Microbial regrowth within building water systems are promoted by water stagnation, low disinfectant residual, high surface-to-volume ratio, amenable growth temperatures, and colonization of drinking water biofilms. Moreover, biofilms provide protection from environmental stresses, access to higher levels of nutrients, and opportunities for symbiotic interactions with other microbes. Disinfectant efficacy information is historically based on inactivation of pathogens in their planktonic, free-floating forms. However, due to the ecological importance of drinking water biofilms for pathogen survival, this study evaluated the efficacy of two common disinfectants, free chlorine and monochloramine, on Legionella pneumophila colonizing mature, drinking water biofilms established on copper and PVC surfaces. Results showed that inactivation was dependent on the disinfectant type and biofilm substratum. Overall, this, and other related research, will provide a better understanding of Legionella ecological stability and survival and aid policy makers in the management of exposure risks to water-based pathogens within building water systems.
KEYWORDS: HPC, biofilm, disinfection, drinking water distribution system, opportunistic pathogen, premise plumbing
ABSTRACT
Building water systems promote the regrowth and survival of opportunistic pathogens, such as Legionella pneumophila, especially within biofilms, where most drinking water microbes reside. However, compared to their planktonic form, disinfection efficacy for the biofilm-associated forms of water-based pathogens is unclear. The aim of this study was to determine the effectiveness of free chlorine and monochloramine in the inactivation of biofilm-associated L. pneumophila strain Philadelphia-1 serogroup 1 (LpP1s1). Mature (1.5- to 2-year-old) drinking water biofilms were developed on copper (Cu) and polyvinyl chloride (PVC) slides within biofilm annular reactors, then colonized with LpP1s1 at approximately 4 log10 CFU cm−2 and exposed to 2 mg liter−1 of free chlorine or monochloramine. Ct (disinfectant concentration × time, expressed as mg min liter−1) inactivation values for 2-, 3-, and 4-log10 reductions of planktonic and biofilm LpP1s1 were determined. For planktonic LpP1s1, free chlorine was more effective at inactivation than was monochloramine treatment, and for biofilm-associated LpP1s1, monochloramine was more effective on Cu biofilms while free chlorine was more effective on PVC biofilms. In contrast to monochloramine, free chlorine treatment of Cu and PVC biofilms, negatively impacted LpP1s1 16S rRNA gene transcript levels and may act synergistically with Cu surfaces to further reduce transcript levels. Moreover, LpP1s1 cells shed from biofilms into the bulk water were more resistant to disinfection than were prepared planktonic LpP1s1 cells. Results from this study indicate that biofilm association, disinfectant type, and substratum play an important role in the survival of Legionella pneumophila in building water systems.
IMPORTANCE Microbial regrowth within building water systems are promoted by water stagnation, low disinfectant residual, high surface-to-volume ratio, amenable growth temperatures, and colonization of drinking water biofilms. Moreover, biofilms provide protection from environmental stresses, access to higher levels of nutrients, and opportunities for symbiotic interactions with other microbes. Disinfectant efficacy information is historically based on inactivation of pathogens in their planktonic, free-floating forms. However, due to the ecological importance of drinking water biofilms for pathogen survival, this study evaluated the efficacy of two common disinfectants, free chlorine and monochloramine, on Legionella pneumophila colonizing mature, drinking water biofilms established on copper and PVC surfaces. Results showed that inactivation was dependent on the disinfectant type and biofilm substratum. Overall, this, and other related research, will provide a better understanding of Legionella ecological stability and survival and aid policy makers in the management of exposure risks to water-based pathogens within building water systems.
INTRODUCTION
Legionella spp. are found in soil and natural water environments where they are subjected to protozoan predation (1). Select members of this genus, e.g., Legionella anisa, L. dumoffii, L. longbeacheae, L. micdadei, and L. pneumophila, can cause Pontiac Fever, a flu-like illness, and Legionnaires’ disease, a potentially fatal form of pneumonia (2). Most clinical isolates are identified as L. pneumophila serogroup (sg) 1 (out of 15 sg’s associated with L. pneumophila) (3). However, underreporting for other disease-causing Legionella strains may be due to diagnostic tests that are only specific for L. pneumophila sg 1 and the infrequent occurrence of those strains in certain environments, which can be further exacerbated by the low probability of causing illness in a susceptible individual (4).
Legionella spp. have been frequently detected within building water systems, or premise plumbing (5), which is defined as the portion within drinking water distribution systems (DWDSs) between the water main and the final point of consumption. These systems are characterized by high surface area-to-volume ratios, water stagnation, the presence of various plumbing materials and cross connections, variable temperatures and water velocities, low to absent disinfectant residuals, and exposure to respirable, drinking water derived aerosols (6). These characteristics have been attributed to the persistence and regrowth of opportunistic pathogens, such as L. pneumophila, Mycobacterium avium, and Pseudomonas aeruginosa and the free-living amoebae, Acanthamoeba spp. and Naegleria fowleri (7). The absence and infrequent detection of Legionella in finished and distributed water (8, 9) and within biofilms collected from distribution pipe sections (10) further supports the hypothesis that conditions within building water systems promote the growth and survival of water-based, opportunistic pathogens.
Selective pressure on Legionella to thrive in low-nutrient environments, such as drinking water, have enabled them to evolve ways to (i) acquire nutrients by residing in nutrient rich biofilms and (ii) avoid predatory digestion by various free-living amoebae (FLA) (11); the latter of which has conferred the ability of certain Legionella species to avoid lysosomal degradation and other bactericidal mechanisms of human lung macrophages (12). This had led to the hypothesis that FLA may be an evolutionary training ground for intracellular pathogens, such as L. pneumophila (13), and positive correlations between the occurrence of FLA and Legionella throughout DWDSs (14–16) suggest a continual selective pressure for survival of pathogenic Legionella in these systems.
Biofilms are highly prevalent in DWDSs with a large proportion of drinking water biomass located within biofilms, including pathogenic microorganisms (17, 18). Chemical disinfection, with free chlorine or monochloramine, is commonly used to inactivate pathogens and control biofilm growth within DWDSs. Chlorine is a potent oxidizer and has been shown to extensively permeabilize bacterial membranes causing leakage of protein and nucleic acid material, as well as lethal DNA damage (19, 20). Monochloramine has been shown to react slowly with DNA and RNA but rapidly with several amino acids, with little damage to bacterial membranes; thus, the lower potency of monochloramine is hypothesized to require several targeted reactions before microbial inactivation can be observed (21). Measurements of free chlorine and monochloramine biofilm penetration confirmed that monochloramine is more effective at penetrating biofilms than is free chlorine; however, the increased penetration did not correlate with greater inactivation of biofilm microorganisms within the same depth as full free chlorine penetration, i.e., ca. 50 to 75 μm (22).
For effective disinfection of drinking water, the World Health Organization (WHO) recommends 5 mg liter−1 for chlorine and 3 mg liter−1 for monochloramine. However, residual concentrations within DWDSs can range from 0.2 to 1 and 0.6 to 5 mg liter−1 for chlorine and monochloramine, respectively (23, 24) with no disinfectant residuals applied in several European countries, e.g., Austria, Germany, the Netherlands, and Switzerland (25). Furthermore, to control the growth of Legionella and other opportunistic pathogens within building water systems, secondary disinfection treatments, such as chlorine dioxide, ozone, UV light, copper-silver ionization, thermal treatment, chlorine, and chloramine, have been utilized (26). In this study, free chlorine and monochloramine concentrations of 2 mg liter−1 were chosen to represent the midrange of the WHO guideline values for effective drinking water treatment, especially given that ≤1.5 mg liter−1 of chlorine and chloramine was shown to be inadequate for microbial control (27–29).
Given the importance of biofilms in pathogen survival within DWDSs, this study examined the effect of free chlorine and monochloramine on culturability and 16S rRNA gene and transcript levels of biofilm-associated L. pneumophila established on copper and polyvinyl chloride surfaces, which are common to building water system plumbing. The goal of this study is to provide a better understanding of Legionella survival within drinking water biofilms and aid in the mitigation of exposure risks to this opportunistic pathogen within building water systems.
RESULTS
Establishment of drinking water biofilms.
One-and-a-half- to two-year-old drinking water biofilms were grown on copper (Cu) and polyvinyl chloride (PVC) surfaces within biofilm annular reactors (BARs) before use in disinfection experiments. Drinking water quality parameters were monitored throughout the period with the means ± standard deviations (SD) being 2.45 ± 0.25 log10 CFU ml−1 for heterotrophic plate count; 8.35 ± 0.18 for pH; 19 ± 3.5°C for temperature; 70 ± 22 mg liter−1 for total alkalinity; 154 ± 17 mg liter−1 for hardness; 0.35 ± 0.95 NTU for turbidity; 0.96 ± 0.18 and 1.07 ± 0.18 mg liter−1 for free and total chlorine, respectively; 0.86 ± 0.23 mg liter−1 for nitrate; 0.15 ± 0.07 mg liter−1 for phosphate; and 0.99 ± 0.77 mg liter−1 for total organic carbon; the results of the elemental analyses are summarized in Table 1 . Seasonal variations for each parameter are reported in Supplemental Information (Tables S1–S6). For each BAR, heterotrophic plate count (HPC), free and total chlorine, and total organic carbon (TOC) was monitored and did not differ significantly between each reactor (P > 0.05, Table 1).
TABLE 1.
Water quality measurements for influent tap water and BARsa
| Water quality parameter | Measurements (mean [SD]) for: |
|||
|---|---|---|---|---|
| Influent tap | BAR1 | BAR2 | BAR3 | |
| HPC (log10 CFU ml−1) | 2.45 (0.25) | 3.58 (0.70) | 3.71 (0.73) | 3.65 (0.79) |
| pH | 8.35 (0.18) | ND | ND | ND |
| Temp (°C) | 19 (3.5) | 20.8 (2.3)b | ||
| Total alkalinity (mg liter−1) | 70 (22) | ND | ND | ND |
| Hardness (mg liter−1) | 154 (17) | ND | ND | ND |
| Turbidity (NTU) | 0.35 (0.95) | ND | ND | ND |
| Free Cl2 (mg liter−1) | 0.96 (0.18) | 0.61 (0.24) | 0.62 (0.21) | 0.54 (0.23) |
| Total Cl2 (mg liter−1) | 1.07 (0.18) | 0.71 (0.24) | 0.72 (0.22) | 0.64 (0.24) |
| NO3 (mg liter−1) | 0.86 (0.23) | ND | ND | ND |
| PO4 (mg liter−1) | 0.15 (0.07) | ND | ND | ND |
| TOC (mg liter−1) | 0.99 (0.77) | 1.11 (1.01) | 1.02 (0.81) | 1.00 (0.62) |
| Essential elementsc | ||||
| Ca | 30.60 (4.80) | |||
| Cu | 0.01 (0.01) | ND | ND | ND |
| Fe | 0.02 (0.04) | ND | ND | ND |
| K | 2.40 (0.71) | ND | ND | ND |
| Mg | 9.08 (1.96) | ND | ND | ND |
| Total N | 0.83 (0.13) | |||
| Na | 26.00 (8.69) | ND | ND | ND |
| Total P | 0.19 (0.08) | ND | ND | ND |
| Zn | 0.02 (0.01) | ND | ND | ND |
| Beneficial elementsd | ||||
| Si | 2.28 (0.69) | ND | ND | ND |
| Toxic elementse | ||||
| Al | 0.05 (0.04) | ND | ND | ND |
Abbreviations: BAR, biofilm annular reactor (three operated in parallel for biofilm development; see Materials and Methods); Cl2, chlorine; HPC, heterotrophic plate count; ND, not detected; NO3, nitrate; NTU, nephelometric turbidity units; PO4, phosphate; SD, standard deviation; TOC, total organic carbon. Other parameters detected, mean mg liter−1 (SD): Ba, 0.04 (0.01); S, 21.18 (4.82); Sr, 0.22 (0.05). Other parameters not detected: Be; NH3, ammonia as N; Sb; and V.
Temperature was measured in BAR3.
Classifications are based on the World Health Organization criteria (65).
Classifications are based on the World Health Organization criteria (65). Mn and Ni were not detected.
Classifications are based on the World Health Organization criteria (65). As, Cd, Li, Pb, and Sn were not detected.
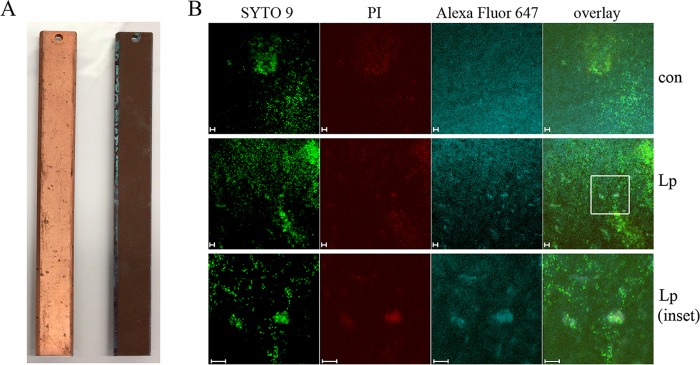
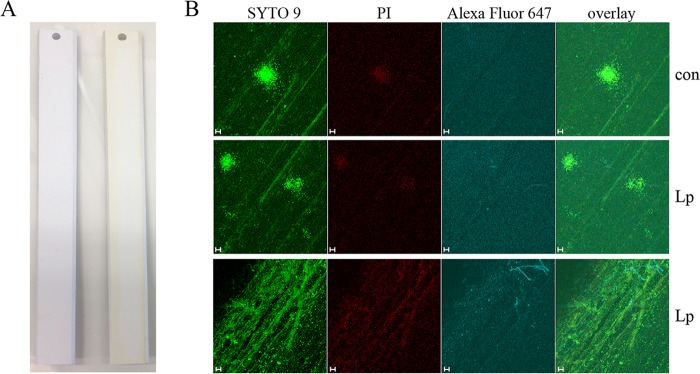
Biofilm material was more visible on the Cu and PVC surfaces than on new slides (Fig. 1A and 2A), with the Cu slides displaying heavy corrosion in the form of white/green copper oxide deposits along the edges of the slide (Fig. 1A, right slide). To visualize biofilm structures via confocal microscopy, Cu and PVC control (con) and LpP1s1-inoculated slides were fluorescently stained with nucleic acid dyes, SYTO 9 and propidium iodide (PI), and a L. pneumophila-specific antibody against strain Philadelphia-1 LPS (Fig. 1B and 2B). SYTO 9 stains all cells (shown in green, representing intact, presumably live cells), whereas PI can only enter cells with compromised membranes and quenches the intracellular SYTO 9 signal (shown in red, representing presumably injured or dead cells).
FIG 1.
Drinking water biofilms on copper (Cu) surfaces. Biofilms were established on Cu BAR slides with uncolonized and colonized slides shown on the left and right, respectively (A). Biofilms were then inoculated with LpP1s1, fluorescently stained, and visualized using confocal microscopy. (B) From left to right, membrane-intact (green, SYTO 9), membrane-permeable (red, propidium iodide [PI]), and L. pneumophila (blue, Alexa Fluor 647) cells were observed by fluorescence microscopy, including an overlay of the signals. The top row shows stained, uninoculated biofilm controls (con), the middle row shows LpP1s1-inoculated biofilms, and the bottom row shows 3×-zoomed-in area of the LpP1s1-inoculated biofilms indicated in the white box. Images are representative of four to six images collected from two independent experiments. Scale bars, 10 μm.
FIG 2.
Drinking water biofilms on polyvinyl chloride surfaces. Biofilms were established on PVC BAR slides with uncolonized and colonized slides shown on the left and right, respectively (A). Biofilms were then inoculated with LpP1s1, fluorescently stained, and visualized using confocal microscopy. (B) From left to right, membrane-intact (green, SYTO 9), membrane-permeable (red, propidium iodide [PI]), and L. pneumophila (blue, Alexa Fluor 647) cells were observed by fluorescence microscopy, including an overlay of the signals. The top row shows stained, uninoculated biofilm controls (con), and the middle and bottom rows show LpP1s1-inoculated biofilms in the middle and edges of the BAR slide, respectively. Images are representative of four to six images collected from two independent experiments. Scale bars, 10 μm.
Cu and PVC biofilms were approximately 30 μm in depth, with no visibly stained LpP1s1 cells present in the control slides (Fig. 1B and 2B, top row). Cu, but not PVC, surfaces were autofluorescent in the same ex/em range as the Alexa Fluor 647-conjugated secondary antibody, which was observed as a uniform blue background. However, LpP1s1-stained cells were clearly visible amid the autofluorescence (Fig. 1B, third column). Cu biofilms were more diffuse with large biofilm colonies observed throughout the slide, where LpP1s1 cells seemed to cluster (Fig. 1B); in contrast, LpP1s1 colonized PVC biofilms in more a punctate pattern (Fig. 2B). Overall, PVC biofilm structures were smaller and more localized, like independent microcolonies, in the middle of the slide, and more diffuse with no large structures visible along the edges (Fig. 2B, middle and bottom rows, respectively). It is not clear if this was correlated to the pattern of prominent brown, biofilm material deposition on the edges of the PVC slide compared to the center portion (Fig. 2A, right slide).
Free chlorine and monochloramine inactivation of planktonic L. pneumophila.
To assess inactivation of planktonic cells, post-exponential-phase LpP1s1 suspensions were prepared as described in Materials and Methods and exposed to initial concentrations (mean ± the SD) of 2.1 ± 0.1 mg free chlorine liter−1 for 0 to 3 min and 2.2 ± 0.1 mg monochloramine liter−1 for 0 to 150 min. Figure 3 displays the log10 CFU ml−1 values at each exposure time during free chlorine and monochloramine inactivation (black and gray bars, respectively) and illustrates the rapid inactivation of LpP1s1 with free chlorine compared to monochloramine. After 1 min of free chlorine exposure, culturable planktonic LpP1s1 cells decreased from 6.3 ± 0.1 to 0.2 ± 0.3 log10 CFU ml−1 compared to the decrease of monochloramine-treated planktonic cells from 6.8 ± 0.0 to 6.5 ± 0.0 log10 CFU ml−1 (Fig. 3). Free chlorine-treated planktonic cells displayed 3- and 4-log10 CFU ml−1 inactivation “concentration times time” (Ct) values (mg min liter−1) of 0.11 and 0.30 compared to the 2-, 3-, and 4-log10 Ct values of 5.35, 6.58, and 7.81 during monochloramine treatment, respectively (Table 2). Due to the rapid inactivation of planktonic cells during free chlorine treatment, the Ct value for 2-log10 reductions could not be determined since that value was beyond the range of linear extrapolation (Table 2).
FIG 3.
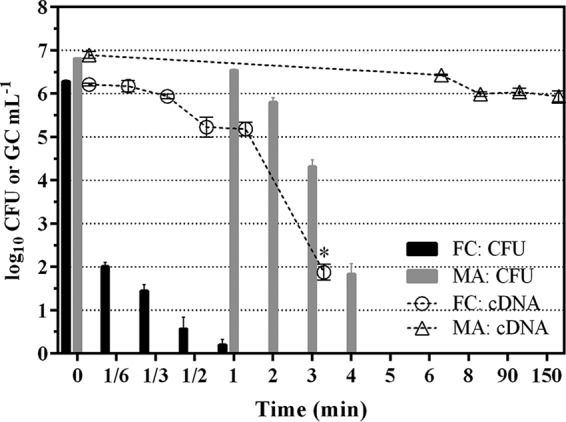
Effect of disinfectants on planktonic L. pneumophila. LpP1s1 suspensions were treated with free chlorine (FC) or monochloramine (MA). Data (mean ± SD) are representative of two independent experiments with three replicates each (n = 6). CFU data are shown as bars and expressed as log10 CFU ml−1. The cDNA qPCR data are shown as open symbols with connecting lines and expressed as log10 genomic copies (GC) ml−1. *, P ≤ 0.01.
TABLE 2.
Ct values for inactivation of planktonic and drinking water biofilm-associated L. pneumophila using free chlorine and monochloraminea
|
Ct value (mg min liter−1) |
||||||
|---|---|---|---|---|---|---|
| Inactivation Ct | Planktonic LpP1s1 |
Drinking water biofilm-associated LpP1s1 |
||||
| Free chlorine | Monochloramine | Lowest | 2nd lowest | 2nd highest | Highest | |
| 2-log10 | ND | 5.35 | Free chlorine PVC, 8.86 | Free chlorine Cu, 13.18 | Monochloramine PVC, 17.16 | Monochloramine Cu, 34.86 |
| 3-log10 | 0.11 | 6.58 | Free chlorine PVC, 36.11 | Free chlorine Cu, 50.83 | Monochloramine Cu, 55.38 | Monochloramine PVC, 62.80 |
| 4-log10 | 0.30 | 7.81 | Free chlorine PVC, 63.67 | Monochloramine Cu, 75.90 | Free chlorine Cu, 88.48 | Monochloramine PVC, 108.44 |
Abbreviations: ND, not determined; Cu, copper; LpP1s1, L. pneumophila strain Philadelphia-1 serogroup 1; PVC, polyvinyl chloride. Ct values (concentration × time) are expressed as mg min liter−1. Data were generated from two independent experiments, except for MA Cu. For planktonic and Cu samples, there were three replicates per experiment (n = 6), except for MA Cu (n = 3). For PVC samples, there were two replicates per experiment (n = 4).
Transcript levels of the 16S rRNA gene, commonly used as a measure of L. pneumophila metabolic activity, was monitored in the free chlorine and monochloramine treated planktonic LpP1s1 cells at each exposure time (Fig. 3, open circles and triangles, respectively). For free chlorine-treated cells, there was a gradual decrease in 16S rRNA transcript levels from 0 to 1 min (1.0 ± 0.4 log10 GC ml−1), followed by a sharp, statistically significant decrease from 1 to 3 min (3.3 ± 0.4 log10 GC ml−1) (Fig. 3, open circles; *, P ≤ 0.01). In contrast, monochloramine-treated planktonic cells displayed a 1.1 ± 0.1 log10 GC ml−1 decrease from 0 to 8 min, followed by no significant changes in 16S rRNA levels from 8 to 90 min (Δ 0.2 ± 0.1 log10 GC ml−1) and 90 to 150 min (Δ 0.3 ± 0.1 log10 GC ml−1) postexposure (Fig. 3, open triangles, P > 0.05).
Free chlorine and monochloramine inactivation of biofilm-associated L. pneumophila.
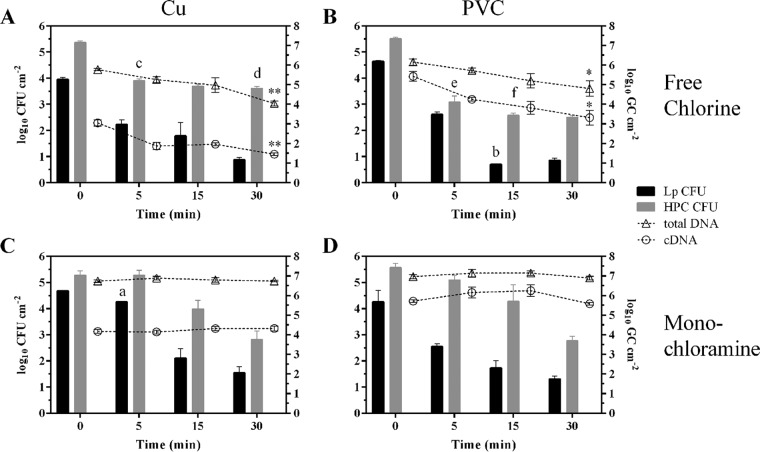
LpP1s1 colonized Cu and PVC drinking water biofilms were exposed to initial concentrations (mean ± the SD) of 2.1 ± 0.1 mg free chlorine liter−1 and 2.2 ± 0.1 mg monochloramine liter−1 for 0 to 30 min as described above. Figure 4 displays the log10 CFU cm−2 values for biofilm LpP1s1 (black bars) levels on Cu and PVC surfaces at each exposure time during inactivation. After 5 min of disinfectant exposure, biofilm LpP1s1 levels (mean log10 CFU cm−2) were reduced by 1.8, 2.0, and 1.7 for free chlorine-treated Cu and PVC biofilms and monochloramine-treated PVC biofilms, respectively (Fig. 4). However, the mean LpP1s1 reduction of 0.4 log10 CFU cm−2 for monochloramine treated Cu biofilms was statistically lower at 5 min than the reduction levels of the other biofilms (P ≤ 0.001). By 15 min, free chlorine-treated PVC biofilms displayed LpP1s1 reduction levels of 3.9 log10 CFU cm−2 which was significantly higher than the reduction levels of 2.6 log10 CFU cm−2 for the other biofilms (P ≤ 0.05). By 30 min, levels of LpP1s1 reduction between the biofilm samples were not statistically different.
FIG 4.
Effect of disinfectants on biofilm-associated bacteria and nucleic acid levels. Cu (A and C) and PVC (B and D) drinking water biofilms, colonized with LpP1s1, were treated with free chlorine (A and B) or monochloramine (C and D). Biofilms were analyzed for LpP1s1 and HPC CFU (shown as bars with corresponding units on the left axis, log10 CFU liter−1) and total DNA and cDNA qPCR (shown as open symbols with connecting lines and units on the right axis, log10 genomic copies [GC] liter−1). Data (mean ± SD) are representative of two independent experiments with three replicates each (n = 6). a, P ≤ 0.001 compared to other biofilms at 5 min. b, P ≤ 0.05 compared to other biofilms at 15 min. c, P ≤ 0.01 compared to monochloramine Cu and PVC at 5 min. d, P ≤ 0.05 compared to free chlorine PVC and monochloramine Cu at 30 min. e, P ≤ 0.001 compared to monochloramine Cu and PVC at 5 min. f, P ≤ 0.01 compared to free chlorine Cu, monochloramine Cu, and PVC at 15 min. *, P ≤ 0.05; **, P ≤ 0.01.
The Ct values for 2-, 3-, and 4-log10 CFU cm−2 reduction levels indicated potential differences in inactivation kinetics based on the disinfectant treatment and biofilm substratum. The samples are listed in order based on the Ct values, from lowest to highest, for 2-log10 reduction—free chlorine PVC, free chlorine Cu, monochloramine PVC, and monochloramine Cu; for 3-log10 reduction—free chlorine PVC, free chlorine Cu, monochloramine Cu, monochloramine PVC; and for 4-log10 reduction—free chlorine PVC, monochloramine Cu, free chlorine Cu, and monochloramine PVC (Table 2).
HPCs were higher than biofilm L. pneumophila levels before exposure and, although decreases were observed, HPCs were generally more resistant to free chlorine and monochloramine, on all surfaces, than L. pneumophila (Fig. 4, gray bars). Notably, after 5 min, HPC log10 CFU cm−2 levels in Cu and PVC biofilms decreased more rapidly during free chlorine treatment, 1.5 and 2.4, compared to 0.0 and 0.5 during monochloramine treatment (P ≤ 0.01). This trend was also observed at 15 min postexposure, where HPC levels in Cu and PVC-free chlorine-treated biofilms decreased 1.7 and 2.9 log10 CFU cm−2, respectively, compared to 1.3 log10 CFU cm−2 for both Cu and PVC monochloramine-treated biofilms (Fig. 4). However, by 30 min, HPC levels in free chlorine-treated Cu biofilms (3.6 ± 0.2) were statistically higher than levels in free chlorine-treated PVC (2.5 ± 0.1) and monochloramine-treated Cu (2.8 ± 0.8) biofilms.
The presence and expression levels of the L. pneumophila 16S rRNA gene were monitored for free chlorine and monochloramine treated Cu and PVC biofilms at each exposure time. Gene transcript (cDNA) levels were lower than total genomic DNA levels for each treatment and surface type (Fig. 4, open circles and triangles, respectively). Free chlorine treatment resulted in a statistically significant decrease in total DNA and cDNA L. pneumophila levels by 30 min for Cu and PVC biofilms (P ≤ 0.01 and P ≤ 0.05, respectively), with no significant differences between starting and ending levels of total DNA and cDNA for monochloramine-treated biofilm samples (Fig. 4).
Analysis of L. pneumophila shed from drinking water biofilms during disinfectant treatment.
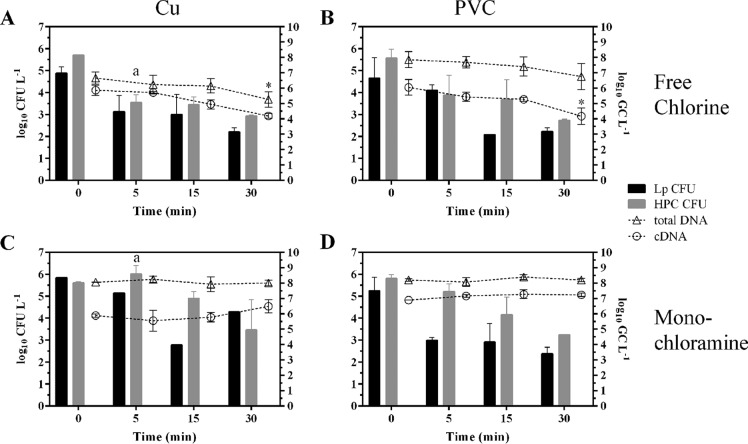
To determine the effect of disinfectant treatment on LpP1s1 shed from drinking water biofilms, beaker water, that contained BAR slides submerged in disinfectant solutions at each exposure time, was processed for LpP1s1 CFU, HPC, total DNA and cDNA analysis as described in Materials and Methods. There were no statistically significant differences in LpP1s1 CFU and HPC levels between free chlorine- and monochloramine-treated Cu and PVC biofilms at each exposure time, except for HPC levels between free chlorine- and monochloramine-treated Cu biofilms at 5 min postexposure (Fig. 5, P ≤ 0.05). As expected, the L. pneumophila 16S rRNA gene total DNA levels were higher than the gene transcript (cDNA) levels for each treatment and surface type (Fig. 5, open triangles and circles, respectively). In concordance with biofilm trends, the total DNA and cDNA levels during free chlorine treatment decreased for Cu and PVC beaker water samples (Fig. 5A and B) compared to the monochloramine-treated samples (Fig. 5C and D). However, statistically significant decreases were only observed for total DNA levels in free chlorine Cu beaker water and cDNA levels in free chlorine PVC beaker water between starting and 30-min postexposure levels (Fig. 5A and B, P ≤ 0.05).
FIG 5.
CFU and nucleic acid levels of biofilm bacteria shed from drinking water biofilms during disinfectant treatment. Beaker water derived from chlorine (A and B) or monochloramine (C and D) treatment of Cu (A and C) and PVC (B and D) drinking water biofilms colonized with LpP1s1 was analyzed for LpP1s1 and HPC CFU (shown as bars with corresponding units on the left axis, log10 CFU liter−1), and total DNA and cDNA qPCR (shown as open symbols with connecting lines and units on the right axis, log10 genomic copies [GC] liter−1). Data (mean ± SD) are representative of two independent experiments (n = 2) for each exposure time. a, P ≤ 0.05 for free chlorine Cu versus monochloramine Cu at 5 min. *, P ≤ 0.05.
DISCUSSION
In this study, mature (1.5- to 2-year-old) drinking water biofilms were developed on Cu and PVC surfaces under conditions typically encountered within building water systems, e.g., turbulent flow with 3 to 9 h periods of stagnation per day (6). Structural differences were observed between Cu and PVC biofilms; notably, larger, more diffuse biofilm colonies observed on Cu surfaces, while the smaller, less diffuse biofilm colonies were on PVC surfaces (Fig. 1B and 2B). The primary goal of this study was to determine efficacy of free chlorine and monochloramine for the inactivation of biofilm-associated L. pneumophila. Disinfectant concentrations of 2 mg liter−1 were chosen to represent the midrange value of the World Health Organization (WHO) guidelines for drinking water treatment (23, 24) and because previous studies had demonstrated that ≤1.5 mg liter−1 chlorine and chloramine was inadequate for microbial control (27–29). To observe and quantify the effectiveness of each disinfectant, e.g., 2-, 3-, and 4-log10 reduction in pathogen concentration posttreatment, the naturally formed Cu and PVC drinking water biofilms were inoculated with L. pneumophila sg 1 strain Philadelphia-1 (LpP1s1), resulting in a biofilm concentration of between 4.0 to 4.7 log10 CFU cm−2.
After colonization, LpP1s1 bacteria clustered within the larger Cu biofilm colonies but displayed a more punctate colonization pattern throughout the PVC surfaces. The maximum thickness of the Cu and PVC biofilms, as measured by Z-stacks, was approximately 30 μm. This depth was similar to that reported for 3-year-old drinking water biofilms grown on stainless steel, where a maximum thickness of 30 μm was observed after 600 days (1.6 year), with an average of 14 μm between 1.4- to 3-year-old biofilms) (30). Martiny et al. observed formation of large, individual microcolonies after 94 days (0.2 year) of biofilm development with a surface coverage of 41%, which progressed to 76% coverage with looser, more spread-out structures from 1.9 to 3 years (30), similar to the large, diffuse structures developed on the Cu surfaces in this study (Fig. 1B).
“Concentration times time” (Ct) values, based on the work of Chick (31) and Watson (32), describes microbial inactivation as a function of disinfectant concentration and contact time with the effectiveness of the disinfectant dependent on the microorganism, disinfectant type and concentration, pH and temperature of the suspension medium (6). In this study, planktonic LpP1s1 Ct (mg min liter−1) values for 4-log10 CFU ml−1 inactivation (99.99% reduction) were 0.3 for free chlorine and 7.8 for monochloramine treatment at pH 8 and 21°C under chlorine demand free conditions (Table 2). In a previous study, the Ct value for 99% inactivation (Ct99) using free chlorine for a L. pneumophila sg 1 isolate was 0.88 at 21°C (pH 6.8 to 7.2) (33), which is higher than the Ct observed in this study, presumably due to the use of sterilized well water as the assay medium, instead of chlorine demand free buffer. Other studies have also evaluated the effectiveness of free chlorine and monochloramine on planktonic L. pneumophila strains, but the use of different strains, buffers, and pH and temperature conditions make comparisons difficult across studies (34–38).
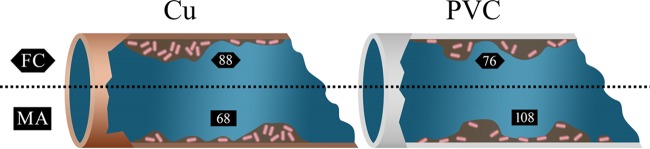
Biofilm L. pneumophila was more quickly inactivated on PVC surfaces using free chlorine than using monochloramine. Free chlorine treatment resulted in a 3.8-log10 reduction after 30 min of exposure compared to 3.0 log10 during monochloramine treatment; thus, for 4-log10 inactivation, more contact time is required with monochloramine than with free chlorine (Fig. 4 and Table 2, 75.90 versus 88.48, respectively). Figure 6 is a summary graphic illustrating the Ct values (mg min liter−1) for 4-log10 or 99.9% reduction of biofilm LpP1s1: 88 for free chlorine-treated Cu biofilms, 63 for free chlorine-treated PVC biofilms, 76 for monochloramine-treated Cu biofilms, and 108 for monochloramine-treated PVC biofilms (Table 2). The interpretation of Ct values for biofilm inactivation should be made cautiously since disinfectant concentration and contact time are not the only parameters affecting the disinfectant’s efficacy. There are many unknowns regarding disinfectant penetration into and microbial aggregation within biofilms, and the potential role of the underlying substratum that could impact disinfectant inactivation.
FIG 6.
Graphical summary of free chlorine and monochloramine disinfection efficacy on biofilm-associated L. pneumophila. Ct inactivation values (mg min liter−1) for 4-log10 CFU cm−2 reduction of biofilm LpP1s1 are shown for free chlorine (FC) and monochloramine (MA) disinfectant treatment of copper (Cu) and polyvinyl chloride (PVC) slides.
Using 1-week-old Pseudomonas aeruginosa and Klebsiella pneumoniae biofilms developed in nutrient rich medium, it was reported that after 30 min of 2.5 mg liter−1 free chlorine exposure, approximately 70% of the bulk free chlorine concentration (∼1.8 mg liter−1) penetrated the biofilms to a depth of 50 μm (39). In contrast, disinfectant penetration was much lower for nitrifying biofilms, where after 30 min of 2.6 to 2.7 mg liter−1 disinfectant exposure, only 8% (∼0.2 mg liter−1) and 15% (∼0.4 mg liter−1) of the bulk water concentration for free chlorine and monochloramine, respectively, penetrated the biofilms to a depth of 50 μm (22). Other studies have also reported the increased penetration of monochloramine within biofilms compared to free chlorine (22, 39), the requirement of higher free chlorine for Cu versus plastic (e.g., polyethylene and PVC) pipes to achieve the same inactivation rates (40, 41), and free chlorine, generally, being more effective than monochloramine at controlling or inactivating L. pneumophila or other biofilm microorganisms within the same study (42–45). Collectively, results from this study suggested that inactivation of biofilm-associated L. pneumophila using monochloramine was more effective on Cu than PVC biofilms, while conversely, free chlorine was more effective on PVC than Cu biofilms.
The heterotrophic plate count (HPC) method is commonly used for enumeration of culturable heterotrophic bacteria in drinking water. Although there have been no conclusive studies indicating that high levels of HPC in drinking water poses a significant health risk, HPC measurements are used as an assessment of drinking water quality (46). There are several studies that correlated HPC levels with Legionella occurrence in drinking water (47–50); however, in this study, there were no similarities or trends in HPC and L. pneumophila levels within the Cu and PVC biofilms during disinfectant treatment. Although free chlorine treatment of Cu and PVC biofilms significantly reduced HPC levels after 5 min of exposure compared to monochloramine treatment, HPC levels remained steady within Cu and PVC biofilms after 15 and 30 min of free chlorine treatment (Fig. 4A and B). In contrast, HPC levels steadily decreased with continual monochloramine exposure (Fig. 4C and D), which may have been due to the better penetration of monochloramine within biofilms and/or the higher disinfectant demand of free chlorine for the biofilms compared to monochloramine.
Notably, the disinfection efficacy of L. pneumophila shed from the Cu and PVC biofilms into the bulk water during treatment, was more like biofilm-associated bacteria than planktonic cells. At each exposure time point, any biofilm material that may have shed and sloughed off from the biofilm slides during disinfection, were collected from the beaker water via filtration and used for CFU enumeration and nucleic acid extraction. Shed biofilm LpP1s1 was culturable from the bulk water at 5 to 30 min of exposure to both disinfectants (Fig. 5), whereas the planktonic LpP1s1 cells were inactivated after 3 to 5 min of exposure (Table 2). Shen et al. also reported higher free chlorine and monochloramine disinfectant resistance of shed L. pneumophila from groundwater PVC biofilms and the accompanying detachment of biofilm material and bacteria during biofilm L. pneumophila shed into the bulk water (51). Interestingly, the same group observed a higher release of biofilm bacteria, as assessed by total 16S rRNA gene copies, from the monochloramine-treated biofilms than from untreated and free chlorine-treated biofilms (51). This trend was observed in this study using the HPC method, as there were higher HPC levels in the beaker water of monochloramine-treated Cu and PVC biofilms at 5 and 10 min postexposure than for free chlorine-treated biofilms (Fig. 5).
The differences in 16S rRNA gene transcript levels of planktonic LpP1s1 during free chlorine and monochloramine treatment did not correlate with the loss culturability. No culturable free chlorine-treated cells were detected at 3 min postexposure, when the significant decline in cDNA levels were observed, and no culturable monochloramine-treated cells were detected at 5 to 150 min postexposure with no significant decreases in cDNA levels observed between 8 and 150 min (Fig. 3). It should be noted that the 16S rRNA gene cDNA quantitative PCR (qPCR) assay cannot differentiate between decreases in transcript levels versus damaged, nonamplifiable transcripts. However, the reported slow reactivity of monochloramine on bacterial RNA (21) suggests the latter may be the case for the differences in transcript levels observed between free chlorine- and monochloramine-treated planktonic LpP1s1 cells.
The cDNA levels of treated Cu biofilms, at 0 and 30 min (Fig. 4A and C), were statistically lower than levels in treated PVC biofilms at the same exposure times (Fig. 4B and D, P ≤ 0.01) indicating that the Cu substratum alone, may be negatively impacting 16S rRNA gene transcript levels. Furthermore, the cDNA levels of free chlorine-treated, compared to monochloramine-treated, Cu biofilms were statistically lower at all time points (Fig. 4A and B, P ≤ 0.0001), indicating a possible synergistic effect of free chlorine and Cu on the reduction transcript levels in LpP1s1 cells. However, the synergistic effect of free chlorine and Cu on transcript levels were less apparent in the beaker water samples since there were no significant differences between free chlorine Cu and PVC beaker water levels by 30 min (Fig. 5A and B, P > 0.05).
This study evaluated the disinfection efficacy for a single L. pneumophila sg 1 strain, the Philadelphia-1 lung tissue isolate, which was previously shown to colonize drinking water biofilms for up to several months after introduction into native drinking water biofilms (52, 53). Similarly, two L. pneumophila drinking water biofilm isolates, sg 1 strain Ads and sg 5 strain 2226A, were reported to stably colonize drinking water biofilms (54, 55), indicating that other L. pneumophila strains may behave similarly in regard to biofilm colonization. Future work will examine the disinfectant susceptibility of other biofilm-associated Legionella strains.
The drinking water used for biofilm development, in this study, is minimally corrosive due to the water utility’s addition of lime and sodium hydroxide to increase pH levels, measured at pH 8.35 ± 0.18 for the BAR influent water (Table 1). However, drinking water pipe material and associated corrosion products have been shown to impact disinfectant efficacy (56, 57). Thus, the chemical interactions between free chlorine, monochloramine, and the Cu and PVC biofilm substratum and their subsequent effect on Legionella inactivation should also be examined in subsequent studies to further elucidate the mechanisms and consequences of various pathogen control strategies employed within building water systems.
L. pneumophila can replicate and reside within various species of free-living amoebae (12, 58), but the abundance, diversity, and distribution of free-living amoebae in building water systems and the proportion of these eukaryotes harboring intracellular pathogens are poorly understood. Vermamoeba vermiformis and various species of the Acanthamoeba genera are commonly used host models to study Legionella-amoeba interactions (58, 59) and can amplify L. pneumophila bacteria to high intracellular concentrations (60). However, V. vermiformis and Acanthamoeba spp. were not detected via qPCR analysis in the biofilm and beaker water DNA samples (data not shown).
Collectively, results from this study indicate there were clear differences between free chlorine and monochloramine treatment and their impact on L. pneumophila 16S rRNA gene transcript levels in planktonic, biofilm-associated, and biofilm shed forms of LpP1s1 (Fig. 6). The combined characteristics of building water systems present favorable growth conditions for opportunistic pathogens, such as Legionella pneumophila. However, their stable colonization within biofilms most likely confers the best survival advantage as biofilms offer protection from environmental stresses, increased access to nutrients, and opportunities for symbiotic interactions with other microbes. Legionella infections can be mitigated with implementation of an effective building water quality management plan (61). Thus, understanding the efficacy of common disinfectants on the ecologically relevant forms of L. pneumophila and their persistence mechanisms within building water systems could aid policy makers in their recommendations to control and eliminate exposure risks to these water-based pathogens.
MATERIALS AND METHODS
Establishment of drinking water biofilms.
Drinking water biofilm communities were established using three bench scale biofilm annular reactor (BAR) model systems (BioSurface Technologies Corporation) and BAR slides (15 cm length by 1.6 cm width by 0.2 mm depth), made of copper alloy C12200 (Cu) and type 2 polyvinyl chloride (PVC). Biofilms were grown under simulated premise plumbing conditions to mimic private residential water conditions with drinking water derived from river water treated by coagulation, flocculation, sedimentation; followed by sand, gravel, and granular activated carbon filtration; and then chlorination. The three BARs were operated, in parallel, at approximately 110 to 120 rpm, resulting in shear stress similar to that of a 0.5- to 1-inch pipe with water flowing at a velocity of 1 foot s−1 (assuming a Hazen-Williams coefficient, measure of head loss due to friction from pipe roughness, of 140 for copper [Cu] and 150 for PVC). The flow rate was set at 100 ml min−1, resulting in a hydraulic residence time of 10 min. A valve under the control of a timer, allowed flow of drinking water into each BAR on weekdays for 2 h in the morning and 4 h at night and on weekends for 2 h in the morning, 2 h midday, and 4 h at night. The valve was connected to a manifold consisting of three outlets which supplied the same water flow rate to the three BARs. In addition, the timer was connected to each of the motor controller so that the drum turned on when there was water flow. The absence of shear force, produced by turning off drum rotation, allowed the simulation of building water system stagnation, as well as operation of BARs at room temperature. The outer cylinders were wrapped to reduce the potential of phototrophic growth within the BARs. Each BAR can accommodate up to 20 slides. Biofilms were developed on 60 Cu slides and then used for disinfection experiments described below. After their removal, new PVC slides were placed into the BARs for biofilm accumulation.
Water quality parameter measurements.
Drinking water, feeding the BARs, was sampled weekly and analyzed for free and total chlorine (N,N-diethyl-p-phenylenediamine [DPD] method; Pocket Colorimeter II; Hach), pH (Accumet AB15; Fisher Scientific), temperature, hardness (EDTA titration method; Hach), turbidity (2100Q portable turbidimeter; Hach), total organic carbon (TOC; EPA method 415.3, rev1.1), trace metals (EPA method 200.7), phosphate (PO4; EPA method 365.1), and nitrate (NO3; EPA method 353.2) by the National Risk Management Research Laboratory at the US Environmental Protection Agency in Cincinnati, OH (Table 1). Water samples were also taken from each BAR and analyzed for TOC and HPC bacteria. HPCs were enumerated by the spread plate method on Reasoner’s 2 A agar (R2A, Difco Laboratories) after incubation at 28°C for 7 days. The limit of detection (LOD) was 0.7 log10 CFU ml−1. A temperature logger (Nomad OM-74; Omega Engineering, Inc.) was used to monitor water temperature in BAR3 during biofilm establishment.
Bacterial culture preparation and enumeration.
Legionella pneumophila sg 1 strain Philadelphia-1 (LpP1s1) is a clinical isolate derived from a human lung (American Type Culture Collection [ATCC 33152]). LpP1s1 cells were grown and enumerated as previously described (62). Briefly, individual frozen stock cultures were stored at −80°C in buffered yeast extract (BYE) broth with 20% (vol/vol) glycerol. A 10-μl aliquot of a thawed suspension was streaked onto buffered charcoal yeast extract (BCYE) agar plates (BD Diagnostics) and incubated at 37°C for 72 h. An LpP1s1 colony was inoculated into 10 ml of BYE broth and grown overnight with continuous shaking at 37°C. Post-exponential-phase cultures (approximately 109 CFU ml−1) were centrifuged (2,420 relative centrifugal force [rcf], 10 min, room temperature) and washed three times with 10 ml UV-light dechlorinated, 0.22-μm-filtered drinking water (dfH2O). LpP1s1 densities, as measured by CFU, were determined by spread plating undiluted and serially diluted bacterial suspensions on BCYE plates incubated for 4 to 6 days at 37°C.
L. pneumophila inoculation of BAR slides.
Cu and PVC slides were aseptically removed from the BARs. Two independent experiments (I and II) were conducted for each disinfectant (free chlorine and monochloramine) with three Cu and two PVC slides per experiment and for each time point evaluated (0, 5, 15, and 30 min). Slides were submerged in 1-liter beakers containing dfH2O and a magnetic stir bar. Beakers were placed on stir plates operated under ambient conditions (21 to 23°C) with continuous stirring. Slides were incubated for 5 days in the presence of LpP1s1 cells at a final concentration of 6.7 ± 0.4 log10 CFU ml−1 (mean ± the SD). On day 5, the LpP1s1 densities in the beaker water were 6.7 ± 0.3 log10 CFU ml−1 (except for monochloramine Cu experiment I, where no culturable LpP1s1 was detected in the beaker water at day 5, but was culturable at day 0). After the 5-day incubation, the BAR slides were gently washed twice with 150 ml of dfH2O to remove loosely adherent bacteria and submerged in fresh 1-liter beakers containing dfH2O and a magnetic stir bar. Beakers were then placed on stir plates operated under ambient conditions (21 to 23°C) with continuous stirring for 24 h prior to use in disinfection experiments.
Disinfectant preparation.
Free chlorine and total chlorine measurements were performed using the DPD colorimetric method (Powder Pillows; Hach). Monochloramine and free ammonia measurements were performed using the indophenol method (method 10200, Powder Pillows, free ammonia chlorinating solution; Hach). Disinfection experiments were all performed under chlorine demand free (CDF) conditions. Disinfectant solutions were prepared using 1:10 diluted CDF (dCDF) buffer (pH 8), prepared as previously described (63). Undiluted CDF buffer contained 50 mg liter−1 of phosphate (PO43−) which negatively impacted the colorimetric readings for monochloramine and free ammonia. The excess phosphate during the indophenol reaction resulted in heavy precipitation within the Hach test vials. Thus, CDF was diluted 10-fold, which maintained the same pH and did not interfere with the indophenol test.
Free chlorine solutions were prepared by diluting 15% reagent-grade sodium hypochlorite solution in dCDF buffer (pH 8) to provide a free residual chlorine concentration of 2 mg liter−1. To prepare a 2-mg liter−1 monochloramine solution, ammonium sulfate [(NH4)2SO4] was dissolved in dCDF, with the addition of chlorine at a 4.5:1 chlorine-to-nitrogen (Cl2:N) mass ratio. All tests were performed under ambient conditions (21°C). Before each experiment, each Hach Pocket Colorimeter II were checked using DPD chlorine and monochloramine/free ammonia secondary gel standards (Hach) to confirm consistent instrument response. The pH, free chlorine, total chlorine, monochloramine, and free ammonia levels were measured at the start and end of each experiment. The average (± the SD) concentrations of free chlorine and monochloramine solutions prepared for all the experiments performed in this study were 2.1 ± 0.1 and 2.2 ± 0.1 mg liter−1, respectively.
Disinfection of planktonic L. pneumophila suspensions and L. pneumophila-colonized biofilms.
Planktonic suspensions of LpP1s1 were exposed to either free chlorine or monochloramine in 50-ml conical tubes at various exposure times with continuous shaking using a vortexer. An aliquot of the washed, overnight LpP1s1 cultures, prepared as described above, was inoculated into the disinfection solution resulting in a concentration of approximately 6 log10 CFU ml−1. A portion of the planktonic suspensions was collected for disinfectant measurements immediately before neutralization by the addition of 0.1 ml of sodium thiosulfate (10% [wt/vol]) at the end of each exposure time. After neutralization, treated planktonic suspensions were enumerated for LpP1s1 CFU and used for cDNA preparation described below. Two independent experiments were performed for each disinfectant with three replicates per exposure time (n = 6).
After the LpP1s1 inoculation period into drinking water biofilms, slide wash, and a 24-h incubation period, as described above, the BAR slides were placed into four separate 1-liter beakers labeled t = 0, 5, 15, and 30 min, indicating disinfectant exposure times, and containing either three Cu or two PVC slides each. Two independent experiments were performed for each disinfectant with two PVC (n = 4) and three Cu (n = 6) replicates per exposure time. The control (t = 0 min) beaker contained 1.25 liters of dCDF. The remaining three beakers contained either 1.25 liters of free chlorine or monochloramine disinfectant solution. Disinfectant measurements of the beaker solution were made immediately before neutralization by the addition of 1 ml of sodium thiosulfate (10% [wt/vol]) at the end of each exposure time. These measurements were used to calculate the Ct (contact time) in mg min liter−1. The log10 CFU ml−1 or cm−2 reduction values were plotted against Ct values at each exposure time, and linear interpolations and extrapolations were used to calculate 2-, 3-, and 4-log10 CFU Ct inactivation values. R2 values for the planktonic LpP1s1 and biofilm LpP1s1 linear plots ranged between 0.9991 and 1 and between 0.8073 and 0.9683, respectively.
To account for zero values, 1 was added to all data points before conversion to the log10 scale [e.g., log10(CFU + 1)]. For planktonic samples, the LOD was 0.0 log10 CFU ml−1. For LpP1s1 and HPC enumeration, the LOD was 2.1 log10 CFU liter−1 for beaker water samples and 0.7 log10 CFU cm−2 for biofilm samples.
Biofilm and beaker water collection.
To recover biofilm material, individual BAR slides were placed into a sterile petri dish (150 by 15 mm) containing 12 ml of dfH2O. A sterile polyester tipped applicator was used to swab the slide surface several times. The slide was then washed three times using a portion of the biofilm suspension in the petri dish. For further extraction and homogenization of biofilm material, the biofilm suspension and BAR slide were transferred to a stomacher bag (380 mm by 510 mm) and placed in a stomacher (Stomacher 3500; Seward, United Kingdom) operated for 4 min at 175 rpm. The biofilm suspension was collected and used for CFU enumeration and nucleic acid extraction. The BAR slide dimensions were 15 × 1.6 × 0.2 cm (length × width × depth), with an approximate surface area of 24 cm2. Calculations from CFU and molecular analyses were expressed as units per cm2.
After disinfectant neutralization, 1 liter of the beaker water at each time point was filtered through a 0.2-μm-pore size polyethersulfone membrane (Supor Membrane; PALL Life Sciences). Filters were placed into 12 ml of dfH2O and vortexed at maximum speed for 1 min. The beaker water concentrate/suspension was collected and used for CFU enumeration and nucleic acid extraction. Two independent experiments were performed for each disinfectant with biofilm slides submerged in one beaker (n = 2) per exposure time. Calculations from CFU and molecular analyses were adjusted and expressed as units per liter.
Total DNA isolation.
DNA was extracted from 1 ml of the biofilm and beaker water suspensions, described in the previous section, using the PowerWater DNA isolation kit (MO BIO Laboratories). The 1-ml biofilm and beaker water suspensions were concentrated by centrifugation (9,391 rcf, 5 min, room temperature), removal of 800 μl of the supernatant, and resuspension of the pellet in the remaining 200 μl of solution. Biofilm swabs or filters were placed into the PowerWater bead tube, along with 200 μl of concentrated biofilm and beaker water suspensions and 1 ml of solution PW1. The remainder of the procedure was performed according to the manufacturer’s protocol.
Complementary DNA preparation.
A PowerSYBR Green Cells-to-CTTM kit (Thermo Fisher Scientific) was used to obtain cDNA from biofilm and beaker water samples according to the manufacturer’s protocol with the following modifications. Then, 1 ml of the biofilm, beaker water, and planktonic suspensions described above, was concentrated by centrifugation (15,000 rcf, 10 min, room temperature) and removal of 990 μl of the supernatant. Next, 50 μl of the lysis buffer was added to the pellet and the remaining 10 μl of solution, followed by incubation for 5 min at room temperature. DNA was removed by adding 4 U of DNase (TURBO DNase; Invitrogen) and incubation at 37°C for 30 min. The removal of DNA was confirmed by the absence of DNA amplification via qPCR using the L. pneumophila-specific primers described below. To convert RNA to cDNA, reverse transcription was performed as described according to the manufacturer’s protocol. Aliquots of the cDNA sample were analyzed by qPCR as described below.
Quantitative PCR.
Biofilm and beaker water total DNA and cDNA and planktonic cDNA samples were analyzed using a Applied Biosystems QuantStudio 6 Flex Fast Real-Time PCR system (Thermo Fisher Scientific). The TaqMan qPCR assay for L. pneumophila detection was performed using the 16S rRNA gene forward (LpneuF1, 5′-CGG AAT TAC TGG GCG TAA AGG-3′) and reverse (LpneuR1, 5′-GAG TCA ACC AGT ATT ATC TGA CCG T-3′) primers and probe (LpneuP1, 5′ FAM-AAG CCC AGG AAT TTC ACA GAT AAC TTA ATC AAC CA-BHQ1 3′) (64). The reaction mixture (20 μl final volume) contained 1× TaqMan Environmental Master Mix 2.0 (Applied Biosystems), 200 nM forward and reverse primers, and 7.5 nM probe. The cycling conditions consisted of a preincubation step at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 10 s and annealing and extension at 60°C for 1 min. Undiluted and 10-fold-diluted experimental samples were analyzed in duplicate with no inhibition observed between the undiluted and diluted samples. Standard curves were generated, on each plate, using a plasmid vector (pUCIDT-AMP; Integrated DNA Technologies, Inc.) containing a cloned 189-bp region of the L. pneumophila Philadelphia-1 16S rRNA gene (NCBI reference sequence NC_002942.5, positions 609325 to 609513). Serial dilutions of eight concentrations ranging from 107 to 1 gene copy were generated and analyzed in triplicate along with duplicate no-template control, for each 96-well plate. Data were expressed as log10 gene copy numbers (GC). The limits of quantification were 2.0 log10 GC ml−1 for planktonic cDNA samples, 3.1 log10 GC liter−1 for beaker water samples, and 1.7 log10 GC cm−2 for biofilm samples.
Fluorescent staining of biofilm samples.
Uninoculated control and LpP1s1-inoculated slides were incubated in a solution containing 10 μM SYTO 9 and 60 μM propidium iodide (PI; Molecular Probes). For L. pneumophila staining, slides were incubated with a mouse IgG monoclonal antibody specific to L. pneumophila Philadelphia-1 strain lipopolysaccharide (Meridian Life Science, Inc.) at a 2-μg ml−1 concentration, followed by incubation with a secondary polyclonal donkey anti-mouse IgG antibody conjugated to Alexa Fluor 647 (Thermo Fisher Scientific) at a concentration of 10 μg ml−1. All fluorescent stains and antibodies were diluted to the target concentrations in staining solution (dfH2O containing 10% bovine serum albumin) and incubated with the BAR slides in the dark for 30 min at room temperature. After each incubation, slides were gently washed twice with the staining solution. Slides were submerged in dfH2O for confocal microscopy.
Confocal microscopy.
BAR slides were imaged on a LSM 510 confocal laser scanning microscope (Zeiss, Germany) with a 40×, 0.8 NA C-Achroplan water-immersion objective lens. Biofilm structures were imaged using confocal reflectance (488-nm laser light scanned across sample with the photomultiplier tube collecting the same 488 nm light which reflects off the sample, and a small confocal pinhole to reduce the optical section thickness to ∼1 μm). Z-stacks were acquired to calculate biofilm thickness. Fluorescence images were acquired using the following excitation (ex) lasers and emission (em) filters for the following dyes indicated as wavelengths (nm): SYTO 9 (ex 488, em 500 to 530), PI (ex 543, em 550 to 600), and Alexa 647 (ex 633, em >650).
Statistical analysis.
Statistical significance was determined for cDNA and total DNA differences using one-way analysis of variance (ANOVA) and Tukey’s multiple-comparison test. Statistical differences between LpP1s1 CFU, HPC, and Ct data were determined using two-way ANOVA and the Bonferroni- Šídák multiple-comparison test. One- and two-way ANOVA was performed using Prism 6 (GraphPad Software). Data in the bar and line graphs are shown as mean with the SD. Water quality parameters were analyzed using the single factor ANOVA function in Microsoft Excel 2016 (Microsoft).
Supplementary Material
ACKNOWLEDGMENTS
We thank Chet Closson at the University of Cincinnati’s Live Microscopy Core for his technical expertise on confocal microscopy and David Wahman at the EPA National Risk Management Research Laboratory for his technical guidance and expertise on monochloramine generation. We also thank our colleagues, Vincent Gallardo and Jingrang Lu, for their critical review of the manuscript.
The U.S. Environmental Protection Agency (EPA) through its Office of Research and Development funded and managed the research described under contract EP-C-11-006 to Pegasus Technical Services, Inc. The manuscript has been subjected to the Agency’s review and has been approved for publication.
The views expressed in this article are those of the authors and do not necessarily represent the views or policies of the Agency. Mention of trade names, commercial products, and/or services does not imply an endorsement or recommendation for use by the U.S. Government or the EPA.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02956-18.
REFERENCES
- 1.Fliermans CB. 1996. Ecology of Legionella: from data to knowledge with little wisdom. Microb Ecol 32:203–228. [DOI] [PubMed] [Google Scholar]
- 2.Diederen BM. 2008. Legionella spp. and Legionnaires’ disease. J Infect 56:1–12. doi: 10.1016/j.jinf.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 3.Yu VL, Plouffe JF, Pastoris MC, Stout JE, Schousboe M, Widmer A, Summersgill J, File T, Heath CM, Paterson DL, Chereshsky A. 2002. Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: an international collaborative survey. J Infect Dis 186:127–128. doi: 10.1086/341087. [DOI] [PubMed] [Google Scholar]
- 4.Fields B, Benson RF, Besser RE. 2002. Legionella and Legionnaires’ disease: 25 years of investigation. Clin Microbiol Rev 15:506–526. doi: 10.1128/CMR.15.3.506-526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buse HY, Schoen ME, Ashbolt NJ. 2012. Legionellae in engineered systems and use of quantitative microbial risk assessment to predict exposure. Water Res 46:921–933. doi: 10.1016/j.watres.2011.12.022. [DOI] [PubMed] [Google Scholar]
- 6.National Research Council. 2006. Drinking water distribution systems: assessing and reducing risks. The National Academy Press, Washington, DC. [Google Scholar]
- 7.Wang H, Edwards MA, Falkinham JO, III, Pruden A. 2013. Probiotic approach to pathogen control in premise plumbing systems? A review. Environ Sci Technol 47:10117–10128. doi: 10.1021/es402455r. [DOI] [PubMed] [Google Scholar]
- 8.King DN, Donohue MJ, Vesper SJ, Villegas EN, Ware MW, Vogel ME, Furlong EF, Kolpin DW, Glassmeyer ST, Pfaller S. 2016. Microbial pathogens in source and treated waters from drinking water treatment plants in the United States and implications for human health. Sci Total Environ 562:987–995. doi: 10.1016/j.scitotenv.2016.03.214. [DOI] [PubMed] [Google Scholar]
- 9.States SJ, Conley LF, Kuchta JM, Oleck BM, Lipovich MJ, Wolford RS, Wadowsky RM, McNamara AM, Sykora JL, Keleti G, Yee RB. 1987. Survival and multiplication of Legionella pneumophila in municipal drinking water systems. Appl Environ Microbiol 53:979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wingender J, Flemming HC. 2004. Contamination potential of drinking water distribution network biofilms. Water Sci Technol 49:277–286. doi: 10.2166/wst.2004.0861. [DOI] [PubMed] [Google Scholar]
- 11.Berry D, Xi C, Raskin L. 2006. Microbial ecology of drinking water distribution systems. Curr Opin Biotechnol 17:297–302. doi: 10.1016/j.copbio.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 12.Lau HY, Ashbolt NJ. 2009. The role of biofilms and protozoa in Legionella pathogenesis: implications for drinking water. J Appl Microbiol 107:368–378. doi: 10.1111/j.1365-2672.2009.04208.x. [DOI] [PubMed] [Google Scholar]
- 13.Molmeret M, Horn M, Wagner M, Santic M, Abu Kwaik Y. 2005. Amoebae as training grounds for intracellular bacterial pathogens. Appl Environ Microbiol 71:20–28. doi: 10.1128/AEM.71.1.20-28.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berk SG, Gunderson JH, Newsome AL, Farone AL, Hayes BJ, Redding KS, Uddin N, Williams EL, Johnson RA, Farsian M, Reid A, Skimmyhorn J, Farone MB. 2006. Occurrence of infected amoebae in cooling towers compared with natural aquatic environments: implications for emerging pathogens. Environ Sci Technol 40:7440–7444. doi: 10.1021/es0604257. [DOI] [PubMed] [Google Scholar]
- 15.Corsaro D, Pages GS, Catalan V, Loret JF, Greub G. 2010. Biodiversity of amoebae and amoeba-associated bacteria in water treatment plants. Int J Hyg Environ Health 213:158–166. doi: 10.1016/j.ijheh.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 16.Marciano-Cabral F, Jamerson M, Kaneshiro ES. 2010. Free-living amoebae, Legionella and Mycobacterium in tap water supplied by a municipal drinking water utility in the USA. J Water Health 8:71–82. doi: 10.2166/wh.2009.129. [DOI] [PubMed] [Google Scholar]
- 17.Camper AK. 1993. Coliform regrowth and biofilm accumulation in drinking water systems: a review, p 91–105. In Geesey GG, Lewandowski Z, Flemming HC (ed), Biofouling and biocorrosion in industrial water systems. Lewis Publishers, Boca Raton, FL. [Google Scholar]
- 18.Wingender J, Flemming HC. 2011. Biofilms in drinking water and their role as reservoir for pathogens. Int J Hyg Environ Health 214:417–423. doi: 10.1016/j.ijheh.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Dukan S, Touati D. 1996. Hypochlorous acid stress in Escherichia coli: resistance, DNA damage, and comparison with hydrogen peroxide stress. J Bacteriol 178:6145–6150. doi: 10.1128/jb.178.21.6145-6150.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Virto R, Mañas P, Álvarez I, Condon S, Raso J. 2005. Membrane damage and microbial inactivation by chlorine in the absence and presence of a chlorine-demanding substrate. Appl Environ Microbiol 71:5022–5028. doi: 10.1128/AEM.71.9.5022-5028.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacangelo JG, Olivieri VP, Kawata K. 1991. Investigating the mechanism of inactivation of Escherichia coli B by monochloramine. J Am Water Works Assoc 83:80–87. doi: 10.1002/j.1551-8833.1991.tb07152.x. [DOI] [Google Scholar]
- 22.Lee WH, Wahman DG, Bishop PL, Pressman JG. 2011. Free chlorine and monochloramine application to nitrifying biofilm: comparison of biofilm penetration, activity, and viability. Environ Sci Technol 45:1412–1419. doi: 10.1021/es1035305. [DOI] [PubMed] [Google Scholar]
- 23.World Health Organization. 2003. Chlorine in drinking-water, p 11 World Health Organization, Geneva, Switzerland. [Google Scholar]
- 24.World Health Organization. 2004. Monochloramine in drinking-water, p 21 World Health Organization, Geneva, Switzerland. [Google Scholar]
- 25.Rosario-Ortiz F, Rose J, Speight V, von Gunten U, Schnoor J. 2016. How do you like your tap water? Science 351:912–914. doi: 10.1126/science.aaf0953. [DOI] [PubMed] [Google Scholar]
- 26.Rhoads WJ, Pruden A, Edwards MA. 2014. Anticipating challenges with in-building disinfection for control of opportunistic pathogens. Water Environ Res 86:540–549. doi: 10.2175/106143014X13975035524989. [DOI] [PubMed] [Google Scholar]
- 27.LeChevallier MW, Babcock TM, Lee RG. 1987. Examination and characterization of distribution system biofilms. Appl Environ Microbiol 53:2714–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neden DG, Jones RJ, Smith JR, Kirmeyer GJ, Foust GW. 1992. Comparing chlorination and chloramination for controlling bacterial regrowth. J Am Water Works Assoc 84:80–88. doi: 10.1002/j.1551-8833.1992.tb07395.x. [DOI] [Google Scholar]
- 29.Wolfe RL, Lieu NI, Izaguirre G, Means EG. 1990. Ammonia-oxidizing bacteria in a chloraminated distribution system: seasonal occurrence, distribution and disinfection resistance. Appl Environ Microbiol 56:451–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martiny AC, Jorgensen TM, Albrechtsen HJ, Arvin E, Molin S. 2003. Long-term succession of structure and diversity of a biofilm formed in a model drinking water distribution system. Appl Environ Microbiol 69:6899–6907. doi: 10.1128/AEM.69.11.6899-6907.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chick H. 1908. An investigation of the laws of disinfection. J Hyg (Lond) 8:92–158. doi: 10.1017/S0022172400006987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watson HE. 1908. A note on the variation of the rate of disinfection with change in the concentration of the disinfectant. J Hyg (Lond) 8:536–542. doi: 10.1017/S0022172400015928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cargill KL, Pyle BH, Sauer RL, McFeters GA. 1992. Effects of culture conditions and biofilm formation on the iodine susceptibility of Legionella pneumophila. Can J Microbiol 38:423–429. doi: 10.1139/m92-071. [DOI] [PubMed] [Google Scholar]
- 34.Domingue EL, Tyndall RL, Mayberry WR, Pancorbo OC. 1988. Effects of three oxidizing biocides on Legionella pneumophila serogroup 1. Appl Environ Microbiol 54:741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dupuy M, Mazoua S, Berne F, Bodet C, Garrec N, Herbelin P, Ménard-Szczebara F, Oberti S, Rodier M-H, Soreau S, Wallet F, Héchard Y. 2011. Efficiency of water disinfectants against Legionella pneumophila and Acanthamoeba. Water Res 45:1087–1094. doi: 10.1016/j.watres.2010.10.025. [DOI] [PubMed] [Google Scholar]
- 36.Kuchta JM, States SJ, McGlaughlin JE, Overmeyer JH, Wadowsky RM, McNamara AM, Wolford RS, Yee RB. 1985. Enhanced chlorine resistance of tap water-adapted Legionella pneumophila as compared with agar medium-passaged strains. Appl Environ Microbiol 50:21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuchta JM, States SJ, McNamara AM, Wadowsky RM, Yee RB. 1983. Susceptibility of Legionella pneumophila to chlorine in tap water. Appl Environ Microbiol 46:1134–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyamoto M, Yamaguchi Y, Sasatsu M. 2000. Disinfectant effects of hot water, ultraviolet light, silver ions and chlorine on strains of Legionella and nontuberculous mycobacteria. Microbios 101:7–13. [PubMed] [Google Scholar]
- 39.De Beer D, Srinivasan R, Stewart PS. 1994. Direct measurement of chlorine penetration into biofilms during disinfection. Appl Environ Microbiol 60:4339–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lehtola MJ, Miettinen IT, Lampola T, Hirvonen A, Vartiainen T, Martikainen PJ. 2005. Pipeline materials modify the effectiveness of disinfectants in drinking water distribution systems. Water Res 39:1962–1971. doi: 10.1016/j.watres.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 41.Zheng M, He C, He Q. 2015. Fate of free chlorine in drinking water during distribution in premise plumbing. Ecotoxicology 24:2151–2155. doi: 10.1007/s10646-015-1544-3. [DOI] [PubMed] [Google Scholar]
- 42.Chauret C, Volk C, Stover L, Dykstra TS, Andrews RC, Gagnon GA. 2005. Effect of disinfectants on microbial ecology in model distribution systems. J Water Health 3:359–369. doi: 10.2166/wh.2005.050. [DOI] [PubMed] [Google Scholar]
- 43.Donlan RM, Forster T, Murga R, Brown E, Lucas C, Carpenter J, Fields B. 2005. Legionella pneumophila associated with the protozoan Hartmannella vermiformis in a model multi-species biofilm has reduced susceptibility to disinfectants. Biofouling 21:1–7. doi: 10.1080/08927010500044286. [DOI] [PubMed] [Google Scholar]
- 44.Loret JF, Robert S, Thomas V, Cooper AJ, McCoy WF, Levi Y. 2005. Comparison of disinfectants for biofilm, protozoa and Legionella control. J Water Health 3:423–433. doi: 10.2166/wh.2005.047. [DOI] [PubMed] [Google Scholar]
- 45.Thomas V, Bouchez T, Nicolas V, Robert S, Loret JF, Levi Y. 2004. Amoebae in domestic water systems: resistance to disinfection treatments and implication in Legionella persistence. J Appl Microbiol 97:950–963. doi: 10.1111/j.1365-2672.2004.02391.x. [DOI] [PubMed] [Google Scholar]
- 46.Allen MJ, Edberg SC, Reasoner DJ. 2004. Heterotrophic plate count bacteria–what is their significance in drinking water? Int J Food Microbiol 92:265–274. doi: 10.1016/j.ijfoodmicro.2003.08.017. [DOI] [PubMed] [Google Scholar]
- 47.Bargellini A, Marchesi I, Righi E, Ferrari A, Cencetti S, Borella P, Rovesti S. 2011. Parameters predictive of Legionella contamination in hot water systems: association with trace elements and heterotrophic plate counts. Water Res 45:2315–2321. doi: 10.1016/j.watres.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 48.Edagawa A, Kimura A, Doi H, Tanaka H, Tomioka K, Sakabe K, Nakajima C, Suzuki Y. 2008. Detection of culturable and nonculturable Legionella species from hot water systems of public buildings in Japan. J Appl Microbiol 105:2104–2114. doi: 10.1111/j.1365-2672.2008.03932.x. [DOI] [PubMed] [Google Scholar]
- 49.Solimini AG, Cottarelli A, Marinelli L, De Giusti M. 2014. Factors influencing persistence of Legionella pneumophila serogroup 1 in laboratory cocultures. BMC Microbiol 14:249. doi: 10.1186/s12866-014-0249-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams K, Pruden A, Falkinham JO III, Edwards M. 2015. Relationship between organic carbon and opportunistic pathogens in simulated glass water heaters. Pathogens 4:355–372. doi: 10.3390/pathogens4020355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen Y, Huang C, Lin J, Wu W, Ashbolt NJ, Liu W-T, Nguyen TH. 2017. Effect of disinfectant exposure on Legionella pneumophila associated with simulated drinking water biofilms: release, inactivation, and infectivity. Environ Sci Technol 51:2087–2095. doi: 10.1021/acs.est.6b04754. [DOI] [PubMed] [Google Scholar]
- 52.Buse HY, Ji P, Gomez-Alvarez V, Pruden A, Edwards MA, Ashbolt NJ. 2017. Effect of temperature and colonization of Legionella pneumophila and Vermamoeba vermiformis on bacterial community composition of copper drinking water biofilms. Microb Biotechnol 10:773–788. doi: 10.1111/1751-7915.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buse HY, Lu J, Struewing IT, Ashbolt NJ. 2014. Preferential colonization and release of Legionella pneumophila from mature drinking water biofilms grown on copper versus unplasticized polyvinylchloride coupons. Int J Hyg Environ Health 217:219–225. doi: 10.1016/j.ijheh.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 54.Lehtola MJ, Torvinen E, Kusnetsov J, Pitkanen T, Maunula L, von Bonsdorff CH, Martikainen PJ, Wilks SA, Keevil CW, Miettinen IT. 2007. Survival of Mycobacterium avium, Legionella pneumophila, Escherichia coli, and caliciviruses in drinking water-associated biofilms grown under high-shear turbulent flow. Appl Environ Microbiol 73:2854–2859. doi: 10.1128/AEM.02916-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moritz MM, Flemming HC, Wingender J. 2010. Integration of Pseudomonas aeruginosa and Legionella pneumophila in drinking water biofilms grown on domestic plumbing materials. Int. J Hyg Environ Health 213:190–197. doi: 10.1016/j.ijheh.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 56.LeChevallier MW, Lowry CD, Lee RG. 1990. Disinfecting biofilms in a model distribution system. J Am Water Works Assoc 82:87–99. doi: 10.1002/j.1551-8833.1990.tb06996.x. [DOI] [Google Scholar]
- 57.Nguyen C, Elfland C, Edwards M. 2012. Impact of advanced water conservation features and new copper pipe on rapid chloramine decay and microbial regrowth. Water Res 46:611–621. doi: 10.1016/j.watres.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 58.Rowbotham TJ. 1986. Current views on the relationships between amoebae, legionellae, and man. Isr J Med Sci 22:678–689. [PubMed] [Google Scholar]
- 59.Hilbi H, Weber SS, Ragaz C, Nyfeler Y, Urwyler S. 2007. Environmental predators as models for bacterial pathogenesis. Environ Microbiol 9:563–575. doi: 10.1111/j.1462-2920.2007.01238.x. [DOI] [PubMed] [Google Scholar]
- 60.Buse HY, Ashbolt NJ. 2012. Counting Legionella cells within single amoeba host cells. Appl Environ Microbiol 78:2070–2072. doi: 10.1128/AEM.07392-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garrison LE, Kunz JM, Cooley LA, Moore MR, Lucas C, Schrag S, Sarisky J, Whitney CG. 2016. Vital signs: deficiencies in environmental control identified in outbreaks of Legionnaires’ disease—North America, 2000–2014. MMWR Morb Mortal Wkly Rep 65:576–584. doi: 10.15585/mmwr.mm6522e1. [DOI] [PubMed] [Google Scholar]
- 62.Buse HY, Ashbolt NJ. 2011. Differential growth of Legionella pneumophila strains within a range of amoebae at various temperatures associated with in-premise plumbing. Lett Appl Microbiol 53:217–224. doi: 10.1111/j.1472-765X.2011.03094.x. [DOI] [PubMed] [Google Scholar]
- 63.Rice EW, Adcock NJ, Sivaganesan M, Rose LJ. 2005. Inactivation of spores of Bacillus anthracis Sterne, Bacillus cereus, and Bacillus thuringiensis subsp. israelensis by chlorination. Appl Environ Microbiol 71:5587–5589. doi: 10.1128/AEM.71.9.5587-5589.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Donohue MJ, O’Connell K, Vesper SJ, Mistry JH, King D, Kostich M, Pfaller S. 2014. Widespread molecular detection of Legionella pneumophila serogroup 1 in cold water taps across the United States. Environ Sci Technol 48:3145–3152. doi: 10.1021/es4055115. [DOI] [PubMed] [Google Scholar]
- 65.World Health Organization. 2005. Nutrients in drinking water. World Health Organization, Geneva, Switzerland. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.