Although computational and reverse metabolic engineering approaches often lead to improved gene deletion mutants for cell factory engineering, the systems level effects of such gene deletions on the production phenotypes have not been extensively studied. Understanding the genetic and molecular function of such gene alterations on production strains will minimize the risk inherent in the development of large-scale fermentation processes, which is a daunting challenge in the field of industrial biotechnology. Therefore, we established a detailed experimental and systems biology approach to uncover the molecular mechanisms of YPL062W deletion in S. cerevisiae, which is shown to improve the production of all terpenoid classes. This study redefines the genetic function of YPL062W, demonstrates a strong correlation between YPL062W and terpenoid production, and provides a useful modification for the creation of terpenoid production platform strains. Further, this study underscores the benefits of detailed and systematic characterization of the metabolic effects of genetic alterations on engineered biosynthetic factories.
KEYWORDS: ALD6, Saccharomyces cerevisiae, YPL062W, terpenoids
ABSTRACT
Saccharomyces cerevisiae is an established cell factory for production of terpenoid pharmaceuticals and chemicals. Numerous studies have demonstrated that deletion or overexpression of off-pathway genes in yeast can improve terpenoid production. The deletion of YPL062W in S. cerevisiae, in particular, has benefitted carotenoid production by channeling carbon toward carotenoid precursors acetyl coenzyme A (acetyl-CoA) and mevalonate. The genetic function of YPL062W and the molecular mechanisms for these benefits are unknown. In this study, we systematically examined this gene deletion to uncover the gene function and its molecular mechanism. RNA sequencing (RNA-seq) analysis uncovered that YPL062W deletion upregulated the pyruvate dehydrogenase bypass, the mevalonate pathway, heterologous expression of galactose (GAL) promoter-regulated genes, energy metabolism, and membrane composition synthesis. Bioinformatics analysis and serial promoter deletion assay revealed that YPL062W functions as a core promoter for ALD6 and that the expression level of ALD6 is negatively correlated to terpenoid productivity. We demonstrate that ΔYPL062W increases the production of all major terpenoid classes (C10, C15, C20, C30, and C40). Our study not only elucidated the biological function of YPL062W but also provided a detailed methodology for understanding the mechanistic aspects of strain improvement.
IMPORTANCE Although computational and reverse metabolic engineering approaches often lead to improved gene deletion mutants for cell factory engineering, the systems level effects of such gene deletions on the production phenotypes have not been extensively studied. Understanding the genetic and molecular function of such gene alterations on production strains will minimize the risk inherent in the development of large-scale fermentation processes, which is a daunting challenge in the field of industrial biotechnology. Therefore, we established a detailed experimental and systems biology approach to uncover the molecular mechanisms of YPL062W deletion in S. cerevisiae, which is shown to improve the production of all terpenoid classes. This study redefines the genetic function of YPL062W, demonstrates a strong correlation between YPL062W and terpenoid production, and provides a useful modification for the creation of terpenoid production platform strains. Further, this study underscores the benefits of detailed and systematic characterization of the metabolic effects of genetic alterations on engineered biosynthetic factories.
INTRODUCTION
Saccharomyces cerevisiae is an attractive platform for heterologous terpenoid production due to its versatile features, including genetic tractability, biosafety, and robustness in industrial fermentation (1–3). In S. cerevisiae, all terpenoids are produced from five-carbon (C5) isopentenyl diphosphate (IPP) biosynthesized by the mevalonate (MVA) pathway from the common precursor acetyl coenzyme A (acetyl-CoA). Terpenoids encompass a vast range of structures that fall into different classes based on the number of C5 IPP precursors used for the synthesis: monoterpenoids, C10; sesquiterpenoids, C15; diterpenoids, C20; triterpenoids, C30 from 2×C15; and tetraterpenoids (carotenoids), C40 from 2×C20 (1). Previous efforts in engineering pathway-relevant genes to increase acetyl-CoA and MVA levels have successfully increased terpenoid yield and productivity in S. cerevisiae (4, 5). However, many of these efforts treat the manipulated metabolic pathways as independent entities, even though they are known to be highly interconnected with the rest of cellular metabolism and therefore tightly regulated (3, 6). This interconnectedness is why seemingly irrelevant genes can have significant and unexpected effects on a given pathway (7–9). With the aid of in silico strategies, 10 genes unrelated to the MVA or terpenoid production pathways were individually deleted and shown to confer 8- to 10-fold increases to amorphadiene production titers in S. cerevisiae (10). Thus, the screening of deletion collections in S. cerevisiae can serve as a rich resource for identifying genetic targets beneficial to terpenoid production (11).
Using a deletion collection, Özaydın et al. (12) identified 24 individual deletions of genes unrelated to carotenoid (C40) production that nonetheless increased production of this class of terpenoids in S. cerevisiae. However, only 3 deletions (ΔROX1, ΔYJL064W, and ΔYPL062W) also increased the production of the sesquiterpenoid bisabolene (C15). Due to the complex nature of genetic interactions and the unique properties of different terpenoid classes, the impacts of modifying different genetic targets on different terpenoid-producing strains have been shown to vary (8). Of the three genes identified by Özaydın et al., deletion of YPL062W resulted in a 4-fold increase in intracellular MVA levels (12). Previously, ΔYPL062W was shown to reduce glycogen accumulation (13, 14). More recently, we found that ΔYPL062W possessed a crucial role in reducing acetate accumulation and elevating acetyl-CoA content, thereby improving lycopene (C40) yield in S. cerevisiae (15). Although computational and genetic approaches for screening and reconstructing such highly effective gene deletion targets are being developed and deployed in metabolic engineering, more detailed studies to uncover the molecular mechanism of such gene deletions have been less forthcoming. Understanding the genetic and molecular function of such gene alterations on production strains will minimize risk inherent in the development of large-scale fermentation processes, which is a daunting challenge in the field of industrial biotechnology.
In this study, we designed a systematic experimental approach incorporating systems biology approaches to elucidate the genetic function of YPL062W and understand how ΔYPL062W improves carbon flow to acetyl-CoA and enhances terpenoid production in S. cerevisiae. The transcriptional effects of YPL062W deletion were analyzed by RNA sequencing (RNA-seq) and revealed an impact on MVA formation, energy metabolism, heterologous gene expression, and cytomembrane composition. Moreover, YPL062W was shown to be nontranscribed, and its deletion decreased the transcription level of the downstream gene ALD6. Serial deletions of conserved domains of ALD6 promoter revealed that YPL062W functions as a core promoter of ALD6. Finally, we demonstrate the correlation between ALD6 transcription level and heterologous terpenoid production in S. cerevisiae.
RESULTS
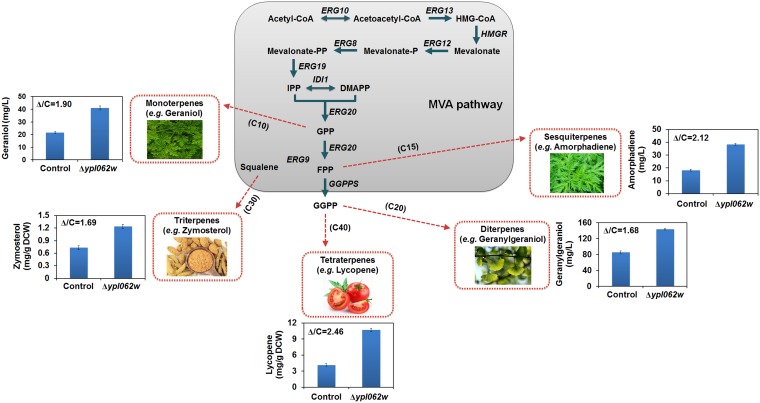
ΔYPL062W can improve the production of all terpenoids.
In our previous study, ΔYPL062W was found to enhance lycopene (C40) production in S. cerevisiae by increasing carbon flux to acetyl-CoA (15). Before conducting detailed experiments on the mechanism of YPL062W deletion, we first examined the generalizability of this gene deletion by assaying its effect on the production of various classes of terpenoids in S. cerevisiae, including (but not limited to) lycopene. SyBE_Sc14C02 (control strain) and SyBE_Sc14C10 (ΔYPL062W strain) were chosen as the host strains for recombinant terpenoid pathways. The biosynthetic pathways of geraniol (monoterpenoid, C10), amorphadiene (sesquiterpenoid, C15), geranylgeraniol (diterpenoid, C20), zymosterol (triterpenoid, C30) and lycopene (tetraterpenoid, C40) were constructed by genomic integration of corresponding terpenogenic modules (see Fig. S1 in the supplemental material). ΔYPL062W significantly increased geraniol, amorphadiene, geranylgeraniol, zymosterol, and lycopene production in S. cerevisiae by 90%, 112%, 68%, 69%, and 146%, respectively (Fig. 1), showing that this deletion benefits the production of all terpenoid classes. Deletion of YPL062W in the control strain did not cause any obvious growth deficiency when utilizing either glucose or ethanol as the sole carbon source (Fig. S2). Therefore, YPLW062W is a promising target for the engineering of S. cerevisiae terpenoid production strains. ΔYPL062W was advanced for detailed characterization of its genetic and molecular effects on terpenoid production.
FIG 1.
The impact of ΔYPL062W on terpenoid production. Recombinant strains expressing terpenoid production pathways were created and assayed to investigate the effect of ΔYPL062W on their corresponding production titers. Geraniol, amorphadiene, geranylgeraniol, zymosterol, and lycopene are representative of monoterpenoid, sesquiterpenoid, diterpenoid, triterpenoid, and tetraterpenoid, respectively. The improvement in production titer is indicated as Δ/C, which is the ratio of terpenoid production titer of the ΔYPL062W strain to that of the control/parent strain.
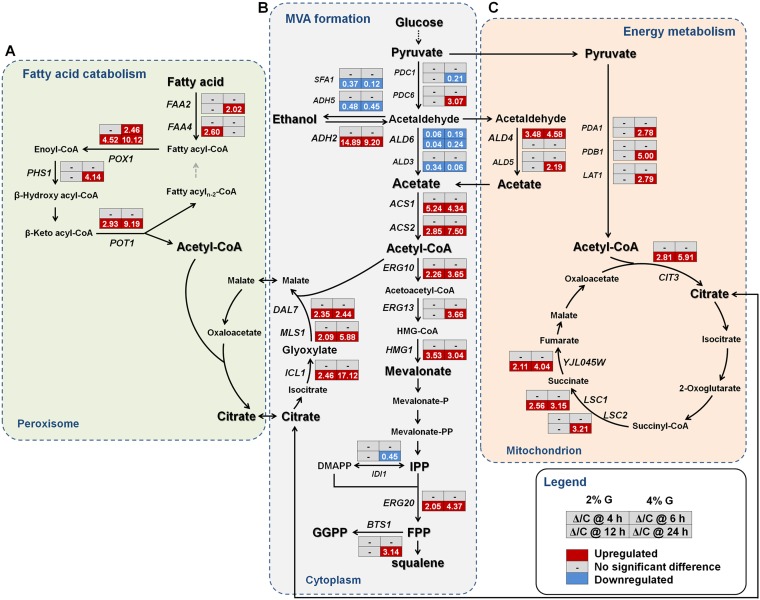
ΔYPL062W upregulates pyruvate dehydrogenase bypass, mevalonate pathway, and energy metabolism.
In order to determine the molecular mechanisms of YPL062W, RNA-seq analysis was applied to investigate the transcriptional effect of YPL062W deletion by comparing SyBE_Sc14C02 (control strain) and SyBE_Sc14C10 (ΔYPL062W strain) during the glucose consumption phase (building up biomass) and the ethanol consumption phase (synthesizing products) (Fig. 2). ALD6 is primarily responsible for cytosolic acetate generation from acetaldehyde. In the central carbon metabolic pathways of the ΔYPL062W strain (Fig. 2B), ALD6 is significantly downregulated during the whole-cell growth stage. Cells lacking ALD6 produce less acetate when assimilating glucose in S. cerevisiae (16, 17). ALD4 encoding a major mitochondrial aldehyde dehydrogenase is upregulated (Fig. 2C) to compensate for the loss of ALD6 (17, 18). In this case, cytosolic acetaldehyde (produced by decarboxylation of pyruvate) is transported into mitochondria to generate acetate (16).
FIG 2.
Transcriptional changes of genes involved in fatty acid catabolism, MVA formation, and energy metabolism by ΔYPL062W, whose reactions mainly occur in the peroxisome (A), cytoplasm (B), and mitochondrion (C), respectively. The key metabolites in each pathway are marked in bold. Transcriptional data are boxed next to each gene. Strain names are abbreviated as follows: C, SyBE_Sc14C10, and Δ, ΔYPL062W strain SyBE_Sc14C02. Strains were cultured under both 2% (mass/vol) (2% G) and 4% (mass/vol) (4% G) glucose conditions. The time reading after the “@” symbol in the legend indicates the sampling time within the glucose consumption phase (i.e., 4 h) or the ethanol consumption phase (i.e., 12 h). The relative transcription level for each gene is indicated as Δ/C, which is the ratio of the transcription level in the ΔYPL062W to that in the control strain. Genes that are significantly upregulated and downregulated and genes without significant transcriptional differences are colored red, blue, and gray, respectively.
When glucose is depleted, cell growth enters the ethanol consumption phase. ADH2, responsible for the oxidization of ethanol to acetaldehyde, is upregulated in strain Δ (Fig. 2B). The genes involved in its reverse reduction reaction (ADH5 and SFA1) are downregulated (Fig. 2B). ALD4 is also upregulated during the ethanol consumption phase (Fig. 2B). The upregulation of ADH2 may be concomitant with upregulation of ALD4, which is reported to increase the reduction flux of NAD+ not only in mitochondria but also in the cytosol (19). The increase in NAD+ reduction flux should improve ATP production and the biosynthetic pathway (19). Moreover, the pyruvate dehydrogenase (PDH) bypass is enhanced by increased expression the genes coding for rate-limiting enzymes ACS1 and ACS2 in strain Δ (Fig. 2B). ACS1 and ACS2 upregulation is necessary for the generation of cytosolic acetyl-CoA from acetate (20, 21). Furthermore, several MVA pathway genes (ERG10, ERG13, HMG1, and ERG20) are also significantly upregulated in the ΔYPL062W strain (Fig. 2B). Among these activated genes, HMG1 and ERG20 encode the major rate-limiting enzymes of MVA pathway (22, 23). The combined above-mentioned changes push ethanol via acetyl-CoA toward MVA, probably leading to the increased intracellular MVA level and increased terpenoid production by ΔYPL062W (24, 25).
In addition to the central carbon metabolism, YPL062W deletion also impacts other types of cell metabolism. The genes responsible for fatty acid β-oxidation (FAA2, FAA4, PHS1, POX1, and POT1) are upregulated in the ΔYPL062W strain, especially during cell growth on nonfermentable carbon sources (i.e., ethanol or acetate) (Fig. 2A). The β-oxidation of fatty acids occurs in peroxisomes and serves as another important source of acetyl-CoA through the glyoxylate cycle (26). The glyoxylate shunt plays an essential role in allowing growth on nonfermentable carbon sources, as it offers the net synthesis of C4 dicarboxylic acid for the tricarboxylic acid (TCA) cycle. The TCA cycle not only generates reducing equivalents for ATP synthesis but also provides metabolic precursors for biomass formation. The genes involved in glyoxylate cycle (ICL1, MLS1, and DAL7 [Fig. 2B]) and TCA cycle (CIT3, LSC1, LSC2, and YJL045W [Fig. 2C]) are significantly upregulated by ΔYPL062W in the ethanol consumption phase, likely benefitting the flux of both the glyoxylate shunt and the mitochondrial TCA cycle (27, 28). Upregulation of the TCA cycle has previously proven to be beneficial for terpenoid production by increasing the supply of both energy and reducing power (29, 30). Therefore, upregulation of glyoxylate cycle in combination with strengthening of the TCA cycle by ΔYPL062W improves energy storage for terpenoid biosynthesis.
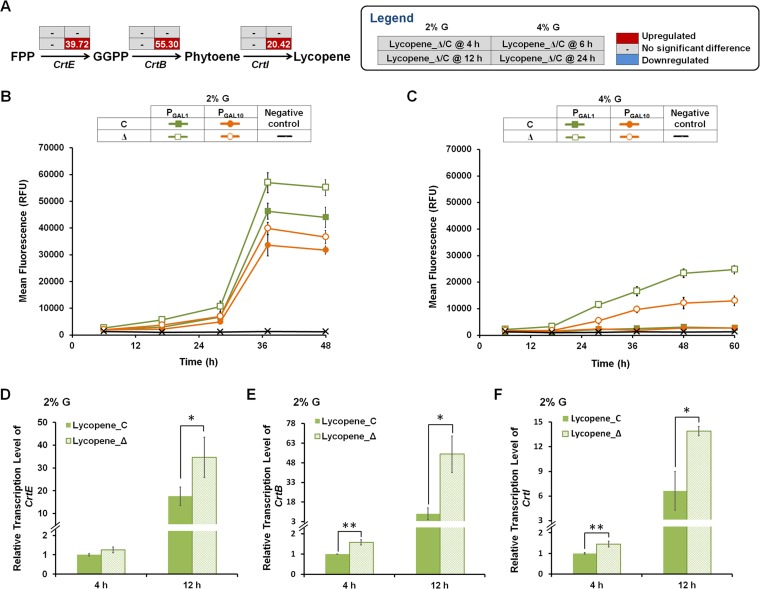
ΔYPL062W improves heterologous expression of GAL promoter-regulated genes.
In order to investigate the influence of YPL062W deletion on the heterologous terpenoid biosynthetic pathway, lycopene-producing strain SyBE_Sc14C07 (control strain; Lycopene_C) and SyBE_Sc14C23 (ΔYPL062W strain; Lycopene_Δ) were selected for transcriptome analysis. In our previous study, we attributed acetate accumulation to the marginal lycopene production of control strain on 4% (mass/vol) glucose (15). In this study, the RNA-seq data demonstrated that the transcription of three heterologous genes responsible for inducible lycopene synthesis (CrtE, CrtB, and CrtI) were significantly upregulated by ΔYPL062W under 4% (mass/vol) glucose, whereas no significant difference was detected between strains Lycopene_C and Lycopene_Δ on 2% (mass/vol) glucose (Fig. 3A). Meanwhile, since CrtE, CrtB, and CrtI are under the control of inducible galactose (GAL) promoters (PGAL1 and PGAL10) (15), the effect of ΔYPL062W on their activities was characterized by promoter fusion with RFP in the host control strain and ΔYPL062W strain. Deletion of YPL062W significantly increased PGAL1 and PGAL10 activities on 4% (mass/vol) glucose (Fig. 3C). However, the impact on GAL promoter activities under 2% (mass/vol) glucose (Fig. 3B) was less statistically robust. Therefore, reverse transcription-PCR (RT-PCR) was conducted to measure the transcription levels of CrtE, CrtB, and CrtI under 2% (mass/vol) glucose. As shown in Fig. 3D to F, the transcription of these three genes was significantly upregulated during the ethanol consumption phase (at 12 h), indicating that ΔYPL062W ubiquitously increases the expression of heterologous genes activated by GAL promoters. The conflicting results between RNA-seq and RT-PCR assays are due to differences in sensitivities of the methods and the fact that differentially expressed genes were defined with a low-resolution threshold (log2 fold change > 1.0). Figure 3B and C show that the overall decreases in promoter activities on 4% (mass/vol) glucose compared to 2% (mass/vol) glucose are likely due to catabolite repression (31). Deletion of YPL062W increases the flux of the carotenoid (C40) pathway by increasing heterologous gene expression and facilitates increased terpenoid production.
FIG 3.
The effect of ΔYPL062W on heterologous gene expression. (A) The transcriptional changes of CrtE, CrtB, and CrtI in lycopene-producing strains revealed by RNA-seq analysis. Strains SyBE_Sc14C07 (control strain, Lycopene_C) and SyBE_Sc14C23 (ΔYPL062W strain, Lycopene_Δ) were cultured under both 2% (mass/vol) (2% G) and 4% (mass/vol) (4% G) glucose conditions. The time reading after the “@” symbol in the legend indicates the sampling time within the glucose consumption (the top line of the table) and ethanol consumption phases (the bottom line of the table). The relative transcription level for each gene is indicated as Lycopene_Δ/C, which is the ratio of the transcription level of the recombinant lycopene pathway in the ΔYPL062W strain to that in the control strain. Genes that are significantly upregulated and downregulated and genes without significant transcriptional differences are colored red, blue, and gray, respectively. (B and C) The effects of ΔYPL062W on PGAL1 and PGAL10 activities when cultured under 2% (mass/vol) glucose (B) or 4% (mass/vol) glucose (C). Promoter activities are represented as relative fluorescence intensities of RFP in the control strain (SyBE_Sc14C02, C) and ΔYPL062W strain (SyBE_Sc14C10, Δ) without lycopene synthesis. (D, E, and F) The transcription level of genes CrtE (D), CrtB (E), and CrtI (F) in lycopene-producing strains (Lycopene_C and Lycopene_Δ) as determined by real-time PCR. Cells were cultured under 2% (mass/vol) glucose, and samples were taken within the glucose consumption phase (i.e., 4 h) or the ethanol consumption phase (i.e., 12 h). The relative transcription level for each gene was determined as 2−ΔΔCT using gene ALG9 for normalization. All data are from at three or more experimental replicates. Statistically significant differences are indicated as follows: *, P < 0.05, and **, P < 0.01 (two-tailed Student t test).
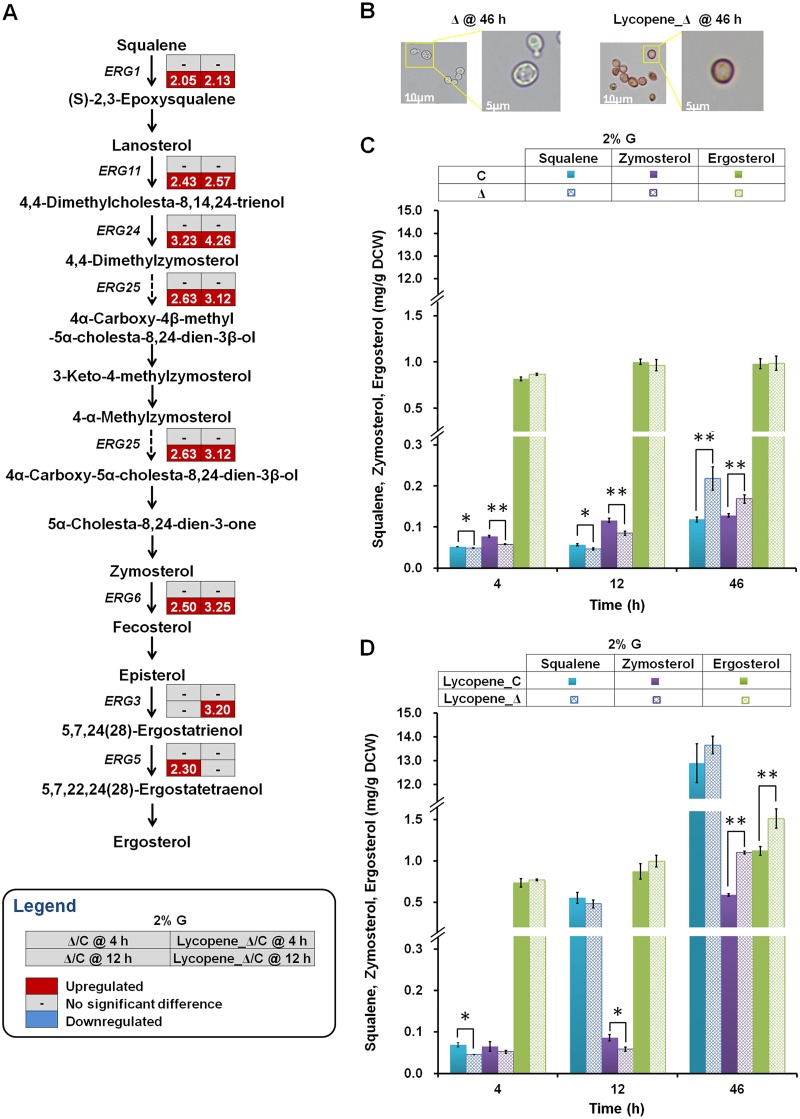
Analysis of the effect of ΔYPL062W on cytomembrane composition.
Since most terpenoid compounds are hydrophobic, they tend to accumulate in the lipophilic cytomembrane and subsequently elicit membrane stress (32, 33). As shown in Fig. 4B, lycopene accumulates in the cell membrane. A high concentration of lycopene increases membrane fluidity and disturbs membrane packing (34, 35). In S. cerevisiae, heterologous carotenoid accumulation leads to reduced levels of intracellular fatty acids and ergosterol. In order to adapt to the stresses, yeast cells change their membrane composition or structure (36–38). In this respect, we hypothesized that the deletion of YPL062W would affect cytomembrane composition. To test the above hypothesis, the expression levels of genes in the membrane biosynthesis pathway as well as the compositions of membrane components were analyzed in host strains (the control strain and the ΔYPL062W strain) and lycopene-producing strains (Lycopene_C and Lycopene_Δ).
FIG 4.
The effect of ΔYPL062W on ergosterol biosynthesis. (A) Transcriptional profiles of ergosterol biosynthesis genes in host strains (SyBE_Sc14C02, C) and (SyBE_Sc14C10, Δ) and lycopene-producing strains (SyBE_Sc14C07, Lycopene_C) and (SyBE_Sc14C23, Lycopene_Δ) on 2% (mass/vol) glucose. Transcriptional data are boxed next to each gene. The time reading after the “@” symbol in the legend indicates the sampling time within the glucose consumption phase (i.e., 4 h) or the ethanol consumption phase (i.e., 12 h). The relative transcription level for each gene is denoted as Lycopene_Δ/C or Δ/C, which are the ratios of the transcription levels in the ΔYPL062W strain to that in the no-deletion control strain for either lycopene-producing recombinant strains or nonproducing parental strains. Genes that are significantly upregulated and downregulated and genes without significant transcriptional differences are colored red, blue, and gray, respectively. (B) Visual microscopic analysis of ΔYPL062W strains with/without lycopene synthesis (Lycopene_Δ and Δ). Cells were cultured under 2% (mass/vol) glucose for 46 h. The contents of squalene, zymosterol, and ergosterol in (C) host strains (C and Δ) and (D) lycopene-producing strains (Lycopene_C and Lycopene_Δ) cultured on 2% (mass/vol) glucose are quantified from experimental triplicates.
In S. cerevisiae, ergosterol and zymosterol constitute the predominant sterols in cell membrane. When YPL062W is knocked out, most of genes involved in ergosterol biosynthesis (ERG1, ERG11, ERG24, ERG25, ERG6, ERG3, and ERG5) are upregulated during the ethanol consumption phase (Fig. 4A). On 2% (mass/vol) glucose fermentation condition, the levels of precursor squalene and zymosterol in the ΔYPL062W strains are significantly lower than those in the control strains at 4 h and 12 h, respectively (Fig. 4C and D). Conversely, ΔYPL062W causes significant increases in squalene and zymosterol contents (by 83.9% and 30.9%, respectively) at 46 h in noncarotenogenic strains (Fig. 4C). No significant difference in ergosterol content was observed in noncarotegenic strains during the whole fermentation process (Fig. 4C).
In contrast, in the lycopene-producing ΔYPL062W strains significant increases zymosterol and ergosterol contents (by 86.7% and 34.5%, respectively) at 46 h of fermentation, while the effect on squalene content was negligible (Fig. 4D). The large amounts of squalene in lycopene-producing strains (Fig. 4D) results from the overexpression of a deregulated tHMG1, the main bottleneck of the early ergosterol pathway (39, 40). These results suggested that YPL062W deletion enhances the intracellular sterol levels during the ethanol consumption phase, which could increase tolerance to these hydrophobic molecules (i.e., lycopene). Indeed, the intracellular ergosterol content has been proven to be correlated with d-limonene tolerance in S. cerevisiae (41).
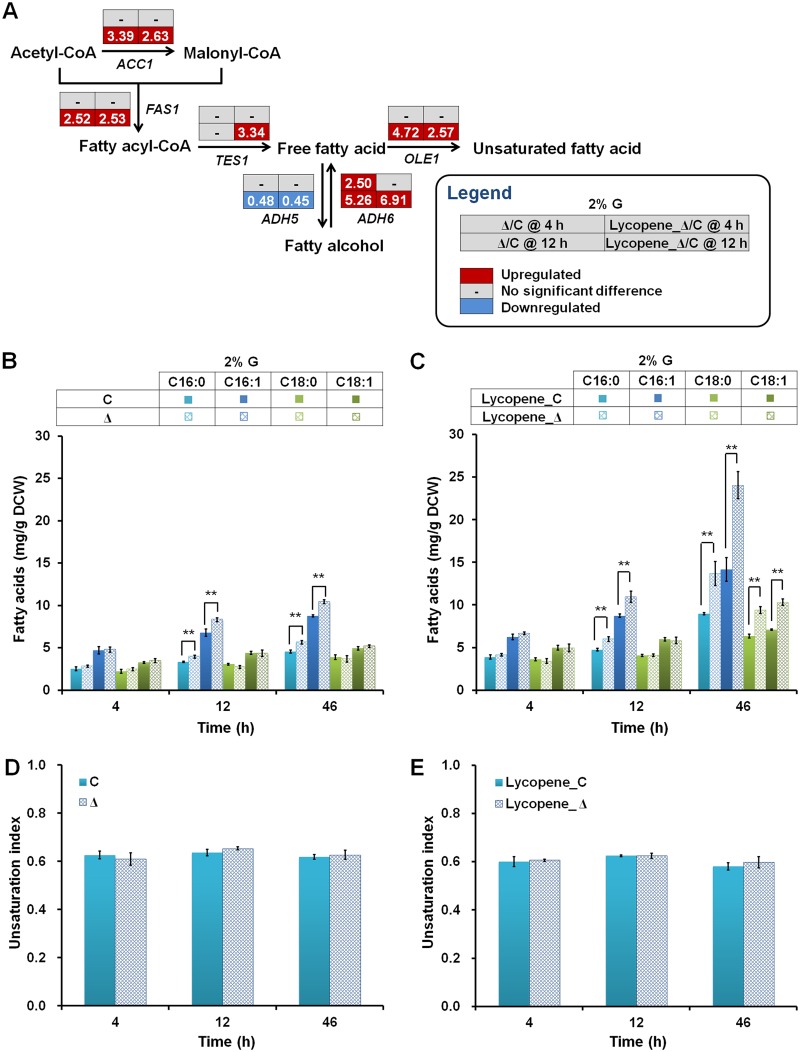
In addition to sterols, fatty acids are key components of the cytomembrane, serving as the backbone of the lipid bilayer. The majority of genes associated with fatty acid biosynthesis (ACC1, FAS1, TES1, ADH6, and OLE1) are transcriptionally upregulated in the YPL062W deletion strain (Fig. 5A). The contents of C16:0 and C16:1 (at 46 h) in the noncarotenogenic ΔYPL062W strain are significantly increased by 21.3% and no significant difference is detected in C18 fatty acid content (Fig. 5B). Total fatty acid contents in ΔYPL062W strains are significantly increased relative to those in control strains (Fig. 5B and C), consistent with upregulation of the fatty acid biosynthesis pathway by ΔYPL062W.
FIG 5.
The effect of ΔYPL062W on fatty acid biosynthesis. (A) Transcriptional profiles of fatty acid biosynthesis genes in host strains control (C, SyBE_Sc14C02) and ΔYPL062W (Δ, SyBE_Sc14C10) and corresponding lycopene-producing strains (Lycopene_C, SyBE_Sc14C07 and Lycopene_Δ, SyBE_Sc14C23) cultured on 2% (mass/vol) glucose. Transcriptional data are boxed next to each gene. The time reading after the “@” symbol in the legend indicates the sampling time within the glucose consumption phase (i.e., 4 h) or the ethanol consumption phase (i.e., 12 h). The relative transcription level for each gene is denoted as Lycopene_Δ/C or Δ/C, which are the ratios of the transcription levels in the ΔYPL062W strain to that in the no-deletion control strain for either lycopene-producing recombinant strains or nonproducing parental strains. Genes that are significantly upregulated and downregulated and genes without significant transcriptional differences are colored red, blue, and gray, respectively. (B and C) Fatty acid contents in host strains (C and Δ [B]) and lycopene-producing strains (Lycopene_C and Lycopene_Δ [C]) cultured on 2% (mass/vol) glucose. are quantified from experimental triplicates. (D and E) Unsaturation index of host strains (C and Δ [D]) and lycopene-producing strains (C and Δ [E]) cultured on 2% (mass/vol) glucose. Unsaturation index was calculated as the ratio of unsaturated fatty acid content to that of total fatty acids.
As for lycopene-producing strains, recovery of the TRP auxotroph gave a significant rise to the overall contents of fatty acids relative to the TRP− host strains (42), as illustrated in Fig. 5B and C. Moreover, the abundances of C18:0 and C18:1 were significantly increased (46 h, increased by 8.4%) by ΔYPL062W as well as C16:0 and C16:1 (46 h, increased by 63.8%) (Fig. 5C). Exogenous unsaturated fatty acids (i.e., oleic acid and linoleic acid) would help to enhance carotenoid yield by preventing against the decreased membrane fluidity caused by carotenoid accumulation (36, 43). However, the unsaturation index, a key factor evaluating yeast membrane fluidity outside of ergosterol content, remained unchanged in all test strains (Fig. 5D and E).
Redefinition and reconstitution of YPL062W.
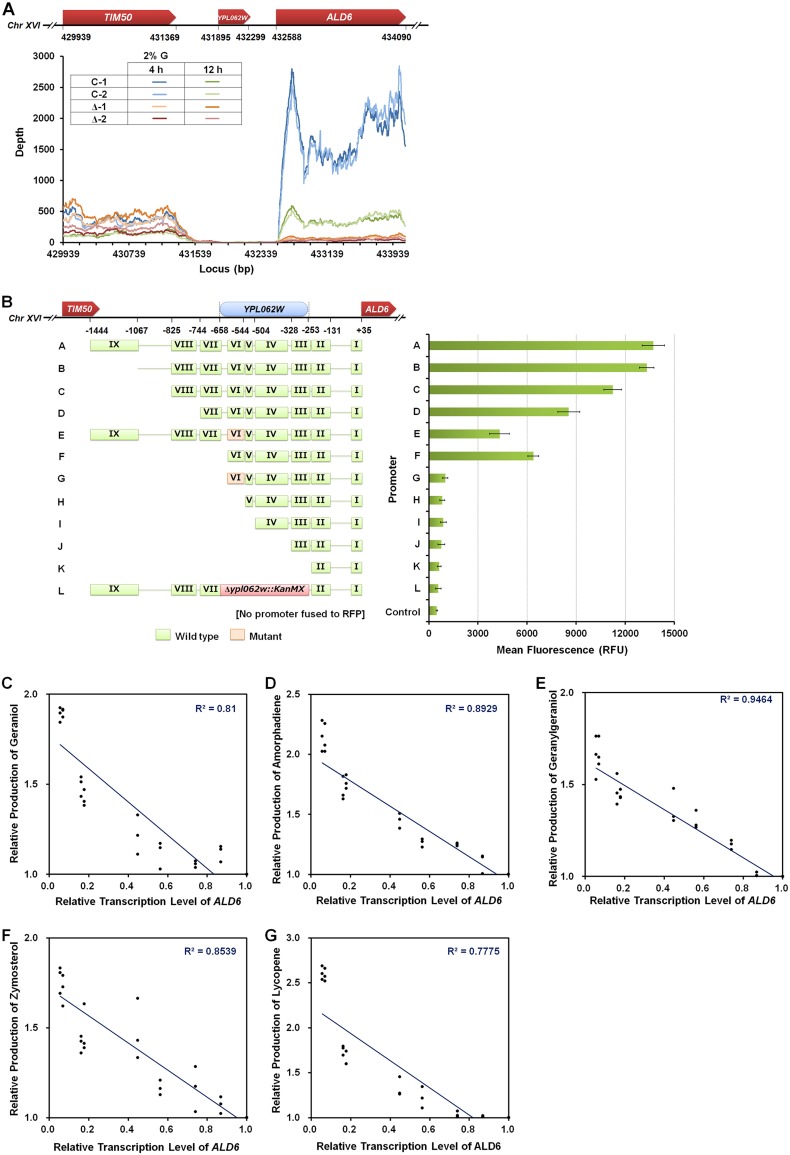
Previous annotation claimed YPL062W as a likely open reading frame (ORF) related to glycogen metabolism and MVA formation (12–14). However, our RNA-seq data show that YPL062W is not actively transcribed (Fig. 6A). Moreover, it was found that the deletion of YPL062W decreases the transcription level of its downstream gene ALD6 without influencing the upstream gene TIM50 (Fig. 6A). When the upstream region of ALD6 (from nucleotide [nt] +35 to −1444 relative to the transcription start site of gene ALD6, Fig. 6B) harboring promoter A (PA; full-length ALD6 promoter) or promoter L (PL; PA with YPL062W deleted) was fused with RFP, it was found that the activity of PL was only 4.2% of PA activity (Fig. 6B), demonstrating that YPL062W is part of the ALD6 promoter and is involved in regulation of ALD6 transcription.
FIG 6.
Redefinition and reconstitution of YPL062W. (A) YPL062W is not expressed, and its deletion negatively impacts ALD6 expression. Transcriptional profiles of Chr XVI (bp 429929 to 434090). Schematic diagram of genome structure spanning YPL062W locus (Chr XVI [bp 429929 to 434090]). Transcriptional profile of gene contexts (from TIM50 through YPL062W to ALD6) in host strains (SyBE_Sc14C02, C and SyBE_Sc14C10, Δ) on 2% (mass/vol) glucose. Cells were sampled at 4 h and 12 h. (B) Characterization of YPL062W as the core promoter driving transcription of ALD6. Various constructs (PA to PL) containing all or part of the sequence spanning the YPL062W locus were fused to RFP, and their activities were measured by relative fluorescence intensities at 48 h of cultivation on 2% (mass/vol) glucose. PA is a full-length promoter. PB to PD, PF, and PH toPK each contain increasingly larger truncations of the conserved region. PE and PG contain transversion mutations in conserved domain VI (marked in orange). The null-expression control PL has YPL062W entirely replaced by a KanMX cassette. The correlation between the transcription levels of ALD6 and terpenoid production titers. Geraniol (monoterpenoid) (C), amorphadiene (sesquiterpenoid) (D), geranylgeraniol (diterpenoid) (E), zymosterol (triterpenoid) (F), and lycopene (tetraterpenoid) (G) were selected to test the correlation between ALD6 transcription levels and terpenoid production. The relative production for each terpenoid was determined as the ratio of product titer in each strain harboring a truncated conserved region to that of the control strain containing and intact PA region. The relative transcription level of ALD6 was determined as the ratio of the transcription level of ALD6 controlled by corresponding truncated promoter to that of ALD6 regulated by the intact PA region.
To characterize the role of YPL062W, we performed serial deletion of the upstream region of ALD6 in the control strain SyBE_Sc14C02, generating strains SyBE_Sc14C96 through SyBE_Sc14C105 (Table 1). Seven conserved domains (IX to III) of ALD6 promoter predicted by evolution conservation (Fig. S3) were serially deleted (obtaining promoters PB to PK) and fused with RFP to identify the core promoter of ALD6 (at 48 h of cultivation in Fig. S4). As illustrated in Fig. 6B, increasing deletions of domains VIII, VII, and VI from the corresponding promoters C (VIII to I), D (VII to I), and F (VI to I) dramatically reduced promoter activities, leaving merely 62.3% (obtaining promoter D), 46.5% (obtaining promoter F), and 6% (obtaining promoter H, V to I) of the full-length promoter (PA, domains I to IX) activity, respectively. Further deletions of the remaining domains (V to III) in PH to PJ exhibit no additional significant decrease in promoter activity (Fig. 6B). These results indicate that domains VIII, VII, and VI form the core promoter of ALD6.
TABLE 1.
S. cerevisiae strains used in this study
| Strain name | Description | Source or reference |
|---|---|---|
| CEN.PK2-1C | MATa ura3-52 trp1-289 leu2-3,112 his3Δ1 MAL2-8C SUC2 | EUROSCARF |
| SyBE_Sc14C02 | CEN.PK2-1C Δgal1 Δgal7 Δgal10::HIS3 | 15 |
| SyBE_Sc14C10 | CEN.PK2-1C Δgal1 Δgal7 Δgal10::HIS3 Δypl062w::KanMX | 15 |
| SyBE_Sc14C07 | SyBE_Sc14C02 trp1::TRP1_TCYC1-BtCrtI-PGAL10-PGAL1-PaCrtB-TPGK1 leu2::LEU2_TACT1-tHMG1-PGAL10-PGAL1-PaCrtE-TGPM1 | 15 |
| SyBE_Sc14C23 | SyBE_Sc14C10 trp1::TRP1_TCYC1-BtCrtI-PGAL10-PGAL1-PaCrtB-TPGK1 leu2::LEU2_TACT1-tHMG1-PGAL10-PGAL1-PaCrtE-TGPM1 | 15 |
| SyBE_Sc14C71_Tri | SyBE_Sc14C02 leu2::LEU2_TACT1-tHMG1-PGAL10 | This study |
| SyBE_Sc14C72_Tri | SyBE_Sc14C10 leu2::LEU2_TACT1-tHMG1-PGAL10 | This study |
| SyBE_Sc14C73_Mono | SyBE_Sc14C02 trp1::TRP1_PGAL1-GES-TPGK1 leu2::LEU2_TACT1-tHMG1-PGAL10 | This study |
| SyBE_Sc14C74_Mono | SyBE_Sc14C10 trp1::TRP1_PGAL1-GES-TPGK1 leu2::LEU2_TACT1-tHMG1-PGAL10 | This study |
| SyBE_Sc14C75_Sesqui | SyBE_Sc14C02 trp1::TRP1_PGAL1-ADS-TPGK1 leu2::LEU2_TACT1-tHMG1-PGAL10 | This study |
| SyBE_Sc14C76_Sesqui | SyBE_Sc14C10 trp1::TRP1_PGAL1-ADS-TPGK1 leu2::LEU2_TACT1-tHMG1-PGAL10 | This study |
| SyBE_Sc14C77_Di | SyBE_Sc14C02 leu2::LEU2_TACT1-tHMG1-PGAL10-PGAL1-TmGGPPS-TGPM1 | This study |
| SyBE_Sc14C78_Di | SyBE_Sc14C10 leu2::LEU2_TACT1-tHMG1-PGAL10-PGAL1-TmGGPPS-TGPM1 | This study |
| SyBE_Sc14C80 | SyBE_Sc14C02 leu2::LEU2_TCYC1-RFP | This study |
| SyBE_Sc14C81 | SyBE_Sc14C02 leu2::LEU2_TCYC1-RFP-PGAL1 | This study |
| SyBE_Sc14C82 | SyBE_Sc14C10 leu2::LEU2_TCYC1-RFP-PGAL1 | This study |
| SyBE_Sc14C83 | SyBE_Sc14C02 leu2::LEU2_TCYC1-RFP-PGAL10 | This study |
| SyBE_Sc14C43 | SyBE_Sc14C10 leu2::LEU2_TCYC1-RFP-PGAL10 | 15 |
| SyBE_Sc14C84 | SyBE_Sc14C02 leu2::LEU2_TCYC1-RFP-PA (−1444_+35) | This study |
| SyBE_Sc14C85 | SyBE_Sc14C02 leu2::LEU2_TCYC1-RFP-PB (−1067_+35) | This study |
| SyBE_Sc14C86 | SyBE_Sc14C02 leu2::LEU2_TCYC1-RFP-PC (−825_+35) | This study |
| SyBE_Sc14C87 | SyBE_Sc14C02 leu2::LEU2_TCYC1-RFP-PD (−744_+35) | This study |
| SyBE_Sc14C88 | SyBE_Sc14C02 leu2::LEU2_TCYC1-RFP-PE (−1444_+35, mutVI) | This study |
| SyBE_Sc14C89 | SyBE_Sc14C02 leu2::LEU2_TCYC1-RFP-PF (−658_+35) | This study |
| SyBE_Sc14C90 | SyBE_Sc14C02 leu2::LEU2_TCYC1-RFP-PG (−658_+35, mutVI) | This study |
| SyBE_Sc14C91 | SyBE_Sc14C02 leu2::LEU2_TCYC1-RFP-PH (−544_+35) | This study |
| SyBE_Sc14C92 | SyBE_Sc14C02 leu2::LEU2_TCYC1-RFP-PI (−504_+35) | This study |
| SyBE_Sc14C93 | SyBE_Sc14C02 leu2::LEU2_TCYC1-RFP-PJ (−328_+35) | This study |
| SyBE_Sc14C94 | SyBE_Sc14C02 leu2::LEU2_TCYC1-RFP-PK (−253_+35) | This study |
| SyBE_Sc14C95 | SyBE_Sc14C02 leu2::LEU2_TCYC1-RFP-PL (−1444_+35, Δypl062w::KanMX) | This study |
| SyBE_Sc14C96 | SyBE_Sc14C02 ALD6::KanMX_PB (−1067_+35) | This study |
| SyBE_Sc14C97 | SyBE_Sc14C02 ALD6::KanMX_PC (−825_+35) | This study |
| SyBE_Sc14C98 | SyBE_Sc14C02 ALD6::KanMX_PD (−744_+35) | This study |
| SyBE_Sc14C99 | SyBE_Sc14C02 ALD6::KanMX_PE (−1444_+35, mutVI) | This study |
| SyBE_Sc14C100 | SyBE_Sc14C02 ALD6::KanMX_PF (−658_+35) | This study |
| SyBE_Sc14C101 | SyBE_Sc14C02 ALD6::KanMX_PG (−658_+35, mutVI) | This study |
| SyBE_Sc14C102 | SyBE_Sc14C02 ALD6::KanMX_PH (−544_+35) | This study |
| SyBE_Sc14C103 | SyBE_Sc14C02 ALD6::KanMX_PI (−504_+35) | This study |
| SyBE_Sc14C104 | SyBE_Sc14C02 ALD6::KanMX_PJ (−328_+35) | This study |
| SyBE_Sc14C105 | SyBE_Sc14C02 ALD6::KanMX_PK (−253_+35) | This study |
| SyBE_Sc14C97_Tri | SyBE_Sc14C02 ALD6::KanMX_PC leu2::LEU2_TACT1-tHMG1-PGAL10 | This study |
| SyBE_Sc14C98_Tri | SyBE_Sc14C02 ALD6::KanMX_PD leu2::LEU2_TACT1-tHMG1-PGAL10 | This study |
| SyBE_Sc14C99_Tri | SyBE_Sc14C02 ALD6::KanMX_PE leu2::LEU2_TACT1-tHMG1-PGAL10 | This study |
| SyBE_Sc14C100_Tri | SyBE_Sc14C02 ALD6::KanMX_PF leu2::LEU2_TACT1-tHMG1-PGAL10 | This study |
| SyBE_Sc14C101_Tri | SyBE_Sc14C02 ALD6::KanMX_PG leu2::LEU2_TACT1-tHMG1-PGAL10 | This study |
| SyBE_Sc14C102_Tri | SyBE_Sc14C02 ALD6::KanMX_PH leu2::LEU2_TACT1-tHMG1-PGAL10 | This study |
| SyBE_Sc14C105_Tri | SyBE_Sc14C02 ALD6::KanMX_PK leu2::LEU2_TACT1-tHMG1-PGAL10 | This study |
| SyBE_Sc14C97_Mono | SyBE_Sc14C02 ALD6::KanMX_PC trp1::TRP1_PGAL1-GES-TPGK1 leu2::LEU2_TACT1-tHMG1-PGAL10 | This study |
| SyBE_Sc14C98_Mono | SyBE_Sc14C02 ALD6::KanMX_PD trp1::TRP1_PGAL1-GES-TPGK1 leu2::LEU2_TACT1-tHMG1-PGAL10 | This study |
| SyBE_Sc14C99_Mono | SyBE_Sc14C02 ALD6::KanMX_PE trp1::TRP1_PGAL1-GES-TPGK1 leu2::LEU2_TACT1-tHMG1-PGAL10 | This study |
| SyBE_Sc14C100_Mono | SyBE_Sc14C02, ALD6::KanMX_PF trp1::TRP1_PGAL1-GES-TPGK1 leu2::LEU2_TACT1-tHMG1-PGAL10 | This study |
| SyBE_Sc14C101_Mono | SyBE_Sc14C02 ALD6::KanMX_PG, trp1::TRP1_PGAL1-GES-TPGK1 leu2::LEU2_TACT1-tHMG1-PGAL10 | This study |
| SyBE_Sc14C102_Mono | SyBE_Sc14C02 ALD6::KanMX_PH trp1::TRP1_PGAL1-GES-TPGK1 leu2::LEU2_TACT1-tHMG1-PGAL10 | This study |
| SyBE_Sc14C105_Mono | SyBE_Sc14C02 ALD6::KanMX_PK trp1::TRP1_PGAL1-GES-TPGK1 leu2::LEU2_TACT1-tHMG1-PGAL10 | This study |
| SyBE_Sc14C97_Sesqui | SyBE_Sc14C02 ALD6::KanMX_PC trp1::TRP1_PGAL1-ADS-TPGK1 leu2::LEU2_TACT1-tHMG1-PGAL10 | This study |
| SyBE_Sc14C98_Sesqui | SyBE_Sc14C02 ALD6::KanMX_PD trp1::TRP1_PGAL1-ADS-TPGK1 leu2::LEU2_TACT1-tHMG1-PGAL10 | This study |
| SyBE_Sc14C99_Sesqui | SyBE_Sc14C02 ALD6::KanMX_PE trp1::TRP1_PGAL1-ADS-TPGK1 leu2::LEU2_TACT1-tHMG1-PGAL10 | This study |
| SyBE_Sc14C100_Sesqui | SyBE_Sc14C02 ALD6::KanMX_PF, trp1::TRP1_PGAL1-ADS-TPGK1 leu2::LEU2_TACT1-tHMG1-PGAL10 | This study |
| SyBE_Sc14C101_Sesqui | SyBE_Sc14C02 ALD6::KanMX_PG trp1::TRP1_PGAL1-ADS-TPGK1 leu2::LEU2_TACT1-tHMG1-PGAL10 | This study |
| SyBE_Sc14C102_Sesqui | SyBE_Sc14C02 ALD6::KanMX_PH, trp1::TRP1_PGAL1-ADS-TPGK1 leu2::LEU2_TACT1-tHMG1-PGAL10 | This study |
| SyBE_Sc14C105_Sesqui | SyBE_Sc14C02 ALD6::KanMX_PK trp1::TRP1_PGAL1-ADS-TPGK1 leu2::LEU2_TACT1-tHMG1-PGAL10 | This study |
| SyBE_Sc14C97_Di | SyBE_Sc14C02 ALD6::KanMX_PC leu2::LEU2_TACT1-tHMG1-PGAL10-PGAL1-TmCrtE-TGPM1 | This study |
| SyBE_Sc14C98_Di | SyBE_Sc14C02 ALD6::KanMX_PD leu2::LEU2_TACT1-tHMG1-PGAL10-PGAL1-TmCrtE-TGPM1 | This study |
| SyBE_Sc14C99_Di | SyBE_Sc14C02 ALD6::KanMX_PE leu2::LEU2_TACT1-tHMG1-PGAL10-PGAL1-TmCrtE-TGPM1 | This study |
| SyBE_Sc14C100_Di | SyBE_Sc14C02 ALD6::KanMX_PF leu2::LEU2_TACT1-tHMG1-PGAL10-PGAL1-TmCrtE-TGPM1 | This study |
| SyBE_Sc14C101_Di | SyBE_Sc14C02 ALD6::KanMX_PG leu2::LEU2_TACT1-tHMG1-PGAL10-PGAL1-TmCrtE-TGPM1 | This study |
| SyBE_Sc14C102_Di | SyBE_Sc14C02 ALD6::KanMX_PH leu2::LEU2_TACT1-tHMG1-PGAL10-PGAL1-TmCrtE-TGPM1 | This study |
| SyBE_Sc14C105_Di | SyBE_Sc14C02 ALD6::KanMX_PK leu2::LEU2_TACT1-tHMG1-PGAL10-PGAL1-TmCrtE-TGPM1 | This study |
| SyBE_Sc14C97_Tetra | SyBE_Sc14C02 ALD6::KanMX_PC trp1::TRP1_TCYC1-BtCrtI-PGAL10-PGAL1-PaCrtB-TPGK1 leu2::LEU2_TACT1-tHMG1-PGAL10-PGAL1-PaCrtE-TGPM1 | This study |
| SyBE_Sc14C98_Tetra | SyBE_Sc14C02 ALD6::KanMX_PD trp1::TRP1_TCYC1-BtCrtI-PGAL10-PGAL1-PaCrtB-TPGK1 leu2::LEU2_TACT1-tHMG1-PGAL10-PGAL1-PaCrtE-TGPM1 | This study |
| SyBE_Sc14C99_Tetra | SyBE_Sc14C02 ALD6::KanMX_PE trp1::TRP1_TCYC1-BtCrtI-PGAL10-PGAL1-PaCrtB-TPGK1 leu2::LEU2_TACT1-tHMG1-PGAL10-PGAL1-PaCrtE-TGPM1 | This study |
| SyBE_Sc14C100_Tetra | SyBE_Sc14C02 ALD6::KanMX_PF trp1::TRP1_TCYC1-BtCrtI-PGAL10-PGAL1-PaCrtB-TPGK1 leu2::LEU2_TACT1-tHMG1-PGAL10-PGAL1-PaCrtE-TGPM1 | This study |
| SyBE_Sc14C101_Tetra | SyBE_Sc14C02 ALD6::KanMX_PG trp1::TRP1_TCYC1-BtCrtI-PGAL10-PGAL1-PaCrtB-TPGK1 leu2::LEU2_TACT1-tHMG1-PGAL10-PGAL1-PaCrtE-TGPM1 | This study |
| SyBE_Sc14C102_Tetra | SyBE_Sc14C02 ALD6::KanMX_PH trp1::TRP1_TCYC1-BtCrtI-PGAL10-PGAL1-PaCrtB-TPGK1 leu2::LEU2_TACT1-tHMG1-PGAL10-PGAL1-PaCrtE-TGPM1 | This study |
| SyBE_Sc14C105_Tetra | SyBE_Sc14C02 ALD6::KanMX_PK trp1::TRP1_TCYC1-BtCrtI-PGAL10-PGAL1-PaCrtB-TPGK1 leu2::LEU2_TACT1-tHMG1-PGAL10-PGAL1-PaCrtE-TGPM1 | This study |
Domain VI (nt −599 to −545 relative to transcription start site) is the only region within the ALD6 core promoter overlapping YPL062W (which covers domains III to VI). To further study the role of this domain, mutations were introduced into PA and PF at domain VI (Fig. 6B). To avoid structural changes to the promoter region, transversion mutations interchanging either two purine nucleotides (A and G) or two pyrimidine nucleotides (C and T) were made according to the method of Makino K et al. (44). Mutation of domain VI within PA (generating PE) decreased promoter activity by 68.7%, an effect roughly equivalent to deletion of domain VI from PF (Fig. 6B). Therefore, domain VI exhibits the strongest ALD6 promoter activity among the nine conserved domains and is the most important region for promoter activity within YPL062W. Since we did not observe transcription of YPL062W, we posit that YPL062W is part of the core promoter regulating the transcription of ALD6.
This observed connection between YPL062W deletion, decreased ALD6 expression, and increased terpenoid production prompted us to explore the correlation between ALD6 transcription and terpenoid production in S. cerevisiae. ALD6 expression was measured by RT-PCR (Fig. S5) in the control and serial deletion strains corresponding to PA through PL. The C10, C15, C20, C30, and C40 terpenoid biosynthesis pathways were introduced into these control and serial deletion strains, and production of the respective terpenoid was quantified (Fig. S5). As shown in Fig. 6C to G, the transcription level of ALD6 is inversely correlated to terpenoid production across all terpenoid classes.
DISCUSSION
Various nonessential genes can impact terpene production in S. cerevisiae in unique and orthogonal ways (12). For example, ΔKEX1, ΔLAC1, ΔSOL1, and ΔYPK9 increase carotenoid production while decreasing bisabolene production, while YPL062W deletion enhances both carotenoid and bisabolene production (12). In this study, it was further demonstrated that ΔYPL062W increases the productivity of geraniol (monoterpenoid), amorphadiene (sesquiterpenoid), geranylgeraniol (diterpenoid), and zymosterol (triterpenoid), as well as lycopene (tetraterpenoid) (Fig. 1), suggesting that ΔYPL062W improves terpenoid production regardless of the class of product. Moreover, deletion of YPL062W does not impair cell growth on either glucose or ethanol as the sole carbon source (Fig. S2). These characteristics make ΔYPL062W a promising and powerful engineering target for the creation of terpenoid-producing S. cerevisiae strains for commercial production.
Although computational and reverse metabolic engineering approaches often lead to improved gene deletion mutants for cell factory engineering (7), the systems level effects of such gene deletions on production phenotypes have not always been extensively studied. Understanding the genetic and molecular function of such gene alterations regarding production will minimize the risks inherent in the development of large-scale fermentation processes, a daunting challenge in the field of industrial biotechnology (45). In a study by Ferreira et al., the creation of multiple deletion mutants enabled the study of fatty acid dynamics in lipid metabolism and generated a platform strain with interesting properties that provided insights into the future development of lipid-related cell factories (46). In an analogous manner, this study established a detailed experimental and systems biology approach to uncover the molecular mechanisms of a key gene deletion (ΔYPL062W) in metabolically engineered yeast. Through transcriptome analysis, it was concluded that (i) the ΔYPL062W strain enhanced pathway expression through upregulated mitochondrial PDH bypass and fatty acid β-oxidation pathways contributing to intracellular acetyl-CoA levels (Fig. 2), upregulation of the MVA pathway for an increased flux from acetyl-CoA to MVA (Fig. 2B), and enhanced expression of heterologous terpenoid genes (Fig. 3); (ii) upregulation of the TCA cycle and glyoxylate shunt increasing the energy resources available for terpenoid biosynthesis (Fig. 2); and (iii) upregulation of fatty acid and ergosterol biosynthesis pathways increased the abundance of fatty acids and sterols (Fig. 4), potentially reflecting a wider dynamic range of membrane composition for better tolerance to terpenoid accumulation stress (36, 41, 43). It is known that adequate precursor and energy supply, high efficiency of the heterologous expression system, and a compatible microenvironment for target compound accumulation are important for maximizing production of natural products in microbes (47). Surprisingly, all of these characteristics critical for highly efficient cell factories are affected by the deletion of YPL062W.
Detailed analysis of YPL062W expression revealed that YPL062W is not transcribed and that its deletion significantly downregulates the downstream gene ALD6 (Fig. 6A). ALD6 contributes to the primary cytosolic acetaldehyde dehydrogenase activity, and cells lacking ALD6 produce less acetic acid during fermentation (16, 17). In this study, deletion of YPL062W directly downregulated ALD6 expression, leading to reduced acetate accumulation. Serial deletions of conserved domains within ALD6 promoter revealed that YPL062W (containing domain VI) acted as a core promoter of ALD6 (Fig. 6B). Although the evolution prediction based on UCSC Genome Browser did not suggest any potential cis-elements binding within domain VI, Walkey et al. (48) reported that the sequence from nt −551 to −556 (GAGGGG) within this region was the binding site of a zinc finger transcriptional activator YML081W. Mutation of this GAGGGG site led to 54% reduction in ALD6 promoter activity (48), which corresponds well with the 68.7% decrease resulting from the base transition mutation of domain VI in our study (Fig. 6B). Therefore, ΔYPL062W might function by disrupting the interaction between YML081W and the ALD6 promoter.
The expression level of ALD6 was negatively correlated to terpenoid production, and this correlation was independent of terpenoid class (Fig. 6C to G). Therefore, the regulation of ALD6 by YPL062W acts as a crucial control element for terpenoid production. The genetic function of YPL062W explains the correlation between YPL062W and terpenoid biosynthesis and provides a powerful engineering platform for yeast terpenoid biosynthesis in general by using ΔYPL062W strains.
This study emphasizes that detailed characterization of genetic alterations is a reliable way to generate knowledge and understanding of the genetic, physiological, metabolic, and phenotypic effects of strain modifications. This understanding and the knowledge that results from it enable more effective and efficient engineering of commercial production strains and minimize the risks associated with commercial strain, process, and scale-up development.
MATERIALS AND METHODS
Strains, media, and culture conditions.
S. cerevisiae SyBE_Sc14C02 and SyBE_Sc14C10 (15), derived from CEN.PK2-1C, were created as the host strains for this work. All yeast strains engineered in this study are listed in Table 1. E. coli DH5α was used for routine cloning procedures. Yeast strains for normal cultivations were cultured in yeast extract-peptone-dextrose (YPD) medium (15) or YPE medium (1% [mass/vol] yeast extract, 2% [mass/vol] peptone, and 2% [vol/vol] ethanol). Synthetic complete (SC) medium (15) was used for yeast recombinant selection. Shake flask batch fermentations for terpenoid production were carried out in YPDG medium according to our previous work (15). The cultivation was maintained for 48 h under 2% (mass/vol) glucose or 60 h under 4% (mass/vol) glucose. For two-phase fermentation, isopropyl myristate (IPM) was added to YPDG medium at a final concentration of 20% (vol/vol).
DNA manipulation.
Integration modules used for yeast homologous recombination were constructed as described previously (15) and performed according to Fig. S1. Oligonucleotides used in this study are listed in Table 2. The genes encoding geraniol synthase (GES) originating from Catharanthus roseus and amorphadiene synthase (ADS) from Artemisia annua were custom-synthesized by Genewiz (Beijing, China) for optimal expression in S. cerevisiae (Fig. S6). The genes encoding geranylgeranyl diphosphate synthase (GGPPS) from Taxus x media, CrtE from Pantoea agglomerans, and Blakeslea trispora-derived CrtB and CrtI were synthesized in our last work (15). Terpenogenic modules included geraniol biosynthetic modules (modules 1 and 4 [Fig. S1]), amorphadiene biosynthetic modules (modules 2 and 4 [Fig. S1]), geranylgeraniol biosynthetic modules (module 5 [Fig. S1]), zymosterol biosynthetic modules (module 4 [Fig. S1]), and lycopene biosynthetic modules (modules 3 and 5 [Fig. S1]).
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide purpose and name | Sequence (5′–3′) |
|---|---|
| For construction of PGAL1-GES/ADS-TPGK1, TRP1 homologous arm | |
| TPR1_LF | GTTTAAACGGAAGAGGAGTAGGGAA |
| TPR1_LR | TACGATGCTGTTCTATTAAATGCT |
| GAL1p_F | AGCATTTAATAGAACAGCATCGTAAGTACGGATTAGAAGCCGC |
| GAL1p_R | TATAGTTTTTTCTCCTTGACGTTA |
| GES_F | TAACGTCAAGGAGAAAAAACTATAATGTCATTACCATTGGCTACACC |
| GES_R | CTATCGATTTCAATTCAATTCAATTTAGAAACAAGGTGTGAAAAATAAAGC |
| ADS_F | TAACGTCAAGGAGAAAAAACTATAATGTCTTTGACTGAAGAAAAGCC |
| ADS_R | CTATCGATTTCAATTCAATTCAATTTAGATAGACATTGGGTAAACCAAC |
| PGK1t_F | ATTGAATTGAATTGAAATCGATAG |
| PGK1t_R | CGTCATAACTGCAAAGTACACATATATAACGAACGCAGAATTTTCG |
| TPR1_RF | ATATATGTGTACTTTGCAGTTATGACG |
| TPR1_RR | GTTTAAACACGCCAACCAAGTATTT |
| PCR verification of PGAL1-GES/ADS-TPGK1, TRP1 homologous arm | |
| TPR1_VF | AGACATGGAGGGCGTTATTA |
| GAL1p_VR | CTTTATTGTTCGGAGCAGTG |
| GAL1p_VF | TGCGTCCTCGTCTTCACCG |
| TPR1_VR | AGTTTGATTCCATTGCGGT |
| For construction of TACT1-tHMG1-PGAL10, LEU2 homologous arm with LEU2 marker | |
| LEU2_LF | GTTTAAACATAACGAGAACACACAGGG |
| LEU2_LR | ATCATTAAAGTAACTTAAGGAGTTAAATTTAAGCAAGGATTTTCTTAACTTC |
| TDH2t_F | ATTTAACTCCTTAAGTTACTTTAATGAT |
| TDH2t_R | GCGAAAAGCCAATTAGTGT |
| ACT1t_F | TCTCTGCTTTTGTGCGC |
| ACT1t_R | ACACTAATTGGCTTTTCGCTACACGGTCCAATGGATAAAC |
| tHMG1_F | GTAAGAATTTTTGAAAATTCAATATAAATGGTTTTAACCAATAAAACAGTC |
| tHMG1_R | GCGCACAAAAGCAGAGATTAGGATTTAATGCAGGTGAC |
| GAL10p_F | TTATATTGAATTTTCAAAAATTCTTAC |
| GAL10p_R | CAAATATCATAAAAAAAGAGAATCTTTAGTGGTTATGCAGCTTTTCCA |
| LEU2_RF | AAAGATTCTCTTTTTTTATGATATTTG |
| LEU2_RR | GTTTAAACTCCATCAAATGGTCAGG |
| PCR verification of TACT1-tHMG1-PGAL10, LEU2 homologous arm with LEU2 marker | |
| LEU2_VF | GGAATACTCAGGTATCGTAAGATGC |
| GAL10p_VR | CTTTATTGTTCGGAGCAGTG |
| GAL10p_VF | CGCTTAACTGCTCATTGCTAT |
| LEU2_VR | CGTTAAGGCCGTTTCTGACA |
| For construction of PGAL1/GAL10/PA-L-RFP-TCYC1, LEU2 homologous arm with LEU2 marker | |
| LEU2_LF | GTTTAAACATAACGAGAACACACAGGG |
| TDH2t_R | GCGAAAAGCCAATTAGTGT |
| CYC1t_RFP_F | CTGGTGGTATGGATGAATTATATAAATAAGGCCGCATCATGTAATTAGTT |
| CYC1t_LEU_R | ACACTAATTGGCTTTTCGCGCAAATTAAAGCCTTCGAGC |
| RFP_F | ATGGTTTCAAAAGGTGAAGAAGAT |
| RFP_R | TTATTTATATAATTCATCCATACCACCAG |
| GAL1p_F | CAAATATCATAAAAAAAGAGAATCTTTAGTACGGATTAGAAGCCGCC |
| GAL1p_R | ATCTTCTTCACCTTTTGAAACCATTATAGTTTTTTCTCCTTGACGTTAAAG |
| GAL10p_F | CAAATATCATAAAAAAAGAGAATCTTTAGTGGTTATGCAGCTTTTCCA |
| GAL10p_R | ATCTTCTTCACCTTTTGAAACCATTTATATTGAATTTTCAAAAATTCTTACTT |
| A/E/L_F | CAAATATCATAAAAAAAGAGAATCTTTACAAAAGAACTATTTAATGTTCATGAAG |
| B_F | CAAATATCATAAAAAAAGAGAATCTTTCTACCACTGCACCTCCTAACAT |
| C_F | CAAATATCATAAAAAAAGAGAATCTTTGAATATAAGGCCGCCGCC |
| D_F | CAAATATCATAAAAAAAGAGAATCTTTACTTTCCGCGGACGCTAA |
| F/G_F | CAAATATCATAAAAAAAGAGAATCTTTATGATAGAATTGGATTATGTAAAAGGTG |
| H_F | CAAATATCATAAAAAAAGAGAATCTTTCTGTTTTTCGACATAAATGAGGG |
| I_F | CAAATATCATAAAAAAAGAGAATCTTTACGTCATTGTTGCATATGGC |
| J_F | CAAATATCATAAAAAAAGAGAATCTTTACCGTTTTGGGCATCGGG |
| K_F | CAAATATCATAAAAAAAGAGAATCTTTCACCGACCATGTGGGCAAA |
| A-L_R | ATCTTCTTCACCTTTTGAAACCATTGTATTCTGATAGTATGTGTTTGTGTATG |
| LEU2_RF | AAAGATTCTCTTTTTTTATGATATTTG |
| LEU2_RR | GTTTAAACTCCATCAAATGGTCAGG |
| PCR verification of PGAL1/GAL10/PA-L-RFP-TCYC1, LEU2 homologous arm with LEU2 marker | |
| LEU2_VF | GGAATACTCAGGTATCGTAAGATGC |
| LEU2_VR | CGTTAAGGCCGTTTCTGACA |
| For construction of PB-K, ALD6 homologous arm with KanMX marker | |
| ALD6_LF | GTTTAAACATTTACGCTGAAAGACTATGTT |
| ALD6_LR | TTAAAATTTAACATAGAAAAAATAAATAGGC |
| KanMX_F | GCCTATTTATTTTTTCTATGTTAAATTTTAATCTGTTTAGCTTGCCTCGTCC |
| KanMX_F | GTTTTCGACACTGGATGGCG |
| B_vivoF | CGCCATCCAGTGTCGAAAACCTACCACTGCACCTCCTAACAT |
| C_vivoF | CGCCATCCAGTGTCGAAAACGAATATAAGGCCGCCGCC |
| D_vivoF | CGCCATCCAGTGTCGAAAACACTTTCCGCGGACGCTAA |
| E_vivoF | CGCCATCCAGTGTCGAAAACACAAAAGAACTATTTAATGTTCATGAAG |
| F/G_vivoF | CGCCATCCAGTGTCGAAAACATGATAGAATTGGATTATGTAAAAGGTG |
| H_vivoF | CGCCATCCAGTGTCGAAAACCTGTTTTTCGACATAAATGAGGG |
| I_vivoF | CGCCATCCAGTGTCGAAAACACGTCATTGTTGCATATGGC |
| J_vivoF | CGCCATCCAGTGTCGAAAACACCGTTTTGGGCATCGGG |
| K_vivoF | CGCCATCCAGTGTCGAAAACCACCGACCATGTGGGCAAA |
| A-L_vivoR | GCAGTGTCAAAGTGTAGCTTAGTCATTGTATTCTGATAGTATGTGTTTGTGTATG |
| ALD6_RF | ATGACTAAGCTACACTTTGACACTGC |
| ALD6_RR | GTTTAAACGGTGTTTTCAGTGGAAGG |
| PCR verification of PB-K, ALD6 homologous arm with KanMX marker | |
| ALD6_VF | CAGCACCAAGTTCCCGCTC |
| ALD6_VR | CTACCCTGACTGGAAGGCGG |
| Primers for real-time PCR | |
| Actin_F | ACCATGTTCCCAGGTATTGC |
| Actin_R | TGGACCACTTTCGTCGTATTC |
| ALD6_F | GAACTTCACCACCTTAGAGCCA |
| ALD6_R | GCAGCGGGTTTCAAGATACA |
| ALG9_F | TAATCCGGGCTGGTTCCATGC |
| ALG9_R | TAGAAGTAGACCCAGTGGACAGATAGCG |
| CrtE_F | TGTTCTCTGCTATGTTGCAGATAGTCGC |
| CrtE_R | TGTTCCTGTCCTTTCCTGTCTCTGG |
| CrtB_F | ACACCTGGTGTAGACATTGCGATG |
| CrtB_R | GGTTAATGCAACTTCTTGGAAAGCGGC |
| CrtI_F | CGAGAGGATAGGAGACCACTTGGAC |
| CrtI_R | ACCTACCGAATCCTAAAGGTCCCTC |
To construct serially deleted ALD6 promoters, the conserved domains of ALD6 promoter (region from nt +35 to −1444 relative to the transcription start site of gene ALD6) were predicted by evolution prediction based on the UCSC Genome Browser (http://genome-asia.ucsc.edu) (49). Serially deleted ALD6 promoters containing different conserved domains were assembled into promoter-activity-characterization modules according to module 6 (Fig. S1) and promoter replacement modules according to module 7 (Fig. S1), respectively. Base transition mutation of conserved domain was introduced through interchanged mutation between either two purine nucleotides (A and G) or two pyrimidine nucleotides (C and T) according to the method of Wang et al. (50) and assembled according to modules 6 and 7 in Fig. S1.
Promoter activity assay.
Promoter activity was characterized by the relative fluorescence intensity of red fluorescent protein (RFP) as previously described (51). The relative fluorescence intensity was the ratio of the fluorescence to optical density at 600 nm (OD600) for each strain during cultivation. The strain SyBE_Sc14C80 without promoter fused to RFP was used as a negative control. Activities of PGAL1 and PGAL10 were determined in the background of strains SyBE_Sc14C02 and SyBE_Sc14C10, respectively. Culturing procedures for the resulting strains (SyBE_Sc14C80-SyBE_Sc14C83, and SyBE_Sc14C43) (Table 1) were the same as terpenoid fermentation in YPDG medium supplemented with 2% (mass/vol) or 4% (mass/vol) glucose, respectively. To determine the activities of PA to PL, the serially deleted ALD6 promoters were fused to RFP in strain SyBE_Sc14C02. The resulting strains (SyBE_Sc14C84 to SyBE_Sc14C95) (Table 1) were cultivated in YPD medium for fluorescence assay.
Microscopic analysis.
For microscopic observation, strain SyBE_Sc14C23 was used to investigate lycopene accumulation after 46 h of shake flask fermentation. Nonproducing strain SyBE_Sc14C10 was used as a control. Cells were harvested by centrifugation, washed, and diluted with sterile water to an OD600 of 5.0. Images were taken with an Olympus CX41 (Olympus, Tokyo, Japan) at magnifications of ×40 and ×100.
Metabolite quantification.
Fatty acids from whole yeast cells were extracted, methyl esterified, and quantified as previously described (52). A fatty acid methyl ester (FAME) standard mixture with acyl chain lengths ranging from C8 to C22 (Sigma) was used for quantification. The fatty acid was calculated as milligrams per gram (dry cell weight [DCW]). Total fatty acid concentrations were calculated as the sum of C16 to C18 (saturated and unsaturated) fatty acids. The unsaturation index was calculated as the ratio of unsaturated fatty acids to total fatty acids. Sterols, including squalene, zymosterol, and ergosterol, were extracted and quantified by gas chromatography-mass spectrometry (GC-MS) as described by Su et al. (53). The contents of sterols were determined by standard curves of squalene, zymosterol, and ergosterol (Sigma), respectively, and expressed as milligrams per gram (DCW). Lycopene quantification was performed as described by our group (15). Production of geraniol, amorphadiene, and geranylgeraniol was carried out as liquid-liquid two-phase fermentation using a 20% (vol/vol) IPM overlay and analyzed by GC-MS as described by Jiang et al. (54), Ro et al. (4), and Tokuhiro et al. (55), respectively. Terpenoid production was quantified by comparison with standard curves of authentic compounds purchased from Sigma.
RNA-sequencing analysis.
Cells were harvested from cultivation under both 2% (mass/vol) and 4% (mass/vol) glucose fermentation conditions according to our previous work (15). In the case of 2% (mass/vol) glucose, cells were sampled at 4 h (glucose consumption phase) and 12 h (ethanol consumption phase). Cells in 4% (mass/vol) glucose were harvested at 6 h (glucose consumption phase) and 24 h (ethanol consumption phase) (15). Total RNA was isolated following the NEBNext Ultra RNA protocol and using the NEBNext poly(A) mRNA magnetic isolation module (New England BioLabs [NEB]) according to the manufacturer’s instructions and was qualified and quantified with an Agilent 2100 bioanalyzer (Agilent Technologies). Sequencing service was performed by Genewiz Inc. on the Illumina HiSeq2500 platform. Image analysis and base calling were conducted by HiSeq control software on the HiSeq instrument. Data were normalized to reads per kilobase per million reads (RPKM) using Htseq software (56). Differentially expressed genes were identified by DESeq2 software (57) with log2 fold change of >1.0 and a corrected P value of <0.05. The R software environment was used for hierarchical clustering analysis. RNA-seq data were calculated from two biological replicates. The Saccharomyces Genome Database (SGD) (58) was used to gather and annotate gene information.
Real-time PCR.
To determine the relative transcriptional level of ALD6, CrtE, CrtB, and CrtI, cells were harvested after 4 h or 12 h of cultivation in YPDG (2% [mass/vol] glucose). Isolation of total RNA was as described in the previous section. Reverse transcription was performed using a PrimeScript RT reagent kit with genomic DNA (gDNA) eraser (TaKaRa Biotech) according to the manufacturer’s instructions. SYBR Premix Ex Taq II (Tli RNaseH Plus) and ROX plus (TaKaRa Biotech) were used for RT-PCR experiments with the primers listed in Table 2. Target gene expression was normalized to actin or ALG9 expression. The relative gene transcription analysis was performed using the threshold cycle (2−ΔΔCT) method (59). RT-PCR data were calculated from three biological replicates. Statistical analysis was performed with SPSS 19.0, with significance levels shown in figures as follows: *, P < 0.05, and **, P < 0.01.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful for the financial support from the National Natural Science Foundation of China (21621004, 21676192, 21676190, and 31600052), the Ministry of Science and Technology of China (“973” Program 2014CB745100), and Innovative Talents and Platform Program of Tianjin (16PTSYJC00050 and 16PTGCCX00140).
We thank Bradley Biggs and Ajikumar Parayil for review and comments on the manuscript. We also thank Ryan Nicholas Philippe for thoroughly proofreading the revised manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01990-18.
REFERENCES
- 1.Ajikumar PK, Tyo K, Carlsen S, Mucha O, Phon TH, Stephanopoulos G. 2008. Terpenoids: opportunities for biosynthesis of natural product drugs using engineered microorganisms. Mol Pharm 5:167–190. doi: 10.1021/mp700151b. [DOI] [PubMed] [Google Scholar]
- 2.Liu L, Redden H, Alper HS. 2013. Frontiers of yeast metabolic engineering: diversifying beyond ethanol and Saccharomyces. Curr Opin Biotechnol 24:1023–1030. doi: 10.1016/j.copbio.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Nielsen J, Keasling JD. 2016. Engineering cellular metabolism. Cell 164:1185–1197. doi: 10.1016/j.cell.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 4.Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, Chang MC, Withers ST, Shiba Y, Sarpong R, Keasling JD. 2006. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 5.Lv X, Wang F, Zhou P, Ye L, Xie W, Xu H, Yu H. 2016. Dual regulation of cytoplasmic and mitochondrial acetyl-CoA utilization for improved isoprene production in Saccharomyces cerevisiae. Nat Commun 7:12851. doi: 10.1038/ncomms12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szappanos B, Kovács K, Szamecz B, Honti F, Costanzo M, Baryshnikova A, Gelius-Dietrich G, Lercher MJ, Jelasity M, Myers CL, Andrews BJ, Boone C, Oliver SG, Pál C, Papp B. 2011. An integrated approach to characterize genetic interaction networks in yeast metabolism. Nat Genet 43:656–662. doi: 10.1038/ng.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alper H, Miyaoku K, Stephanopoulos G. 2005. Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nat Biotechnol 23:612–616. doi: 10.1038/nbt1083. [DOI] [PubMed] [Google Scholar]
- 8.Alper H, Jin YS, Moxley JF, Stephanopoulos G. 2005. Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metab Eng 7:155–164. doi: 10.1016/j.ymben.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Trikka FA, Nikolaidis A, Athanasakoglou A, Andreadelli A, Ignea C, Kotta K, Argiriou A, Kampranis SC, Makris AM. 2015. Iterative carotenogenic screens identify combinations of yeast gene deletions that enhance sclareol production. Microb Cell Fact 14:60. doi: 10.1186/s12934-015-0246-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Z, Meng H, Li J, Wang J, Li Q, Wang Y, Zhang Y. 2014. Identification of novel knockout targets for improving terpenoids biosynthesis in Saccharomyces cerevisiae. PLoS One 9:e112615. doi: 10.1371/journal.pone.0112615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M'Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Veronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 12.Özaydın B, Burd H, Lee TS, Keasling JD. 2013. Carotenoid-based phenotypic screen of the yeast deletion collection reveals new genes with roles in isoprenoid production. Metab Eng 15:174–183. doi: 10.1016/j.ymben.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 13.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian K-D, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Güldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kötter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang C-y, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 14.Wilson WA, Wang Z, Roach PJ. 2002. Systematic identification of the genes affecting glycogen storage in the yeast Saccharomyces cerevisiae: implication of the vacuole as a determinant of glycogen level. Mol Cell Proteomics 1:232–242. doi: 10.1074/mcp.M100024-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y, Xiao W, Wang Y, Liu H, Li X, Yuan Y. 2016. Lycopene overproduction in Saccharomyces cerevisiae through combining pathway engineering with host engineering. Microb Cell Fact 15:113. doi: 10.1186/s12934-016-0509-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Remize F, Andrieu E, Dequin S. 2000. Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae: role of the cytosolic Mg2+ and mitochondrial K+ acetaldehyde dehydrogenases Ald6p and Ald4p in acetate formation during alcoholic fermentation. Appl Environ Microbiol 66:3151–3159. doi: 10.1128/AEM.66.8.3151-3159.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saint-Prix F, Bonquist L, Dequin S. 2004. Functional analysis of the ALD gene family of Saccharomyces cerevisiae during anaerobic growth on glucose: the NADP+-dependent Ald6p and Ald5p isoforms play a major role in acetate formation. Microbiology 150:2209–2220. doi: 10.1099/mic.0.26999-0. [DOI] [PubMed] [Google Scholar]
- 18.Boubekeur S, Bunoust O, Camougrand N, Castroviejo M, Rigoulet M, Guerin B. 1999. A mitochondrial pyruvate dehydrogenase bypass in the yeast Saccharomyces cerevisiae. J Biol Chem 274:21044–21048. doi: 10.1074/jbc.274.30.21044. [DOI] [PubMed] [Google Scholar]
- 19.Boubekeur S, Camougrand N, Bunoust O, Rigoulet M, Guerin B. 2001. Participation of acetaldehyde dehydrogenases in ethanol and pyruvate metabolism of the yeast Saccharomyces cerevisiae. Eur J Biochem 268:5057–5065. doi: 10.1046/j.1432-1033.2001.02418.x. [DOI] [PubMed] [Google Scholar]
- 20.Chen F, Zhou J, Shi Z, Liu L, Du G, Chen J. 2010. Effect of acetyl-CoA synthase gene overexpression on physiological function of Saccharomyces cerevisiae. Wei Sheng Wu Xue Bao 50:1172–1179. (In Chinese.) [PubMed] [Google Scholar]
- 21.Ding J, Holzwarth G, Penner MH, Patton-Vogt J, Bakalinsky AT. 2015. Overexpression of acetyl-CoA synthetase in Saccharomyces cerevisiae increases acetic acid tolerance. FEMS Microbiol Lett 362:1–7. doi: 10.1093/femsle/fnu042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai Z, Liu Y, Huang L, Zhang X. 2012. Production of miltiradiene by metabolically engineered Saccharomyces cerevisiae. Biotechnol Bioeng 109:2845–2853. doi: 10.1002/bit.24547. [DOI] [PubMed] [Google Scholar]
- 23.Basson ME, Thorsness M, Finer-Moore J, Stroud RM, Rine J. 1988. Structural and functional conservation between yeast and human 3-hydroxy-3-methylglutaryl coenzyme A reductases, the rate-limiting enzyme of sterol biosynthesis. Mol Cell Biol 8:3797–3808. doi: 10.1128/MCB.8.9.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y, Daviet L, Schalk M, Siewers V, Nielsen J. 2013. Establishing a platform cell factory through engineering of yeast acetyl-CoA metabolism. Metab Eng 15:48–54. doi: 10.1016/j.ymben.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 25.Lv X, Xie W, Lu W, Guo F, Gu J, Yu H, Ye L. 2014. Enhanced isoprene biosynthesis in Saccharomyces cerevisiae by engineering of the native acetyl-CoA and mevalonic acid pathways with a push-pull-restrain strategy. J Biotechnol 186:128–136. doi: 10.1016/j.jbiotec.2014.06.024. [DOI] [PubMed] [Google Scholar]
- 26.van Roermund CW, Waterham HR, Ijlst L, Wanders RJ. 2003. Fatty acid metabolism in Saccharomyces cerevisiae. Cell Mol Life Sci 60:1838–1851. doi: 10.1007/s00018-003-3076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y, Siewers V, Nielsen J. 2012. Profiling of cytosolic and peroxisomal acetyl-CoA metabolism in Saccharomyces cerevisiae. PLoS One 7:e42475. doi: 10.1371/journal.pone.0042475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee YJ, Jang JW, Kim KJ, Maeng PJ. 2011. TCA cycle-independent acetate metabolism via the glyoxylate cycle in Saccharomyces cerevisiae. Yeast 28:153–166. doi: 10.1002/yea.1828. [DOI] [PubMed] [Google Scholar]
- 29.Du W, Song Y, Liu M, Yang H, Zhang Y, Fan Y, Luo X, Li Z, Wang N, He H, Zhou H, Ma W, Zhang T. 2016. Gene expression pattern analysis of a recombinant Escherichia coli strain possessing high growth and lycopene production capability when using fructose as carbon source. Biotechnol Lett 38:1571–1577. doi: 10.1007/s10529-016-2133-0. [DOI] [PubMed] [Google Scholar]
- 30.Zhao J, Li Q, Sun T, Zhu X, Xu H, Tang J, Zhang X, Ma Y. 2013. Engineering central metabolic modules of Escherichia coli for improving beta-carotene production. Metab Eng 17:42–50. doi: 10.1016/j.ymben.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 31.Napp SJ, Da Silva NA. 1994. Catabolite repression and induction time effects for a temperature-sensitive GAL-regulated yeast expression system. J Biotechnol 32:239–248. doi: 10.1016/0168-1656(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 32.Brennan TC, Turner CD, Kromer JO, Nielsen LK. 2012. Alleviating monoterpene toxicity using a two-phase extractive fermentation for the bioproduction of jet fuel mixtures in Saccharomyces cerevisiae. Biotechnol Bioeng 109:2513–2522. doi: 10.1002/bit.24536. [DOI] [PubMed] [Google Scholar]
- 33.Ro DK, Ouellet M, Paradise EM, Burd H, Eng D, Paddon CJ, Newman JD, Keasling JD. 2008. Induction of multiple pleiotropic drug resistance genes in yeast engineered to produce an increased level of anti-malarial drug precursor, artemisinic acid. BMC Biotechnol 8:83. doi: 10.1186/1472-6750-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gruszecki WI, Strzałka K. 2005. Carotenoids as modulators of lipid membrane physical properties. Biochim Biophys Acta 1740:108–115. doi: 10.1016/j.bbadis.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 35.Xia S, Tan C, Zhang Y, Abbas S, Feng B, Zhang X, Qin F. 2015. Modulating effect of lipid bilayer-carotenoid interactions on the property of liposome encapsulation. Colloids Surf B Biointerfaces 128:172–180. doi: 10.1016/j.colsurfb.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 36.Liu P, Sun L, Sun Y, Shang F, Yan G. 2016. Decreased fluidity of cell membranes causes a metal ion deficiency in recombinant Saccharomyces cerevisiae producing carotenoids. J Ind Microbiol Biotechnol 43:525–535. doi: 10.1007/s10295-015-1728-0. [DOI] [PubMed] [Google Scholar]
- 37.Parveen M, Hasan MK, Takahashi J, Murata Y, Kitagawa E, Kodama O, Iwahashi H. 2004. Response of Saccharomyces cerevisiae to a monoterpene: evaluation of antifungal potential by DNA microarray analysis. J Antimicrob Chemother 54:46–55. doi: 10.1093/jac/dkh245. [DOI] [PubMed] [Google Scholar]
- 38.Wriessnegger T, Pichler H. 2013. Yeast metabolic engineering—targeting sterol metabolism and terpenoid formation. Prog Lipid Res 52:277–293. doi: 10.1016/j.plipres.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 39.Donald KA, Hampton RY, Fritz IB. 1997. Effects of overproduction of the catalytic domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase on squalene synthesis in Saccharomyces cerevisiae. Appl Environ Microbiol 63:3341–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Polakowski T, Stahl U, Lang C. 1998. Overexpression of a cytosolic hydroxymethylglutaryl-CoA reductase leads to squalene accumulation in yeast. Appl Microbiol Biotechnol 49:66–71. doi: 10.1007/s002530051138. [DOI] [PubMed] [Google Scholar]
- 41.Liu J, Zhu Y, Du G, Zhou J, Chen J. 2013. Exogenous ergosterol protects Saccharomyces cerevisiae from D-limonene stress. J Appl Microbiol 114:482–491. doi: 10.1111/jam.12046. [DOI] [PubMed] [Google Scholar]
- 42.Rodríguez-Vargas S, Sánchez-García A, Martínez-Rivas JM, Prieto JA, Randez-Gil F. 2007. Fluidization of membrane lipids enhances the tolerance of Saccharomyces cerevisiae to freezing and salt stress. Appl Environ Microbiol 73:110–116. doi: 10.1128/AEM.01360-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun Y, Sun L, Shang F, Yan G. 2016. Enhanced production of β-carotene in recombinant Saccharomyces cerevisiae by inverse metabolic engineering with supplementation of unsaturated fatty acids. Process Biochem 51:568–577. doi: 10.1016/j.procbio.2016.02.004. [DOI] [Google Scholar]
- 44.Makino K, Amemura M, Kawamoto T, Kimura S, Shinagawa H, Nakata A, Suzuki M. 1996. DNA binding of PhoB and its interaction with RNA polymerase. J Mol Biol 259:15–26. doi: 10.1006/jmbi.1996.0298. [DOI] [PubMed] [Google Scholar]
- 45.Jenjaroenpun P, Wongsurawat T, Pereira R, Patumcharoenpol P, Ussery DW, Nielsen J, Nookaew I. 2018. Complete genomic and transcriptional landscape analysis using third-generation sequencing: a case study of Saccharomyces cerevisiae CEN.PK113-7D. Nucleic Acids Res 46:e38. doi: 10.1093/nar/gky014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferreira R, Teixeira PG, Siewers V, Nielsen J. 2018. Redirection of lipid flux toward phospholipids in yeast increases fatty acid turnover and secretion. Proc Natl Acad Sci U S A 115:1262–1267. doi: 10.1073/pnas.1715282115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen X, Gao C, Guo L, Hu G, Luo Q, Liu J, Nielsen J, Chen J, Liu L. 2018. DCEO biotechnology: tools to design, construct, evaluate, and optimize the metabolic pathway for biosynthesis of chemicals. Chem Rev 118:4–72. doi: 10.1021/acs.chemrev.6b00804. [DOI] [PubMed] [Google Scholar]
- 48.Walkey CJ, Luo Z, Madilao LL, van Vuuren HJ. 2012. The fermentation stress response protein Aaf1p/Yml081Wp regulates acetate production in Saccharomyces cerevisiae. PLoS One 7:e51551. doi: 10.1371/journal.pone.0051551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tyner C, Barber GP, Casper J, Clawson H, Diekhans M, Eisenhart C, Fischer CM, Gibson D, Gonzalez JN, Guruvadoo L, Haeussler M, Heitner S, Hinrichs AS, Karolchik D, Lee BT, Lee CM, Nejad P, Raney BJ, Rosenbloom KR, Speir ML, Villarreal C, Vivian J, Zweig AS, Haussler D, Kuhn RM, Kent WJ. 2017. The UCSC Genome Browser database: 2017 update. Nucleic Acids Res 45:D626–D634. doi: 10.1093/nar/gkw1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Cen XF, Zhao GP, Wang J. 2012. Characterization of a new GlnR binding box in the promoter of amtB in Streptomyces coelicolor inferred a PhoP/GlnR competitive binding mechanism for transcriptional regulation of amtB. J Bacteriol 194:5237–5244. doi: 10.1128/JB.00989-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cao YX, Xiao WH, Liu D, Zhang JL, Ding MZ, Yuan YJ. 2015. Biosynthesis of odd-chain fatty alcohols in Escherichia coli. Metab Eng 29:113–123. doi: 10.1016/j.ymben.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 52.Runguphan W, Keasling JD. 2014. Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid-derived biofuels and chemicals. Metab Eng 21:103–113. doi: 10.1016/j.ymben.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 53.Su W, Xiao WH, Wang Y, Liu D, Zhou X, Yuan YJ. 2015. Alleviating redox imbalance enhances 7-dehydrocholesterol production in engineered Saccharomyces cerevisiae. PLoS One 10:e0130840. doi: 10.1371/journal.pone.0130840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang GZ, Yao MD, Wang Y, Zhou L, Song TQ, Liu H, Xiao WH, Yuan YJ. 2017. Manipulation of GES and ERG20 for geraniol overproduction in Saccharomyces cerevisiae. Metab Eng 41:57–66. doi: 10.1016/j.ymben.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 55.Tokuhiro K, Muramatsu M, Ohto C, Kawaguchi T, Obata S, Muramoto N, Hirai M, Takahashi H, Kondo A, Sakuradani E, Shimizu S. 2009. Overproduction of geranylgeraniol by metabolically engineered Saccharomyces cerevisiae. Appl Environ Microbiol 75:5536–5543. doi: 10.1128/AEM.00277-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 57.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hitz BC, Karra K, Krieger CJ, Miyasato SR, Nash RS, Park J, Skrzypek MS, Simison M, Weng S, Wong ED. 2012. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res 40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.