This study established an in-frame gene deletion method in the alphaproteobacterium Labrenzia aggregata LZB033 and generated an rpoN gene mutant. A comparison of the transcriptomes and phenotypic characteristics between the mutant and wild-type strains confirmed the role of RpoN in L. aggregata LZB033 flagellar formation, motility, biofilm formation, and carbon usage. Most importantly, RpoN is a key factor for survival under different environmental challenge conditions. Furthermore, the ability to synthesize and metabolize dimethylsulfoniopropionate (DMSP) was related to RpoN. These features revealed RpoN to be an important regulator of stress resistance and survival for L. aggregata LZB033 in marine environments.
KEYWORDS: Labrenzia aggregata, RNA sequencing, RpoN, environmental adaptation, in-frame gene knockout
ABSTRACT
Labrenzia aggregata LZB033 (Rhodobacteraceae), which produces dimethylsulfoniopropionate (DMSP) and reduces nitrate to nitrogen, was isolated from seawater of the East China Sea. Its genome encodes a large number of transcriptional regulators which may be important for its adaptation to diverse marine environments. The alternative σ54 factor (RpoN) is a central regulator of many bacteria, regulating the transcription of multiple genes and controlling important cellular functions. However, the exact role of RpoN in Labrenzia spp. is unknown. In this study, an in-frame rpoN deletion mutant was constructed in LZB033, and the function of RpoN was determined. To systematically identify RpoN-controlled genes, we performed a detailed analysis of gene expression differences between the wild-type strain and the ΔrpoN mutant using RNA sequencing. The expression of 175 genes was shown to be controlled by RpoN. Subsequent phenotypic assays showed that the ΔrpoN mutant was attenuated in flagellar biosynthesis and swimming motility, utilized up to 13 carbon substrates differently, lacked the ability to assimilate malic acid, and displayed markedly decreased biofilm formation. In addition, stress response assays showed that the ΔrpoN mutant was impaired in the ability to survive under different challenge conditions, including osmotic stress, oxidative stress, temperature changes, and acid stress. Moreover, both the DMSP synthesis and catabolism rates of LZB033 decreased after rpoN was knocked out. Our work provides essential insight into the regulatory function of RpoN, revealing that RpoN is a key determinant for LZB033 flagellar formation, motility, biofilm formation, and environmental fitness, as well as DMSP production and degradation.
IMPORTANCE This study established an in-frame gene deletion method in the alphaproteobacterium Labrenzia aggregata LZB033 and generated an rpoN gene mutant. A comparison of the transcriptomes and phenotypic characteristics between the mutant and wild-type strains confirmed the role of RpoN in L. aggregata LZB033 flagellar formation, motility, biofilm formation, and carbon usage. Most importantly, RpoN is a key factor for survival under different environmental challenge conditions. Furthermore, the ability to synthesize and metabolize dimethylsulfoniopropionate (DMSP) was related to RpoN. These features revealed RpoN to be an important regulator of stress resistance and survival for L. aggregata LZB033 in marine environments.
INTRODUCTION
Alphaproteobacteria is one of the most abundant groups of marine bacteria and includes the family Rhodobacteraceae, which consists of >100 genera and >300 species (1). Rhodobacteraceae representatives show a very versatile physiology by dwelling in greatly varied marine habitats and are key players in biogeochemical cycling (1). Recently, we isolated a Rhodobacteraceae member, Labrenzia aggregata LZB033, from surface seawater of the East China Sea and found that it can utilize and produce dimethylsulfoniopropionate (DMSP), which is one of Earth’s most abundant organosulfur molecules (2). The site (122.58°E, 28.73°N) from which LZB033 was isolated is not far from the coast and is near the Yangtze River and the Qiantang River; hence, it could receive a large amount of terrigenous materials from these rivers. In addition, the site was also influenced by the Taiwan Warm Current (TWC) and the Zhejiang Fujian Coastal Current (ZFCC) and thus showed fluctuating properties, as follows: in winter, the ZFCC, carrying the Yangtze’s brackish water and sediment discharge southward, intensifies, and in summer, under the prevailing southeast monsoon, the northward TWC, carrying warm and saline middle-deep water, intensifies. By searching the genome of L. aggregata LZB033, we found that this isolate has a complex regulatory network that may help this bacterium adapt to complex environments under stressful conditions; this network includes components such as quorum-sensing regulators, two-component regulators, and sigma factors. Sigma factors are a variable subunit that combines with five constant subunits to form the polymerase holoenzyme, which regulates the transcription of many bacterial genes (3). The sigma 54 factor, alternatively named RpoN, is the only member of the sigma 54-type family. The transcription of RpoN is distinct from that of other sigma factors in that it requires unusually diverse regulons and genes that function in a variety of cellular processes (3, 4). RpoN was historically known for its role in nitrogen assimilation (5), but it has also been shown to be involved in regulating other important lifestyle-associated functions in bacteria, such as flagellar motility, type III and type VI protein secretion systems, biosynthesis of exopolysaccharides, and biofilm formation in pathogenic bacteria (6–9). Overexpression of rpoN showed that mucoid Pseudomonas aeruginosa cells become nonmucoid (10). Additionally, it was determined by knocking out the rpoN gene that RpoN has a regulatory effect on P. aeruginosa virulence by modulating the function of the PqsR quorum-sensing regulator (11).
Although the mechanism through which RpoN regulates target gene expression is documented in other bacteria, the exact role of RpoN in L. aggregata is unclear. Here, our goal was to identify the function of RpoN in strain L. aggregata LZB033 during its life cycle in seawater, with the hope of better understanding its environmental adaptation. In this study, we constructed a mutant of rpoN using an in-frame gene knockout method in the genus Labrenzia and investigated the global effects of RpoN in L. aggregata LZB033 using RNA sequencing (RNA-seq) technology. Subsequent experiments were used to determine the involvement of RpoN in growth, flagellar biosynthesis, swimming motility, carbon utilization, biofilm formation, stress response, DMSP synthesis, and DMSP-dependent dimethyl sulfide (DMS) production in L. aggregata LZB033.
RESULTS
Construction of mutant strain.
Using the double-selection strategy of allelic exchange mutagenesis with the suicide vector pK18mobsacB, we deleted a 1,515-bp (from bp 28 to 1542) core portion from the 1,551-bp rpoN gene and obtained a ΔrpoN mutant (see Fig. S1 in the supplemental material). We tried to construct the complementary strain of this mutant but failed, probably because of the use of inappropriate plasmid or method.
RpoN is related to bacterial growth.
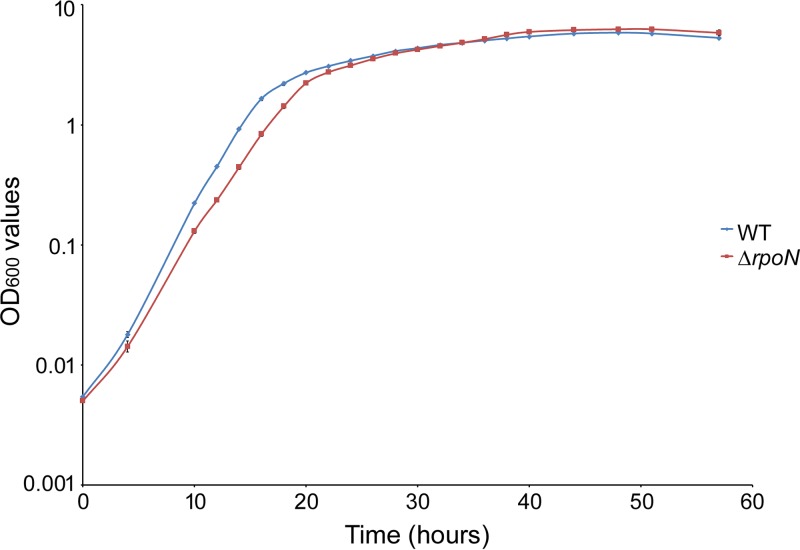
To test whether the ΔrpoN mutant had growth defects, the growth proficiency of the ΔrpoN mutant strain was compared with that of the wild-type strain LZB033 using LB-sea salt (LBSS) medium. When cultured on LBSS agar plates without any antibiotics, the colonies of the ΔrpoN mutant strain were smaller than those of the LZB033 strain (Fig. 1A), which was easily observed after 2 days. Meanwhile, the growth curves of these two bacteria showed that the ΔrpoN mutant grew slower than did LZB033 during the first 28 h, while there was no significant difference from 30 h onward (Fig. 2).
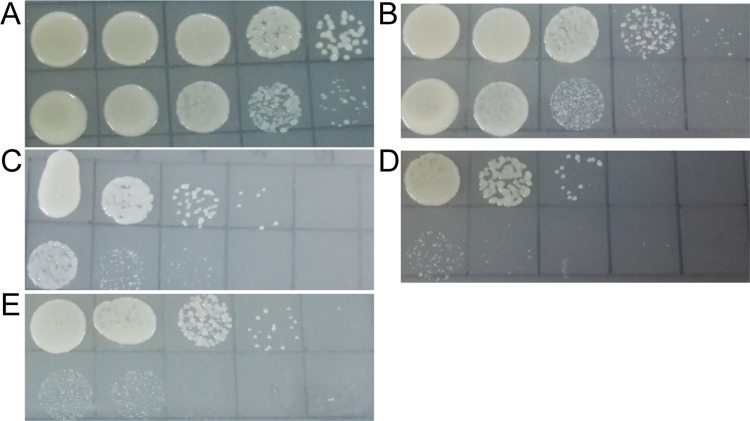
FIG 1.
The viability of L. aggregata wild-type and rpoN mutant strains under different conditions. (A) Control, growth of the strains in LBSS medium without any change. (B) Growth of the strains in LBSS medium supplemented with 2.5 M NaCl and cultured at 28°C for 3 h. (C) Growth of the strains in LBSS medium supplemented with 20 mM H2O2 and cultured at 28°C for 10 min. (D) Growth of the strains in LBSS medium and cultured at 50°C for 30 min. (E) Growth of the strains in LBSS medium (pH 2.5) and cultured at 28°C for 15 min. Cultures were serially diluted 10-fold, spotted at a 10-μl volume in rows on LBSS plates, and incubated at 28°C for 48 h. The leftmost column is 10−1 dilutions, and the rightmost column is 10−5 dilutions. Top row, wild-type strain LZB033; bottom row, ΔrpoN mutant strain.
FIG 2.
Growth curves of L. aggregata strains LZB033 and ΔrpoN mutant cultured in LBSS medium. Overnight cultures were adjusted to an OD600 of 0.5, diluted 1:100 into 200 ml fresh LBSS medium, and cultivated at 28°C with constant shaking. The optical absorbance at 600 nm was measured at 2-h intervals until the bacterial growth reached the stationary phase. y axis, log10 of OD600 values of L. aggregata; x axis, incubation time (in hours). The values of OD600 are shown as the means ± standard deviation (SD); n = 3.
Overview of gene expression profiles.
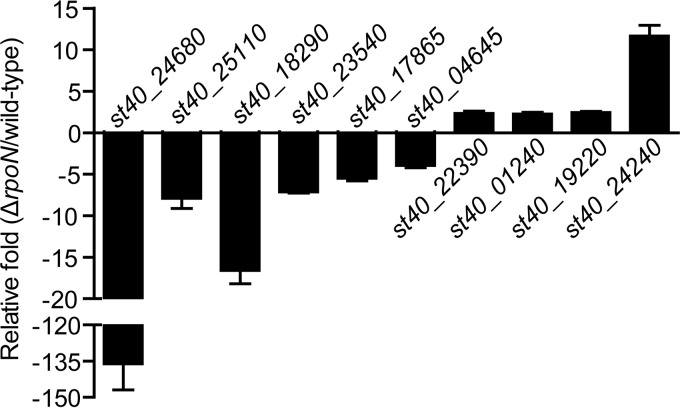
RpoN regulation at the global level during the exponential phase was analyzed using RNA-seq analysis of the wild-type and rpoN mutant L. aggregata strains. After dirty raw reads were filtered, a total of 13,085,602 and 14,869,680 reads were obtained for wild-type and ΔrpoN mutant cells, respectively. Approximately 98.97% of the total reads for both samples were uniquely mapped to the reference genome. According to the screening criteria for differentially expressed genes, the transcription of 175 genes, including 106 downregulated genes and 69 upregulated genes, was found to be associated with RpoN (Table S1), indicating that RpoN has both positive and negative regulatory roles in transcription. The RNA-seq results were further confirmed by quantitative reverse transcription-PCR (qRT-PCR) analysis of 10 randomly selected differentially expressed genes under the same culture conditions used for the RNA-seq experiments. Although some of the fold change values were different, the qRT-PCR results agreed with the RNA-seq results, indicating that the RNA-seq data were robust and valid (Fig. 3).
FIG 3.
Confirmation of the RNA-seq results by qRT-PCR analysis. The data are presented as the means ± SD; n = 3.
RpoN is required for flagellar formation and swimming motility.
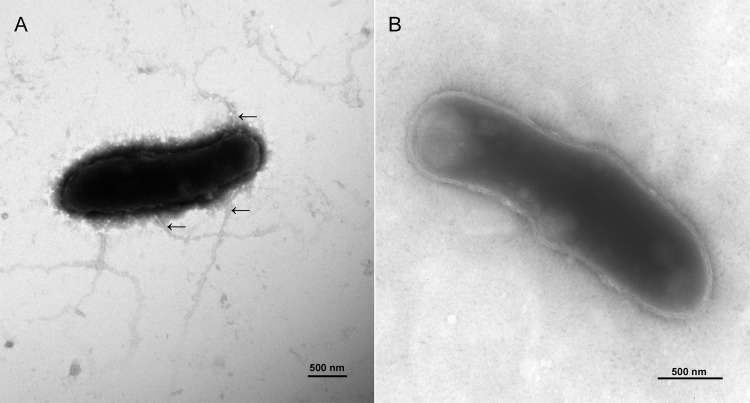
In ΔrpoN mutant cells, 8 out of the top 10 most downregulated genes are annotated as encoding flagellar proteins that participate in flagellar assembly, such as FlaE, FlgH, FlgG, and FlgA. By transmission electron microscopy (TEM) analysis, we found that L. aggregata wild-type strain LZB033 is peritrichously flagellated (Fig. 4A); however, after the rpoN gene was knocked out, no flagella were observed after cultivation for 24 h (Fig. 4B). In the ΔrpoN mutant, the amounts of two genes for chemotaxis, i.e., st40_12110 and st40_25185, were decreased by 3.35- and 2.16-fold, respectively. A swimming motility assay was performed on semisolid agar plates, and the motility levels of the strains were compared based on the measurement of the bacterial halo diameter formed after 48 h. The diameter of the LZB033 halo was 14 ± 0 mm, while that of the ΔrpoN mutant halo was 8.25 ± 0.5 mm (P < 0.01), suggesting that the rpoN mutant is deficient in swimming motility.
FIG 4.
Transmission electron microscopy of L. aggregata LZB033 (A) and ΔrpoN mutant (B) strains.
RpoN regulates carbon usage.
The transcription of genes involved in substrate transport, including the transport of sugars, amino acids, peptides, C4-dicarboxylates, molybdenum, and urea, was changed in the mutant cells. To test the function of rpoN in the usage of carbon sources, Biolog GN2 microplates were used. When the optical density of a well was 0.2 units greater than that of A1, the well was treated as positive. Among the 95 carbon sources and compared with the capability of wild-type strain LZB033, the capability of the ΔrpoN mutant to use four substrates, lactulose, l-asparagine, d,l-carnitine, and d,l-α-glycerol phosphate, increased. In contrast, nine carbon substrates were used less in the ΔrpoN mutant, as follows: N-acetyl-d-galactosamine, N-acetyl-d-glucosamine, l-arabinose, itaconic acid, α-keto glutaric acid, d-saccharic acid, succinic acid, bromosuccinic acid, and hydroxy-l-proline (Tables S2 and S3). In addition, in the API 20NE strip assay, there was no difference between the performances of LZB033 and the ΔrpoN mutant except that the ΔrpoN mutant lacked the ability to assimilate malic acid.
Biofilm formation.
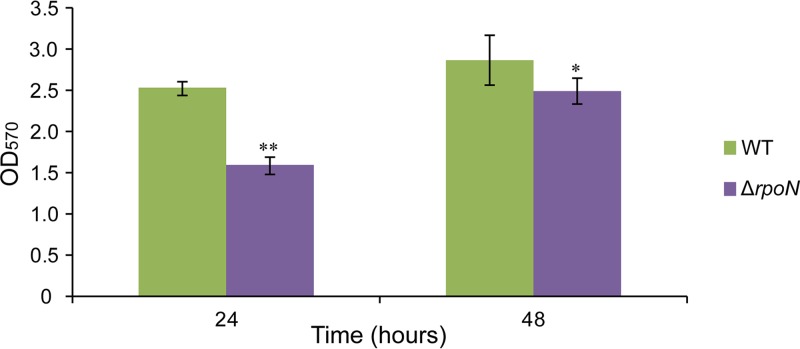
After incubation for 24 h and staining with crystal violet, an obvious purple ring appeared in the gas-liquid interface of the LZB033 culture, contrasting with the lightly colored ring of the ΔrpoN mutant (Fig. S2A). Quantification by an enzyme-linked immunosorbent assay (ELISA) reader indicated that the ΔrpoN mutant produced a significantly thinner (37.2% thinner) biofilm than did the wild-type LZB033 (Fig. 5). After incubation for 48 h, both of the cultures showed obvious purple rings at the gas-liquid interface (Fig. S2B), and the amount of biofilm formed by the ΔrpoN mutant was 13.2% less than that of LZB033 (Fig. 5).
FIG 5.
Quantification of biofilm formed by L. aggregata strains. Overnight cultures were adjusted to an OD600 of 0.5, vortexed, diluted 1:20 in fresh LBSS, transferred into a 96-well microtiter plate, and incubated statically for 48 h. Wells containing LBSS were used as a negative control. The adherent cells were stained with 2% crystal violet and quantified at 570 nm. The values of OD570 are shown as the means ± SD of the results from three individual experiments. Significance was analyzed with Tukey’s test, *, P < 0.05; **, P < 0.01.
RpoN is essential for stress resistance.
In ΔrpoN mutant cells, the expression of two peroxidases was reduced, and the abundance of cytochrome c was decreased by 2.37-fold. Both peroxidases and cytochrome c play a role in resistance to oxidative stress, and we tested the influence of rpoN knockout on bacterial stress resistance. When treated with a high concentration of NaCl (2.5 M) for 3 h, a high concentration of hydrogen peroxide (20 mM) for 10 min, high temperature (50°C) for 30 min, or a solution with a low pH value (pH 2.5) for 15 min, the number of rpoN mutant colonies on plates was less than that of the wild-type strain after incubation for 2 days (Fig. 1B to E). These results suggested that the rpoN mutant suffers more osmotic, oxidative, heat, and acid stress than does the wild type under the same stress conditions.
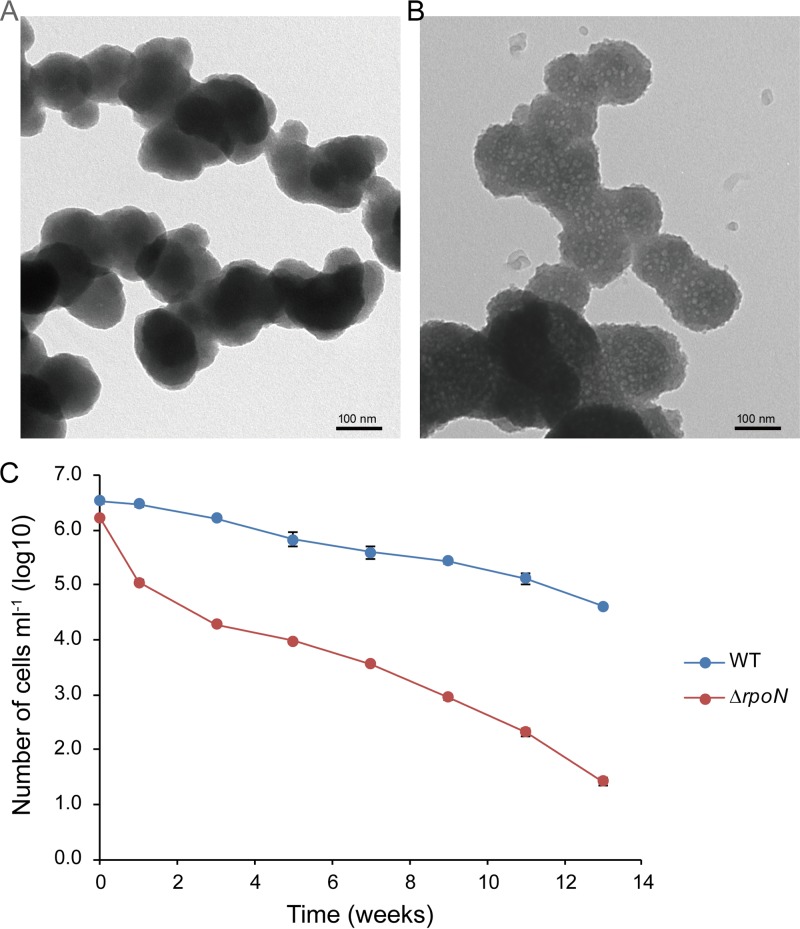
Scanning electron microscopy of the normal cells showed that they were rods and more than 2 μm long (Fig. 4), while after culturing in seawater at 4°C for 13 weeks, the cells gradually changed from rods to coccoid (Fig. 6A and B). Furthermore, the culturable cell counts of LZB033 declined by two orders of magnitude (from 3.42 × 106 CFU/ml to 4.18 × 104 CFU/ml), while the ΔrpoN mutant cell counts rapidly declined by five orders of magnitude (from 1.67 × 106 CFU/ml to 2.62 × 10 CFU/ml) after cultivation for 13 weeks (Fig. 6C).
FIG 6.
Morphology of L. aggregata LZB033 (A) and ΔrpoN mutant (B) strains and the cell counts of these strains (C) after culturing in seawater at 4°C for 13 weeks. Bacteria were cultured in artificial seawater and maintained at 4°C without shaking. Culturability was determined by spread plate count on LBSS plates at 0, 1, 3, 5, 7, 9, 11, and 13 weeks.
DMSP synthesis and catabolism.
The expression of both the DMSP synthesis gene dysB (st40_10620) and the DMSP lyse genes dddP (st40_19850) and dddL (st40_24780) (2), which encode the key methyltransferase enzyme of DMSP synthesis pathway and lyase converting DMSP to acrylate plus DMS, respectively, decreased around 0.47-, 0.34-, and 0.34-fold in the ΔrpoN mutant compared to the wild-type strain, respectively, with no statistical significance (q > 0.005).
The DMSP production results showed that the ΔrpoN mutant had a significantly lower level of DMSP production (10.92 ± 1.21 pmol DMSP/[μg protein] · h) than wild-type strain (33.89 ± 3.39 pmol DMSP/[μg protein] · h) (P < 0.01) under the same incubation conditions. Similarly, when detecting the DMSP-dependent-DMS production, the DMS production in the ΔrpoN mutant was 128.58 ± 3.39 pmol DMS/(μg protein) · h, while that in wild-type strain was 172.4 ± 5.53 pmol DMS/(μg protein) · h (P < 0.01). The results suggested that both DMSP synthesis and catabolism pathways were negatively regulated by RpoN.
DISCUSSION
The genome of L. aggregata LZB033 encodes a large number of transcriptional regulators which may be important for this bacterium to adapt to diverse environments during its life cycle. The role of RpoN (σ54 or also σN) was historically uncovered by a study of the regulation of nitrogen metabolism, but RpoN has been subsequently found to be involved in many other biological activities in diverse proteobacterial bacteria, which implicates RpoN as a central player that controls processes involved in the physical interaction of an organism and its environment (5–9).
The development of next-generation sequencing technologies provides a revolutionary tool for studying the interaction between transcriptional factors and target DNA. In comparison with microarray analysis, RNA-seq analyses provide much increased sensitivity in revealing transcriptional units. Here, we present RNA-seq data to study the transcriptome controlled by the transcription factor RpoN. We identified 175 genes that might be regulated by RpoN. This result is comparable with those from previous studies, where RpoN was found to control the expression of a large number of genes in other strains such as those of Vibrio cholerae (12), Escherichia coli (13), and Pseudomonas protegens (9).
Regulation by RpoN is clearly linked to central nitrogen metabolism (14) and plays a major role in the response of bacteria to nitrogen-limiting conditions. In E. coli, the transcription of at least 14 operons/regulators in the nitrogen regulatory response is directed by RpoN. Furthermore, it was also reported that RpoN is involved in carbon source utilization and certain fermentation pathways (14). In this study, genes that participate in nitrate and nitrous oxide reduction were downregulated, while those associated with sulfite reduction and sulfur transfer were upregulated in the ΔrpoN mutant strain, indicating that rpoN enhances nitrogen utilization and inhibits sulfur utilization. Additionally, the ΔrpoN mutant formed smaller colonies than did the wild-type strain after 48 h of cultivation on LBSS solid medium (Fig. 1A), but the numbers of colonies were approximately the same between the two bacteria, which ensures the reliability of results from experiments on stress. Meanwhile, knocking out the rpoN gene resulted in the mutant strain using 13 particular carbon sources differently from the wild type, confirming that RpoN is a crucial factor for carbon source utilization in L. aggregata.
RpoN has been implicated in the regulation of flagellar and motility genes in many species. A range of comparative genome analyses showed that there was an evolutionary relationship between RpoN and the presence of lipopolysaccharides and the biosynthesis of flagella (15). Biosynthesis of the bacterial flagellum is a complex process that requires the concerted expression of dozens of genes. Studies have noted that RpoN can regulate the transcription of flhDC and fliA in the flagellar hierarchy (13, 16). Moreover, it was found that RpoN not only controls flagellin expression but also modulates flagellum assembly by regulating the expression of the putative glycosyltransferase Jhp0106, thus affecting the posttranslational modification of flagellin (17). In our study, the abundance of flagellar genes was decreased by up to 211.88-fold in the ΔrpoN mutant, and the ΔrpoN mutant was deficient in the synthesis of flagella. After culturing for 48 h in semisolid agar plates, the mutant strain had significantly less motility than did the wild-type strain. This phenotypic analysis supported the view that RpoN is involved in regulating flagellar synthesis and swimming motility in the marine bacterium L. aggregata. Additionally, a previous study revealed that rpoN in Pseudomonas syringae strongly activated the vast majority of genes involved in flagellar synthesis and motility (18), as expected based on the critical relation of RpoN to most flagellar synthesis genes in P. aeruginosa (19).
Biofilms are defined as an assemblage of microbial cells that are associated with a surface and enclosed in an extracellular matrix principally composed of polysaccharide material; they typically help bacteria against antibiotics, native predators, and other stresses. Poly-β-1,6-N-acetyl-d-glucosamine, a polymer of N-acetyl-d-glucosamine, is a key component of bacterial biofilms (20). As mentioned above, when the rpoN gene was knocked out, the mutant cells decreased the usage of N-acetyl-d-glucosamine, which is a change that may be related to biofilm formation. By comparing biofilm formation at 24 h and 48 h between the wild-type and mutant strains, we found that the biofilm-forming ability was initially significantly impaired, which was consistent with the low growth rate of the mutant in the first 28 h. This difference suggested that genes regulated by RpoN are involved in biofilm-related metabolic pathways and that rpoN is essential for initial colonization of LZB033 on the surface. In addition, flagellar and flagellum-mediated motility were believed to be critical for both initial surface attachment and subsequent biofilm formation, and motility can affect biofilm architecture (21, 22). In this study, with the same initial bacterial count, fewer cells of the motility-impaired strain than the wild-type strain attached to polyvinyl chloride (PVC) (Fig. S2), and the amount of biofilm produced by the motility-impaired strain was significantly lower than that of the wild-type strain during the first 24 h after inoculation, which suggests that flagella and flagellum-mediated motility are important for biofilm formation in L. aggregata.
It is of vital importance for microorganism survival that they respond rapidly to environmental changes. The ecological niche where L. aggregata LZB033 lives is complex because the temperature, salinity, nutrients, and other environmental factors vary with time, and bacteria living there must evolve to withstand challenges. The RpoN-dependent gene expression paradigm is widely distributed among bacteria and is responsible for the expression of stress-induced genes, which are important in many bacterial survival processes. The challenge experiments illustrated that the RpoN mutation in L. aggregata significantly impaired its ability to resist various stresses, i.e., hyperosmolarity, oxidative, heat, low pH, cold, and starvation. Moreover, RpoN has been shown to be important for stress resistance in bacteria other than L. aggregata. For example, inactivation of rpoN in Listeria monocytogenes affected its ability to grow under osmotic stress (23). In addition, the inactivation of rpoN in E. coli O157:H7 led to increased acid resistance (24), whereas in our experiments, the inactivation resulted in decreased acid resistance. This result implied that the regulatory roles of RpoN in responses of different bacteria to the same environmental stresses might be different, which may be due to the interactions of different genes involved in stress resistance. Biofilms exhibit greater resistance to unfavorable environments than free-living bacterial cells, and the decrease in the biofilm formation ability of the ΔrpoN mutant may reduce their stress tolerance. Furthermore, by generating and screening a transposon-based mutant library of Campylobacter jejuni, Reid et al. (25) found that cell surface components such as flagella, the outer membrane, capsular polysaccharides, and lipooligosaccharides played an important role. From our data, we suggest that the increased sensitivity of the ΔrpoN mutant may result from reduced flagellar synthesis and swimming motility.
As a representative marine bacterium, L. aggregata LZB033 has been known to produce and catabolize DMSP, a ubiquitous organic sulfur-containing compound. Although almost all types of organisms from marine and estuary environments can participate in DMSP cycling (26–32), the molecular regulatory mechanisms are barely known. We found that the DMSP-producing and -catabolizing abilities of strain LZB033 decreased significantly after rpoN was knocked out; however, the transcriptomic analysis showed no significant downregulation of related genes. It was implied that the regulation might also affect its posttranscription, translation processing, or posttranslational modification. In addition, gene expression could be regulated by more than one regulator and vice versa in a gene regulatory network. More molecular evidence is need to unveil the regulation mechanism of rpoN for DMSP and DMS production in marine bacteria.
In conclusion, this study generated the rpoN mutant of L. aggregata LZB033, comparing the transcriptome and phenotype of the mutant strain with those of the wild-type strain; described the contribution of RpoN to bacterial growth, flagellum formation, swimming motility, biofilm formation, stress resistance, and DMSP synthesis and catabolism; and revealed RpoN to be an important regulator of stress resistance and survival in L. aggregata LZB033 in a marine environment.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The relevant characteristics of the bacterial strains and plasmids used in this work are described in Table 1. If there were no special instructions, Escherichia coli was cultured in Luria-Bertani (LB) medium at 37°C, and L. aggregata was cultured in LBSS medium (LB medium supplied with 8.5 g/liter sea salt; Sigma, St. Louis, MO, USA) at 28°C. Seawater was filtered through a 0.22-μm-pore-size filter (Millipore, Bedford, MA, USA). When required, the antibiotics ampicillin (Amp) and kanamycin (Kan) (Sigma) were added to final concentrations of 100 and 50 μg/ml, respectively. pRK2013, pK18mobsacB, and E. coli 803 were kindly provided by Jonathan D. Todd (University of East Anglia, Norwich, UK).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| Labrenzia aggregata | ||
| LZB033 | Wild-type strain isolated from the East China Sea, Ampr PmBr | 2 |
| ΔrpoN mutant | LZB033 mutant, in-frame deletion of rpoN | This study |
| Escherichia coli | ||
| JM109 | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 Δ(lac-proAB/F) [traD36 proAB+ lacIq lacZΔM15] | 41 |
| 803 | Met−; used as host for transformation with large plasmids | 42 |
| Plasmids | ||
| pRK2013 | RK2-derivative helper plasmid used in triparental matings, tra oriT colE1 immune, Kanr | 43 |
| pK18mobsacB | RP4-IncP suicide vector, lacZα sacB oriT oriV, Kanr | 44 |
PmBr, polymyxin B resistant.
Construction of the rpoN in-frame deletion mutant.
Genome annotation of L. aggregata LZB033 indicated that one open reading frame (st40_00765) was rpoN, and an in-frame deletion mutant (ΔrpoN) was constructed by double-crossover allelic exchange. Briefly, the flanking region of the gene was amplified by overlapping PCR, cloned into the pK18mobsacB suicide vector, and transformed into LZB033 by triparental mating with the help of pRK2013. Transconjugants with a single-crossover insertion in the LZB033 chromosome were obtained by screening on LBSS plates containing Amp and Kan. Allelic exchange between the chromosomal gene and the mutagenized copy was achieved in a second crossover event, which was counterselected on LBSS containing 10% (wt/vol) sucrose to determine the excision of pK18mobsacB from the chromosome. The resultant mutant, ΔrpoN, was selected by antibiotic sensitivity (Kan sensitive [Kans] and Amp resistant [Ampr]) and confirmed by PCR assay followed by nucleotide sequence analysis. The primers used in this study are listed in Table 2.
TABLE 2.
Primers used in this study
| Primer by function | Sequence (5′–3′)a | Product size (bp) |
|---|---|---|
| In-frame mutation | ||
| rpoN-UO | CAGAATTCCGAGTATCGCCCTTTCC | |
| rpoN-UI | TTAAAGTCGCACCATCTCCAGCTTGGTTG | 462 |
| rpoN-DI | GTGCGACTTTAAGTCCTGATGTG | |
| rpoN-DO | GGTCTAGAAGGCTGCATTCAGACTTG | 475 |
| qRT-PCR | ||
| st40_24680-S | CGGGCGACGGCTTTACAATCTC | 226 |
| st40_24680-A | GGACGCTGCTGGTTGTTGGAAT | |
| st40_25110-S | CGGCATGATGACAGCGAAGGAA | 207 |
| st40_25110-A | GTGGTCGGTGCATTGGCATTCT | |
| st40_18290-S | AAGTTCCAGGTGCCGAGTGTCT | 220 |
| st40_18290-A | CGATCTCCAGCCATTGCGTCTG | |
| st40_23540-S | AACGACGAGATGAACGGCAAGG | 156 |
| st40_23540-A | CAGGCGGAACTTCAGCGTGTAG | |
| st40_17865-S | AGACCGTGTCCGTTGCCATCA | 246 |
| st40_17865-A | TGCCGAGATAACCGACCATCCA | |
| st40_04645-S | GCACACGACATCCACGGCAT | 141 |
| st40_04645-A | GGCGGTGAACAGGCAGGAATAC | |
| st40_22390-S | TGAAGGTCCTGTGCGGCTATCA | 229 |
| st40_22390-A | CGGCGAAGTTTCCTGTCGTTGA | |
| st40_01240-S | ACAGTGCCTACATGCGGACCAT | 198 |
| st40_01240-A | AGGGATTGTTCGGACTGCGGAT | |
| st40_19220-S | CCAGCGTACCGAGCAGTTCAAG | 297 |
| st40_19220-A | TCGACTTCTGCCAGCTCTTCCA | |
| st40_24240-S | CCGAAGCCAACGAAGTCACCAT | 249 |
| st40_24240-A | TGATGAAGACAACGCCGATGCC | |
| 16S rRNA-S | CGCAGAACCTTACCAGCCCTTG | 283 |
| 16S rRNA-A | CCACTGTCACCGCCATTGTAGC | |
Nucleotides in bold represent restriction enzyme sites added to the 5′ region of the primers. Underlined nucleotides represent overlap sequences.
Growth determination.
Overnight cultures were adjusted to an optical density at 600 nm (OD600) of 0.5, diluted 1:100 into 200 ml fresh LBSS medium, and cultivated at 28°C with constant shaking. The optical absorbance at 600 nm was measured at 2-h intervals until the bacterial growth reached stationary phase. Three independent experiments were performed, and mean optical absorbance values at each time point were used to determine the growth rate.
Isolation and purification of bacterial RNA.
Cultures of wild-type and ΔrpoN mutant L. aggregata were grown in LBSS to exponential phase and harvested by centrifugation at 4°C. RNA was extracted using the TRIzol LS reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions and purified using the RNeasy minikit (Qiagen, Valencia, CA, USA), followed by treatment with RNase-free DNase I (Qiagen, Hilden, Germany) to eliminate genomic DNA contamination. DNA removal was confirmed using PCR. RNA size, integrity, and total amount were determined using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, MA, USA). Three independent experiments were performed.
Illumina high-throughput transcriptome sequencing and data analysis.
cDNA was prepared and modified for Illumina HiSeq paired-end 150-bp (PE150) sequencing according to the manufacturer’s protocol. Three independent libraries were prepared for each of the RNA-seq samples. After the dirty raw reads were filtered, the clean reads were mapped to the L. aggregata LZB033 genome (GenBank accession number JXMT00000000) (2). The gene expression level was presented in reads per kilobase per million (RPKM) values (33). We used a 2-fold change in average gene expression and a q value (adjusted P value) of <0.005 as criteria to identify RpoN-controlled genes (34).
Quantitative RT-PCR analysis.
Primers for qRT-PCR for 10 randomly selected differentially expressed genes as well as the 16S rRNA gene were designed using Primer Premier 6.0 and are listed in Table 2. The amplification efficiencies of all primer pairs were tested using standard dilution procedures. cDNA was synthesized using the PrimeScript first-strand cDNA synthesis kit (TaKaRa, Kusatsu, Japan), according to the manufacturer’s instructions. Samples were run in triplicate, and qRT-PCR analyses were conducted using an ABI 7500 sequence detection system (Applied Biosystems, CA, USA) with SYBR green fluorescent dye. The 16S rRNA gene was used as a reference control for sample normalization.
Electron microscopy.
Active bacterial cultures grown on LBSS semisolid medium or cultured in artificial seawater (ASW) for 13 weeks were collected directly, negatively stained with 1% phosphotungstic acid (pH 7.4) on a Formvar carbon-coated grid, and observed with a transmission electron microscope (TEM-1200EX; Japan) (35).
Swimming motility assay.
To measure swimming motility, L. aggregata strains were cultured on semisolid agar plates following a procedure described previously (36). Briefly, fresh cultures of L. aggregata strains were spotted on the LBSS plates containing 0.3% (wt/vol) agar and cultured for 48 h. Bacteria proficient for swimming motility move radially in all directions, and the distinct radial zone was measured. Bacteria were considered nonmotile when diffused growth around the colony was not observed.
Carbon usage.
The GN2 MicroPlate kit (Biolog) and API 20NE strip (bioMérieux, France) were used according to the manufacturers’ instructions, except that sterile seawater was used to prepare the inocula.
Quantitative biofilm formation.
Quantification of biofilms was performed as described previously (37), with some modification. Briefly, overnight cultures of L. aggregata were adjusted to an OD600 of 0.5. Then, the cultures were vortexed, diluted 1:20 in fresh LBSS, and transferred into a 96-well microtiter plate before being incubated statically for 48 h. Wells containing LBSS were used as a negative control. The adherent cells were stained with 2% crystal violet and quantified by an ELISA reader (Sunrise; Tecan Group Ltd., Männedorf, Switzerland) at 570 nm.
Stress response assays.
Exponential-phase cells (OD600, 0.5) cultivated in LBSS were harvested by centrifugation for 10 min at 4,000 rpm. The supernatant was removed as much as possible, and the pellet was washed in phosphate-buffered saline and resuspended in appropriate medium before being treated with different challenges. At the indicated time points of the challenge conditions described below, the bacterial cultures were washed, resuspended in LBSS medium, serially diluted 10-fold, and spotted at 10-μl volumes in rows on LBSS plates using the drop plate method (38). The plates were incubated at 28°C for 48 h and observed. The challenge conditions were as follows: for osmotic and oxidative challenges, the pellet was resuspended in LBSS medium supplemented with 2.5 M NaCl and 20 mM H2O2 and cultured at 28°C for 3 h and 10 min, respectively; for the high-temperature challenge, the pellet was resuspended in LBSS medium and cultured at 50°C for 30 min; and for the acid challenge, the pellet was resuspended in LBSS medium (pH 2.5) for 15 min. Pellets resuspended in LBSS medium without any treatment were used as a control.
To measure the response to low temperature and nutrition-deficient conditions, ASW, which contained 40 g/liter sea salt and was sterilized by passage through a 0.22-μm membrane filter, was used. After the L. aggregata strains were cultivated, harvested, and washed as described above, the pellets were resuspended in ASW and maintained at 4°C without shaking. Culturability was determined by a spread plate count on LBSS plates at 0, 1, 3, 5, 7, 9, 11, and 13 weeks.
DMS and DMSP production assays.
Quantification of DMS and DMSP was performed as described previously (39). To measure the DMSP synthesis ability, L. aggregata strains were grown overnight, washed twice with marine basal minimal (MBM) medium (40), diluted to OD600 of 0.3, inoculated into MBM medium containing a mixed carbon source and 0.5 mM Met before being incubated at 28°C for 26 h. When assayed, 100 μl of 10 M NaOH was added to 200 μl of culture, cap sealed, and stored in the dark at room temperature for at least 16 h. Abiotic medium controls of MBM medium supplied with the same concentrations of carbon sources and Met were set up and incubated under the same conditions. For DMS production, the assay was same as described above, expect that the bacteria were inoculated into gas chromatography (GC) vials containing MBM medium supplied with a mixed carbon source and 0.5 mM DMSP. The directly bacterial DMSP-dependent DMS production and alkaline lysis production DMS from bacterial synthesized DMSP were measured by GC (Agilent 7890A). Total protein concentration in the cells was estimated using Bradford assays (Bio-Rad). The rates of DMS and DMSP production were expressed in picomoles DMS or DMSP per milligram of protein per hour.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jonathan D. Todd (University of East Anglia, Norwich, UK) for kindly engaging in useful discussion and for providing pRK2013, pK18mobsacB, and E. coli 803.
This work was funded by the National Natural Science Foundation of China (grants 91751202 and 41521064) and the National Key Research and Development Program of China (grant 2016YFA0601303).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02844-18.
REFERENCES
- 1.Pujalte MJ, Lucena T, Ruvira MA, Arahal DR, Macián MC. 2014. The family Rhodobacteraceae, p 545–577. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes: Alphaproteobacteria and Betaproteobacteria, 4th ed Springer, Berlin, Germany. [Google Scholar]
- 2.Curson AR, Liu J, Bermejo Martinez A, Green RT, Chan Y, Carrion O, Williams BT, Zhang SH, Yang GP, Bulman Page PC, Zhang X-H, Todd JD. 2017. Dimethylsulfoniopropionate biosynthesis in marine bacteria and identification of the key gene in this process. Nat Microbiol 2:17009. doi: 10.1038/nmicrobiol.2017.9. [DOI] [PubMed] [Google Scholar]
- 3.Gruber TM, Gross CA. 2003. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol 57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 4.Bush M, Dixon R. 2012. The role of bacterial enhancer binding proteins as specialized activators of σ54-dependent transcription. Microbiol Mol Biol Rev 76:497–529. doi: 10.1128/MMBR.00006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kustu S, Santero E, Keener J, Popham D, Weiss D. 1989. Expression of sigma 54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev 53:367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hao B, Mo Z-L, Xiao P, Pan H-J, Lan X, Li G-Y. 2013. Role of alternative sigma factor 54 (RpoN) from Vibrio anguillarum M3 in protease secretion, exopolysaccharide production, biofilm formation, and virulence. Appl Microbiol Biotechnol 97:2575–2585. doi: 10.1007/s00253-012-4372-x. [DOI] [PubMed] [Google Scholar]
- 7.Ray SK, Kumar R, Peeters N, Boucher C, Genin S. 2015. rpoN1, but not rpoN2, is required for twitching motility, natural competence, growth on nitrate, and virulence of Ralstonia solanacearum. Front Microbiol 6:229. doi: 10.3389/fmicb.2015.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisher MA, Grimm D, Henion AK, Elias AF, Stewart PE, Rosa PA, Gherardini FC. 2005. Borrelia burgdorferi σ54 is required for mammalian infection and vector transmission but not for tick colonization. Proc Natl Acad Sci U S A 102:5162–5167. doi: 10.1073/pnas.0408536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, Shi H, Wang Z, Huang X, Zhang X. 2018. Pleiotropic control of antibiotic biosynthesis, flagellar operon expression, biofilm formation, and carbon source utilization by RpoN in Pseudomonas protegens H78. Appl Microbiol Biotechnol 102:9719–9730. doi: 10.1007/s00253-018-9282-0. [DOI] [PubMed] [Google Scholar]
- 10.Al Ahmar R, Kirby B, Yu H. 2018. Pyrimidine biosynthesis regulates small-colony variant and mucoidy in Pseudomonas aeruginosa through sigma factor competition. J Bacteriol 201:e00575-18. doi: 10.1128/JB.00575-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai Z, Liu Y, Chen Y, Yam JKH, Chew SC, Chua SL, Wang K, Givskov M, Yang L. 2015. RpoN regulates virulence factors of Pseudomonas aeruginosa via modulating the PqsR quorum sensing regulator. Int J Mol Sci 16:28311–28319. doi: 10.3390/ijms161226103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong TG, Mekalanos JJ. 2012. Characterization of the RpoN regulon reveals differential regulation of T6SS and new flagellar operons in Vibrio cholerae O37 strain V52. Nucleic Acids Res 40:7766–7775. doi: 10.1093/nar/gks567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao K, Liu M, Burgess RR. 2010. Promoter and regulon analysis of nitrogen assimilation factor, σ54, reveal alternative strategy for E. coli MG1655 flagellar biosynthesis. Nucleic Acids Res 38:1273–1283. doi: 10.1093/nar/gkp1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reitzer L, Schneider BL. 2001. Metabolic context and possible physiological themes of σ54-dependent genes in Escherichia coli. Microbiol Mol Biol Rev 65:422–444. doi: 10.1128/MMBR.65.3.422-444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francke C, Groot Kormelink T, Hagemeijer Y, Overmars L, Sluijter V, Moezelaar R, Siezen RJ. 2011. Comparative analyses imply that the enigmatic sigma factor 54 is a central controller of the bacterial exterior. BMC Genomics 12:1. doi: 10.1186/1471-2164-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong T, Yu R, Schellhorn H. 2011. Antagonistic regulation of motility and transcriptome expression by RpoN and RpoS in Escherichia coli. Mol Microbiol 79:375–386. doi: 10.1111/j.1365-2958.2010.07449.x. [DOI] [PubMed] [Google Scholar]
- 17.Kao CY, Chen JW, Wang S, Sheu BS, Wu JJ. 2017. The Helicobacter pylori J99 jhp0106 gene, under the control of the CsrA/RpoN regulatory system, modulates flagella formation and motility. Front Microbiol 8:483. doi: 10.3389/fmicb.2017.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu X, Lund SP, Greenwald JW, Records AH, Scott RA, Nettleton D, Lindow SE, Gross DC, Beattie GA. 2014. Transcriptional analysis of the global regulatory networks active in Pseudomonas syringae during leaf colonization. mBio 5:e01683-14. doi: 10.1128/mBio.01683-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nandini D, Wolfgang MC, Goodman AL, Arora SK, Jyot J, Lory S, Ramphal R. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol Microbiol 50:809–824. doi: 10.1046/j.1365-2958.2003.03740.x. [DOI] [PubMed] [Google Scholar]
- 20.Izano EA, Amarante MA, Kher WB, Kaplan JB. 2008. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl Environ Microbiol 74:470–476. doi: 10.1128/AEM.02073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedlander RS, Vogel N, Aizenberg J. 2015. Role of flagella in adhesion of Escherichia coli to abiotic surfaces. Langmuir 31:6137–6144. doi: 10.1021/acs.langmuir.5b00815. [DOI] [PubMed] [Google Scholar]
- 22.Wood TK, Gonzalez Barrios AF, Herzberg M, Lee J. 2006. Motility influences biofilm architecture in Escherichia coli. Appl Microbiol Biotechnol 72:361–367. doi: 10.1007/s00253-005-0263-8. [DOI] [PubMed] [Google Scholar]
- 23.Okada Y, Okada N, Makino S, Asakura H, Yamamoto S, Igimi S. 2006. The sigma factor RpoN (σ54) is involved in osmotolerance in Listeria monocytogenes. FEMS Microbiol Lett 263:54–60. doi: 10.1111/j.1574-6968.2006.00405.x. [DOI] [PubMed] [Google Scholar]
- 24.Riordan JT, Tietjen JA, Walsh CW, Gustafson JE, Whittam TS. 2010. Inactivation of alternative sigma factor 54 (RpoN) leads to increased acid resistance, and alters locus of enterocyte effacement (LEE) expression in Escherichia coli O157 : H7. Microbiology 156:719–730. doi: 10.1099/mic.0.032631-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reid AN, Pandey R, Palyada K, Whitworth L, Doukhanine E, Stintzi A. 2008. Identification of Campylobacter jejuni genes contributing to acid adaptation by transcriptional profiling and genome-wide mutagenesis. Appl Environ Microbiol 74:1598–1612. doi: 10.1128/AEM.01508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnston AW, Green RT, Todd JD. 2016. Enzymatic breakage of dimethylsulfoniopropionate-a signature molecule for life at sea. Curr Opin Chem Biol 31:58–65. doi: 10.1016/j.cbpa.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 27.Moran MA, Reisch CR, Kiene RP, Whitman WB. 2012. Genomic insights into bacterial DMSP transformations. Annu Rev Mar Sci 4:523–542. doi: 10.1146/annurev-marine-120710-100827. [DOI] [PubMed] [Google Scholar]
- 28.Curson ARJ, Williams BT, Pinchbeck BJ, Sims LP, Martinez AB, Rivera PPL, Kumaresan D, Mercade E, Spurgin LG, Carrion O, Moxon S, Cattolico RA, Kuzhiumparambil U, Guagliardo P, Clode PL, Raina JB, Todd JD. 2018. DSYB catalyses the key step of dimethylsulfoniopropionate biosynthesis in many phytoplankton. Nat Microbiol 3:430–439. doi: 10.1038/s41564-018-0119-5. [DOI] [PubMed] [Google Scholar]
- 29.Kageyama H, Tanaka Y, Shibata A, Waditee-Sirisattha R, Takabe T. 2018. Dimethylsulfoniopropionate biosynthesis in a diatom Thalassiosira pseudonana: identification of a gene encoding MTHB-methyltransferase. Arch Biochem Biophys 645:100–106. doi: 10.1016/j.abb.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 30.Reed RH. 1983. Measurement and osmotic significance of beta-dimethylsulphoniopropionate in marine macroalgae. Mar Biol Lett 4:173–181. [Google Scholar]
- 31.Otte ML, Wilson G, Morris JT, Moran BM. 2004. Dimethylsulphoniopropionate (DMSP) and related compounds in higher plants. J Exp Bot 55:1919–1925. doi: 10.1093/jxb/erh178. [DOI] [PubMed] [Google Scholar]
- 32.Raina JB, Tapiolas DM, Foret S, Lutz A, Abrego D, Ceh J, Seneca FO, Clode PL, Bourne DG, Willis BL, Motti CA. 2013. DMSP biosynthesis by an animal and its role in coral thermal stress response. Nature 502:677–680. doi: 10.1038/nature12677. [DOI] [PubMed] [Google Scholar]
- 33.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 34.Wang QQ, Liu F, Chen XS, Ma XJ, Zeng HQ, Yang ZM. 2010. Transcriptome profiling of early developing cotton fiber by deep-sequencing reveals significantly differential expression of genes in a fuzzless/lintless mutant. Genomics 96:369–376. doi: 10.1016/j.ygeno.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 35.Givaudan A, Lanois A. 2000. flhDC, the flagellar master operon of Xenorhabdus nematophilus: requirement for motility, lipolysis, extracellular hemolysis, and full virulence in insects. J Bacteriol 182:107–115. doi: 10.1128/JB.182.1.107-115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu T, Su Y, Xu Y, He Y, Wang B, Dong X, Li Y, Zhang X-H. 2014. Mutations of flagellar genes fliC12, fliA and flhDC of Edwardsiella tarda attenuated bacterial motility, biofilm formation and virulence to fish. J Appl Microbiol 116:236–244. doi: 10.1111/jam.12357. [DOI] [PubMed] [Google Scholar]
- 37.Sheikh J, Hicks S, Dall’Agnol M, Phillips AD, Nataro JP. 2001. Roles for Fis and YafK in biofilm formation by enteroaggregative Escherichia coli. Mol Microbiol 41:983–997. [DOI] [PubMed] [Google Scholar]
- 38.Hoben H, Somasegaran P. 1982. Comparison of the pour, spread, and drop plate methods for enumeration of Rhizobium spp. in inoculants made from presterilized peat. Appl Environ Microbiol 44:1246–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J, Liu J, Zhang S, Liang J, Lin H, Song D, Yang G, Todd JD, Zhang X-H. 2018. Novel insights into bacterial dimethylsulfoniopropionate catabolism in the East China Sea. Front Microbiol 9:3206. doi: 10.3389/fmicb.2018.03206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baumann P, Baumann L. 1981. The marine gram-negative eubacteria: genera Photobacterium, Beneckea, Alteromonas, Pseudomonas, and Alcaligenes, p 1302–1331. In Starr MP, Stolp H, Trüper HG, Balows A, Schlegel HG (ed), The prokaryotes: a handbook on habitats, isolation and identification of bacteria, 1st ed Springer, Berlin, Germany. [Google Scholar]
- 41.Yanisch-Perron C, Vieira J, Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mpl8 and pUC19 vectors. Gene 33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 42.Wood WB. 1966. Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J Mol Biol 16:118–133. doi: 10.1016/S0022-2836(66)80267-X. [DOI] [PubMed] [Google Scholar]
- 43.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.