (Table S3). In addition, the TOM content in sediments was also significantly positively correlated with the ANME archaea OTU number (P < 0.01) and the Chao 1 index (P < 0.05).
ANME-1, ANME-2c, and ANME-3 archaea were detected in all sediments samples by qPCR. ANME-2c archaea were dominant in sampling points collected in spring, summer and autumn (excluding AS), with the relative abundance ranging from 48.56 to 79.60% (Fig. S4). Furthermore, the relative abundance of ANME-2c archaea was highest in the low-tide zone except in the spring. ANME-3 archaea were the dominant ANME archaea in the AS and winter sampling points, accounting for 62.13 to 91.91% of the total ANME archaea. ANME-1 archaea occupied the lowest percentage of ANMEs archaea, only ranging from 2.48 to 16.26%. The community structure and relative abundance of ANME archaea revealed by high-throughput sequencing were consistent with the qPCR results. Redundancy analysis (RDA) results showed that among all the physicochemical factors tested, the TOM and SO42– content had a significant effect on the community structure of ANME archaea (P < 0.05), while TP and salinity exerted an insignificant influence (0.05 < P < 0.10) (Fig. S5).
Potential S-AOM activities in simulated columns.
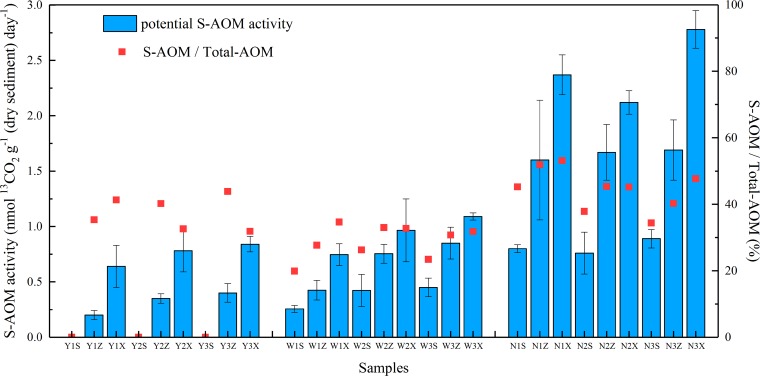
The contribution rates of each methane oxidation process to total methane oxidation equal the ratios of its value to the sum of aerobic methane oxidation, D-AOM, S-AOM, and iron- and manganese-dependent AOM. Compared to the aerobic methane oxidation process, the contribution rates of S-AOM process and D-AOM process to total methane removal were relatively low (Table S4). The methane removed by D-AOM process took up 1.17 to 43.34% of the total methane oxidation, with the potential D-AOM activity ranging from 0.34 to 3.05 nmol 13CO2 g−1 (dry sediment) day−1. The potential S-AOM activity was slightly lower than the potential D-AOM activity in simulated column sediments, varying from 0.00 to 2.78 nmol 13CO2 g−1 (dry sediment) day−1, which accounted for 0.00 to 49.10% of the total methane removal (Fig. 4). Overall, S-AOM activities increased slightly during the incubation. It is remarkable that the S-AOM activities increased in order of column Y, column W, and column N. In addition, S-AOM activities also rose apparently with the depth. Specifically, no S-AOM activity was detected in three surface sampling points of the aerobic soil column samples Y1S, Y2S, and Y3S. These results indicated that oxygen inhibited S-AOM activities considerably, which was subsequently demonstrated by the negative correlation between oxidation-reduction potential (ORP) and S-AOM activity using Pearson correlation analysis (Table S5).
FIG 4.
Potential S-AOM activity and ratio of potential S-AOM activity to total-AOM activity in the samples collected from the simulated columns. Red squares represent the contribution ratios of the S-AOM process to the total anaerobic methane oxidation. Error bars represent the standard deviations.
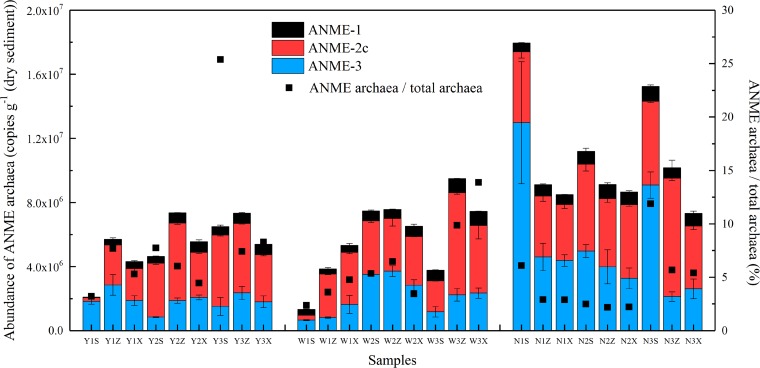
Abundance of ANME archaea in simulated columns.
Through the qPCR experiment, ANME-1, ANME-2c, and ANME-3 archaea were successfully detected in simulated column sediments (Fig. 5). Among all the detected ANME archaea, the average abundance of ANME-2c archaea was the highest, ranging from 2.17 × 105 to 7.39 × 106 copies g−1 (dry sediment). ANME-3 archaea ranked second and showed an approximate abundance to ANME-2c archaea in column N. ANME-1 archaea were the fewest, with an average copy number of 6.15 × 105 copies g−1 (dry sediment). The 16S rRNA gene copy number of the total ANME archaea consisting of ANME-1, ANME-2c, and ANME-3 archaea increased in order of column Y, column W, and column N. In addition, the average ratios of ANME archaea to total archaea in column Y, column W, and column N were 8.40, 6.08, and 4.64%, respectively. This indicated that ANME archaea accounted for a considerable part of the microbial phase in simulated columns.
FIG 5.
Abundance of ANMEs archaea in samples collected from the simulated columns. Black squares represent the ratios of ANME archaea abundance to the total archaea detected by qPCR. Error bars represent the standard deviations.
Pearson correlation analysis revealed that temperature was significantly positively correlated with the abundance of ANME-1, ANME-2c archaea (P < 0.01), and S-AOM activity (P < 0.05) (Table S5). There was also a significant positive correlation between sulfate and ANME-1 archaea (P < 0.05) and potential S-AOM activity (P < 0.01). TOM in the simulated column sediments was significantly positively correlated with the number of ANME-1, ANME-2c archaea, and S-AOM activity (P < 0.01).
Diversity and community structure of ANME archaea in simulated columns.
A total of 37,854 ANME archaeal 16S rRNA gene sequences were obtained in 27 simulated column samples (excluding ANME-2d archaea) (Table S6). The OTU number of ANME archaea was 38 to 89, with a cutoff 3% sequence difference. The largest OTU number of ANME archaea appeared in sample W1Z of column W. The OTU number of ANME archaea varied from different simulated columns, with an average value of 68 in column W, 64 in column Y, and 47 in column N. The Shannon index and the Chao 1 index of the soil column samples showed a similar change in OTU number. Pearson correlation analysis showed that there was a significant negative correlation between ORP and the OTU number (P < 0.05), while SO42– was significantly positively correlated with the OTU number, the Shannon index, and the Chao 1 index (P < 0.01) (Table S7).
Using qPCR, it was found that methane anaerobic oxidation archaea were mainly comprised of ANME-1, ANME-2c, ANME-3, and ANME-2d archaea (Fig. S6). ANME-2c archaea accounted for the largest proportion, while ANME-3 archaea were the dominant clade in some samples. We speculated that these two clades were the main force of S-AOM process. The ANME-2d archaea responsible for nitrate-AOM showed the lowest relative abundance in all the samples. In order of column Y, column W, and column N, the decreasing trend of ANME-2c archaea and the increasing trend of ANME-3 archaea were consistent with the qPCR results (Fig. 5). RDA showed that SO42– and TOM were strongly correlated with the community structure of ANME archaea (P < 0.01, Monte Carlo test, 1,000 times; Fig. S7).
DISCUSSION
The previous research illustrated that ANME-1, ANME-2a/b, ANME-2c, and ANME-3 archaea were involved in S-AOM process (18). However, only ANME-1, ANME-2c, and ANME-3 archaea were detected by qPCR in the present study. It was reported that ANME archaeal 16S rRNA gene copy numbers were lower than the detection limit of the primers used in the experiment in natural gas fields and freshwater lake water columns (31, 33). However, the 16S rRNA gene copy numbers of ANME-1, ANME-2c, and ANME-3 archaea in Sonora Margin cold seep sediments from Maas Basin of California Bay were up to 1010 to 1011 copies g−1 (dry sediment) (24). By using specific primers based on the ANME archaeal mcrA gene, it was found that the mcrA gene copy number of ANME archaea was 107 to 108 copies g−1 (dry sediment) in coastal reedbed sediments (34). We summarized the abundance of ANME archaea detected in different natural habitats in Table 2. This study further proved that the abundance of ANME archaea varied noticeably in different habitats.
TABLE 2.
Comparison of sulfate-dependent AOM rates and abundance of ANME archaea in natural habitats
| Habitat | AOM rate (nmol cm−3 day−1) | Abundance of ANME archaea | Source or reference(s) |
|---|---|---|---|
| Sonora margin cold seep sediments | NAa | 1010–1011 16S rRNA gene copies g−1 (dry sediment) | 24 |
| Offshore Joetsu methane-seep sediments | NA | 105–109 mcrA gene copies g−1 (dry sediment) | 36 |
| Coastal reedbed sediments | NA | 107–108 mcrA gene copies g−1 (dry sediment) | 34 |
| Haima cold seep sediments | NA | 103–107 mcrA gene copies g−1 (dry sediment) | 68 |
| Natural gas fields | NA | <105 16S rRNA gene copies ml−1 | 31 |
| Freshwater lake water column | NA | Less than the detection limit | 33 |
| Black Sea microbial mats and reefs | 1,000–20,000 | NA | 16, 41 |
| Cold seep sediments | 100–5,000 | NA | 27, 38 |
| Mud volcano | 10–500 | NA | 26, 48 |
| Coastal SMTZ | 1–200 | NA | 28, 46, 49 |
| Lake sediments | 0.01–5 | NA | 50, 51 |
| Subsurface SMTZ | 0.001–1 | NA | 30 |
| Black Sea water column | 0.0001–0.01 | NA | 52 |
| Zhoushan archipelago intertidal zone sediments | 0–0.77 nmol g−1 (dry sediment) day−1 | 1.64 × 106 to 9.99 × 106 16S rDNA copies g−1 (dry sediment) | This study |
NA, not applicable
Usually, ANME-1 archaea exist in strictly anaerobic sediments rich in sulfate (35). The low abundance of ANME-1 archaea in the intertidal sediments corroborated that the intertidal ecosystem was not a suitable habitat for ANME-1 archaea. ANME-2c archaea were the dominant ANME archaea in the tested area and might be the major functional microorganisms for S-AOM process. However, previous studies indicated that ANME-2c archaea were mainly distributed in deep marine sediments (36, 37) and estuarine sediments (38). However, this study verified that ANME-2c archaea could also exist in the intertidal surface sediments with large abundance. Notably, the abundance of ANME-1 and ANME-2c archaea fluctuated less in four seasons, whereas ANME-3 archaea abundance upsurged and became the dominant ANME archaea in the winter samples. ANME-3 clade is closely related to the genus Methanococcoides, clustered within Methanosarcinales. In addition, the growth temperature range of the genus Methanococcoides is 1.7 to 35°C (39). Previous studies demonstrated that ANME-3 archaea were mainly found in mud volcanoes and gas seep sediments, which were characterized by a high methane content and low temperature (26, 40). The average temperature of winter sediments was around 9.0°C, which was much lower than the temperatures of sediments in the other three seasons. In addition, the sulfate and TOM content was highest in the winter sediments, which provided sufficient electron acceptors and methane for S-AOM process and was beneficial to the growth of ANME archaea. Therefore, lower temperatures and a higher sulfate and TOM content might be responsible for a large quantity of ANME-3 archaea in winter sediments. However, low temperature resulted not only in the high abundance of ANME-3 archaea but also in the reduction of the activities of all ANME archaea. This could explain why there was a contrary trend between activity and abundance (Fig. 1 and 2).
Quantitative PCR and 16S rRNA gene high-throughput sequencing showed a similar pattern in the community structure of ANME archaea both in situ and in simulated columns. Some studies pointed out that only one single dominant clade of ANME archaea was discovered in one habitat (22–24, 40–42). For example, ANME-1 archaea were dominant in deep anaerobic sediments (ANME-1b) or high-temperature sediments (ANME-1a) (24, 37, 43), while ANME-2c archaea played a leading role in a variety of habitats rich in sulfate (44). ANME-3 archaea were found to be dominant in cold seeps and mud volcanic sediments with high methane partial pressure and low temperatures, such as the Haakon Mosby mud volcano 1,250 m below sea level (26), the surface sediments in Guaymas basin 1,500 m below sea level (24), and the Kazan mud volcano 1,700 m below sea level (32, 45). However, diverse ANME archaea were able to coexist in a particular habitat (46). In this study, ANME-1, ANME-2c, and ANME-3 archaea were simultaneously detected in the intertidal ecosystem, which showed a high diversity. We speculated that the variable environmental conditions in intertidal zones provided different ANME archaea with different habitats suitable for their growth. However, the abundance of ANME-3 clade noted here was an uncertainty because of the primers’ specificity, and this should be resolved by using more specific primers in the future.
Previous work observed significant nitrate production in the surface layer in an intertidal area with bulrush as the dominant vegetation, which could provide substrate for D-AOM process (47). This could also explain the higher D-AOM rates than S-AOM rates observed here. The potential S-AOM activity detected in the present study was far lower than the reported maximum. The potential S-AOM rate of microbial mats and reefs of the anoxic Black Sea was up to 1,000 to 20,000 nmol 13CO2 cm−3 day−1 (16, 41). The S-AOM rates detected in different habitats varied widely, with the rates of 100 to 5,000 nmol 13CO2 cm−3 day−1 in the cold seep sediments of the Gulf of Mexico (27, 38), 10 to 500 nmol 13CO2 cm−3 day−1 in the mud volcanic sediments (26, 48), and 1 to 200 nmol 13CO2 cm−3 day−1 in coastal SMTZs (28, 46, 49). However, the potential S-AOM activity detected in the intertidal sediments was approximate to that in lake sediments and Black seawater column (0.0001 to 1 nmol 13CO2 cm−3 day−1) (50–52). The activities of S-AOM in different natural habitats are summarized in Table 2. The availability of sulfate and methane not only affected the rate of S-AOM process but also the growth of ANME archaea (53, 54). Compared to the concentration of sulfate and methane in the habitats such as seabed microbial mats, cold seep sediments, and mud volcanic sediments, those in the intertidal zone were far from the threshold for the high-rate S-AOM process. Furthermore, the S-AOM activity was also lower than the D-AOM activity for this site. However, the average potential contribution rate of S-AOM to total AOM was 34.5%, which implied that S-AOM process was still an important methane sink that cannot be ignored in the intertidal ecosystem.
Temperature, salinity, sulfate, and methane content could exert impacts on the diversity and niche of ANME archaea (55). In the present study, temperature was found to be significantly positively correlated with the activity of ANME archaea both in situ and in simulated columns (P < 0.05). Sulfate concentration was significantly positively correlated with the OTU number, the Shannon index, and the Chao1 index of ANME archaea in simulated columns (P < 0.01). TOM also showed a significantly positive correlation with the Shannon index and the Chao 1 index of ANME archaea (P < 0.05), which could provide methane for S-AOM process under anaerobic conditions. Moreover, TOM had a significant effect on the community structure of ANME archaea. These results show that temperature, sulfate, and TOM/methane were vital environmental factors influencing the S-AOM process and the ANME archaea.
ORP was proportional to the logarithm of the dissolved oxygen concentration (56). The availability of oxygen in soil was a limiting factor for the control of methanogens and methanotrophs (57). Previous researches proved that Euryarchaeota archaea prefer anaerobic conditions in natural habitats (58, 59). Our work found that the abundance and the activity of ANME archaea showed an obvious increasing trend according to the order of the mid-tide zone, the low-tide zone, and the subtidal zone. The results suggested that ANME archaea preferred a stable anaerobic environment such as the flooded subtidal zone. Considering that the availability of oxygen in soils was primarily affected by such factors as moisture, light conditions, and plants species (5), we conducted the simulated column experiments to further explore the effect of oxygen on ANME archaea by adding different amounts of water to different simulated columns. The ORP value decreased in order of aerobic column Y, microaerobic column W, and anoxic column N, while the abundance and the activity of ANME archaea showed a reverse trend. The results reinforced the view that ANME archaea were extremely sensitive to oxygen.
The majority of ANME archaea were found in habitats that were difficult to simulate in the laboratory (43, 60). Some researchers have probed into the enrichment and culture of ANME archaea under atmospheric pressure (61). The discovery of ANME archaea in intertidal sediments, together with the incubation, in simulated columns provided new insights into inoculum screening and enrichment of anaerobic methanotrophic archaea under laboratory conditions.
MATERIALS AND METHODS
Sample collection.
Sediments were collected from the intertidal zone of Zhoushan West Wharf (N30°06′56.20″, E122°08′22.09″), which is located at the Qiangtang River estuary (Fig. S8). The samples were collected from the mid-tide zone, the low-tide zone, and the subtidal zone, respectively, in the spring (May 2015), summer (August 2015), autumn (November 2015), and winter (February 2016). The samples collected in the spring were named SpM (spring mid-tide zone), SpL (spring low-tide zone), and SpS (spring subtidal zone); the samples collected in summer were named SM, SL, and SS; the samples collected in autumn were named AM, AL, and AS; and the samples collected in winter were named WM, WL, and WS. The top 5 cm of sediments were collected carefully using box cores with a total volume of >1 liter for each sample. The sediments were well mixed and then put into sterile containers, sealed, and transported to the laboratory. Each sample was divided into three parts: one was used for immediate potential methane oxidation activity tests, one was stored at 4°C for physicochemical analysis, and one was stored at –20°C for molecular analysis.
Simulated column experiments.
Extra 50-liter sediments were collected in summer for the simulated column experiments. The sediments were the mixture of sediments from three different parts. The simulated columns were made of polymethyl methacrylate. The main body was 70 cm in height and 10 cm in inner diameter. Each column was filled with well-mixed sediments with a filling height of 50 cm (Fig. S9). The samples were collected at three different depths: 5 cm (S), 25 cm (Z), and 45 cm (X). Three different groups were set for the simulated column experiments (column Y, column W, and column N). A 50-ml portion of filter-sterilized seawater was added to column Y every 10 days during cultivation in order to keep it aerobic. Column W was semisubmerged, and 100 ml of sterilized seawater was added every 2 days. Column N was kept flooded during the whole incubation process, and the overlying water depth was set at 10 to 15 cm. All columns were prepared in triplicate. The incubation lasted for 3 months, and the samples at three different depths were collected monthly. For example, the sample named W2Z represented the sample collected at the 25-cm depth from the column W in the second month.
Physical and chemical analysis.
A pH meter (IQ150; IQ Scientific Instruments, Inc., Carlsbad, CA) and a portable ORP meter (Orion 320P-84; Thermo Fisher Scientific, Massachusetts, MA) were used to measure pH, temperature, and redox potential of the sediments in situ. A conductivity-temperature-depth system (CTD) was used to measure the salinity of surface water. The total nitrogen, total phosphorus, ammonium, nitrite, nitrate, and sulfate of the sediments were measured by spectrophotometry (62). The total iron and manganese contents were measured by flame atomic absorption spectrometry. The physicochemical characteristics are shown in Tables S8 and S9, and the chemical profiles for this region were reported previously (63).
Activity tests.
Portions (10 g) of sediments for each sample and 35 ml of filtered seawater were transferred to a 75-ml serum bottle. A BaCl2 solution was added to each sample according to the SO42– concentration in the serum bottle in order to remove SO42–. Serum bottles were then aerated with Ar gas and placed in a shaker at 30°C with a rotation speed of 150 rpm min−1 for 48 h in order to remove residual oxygen, NOx–, and other electron acceptors in sediments. By adding 13CH4 and different electron acceptors (O2, NO3–, SO42–, ferrihydrite, and birnessite), the aerobic methane oxidation, D-AOM, S-AOM, and iron- and manganese-dependent AOM rates were measured, respectively, by using the 13C stable isotope tracing method, with three replicates in each group. The serum bottles were incubated in a shaker at 30°C in dark, rotated at 150 rpm min−1, and sampled every 24 h. The 13CH4 consumed and 13CO2 produced in the serum bottle were determined by gas chromatography-mass spectrometry (Agilent 7890A inert MSD; Agilent) (64). The dry weight of the sediments in the serum bottle was measured after the activity test was completed. Information about the treatment and the calculation formulas can be found in Table S10.
DNA extraction and Illumina-based 16S rRNA gene sequencing.
DNA extraction was performed using the Power Soil DNA kit (Mo Bio Laboratories, Carlsbad, CA) according to the manufacturer’s instructions. The archaeal 16S rRNA genes (V4-V5 region) were amplified by the primers Arc519F/Arc915R (65). Sequencing of archaeal 16S rRNA genes was performed on the Illumina-MiSeq platform (Shanghai Personal Biotechnology Co., Ltd.). The raw data were submitted to NCBI Sequence Read Archive database under accession numbers SRR7772563 to SRR7772574. The rarefaction curves for the sequencing are shown in Fig. S10. Specific information for the primers used in this study is shown in Table 3.
TABLE 3.
Primers used in this study
| Primer | Sequence (5′–3′) | Target gene | Reference |
|---|---|---|---|
| Arc519F | CAGCCGCCGCGGTAA | Archaeal 16S rRNA gene | 65 |
| Arc915R | GTGCTCCCCCGCCAATTCCT | Archaeal 16S rRNA gene | 65 |
| ANME1-395F | AAC TCT GAG TGC CTC CTA | ANME-1 16S rRNA gene | 31 |
| AAC TCT GAG TGC CTC CAA | 31 | ||
| AAC TCT GAG TGC CCC CTA | 31 | ||
| ANME1-1417R | CCT CAC CTA AAC CCC ACT | ANME-1 16S rRNA gene | 31 |
| CCT CAC CTA AAT CCC ACT | 31 | ||
| ANME-2cF | TCG TTT ACG GCT GGG ACT AC | ANME-2 16S rRNA gene | 24 |
| ANME-2cR | TCC TCT GGG AAA TCT GGT TG | ANME-2 16S rRNA gene | 24 |
| ANME-3F | GGATTGGCATAACACCGG | ANME-3 16S rRNA gene | 24 |
| ANME-3R | TATGCTGGCACTCAGTGTCC | ANME-3 16S rRNA gene | 24 |
| DP397F | TGGCTGTCCAGCTRTYC | ANME-2d 16S rRNA gene | 69 |
| DP569R | GRACGCCTGACGATTRAG | ANME-2d 16S rRNA gene | 69 |
Quantitative PCR.
Specific primers Arc519F/Arc915R were used to determine the abundance of total archaea in sediments by qPCR (65). The specific primers ANME1-395F/ANME1-1417R, ANME-2cF/ANME-2cR (31), and ANME-3F/ANME-3R (24) were used to determine the abundance of ANME-1, ANME-2c, and ANME-3, respectively. The standard curves of qPCR were, respectively, constructed with a series of 10-fold dilutions of plasmid DNA with a known copy number (amplification efficiency [εC] > 0.91).
Phylogenetic and statistical analysis.
ANME archaeal 16S rRNA gene sequences were classified into OTU by using QIIME (www.qiime.org), with a cutoff 3% sequence difference. OTU belonging to ANME archaea with a relative abundance of >0.001% were selected for phylogenetic analysis. The representative sequences of the selected OTUs, as well as the reference sequences retrieved from the National Center for Biotechnology Information (NCBI), were aligned in MEGA 7.0 using the ClustalW algorithm. Phylogenetic trees were reconstructed using the neighbor-joining method in MEGA 7.0 (66).
QIIME was also used to analyze the diversity (Shannon index and Chao1 index) of the obtained OTU. The correlations between the environmental factors and the community structure of ANME archaea were determined by using CANOCO software redundancy analysis (67). Pearson correlation analysis of SPSS 19.0 (SPSS 2010) was used to determine the effect of environmental factors on the abundance, diversity, and activity of ANME archaea.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by the National Natural Science Foundation of China (51478415, 41641031, and 41773074). This study was also funded by the Open Project of State Key Laboratory of Urban Water Resource and Environment, Harbin Institute of Technology (QAK201714) and the Research Funds for Central Universities (2017xzzx010-03).
We thank Fumin Fang for assistance.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02638-18.
REFERENCES
- 1.Core Writing Team, Pachauri RK, Meyer LA. 2014. Climate change 2014: synthesis report—contribution of working groups I, II, and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. IPCC, Geneva, Switzerland. [Google Scholar]
- 2.Tamai N, Takenaka C, Ishizuka S. 2007. Water-soluble Al inhibits methane oxidation at atmospheric concentration levels in Japanese forest soil. Soil Biol Biochem 39:1730–1736. doi: 10.1016/j.soilbio.2007.01.029. [DOI] [Google Scholar]
- 3.Whalen SC. 2005. Biogeochemistry of methane exchange between natural wetlands and the atmosphere. Environ Eng Sci 22:73–94. doi: 10.1089/ees.2005.22.73. [DOI] [Google Scholar]
- 4.Nisbet EG, Dlugokencky EJ, Bousquet P. 2014. Methane on the rise—again. Science 343:493–495. doi: 10.1126/science.1247828. [DOI] [PubMed] [Google Scholar]
- 5.Malyan SK, Bhatia A, Kumar A, Gupta DK, Singh R, Kumar SS, Tomer R, Kumar O, Jain N. 2016. Methane production, oxidation and mitigation: a mechanistic understanding and comprehensive evaluation of influencing factors. Sci Total Environ 572:874–896. doi: 10.1016/j.scitotenv.2016.07.182. [DOI] [PubMed] [Google Scholar]
- 6.Raghoebarsing AA, Pol A, van de Pas-Schoonen KT, Smolders AJP, Ettwig KF, Rijpstra WIC, Schouten S, Damsté JSS, Op den Camp HJM, Jetten MSM, Strous M. 2006. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440:918. doi: 10.1038/nature04617. [DOI] [PubMed] [Google Scholar]
- 7.Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MMM, Schreiber F, Dutilh BE, Zedelius J, de Beer D, Gloerich J, Wessels HJCT, van Alen T, Luesken F, Wu ML, van de Pas-Schoonen KT, Op den Camp HJM, Janssen-Megens EM, Francoijs K-J, Stunnenberg H, Weissenbach J, Jetten MSM, Strous M. 2010. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 8.Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P, Yuan Z, Tyson GW. 2013. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500:567–570. doi: 10.1038/nature12375. [DOI] [PubMed] [Google Scholar]
- 9.Knittel K, Boetius A. 2009. Anaerobic oxidation of methane: progress with an unknown process. Annu Rev Microbiol 63:311–334. doi: 10.1146/annurev.micro.61.080706.093130. [DOI] [PubMed] [Google Scholar]
- 10.Ettwig KF, Zhu B, Speth D, Keltjens JT, Jetten MSM, Kartal B. 2016. Archaea catalyze iron-dependent anaerobic oxidation of methane. Proc Natl Acad Sci U S A 113:12792–12796. doi: 10.1073/pnas.1609534113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He Z, Zhang Q, Feng Y, Luo H, Pan X, Gadd GM. 2018. Microbiological and environmental significance of metal-dependent anaerobic oxidation of methane. Sci Total Environ 610-611:759–768. doi: 10.1016/j.scitotenv.2017.08.140. [DOI] [PubMed] [Google Scholar]
- 12.Barnes RO, Goldberg ED. 1976. Methane production and consumption in anoxic marine sediments. Geol 4:297–300. doi:. [DOI] [Google Scholar]
- 13.Hinrichs KU, Hayes JM, Sylva SP, Brewer PG, Delong EF. 1999. Methane-consuming archaebacteria in marine sediments. Nature 398:802–805. doi: 10.1038/19751. [DOI] [PubMed] [Google Scholar]
- 14.Boetius A, Ravenschlag K, Schubert CJ, Rickert D, Widdel F, Gieseke A, Amann R, Jørgensen BB, Witte U, Pfannkuche O. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623–626. doi: 10.1038/35036572. [DOI] [PubMed] [Google Scholar]
- 15.Orphan VJ, House CH, Hinrichs KU, McKeegan KD, DeLong EF. 2001. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293:484–487. doi: 10.1126/science.1061338. [DOI] [PubMed] [Google Scholar]
- 16.Michaelis W, Seifert R, Nauhaus K, Treude T, Thiel V, Blumenberg M, Knittel K, Gieseke A, Peterknecht K, Pape T, Boetius A, Amann R, Jørgensen BB, Widdel F, Peckmann J, Pimenov NV, Gulin MB. 2002. Microbial reefs in the Black Sea fueled by anaerobic oxidation of methane. Science 297:1013–1015. doi: 10.1126/science.1072502. [DOI] [PubMed] [Google Scholar]
- 17.Hallam SJ, Putnam N, Preston CM, Detter JC, Rokhsar D, Richardson PM, Delong EF. 2004. Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 305:1457–1462. doi: 10.1126/science.1100025. [DOI] [PubMed] [Google Scholar]
- 18.Timmers PHA, Welte CU, Koehorst JJ, Plugge CM, Jetten MSM, Stams AJM. 2017. Reverse methanogenesis and respiration in methanotrophic archaea. Archaea 2017:1–22. doi: 10.1155/2017/1654237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGlynn SE. 2017. Energy metabolism during anaerobic methane oxidation in ANME archaea. Microbes Environ 32:5–13. doi: 10.1264/jsme2.ME16166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wegener G, Krukenberg V, Riedel D, Tegetmeyer HE, Boetius A. 2015. Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature 526:587–590. doi: 10.1038/nature15733. [DOI] [PubMed] [Google Scholar]
- 21.Mcglynn SE, Chadwick GL, Kempes CP, Orphan VJ. 2015. Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 526:531–535. doi: 10.1038/nature15512. [DOI] [PubMed] [Google Scholar]
- 22.Knittel K, Lösekann T, Boetius A, Kort R, Amann R. 2005. Diversity and distribution of methanotrophic archaea at cold seeps. Appl Environ Microbiol 71:467–479. doi: 10.1128/AEM.71.1.467-479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orphan VJ, House CH, Hinrichs KU, Mckeegan KD, Delong EF. 2002. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc Natl Acad Sci U S A 99:7663–7668. doi: 10.1073/pnas.072210299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vigneron A, Cruaud P, Pignet P, Caprais JC, Cambon-Bonavita MA, Godfroy A, Toffin L. 2013. Archaeal and anaerobic methane oxidizer communities in the Sonora Margin cold seeps, Guaymas Basin (Gulf of California). ISME J 7:1595. doi: 10.1038/ismej.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pernthaler A, Dekas AE, Brown CT, Goffredi SK, Embaye T, Orphan VJ. 2008. Diverse syntrophic partnerships from deep-sea methane vents revealed by direct cell capture and metagenomics. Proc Natl Acad Sci U S A 105:7052–7057. doi: 10.1073/pnas.0711303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niemann H, Lösekann T, de Beer D, Elvert M, Nadalig T, Knittel K, Amann R, Sauter EJ, Schlüter M, Klages M, Foucher JP, Boetius A. 2006. Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink. Nature 443:854–858. doi: 10.1038/nature05227. [DOI] [PubMed] [Google Scholar]
- 27.Joye SB, Boetius A, Orcutt BN, Montoya JP, Schulz HN, Erickson MJ, Lugo SK. 2004. The anaerobic oxidation of methane and sulfate reduction in sediments from Gulf of Mexico cold seeps. Chem Geol 205:219–238. doi: 10.1016/j.chemgeo.2003.12.019. [DOI] [Google Scholar]
- 28.Treude T, Krüger M, Boetius A, Jørgensen BB. 2005. Environmental control on anaerobic oxidation of methane in the gassy sediments of Eckernförde Bay (German Baltic). Limnol Oceanogr 50:1771–1786. doi: 10.4319/lo.2005.50.6.1771. [DOI] [Google Scholar]
- 29.Reeburgh WS. 2007. Oceanic methane biogeochemistry. Chem Rev 107:486–513. doi: 10.1021/cr050362v. [DOI] [PubMed] [Google Scholar]
- 30.Sivan O, Schrag DP, Murray RW. 2007. Rates of methanogenesis and methanotrophy in deep-sea sediments. Geobiology 5:141–151. doi: 10.1111/j.1472-4669.2007.00098.x. [DOI] [Google Scholar]
- 31.Miyashita A, Mochimaru H, Kazama H, Ohashi A, Yamaguchi T, Nunoura T, Horikoshi K, Takai K, Imachi H. 2009. Development of 16S rRNA gene-targeted primers for detection of archaeal anaerobic methanotrophs (ANMEs). FEMS Microbiol Lett 297:31–37. doi: 10.1111/j.1574-6968.2009.01648.x. [DOI] [PubMed] [Google Scholar]
- 32.Pachiadaki MG, Lykousis V, Stefanou EG, Kormas KA. 2010. Prokaryotic community structure and diversity in the sediments of an active submarine mud volcano (Kazan mud volcano, East Mediterranean Sea). FEMS Microbiol Lett 72:429–444. doi: 10.1111/j.1574-6941.2010.00857.x. [DOI] [PubMed] [Google Scholar]
- 33.Zigah PK, Oswald K, Brand A, Dinkel C, Wehrli B, Schubert CJ. 2015. Methane oxidation pathways and associated methanotrophic communities in the water column of a tropical lake. Limnol Oceanogr 60:553–572. doi: 10.1002/lno.10035. [DOI] [Google Scholar]
- 34.Zhou Z, Han P, Gu JD. 2014. New PCR primers based on mcrA gene for retrieving more anaerobic methanotrophic archaea from coastal reedbed sediments. Appl Microbiol Biotechnol 98:4663. doi: 10.1007/s00253-014-5599-5. [DOI] [PubMed] [Google Scholar]
- 35.Biddle JF, Cardman Z, Mendlovitz H, Albert DB, Lloyd KG, Boetius A, Teske A. 2012. Anaerobic oxidation of methane at different temperature regimes in Guaymas Basin hydrothermal sediments. ISME J 6:1018. doi: 10.1038/ismej.2011.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yanagawa K, Sunamura M, Lever MA, Morono Y, Hiruta A, Ishizaki O, Matsumoto R, Urabe T, Inagaki F. 2011. Niche separation of methanotrophic archaea (ANME-1 and -2) in methane-seep sediments of the Eastern Japan Sea offshore Joetsu. Geomicrobiol J 28:118–129. doi: 10.1080/01490451003709334. [DOI] [Google Scholar]
- 37.Nunoura T, Oida H, Toki T, Ashi J, Takai K, Horikoshi K. 2006. Quantification of mcrA by quantitative fluorescent PCR in sediments from methane seep of the Nankai Trough. FEMS Microbiol Ecol 57:149–157. doi: 10.1111/j.1574-6941.2006.00101.x. [DOI] [PubMed] [Google Scholar]
- 38.Treude T, Boetius A, Knittel K, Wallmann K, Barker Jørgensen B. 2003. Anaerobic oxidation of methane above gas hydrates at Hydrate Ridge, NE Pacific Ocean. Mar Ecol Prog Ser 264:1–14. doi: 10.3354/meps264001. [DOI] [Google Scholar]
- 39.Sowers KR. 2015. Methanococcoides In Bergey’s manual of systematics of Archaea and Bacteria. John Wiley & Sons, Ltd, London, United Kingdom. doi: 10.1002/9781118960608.gbm00514. [DOI] [Google Scholar]
- 40.Losekann T, Knittel K, Nadalig T, Fuchs B, Niemann H, Boetius A, Amann R. 2007. Diversity and abundance of aerobic and anaerobic methane oxidizers at the Haakon Mosby mud volcano, Barents Sea. Appl Environ Microbiol 73:3348–3362. doi: 10.1128/AEM.00016-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Treude T, Orphan V, Knittel K, Gieseke A, House CH, Boetius A. 2007. Consumption of methane and CO2 by methanotrophic microbial mats from gas seeps of the anoxic Black Sea. Appl Environ Microbiol 73:2271–2283. doi: 10.1128/AEM.02685-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wegener G, Shovitri M, Knittel K, Niemann H, Hovland M, Boetius A. 2008. Biogeochemical processes and microbial diversity of the Gullfaks and Tommeliten methane seeps (Northern North Sea). Biogeosciences 5:1127–1144. doi: 10.5194/bg-5-1127-2008. [DOI] [Google Scholar]
- 43.Ruff SE, Biddle JF, Teske AP, Knittel K, Boetius A, Ramette A. 2015. Global dispersion and local diversification of the methane seep microbiome. Proc Natl Acad Sci U S A 112:4015–4020. doi: 10.1073/pnas.1421865112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Felden J, Ruff SE, Ertefai T, Inagaki F, Hinrichs KU, Wenzhöfer F. 2014. Anaerobic methanotrophic community of a 5,346-m-deep vesicomyid clam colony in the Japan Trench. Geobiology 12:183. doi: 10.1111/gbi.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heijs SK, Haese RR, van der Wielen PWJJ, Forney LJ, van Elsas JD. 2007. Use of 16S rRNA gene based clone libraries to assess microbial communities potentially involved in anaerobic methane oxidation in a Mediterranean cold seep. Microb Ecol 53:384–398. doi: 10.1007/s00248-006-9172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parkes RJ, Cragg BA, Banning N, Brock F, Webster G, Fry JC, Hornibrook E, Pancost RD, Kelly S, Knab N, Jørgensen BB, Rinna J, Weightman AJ. 2007. Biogeochemistry and biodiversity of methane cycling in subsurface marine sediments (Skagerrak, Denmark). Environ Microbiol 9:1146–1161. doi: 10.1111/j.1462-2920.2006.01237.x. [DOI] [PubMed] [Google Scholar]
- 47.Van Der Nat F, De Brouwer J, Middelburg JJ, Laanbroek HJ. 1997. Spatial distribution and inhibition by ammonium of methane oxidation in intertidal freshwater marshes. Appl Environ Microbiol 63:4734–4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Omoregie EO, Mastalerz V, de Lange G, Straub KL, Kappler A, Roy H, Stadnitskaia A, Foucher JP, Boetius A. 2008. Biogeochemistry and community composition of iron- and sulfur-precipitating microbial mats at the Chefren mud volcano (Nile Deep Sea Fan, Eastern Mediterranean). Appl Environ Microbiol 74:3198–3215. doi: 10.1128/AEM.01751-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knab NJ, Dale AW, Lettmann K, Fossing H, Bo BJ. 2008. Thermodynamic and kinetic control on anaerobic oxidation of methane in marine sediments. Geochim Cosmochim Ac 72:3746–3757. doi: 10.1016/j.gca.2008.05.039. [DOI] [Google Scholar]
- 50.Takeuchi M, Yoshioka H, Seo Y, Tanabe S, Tamaki H, Kamagata Y, Takahashi HA, Igari S, Mayumi D, Sakata S. 2011. A distinct freshwater-adapted subgroup of ANME-1 dominates active archaeal communities in terrestrial subsurfaces in Japan. Environ Microbiol 13:3206–3218. doi: 10.1111/j.1462-2920.2011.02517.x. [DOI] [PubMed] [Google Scholar]
- 51.Schubert CJ, Vazquez F, Losekann-Behrens T, Knittel K, Tonolla M, Boetius A. 2011. Evidence for anaerobic oxidation of methane in sediments of a freshwater system (Lago di Cadagno). FEMS Microbiol Ecol 76:26–38. doi: 10.1111/j.1574-6941.2010.01036.x. [DOI] [PubMed] [Google Scholar]
- 52.Schubert CJ, Durisch-Kaiser E, Holzner CP, Klauser L, Wehrli B, Schmale O, Greinert J, Mcginnis DF, Batist MD, Kipfer R. 2013. Methanotrophic microbial communities associated with bubble plumes above gas seeps in the Black Sea. Geochem Geophy Geosy 7. [Google Scholar]
- 53.Girguis PR, Cozen AE, Delong EF. 2005. Growth and population dynamics of anaerobic methane-oxidizing archaea and sulfate-reducing bacteria in a continuous-flow bioreactor. Appl Environ Microbiol 71:3725. doi: 10.1128/AEM.71.7.3725-3733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nauhaus K, Albrecht M, Elvert M, Boetius A, Widdel F. 2007. In vitro cell growth of marine archaeal-bacterial consortia during anaerobic oxidation of methane with sulfate. Environ Microbiol 9:187–196. doi: 10.1111/j.1462-2920.2006.01127.x. [DOI] [PubMed] [Google Scholar]
- 55.Bhattarai S, Cassarini C, Gonzalez-Gil G, Egger M, Slomp CP, Zhang Y, Esposito G, Lens PNL. 2017. Anaerobic methane-oxidizing microbial community in a coastal marine sediment: anaerobic methanotrophy dominated by ANME-3. Microb Ecol 74:608–622. doi: 10.1007/s00248-017-0978-y. [DOI] [PubMed] [Google Scholar]
- 56.Peddie CC, Mavinic DS, Jenkins CJ. 1990. Use of ORP for monitoring and control of aerobic sludge digestion. J Environ Eng 116:461–471. doi: 10.1061/(ASCE)0733-9372(1990)116:3(461). [DOI] [Google Scholar]
- 57.Bender M, Conrad R. 1993. Kinetics of methane oxidation in oxic soils. Chemosphere 26:687–696. doi: 10.1016/0045-6535(93)90453-C. [DOI] [Google Scholar]
- 58.Lehours AC, Bardot C, Thenot A, Debroas D, Fonty G. 2005. Anaerobic microbial communities in Lake Pavin, a unique meromictic lake in France. Appl Environ Microbiol 71:7389–7400. doi: 10.1128/AEM.71.11.7389-7400.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Llirós M, Gich F, Plasencia A, Auguet JC, Darchambeau F, Casamayor EO, Descy JP, Borrego C. 2010. Vertical distribution of ammonia-oxidizing crenarchaeota and methanogens in the epipelagic waters of Lake Kivu (Rwanda-Democratic Republic of the Congo). Appl Environ Microbiol 76:6853–6863. doi: 10.1128/AEM.02864-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Orphan VJ, Ussler W, Naehr TH, House CH, Hinrichs K-U, Paull CK. 2004. Geological, geochemical, and microbiological heterogeneity of the seafloor around methane vents in the Eel River Basin, offshore California. Chem Geol 205:265–289. doi: 10.1016/j.chemgeo.2003.12.035. [DOI] [Google Scholar]
- 61.Meulepas RJW, Jagersma CG, Gieteling J, Buisman CJN, Stams AJM, Lens PNL. 2009. Enrichment of anaerobic methanotrophs in sulfate-reducing membrane bioreactors. Biotechnol Bioeng 104:458–470. doi: 10.1002/bit.22412. [DOI] [PubMed] [Google Scholar]
- 62.Shen L-D, Liu S, Huang Q, Lian X, He Z-F, Geng S, Jin R-C, He Y-F, Lou L-P, Xu X-Y, Zheng P, Hu B-L. 2014. Evidence for the cooccurrence of nitrite-dependent anaerobic ammonium and methane oxidation processes in a flooded paddy field. Appl Environ Microbiol 80:7611–7619. doi: 10.1128/AEM.02379-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He Z, Wang J, Hu J, Yu H, Jetten MSM, Liu H, Cai C, Liu Y, Ren H, Zhang X, Hua M, Xu X, Zheng P, Hu B. 2019. Regulation of coastal methane sinks by a structured gradient of microbial methane oxidizers. Environ Pollut 244:228–237. doi: 10.1016/j.envpol.2018.10.057. [DOI] [PubMed] [Google Scholar]
- 64.Hu B-L, Shen L-D, Lian X, Zhu Q, Liu S, Huang Q, He Z-F, Geng S, Cheng D-Q, Lou L-P, Xu X-Y, Zheng P, He Y-F. 2014. Evidence for nitrite-dependent anaerobic methane oxidation as a previously overlooked microbial methane sink in wetlands. Proc Natl Acad Sci U S A 111:4495–4500. doi: 10.1073/pnas.1318393111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Delong EF. 1992. Archaea in coastal marine environments. Proc Natl Acad Sci U S A 89:5685. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ter Braak CJ, Smilauer P. 2002. CANOCO reference manual and CanoDraw for Windows user’s guide: software for canonical community ordination (version 4.5). Microcomputer Power, New York, New York. [Google Scholar]
- 68.Niu M, Fan X, Zhuang G, Liang Q, Wang F. 2017. Methane-metabolizing microbial communities in sediments of the Haima cold seep area, northwest slope of the South China Sea. FEMS Microbiol Ecol 93:fix101. doi: 10.1093/femsec/fix101. [DOI] [PubMed] [Google Scholar]
- 69.Ding J, Ding Z, Fu L, Lu Y, Cheng SH, Zeng RJ. 2015. New primers for detecting and quantifying denitrifying anaerobic methane oxidation archaea in different ecological niches. Appl Microbiol Biotechnol 99:9805–9812. doi: 10.1007/s00253-015-6893-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.