The ability to generate chromosomal mutations is fundamental to microbiology. Historically, however, GBS pathogenesis research has been made challenging by the relative genetic intractability of the organism. Generating a single knockout in GBS using traditional techniques can take many months, with highly variable success rates. Furthermore, traditional methods do not offer a straightforward way to generate single-base-pair polymorphisms or other subtle changes, especially to noncoding regions of the chromosome. We have developed a new sucrose counterselection-based method that permits rapid, efficient, and flexible GBS mutagenesis. Our technique requires no additional equipment beyond what is needed for traditional approaches. We believe that it will catalyze rapid advances in GBS genetics research by significantly easing the path to generating mutants.
KEYWORDS: counterselection, genome editing, group B streptococcus, mutagenesis, sacB
ABSTRACT
Streptococcus agalactiae (group B Streptococcus [GBS]) is a cause of severe infections, particularly during the newborn period. While methods exist for generating chromosomal mutations in GBS, they are cumbersome and inefficient and present significant challenges if the goal is to study subtle mutations, such as single-base-pair polymorphisms. To address this problem, we have developed an efficient and flexible GBS mutagenesis protocol based on sucrose counterselection against levansucrase (SacB) expressed from a temperature-selective shuttle vector. GBS containing the SacB expression cassette demonstrates lethal sensitivity to supplemental sucrose whether the plasmid DNA is replicating outside of the chromosome or has been integrated during a crossover event. Transmission electron microscopy shows that SacB-mediated lethal sucrose sensitivity results from the accumulation of inclusion bodies that eventually lead to complete degradation of normal cellular architecture and subsequent lysis. We used this new mutagenesis technique to generate an in-frame, allelic exchange knockout of the GBS sortase gene srtA, demonstrating that >99% of colonies that emerge from our protocol had the expected knockout phenotype and that among a subset tested by sequencing, 100% had the correct genotype. We also generated barcoded nonsense mutations in the cylE gene in two GBS strains, showing that the approach can be used to make small, precise chromosomal mutations.
IMPORTANCE The ability to generate chromosomal mutations is fundamental to microbiology. Historically, however, GBS pathogenesis research has been made challenging by the relative genetic intractability of the organism. Generating a single knockout in GBS using traditional techniques can take many months, with highly variable success rates. Furthermore, traditional methods do not offer a straightforward way to generate single-base-pair polymorphisms or other subtle changes, especially to noncoding regions of the chromosome. We have developed a new sucrose counterselection-based method that permits rapid, efficient, and flexible GBS mutagenesis. Our technique requires no additional equipment beyond what is needed for traditional approaches. We believe that it will catalyze rapid advances in GBS genetics research by significantly easing the path to generating mutants.
INTRODUCTION
Streptococcus agalactiae (group B Streptococcus [GBS]) is the most common cause of neonatal sepsis and meningitis (1–3). It can also cause serious infections in adults (4, 5) and in several animal species, including fish, which can be a source of zoonotic transmission (6–9).
GBS is not naturally competent under laboratory conditions (10) and exhibits low rates of spontaneous genetic recombination (11). Genetic studies of GBS have mostly relied on the generation of allelic exchange knockouts using mutagenesis cassettes cloned into a temperature-sensitive shuttle vector (12–14). The mutagenesis cassette typically consists of an antibiotic resistance marker with upstream and downstream homology arms matching the chromosomal regions adjacent to the target gene. A second antibiotic resistance marker on the plasmid, outside of the mutagenesis cassette, confers dual resistance to transformed cells.
After electroporation and transformation of competent GBS cells with the mutagenesis vector, transformed clones are initially grown at a permissive temperature, which allows extrachromosomal replication of the plasmid. A subsequent shift to a higher, nonpermissive temperature selects against extrachromosomal plasmid replication, leaving only cells where a crossover event at one of the homology arms has resulted in plasmid integration into the chromosome.
In order to achieve allelic exchange, a second crossover event must then occur at the other homology arm, followed by plasmid expulsion from the cell. Successful completion of these steps is detected by screening individual colonies for a specific antibiotic resistance phenotype, that is, retained antibiotic resistance from the mutagenesis cassette marker with sensitivity to the second antibiotic due to loss of the plasmid during growth at the nonpermissive temperature (12).
Without effective counterselection against the plasmid, however, detection of the second crossover event—a stochastic and often rare occurrence—is inefficient. Particularly if the desired mutant has a fitness disadvantage compared to the wild type (WT), identification of an allelic exchange mutant may require manual screening of hundreds or thousands of individual colonies (15). Moreover, small-scale and unmarked alterations, such as single nucleotide polymorphisms (SNPs) or subtle mutations to noncoding regions, can be very difficult to obtain.
Levansucrase (sucrose:2,6-β-d-fructan 2,6-β-d-fructosyltransferase) is an enzyme present in multiple bacterial species and has been extensively studied in Bacillus subtilis (16–21). Encoded by the sacB gene, secreted B. subtilis levansucrase polymerizes sucrose into the branched fructan polymer known as levan (16, 19). While the exact function of levansucrase in B. subtilis is unknown, the levan that it generates is believed to serve a structural or nutrient role for the cell (17).
In other bacterial species, expression of B. subtilis sacB confers lethal sensitivity to sucrose (22, 23). The mechanism of sucrose toxicity is believed to be from either intracellular or extracellular accumulation of levan, with resultant disruption of normal cellular processes. Sucrose sensitivity from sacB expression has been used as counterselection in conjunction with plasmid-based mutagenesis systems to isolate mutants in several bacterial species. This approach has been widely used in Gram-negative organisms for decades, including Escherichia coli, Agrobacterium tumefaciens, and Xanthomonas campestris (24–26). Its use in Gram-positive bacteria has been more limited, but successful applications have been reported in several species, including Streptococcus zooepidemicus, Streptococcus pneumoniae, and Mycobacterium spp. (22, 23, 27–29). To date, however, the technique has not been described in GBS.
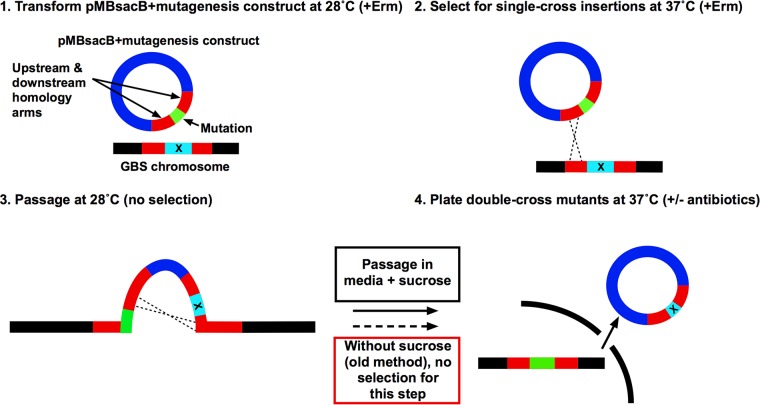
Here, we report the development and validation of a flexible and efficient counterselection system to make targeted mutations in GBS using a sacB-containing, temperature-sensitive, broad-host-range plasmid. A schematic of this new system is presented in Fig. 1. We show that the system can be used to generate marked and unmarked mutations in multiple GBS strains. In our experience, use of this technique dramatically decreases the labor and time required to generate mutants, allowing accelerated discovery.
FIG 1.
Schematic of pMBsacB-mediated GBS mutagenesis. Following insertion of a mutagenesis cassette into pMBsacB and transformation of GBS at 28°C (1), the transformed strain is used to seed a 37°C culture, selecting for single-crossover events at one of the homology arms (2). After removal of antibiotic selection and growth at 28°C to promote a second crossover event and plasmid expulsion (3), sucrose is added as counterselection to isolate the desired allelic exchange clones (4). In the absence of sucrose counterselection, step 4 resembles traditional mutagenesis techniques (red box), in which identification of the allelic exchange depends on chance discovery of a clone with spontaneous loss of plasmid-based antibiotic resistance.
RESULTS
Construction of pMBsacB, a temperature-sensitive, sucrose-counterselectable mutagenesis shuttle vector.
Plasmid pMBsacB is derived from pHY304, a widely used mutagenesis shuttle vector with the temperature-sensitive broad-host-range origin of replication from pWV01 (12, 30).
The B. subtilis sacB coding sequence, complete with signal peptide sequence, was amplified from strain 168 purified genomic DNA and cloned into plasmid pORI23, placing the sacB gene downstream of the p23 promoter. This plasmid is designated pSacB23 and was used in initial experiments to test sacB functionality in GBS (see next section).
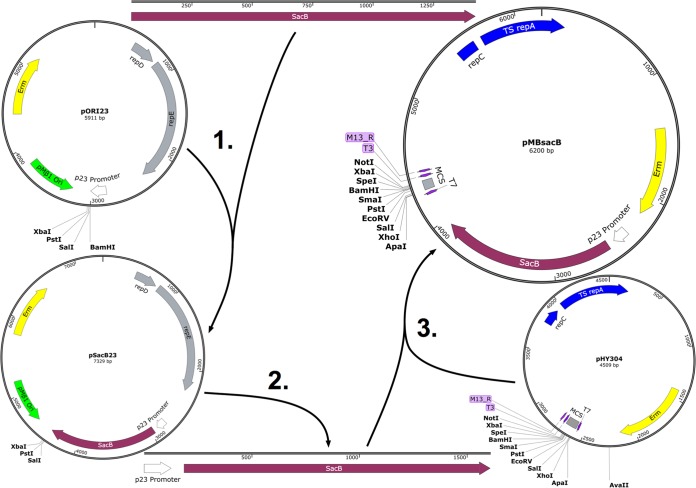
We subsequently generated pMBsacB by amplifying the p23 promoter and sacB coding sequence as a single expression cassette and then subcloning it into pHY304. Figure 2 shows the steps involved in developing pMBsacB.
FIG 2.
Development of pMBsacB. The sacB coding sequence was amplified from B. subtilis 168 and inserted at the BamHI site in pORI23, placing the gene adjacent to the p23 promoter in pSacB23 (1). The promoter-gene cassette was then amplified from pSacB23 (2) and cloned into the AvaII site of the broad-host-range, temperature-sensitive plasmid pHY304 (3), generating pMBsacB.
GBS transformed with sacB-bearing plasmids shows lethal sucrose sensitivity.
Before transforming GBS with sacB-bearing plasmids pSacB23 and pMBsacB, we developed an electroporation protocol that did not include the use of sucrose, which is typically used as an osmoprotectant to prevent bacterial death during transformation (30, 31). We had observed very low rates of successful transformation during early trials with sucrose osmoprotection (data not shown), presumably due to sucrose-mediated toxicity.
We initially replaced sucrose with maltose, a structurally related disaccharide that we hoped would not be lethal to cells transformed with sacB plasmids. However, there was no significant increase in transformation efficiency with maltose osmoprotection (data not shown), which we attributed to SacB-nonspecific reactivity with maltose, likely generating maltosylfructose (32, 33).
Our transformation efficiency returned to expected levels (10−4 to 10−5 per μg plasmid DNA for GBS strain CNCTC 10/84) with replacement of sucrose osmoprotectant with 25% (mass/mass) polyethylene glycol (average molecular weight [MW], 6,000 Da; polyethylene glycol 6000 [PEG 6000]), which was dissolved in rich transformation medium for competent cell outgrowth and in the wash solution in which we store and electroporate competent GBS.
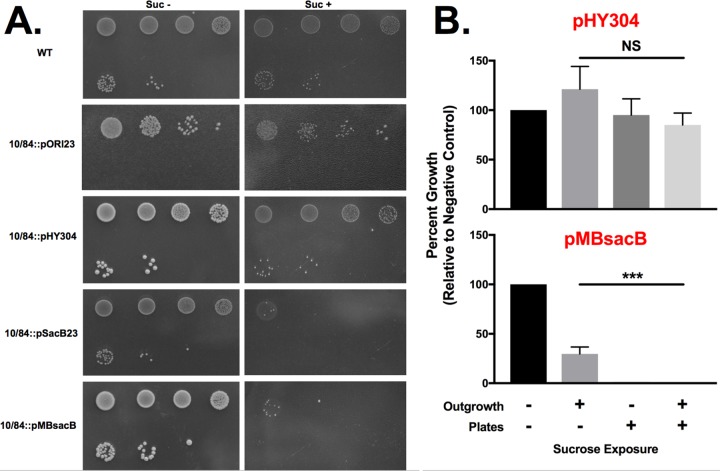
GBS strain 10/84 transformed with pSacB23 or pMBsacB showed significant growth defects on solid medium with a supplemental 0.75 M sucrose (Fig. 3A). We also tested the sensitivity of planktonic 10/84::pMBsacB to sucrose added to liquid medium and found significant growth impairment. Planktonic and solid media exposure could also be combined (Fig. 3B). Strain 10/84 transformed with pHY304, by contrast, did not demonstrate significant sucrose sensitivity.
FIG 3.
sacB confers lethal sucrose sensitivity in GBS. (A) Stationary-phase WT GBS 10/84 or 10/84 transformed with plasmids used in this study was serially diluted 10-fold and plated on TS agar plates with appropriate antibiotic selection with or without supplemental 0.75 M sucrose. The first dilution shown is 10−1. (B) pMBsacB-based sucrose sensitivity functions in liquid culture and on solid medium, whereas pHY304 does not affect GBS survival in sucrose in either growth condition (***, P < 0.0001, analysis of variance [ANOVA]).
In order to directly visualize the phenotypic effect of sucrose exposure on GBS expressing SacB, we performed transmission electron microscopy on 10/84 transformed with pMBsacB or pHY304 and exposed to sucrose or control conditions. As shown in Fig. 4A to C, sucrose exposure of 10/84 with pMBsacB resulted in abnormal cellular morphology, with apparent intracellular accumulation of inclusion bodies, eventually resulting in complete degradation of normal cellular architecture and eventual lysis. Apart from expected cell shrinkage from sucrose-mediated osmosis, GBS transformed with the control plasmid pHY304 showed normal architecture regardless of sucrose exposure (Fig. 4D and E). SacB expression in the absence of sucrose had no apparent effect on morphology (Fig. 4F).
FIG 4.
Transmission electron microscopy reveals that SacB expression under sucrose counterselection results in destructive intracellular inclusions. (A to C) Early log-phase GBS strain 10/84 transformed with pMBsacB and exposed to 0.75 M sucrose for 3 h shows intracellular inclusion bodies that lead to degraded architecture and eventual lysis. GBS transformed with the control plasmid pHY304 shows normal architecture under baseline growth conditions (D) and only expected osmotic effects when grown with supplemental sucrose (E). (F) SacB expression in the absence of sucrose counterselection has no apparent effect on GBS morphology.
Sucrose counterselection improves the efficiency of allelic exchange mutagenesis.
To test whether sucrose counterselection could be used in GBS to produce allelic exchange mutants, we applied our system to deleting a gene that we and others had experience knocking out, the sortase gene srtA (34). This provided a benchmark against which the efficiency of counterselection-assisted mutagenesis could be measured.
We used overlap extension PCR to generate an srtA mutagenesis cassette (ΔsrtA) in which approximately 800-bp homology arms flank an in-frame chloramphenicol acetyltransferase gene (cat), which confers chloramphenicol resistance, replacing the srtA coding sequence. This cassette was subcloned into pHY304 and pMBsacB and then used for transformation of 10/84 using PEG 6000 osmoprotection.
After transformation with either pHY304-ΔsrtA or pMBsacB-ΔsrtA, single-cross intermediate strains were obtained by changing the growth temperature of the liquid cultures from 28°C to 37°C in the presence of erythromycin selection. Once chromosomal insertion was confirmed by PCR, the pHY304 and pMBsacB single-cross strains were serially passaged at 28°C with no antibiotics or counterselection. At that point, each of the two cultures was used to seed two new cultures with chloramphenicol at 37°C, one containing 0.75 M sucrose and the other containing a control lacking sucrose.
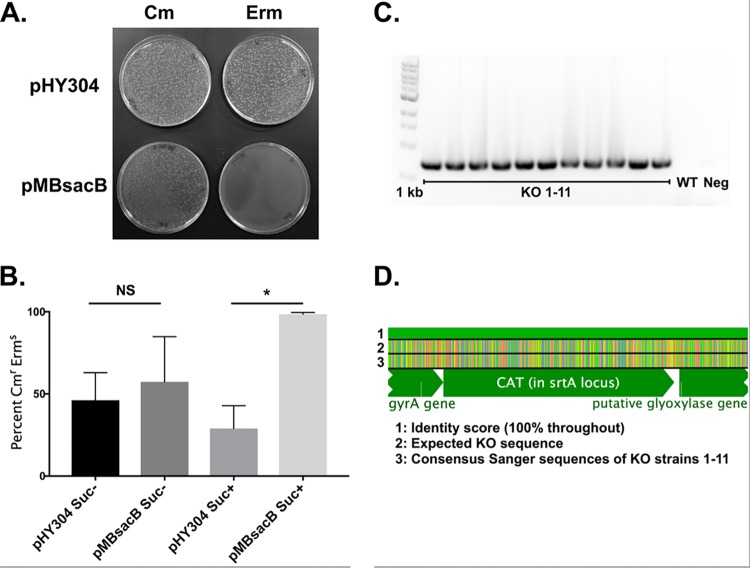
Each of the four cultures (10/84::pHY304-ΔsrtA and 10/84::pMBsacB-ΔsrtA, with and without sucrose) was passaged three times at 37°C with chloramphenicol selection. Serial dilutions were then plated on chloramphenicol-containing solid media with or without erythromycin selection. While the overall CFU concentration did not differ significantly between conditions—as indicated by equal growth on the nonselective medium—there was a notable difference between 10/84::pMBsacB-ΔsrtA grown in the presence of sucrose and the other three outgrowth conditions. Exposure of the pMBsacB single-cross strain to sucrose eliminated virtually all erythromycin-resistant survival, suggesting successful counterselection (Fig. 5A and B). PCR amplification of the cat gene generated the expected 660-bp gel electrophoresis bands when genomic DNA from colonies that survived counterselection was used as template but not when wild-type GBS DNA was used (Fig. 5C). Sanger sequencing of DNA amplified using PCR primers that bind outside of the ΔsrtA homology arms confirmed that the srtA gene had been replaced by cat as intended (Fig. 5D). The pHY304 single-cross strain was not responsive to sucrose, and the pMBsacB erythromycin-resistant CFU survived well in the absence of sucrose exposure.
FIG 5.
Allelic exchange mutagenesis of the srtA gene is made more efficient by using pMBsacB. (A) Sucrose exposure of the single-cross pMBsacB-ΔsrtA knockout intermediates results in selection against erythromycin-resistant clones, which is not seen with the pHY304-ΔsrtA single-cross strain. (B) Recovery of phenotypically correct knockout clones is significantly more efficient using pMBsacB than when mutagenesis is performed with pHY304 (*, P < 0.05, t test). (C) PCR of cat results in successful amplification of genomic DNA from 11 knockout (KO) candidates but not from wild-type (WT) or template-negative (Neg) controls. (D) Sanger sequences of the srtA region amplified using primers outside of the mutagenesis cassette were combined to generate a consensus sequence, which was aligned to the expected knockout template. Erm, erythromycin; Cm, chloramphenicol; Suc, sucrose.
The pMBsacB mutagenesis system permits efficient generation of unmarked mutations in multiple GBS strains.
An effective counterselection system could facilitate generation of unmarked mutations, opening the possibility of performing sequential genome edits to produce complex mutants.
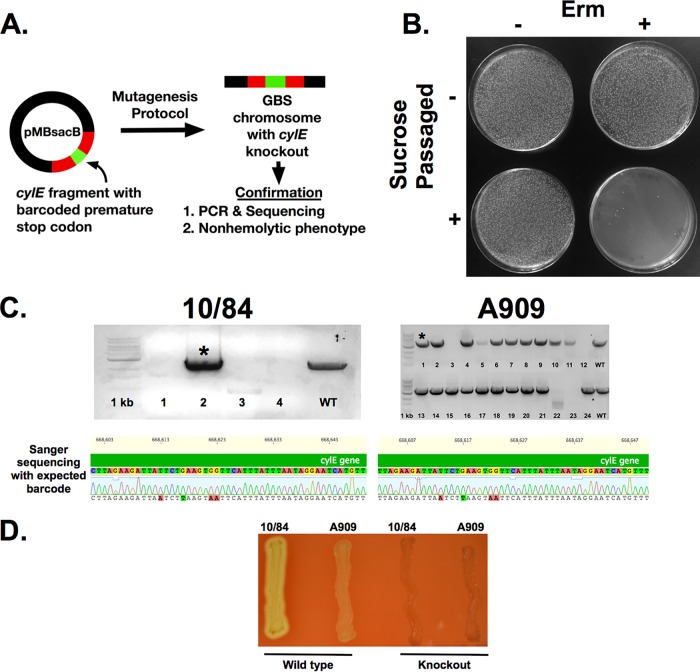
To explore whether pMBsacB could accelerate the generation of unmarked mutations, we developed a mutagenesis cassette against the cylE gene, which encodes a key biosynthetic enzyme responsible for production of the GBS pigmented hemolytic toxin β-hemolysin/cytolysin (35–37). The mutagenesis cassette features a premature stop codon surrounded by a set of silent mutation SNPs, establishing a unique barcode that would not likely arise through spontaneous mutation. The barcoded stop codon is flanked by 500-bp upstream and downstream homology arms (Fig. 6A).
FIG 6.
pMBsacB permits efficient generation of unmarked mutations in the cylE gene. (A) After generation of a cylE mutagenesis cassette bearing a premature stop codon barcoded by additional silent SNPs, the cassette was cloned into pMBsacB and used to knockout β-hemolysin/cytolysin expression in GBS 10/84 and A909. (B) Sucrose counterselection resulted in near-complete elimination of erythromycin-resistant CFU. (C) Clones that survived counterselection were screened for the barcoded mutation by performing PCR with cylE_farout_F/R primers and Sanger sequencing of the amplified genomic region of interest with cylEmut_Conf_F. Representative traces are shown for the bands marked by asterisks. (D) The recovered knockout strains had the expected nonhemolytic phenotype when plated on 5% sheep’s blood agar. Erm, erythromycin.
We cloned this cassette into pMBsacB, generating pMBsacB-cylEmut, which was then used to transform two GBS strains, 10/84 and A909. After generation of the single-cross intermediate, which was nonhemolytic (data not shown), we used sucrose counterselection to isolate putative double-cross GBS CFU, which could be either wild type—due to autoexcision of the plasmid at the same homology arm as that crossed over during insertion—or a cylE knockout.
Counterselection against the single-cross strain again resulted in near-complete elimination of erythromycin-resistant CFU (Fig. 6B).
When plated on solid medium without antibiotic selection, the counterselection culture yielded an equal mix of pigmented and nonpigmented colonies, supporting the concept that the plasmid could autoexcise in one of two ways, only one of which would result in the nonpigmented phenotype expected of the knockout. If not exposed to sucrose, the single-cross intermediate retained erythromycin resistance and the colonies were uniformly nonpigmented, reflecting the fact that the population consisted almost entirely of unchanged, single-cross CFU.
In the 10/84 experiment, we selected four nonpigmented colonies from the nonselective plate on which the sucrose-exposed culture had been grown. These were used for genomic DNA purification, followed by amplification of the cylE coding sequence and Sanger sequencing. One of the four had the correct barcoded sequence, whereas the other three did not properly amplify during PCR, suggesting that the plasmid autoexcised in a manner that left a partial sequence deletion (Fig. 6C).
In the case of strain A909, 20 out of 24 nonpigmented isolates properly amplified by PCR. We sequenced 10 of these, 9 of which had the expected barcoded premature stop codon (Fig. 6C). Both 10/84 and A909 cylE knockout strains generated using pMBsacB-cylEmut showed the anticipated nonpigmented, nonhemolytic phenotype (Fig. 6D).
Unintended deletions during the final plasmid excision are a phenomenon that we have subsequently observed in other mutagenesis experiments, suggesting that all mutants generated with pMBsacB must be confirmed by some combination of PCR and sequencing to ensure the desired genotype.
DISCUSSION
Reliable methods for creating specific mutations are central to microbiological discovery. Existing methods for doing so in GBS have been suboptimal in multiple respects. Without counterselection, the final screening step for plasmid autoexcision and curing is unreliable and inefficient since it depends on random identification of a low-probability biological event. An earlier counterselection-based approach to GBS mutagenesis was limited by the fact that it required an already mutated background strain, which is not ideal for pathogenesis work (15). Together, these barriers make isolation of complex mutants with multiple, subtle chromosomal changes infeasible.
Our sacB-mediated counterselection system overcomes these limitations. The method is simple and does not require any additional equipment or experience beyond what is required for traditional approaches. While we have not attempted mutagenesis of other Gram-positive species using pMBsacB in this study, the possibility that it would be an effective tool in other organisms is an intriguing line of inquiry that we intend to explore.
We validated our system by generating two knockouts, one (srtA) involved allelic exchange with a chloramphenicol resistance marker, while the other (cylE) demonstrated the ability of our technique to produce small chromosomal changes at the single nucleotide level. In order to confirm that the system works in multiple GBS strains from different serotypes (which can show phenotypic variability under the same growth conditions), we generated the same cylE mutation in A909 (serotype Ia) and 10/84 (serotype V). We noted that A909 grew less robustly on 0.75 M sucrose than 10/84; so for the A909 cylE mutation, we used 0.5 M sucrose counterselection. When using this system in different strains, it is important to optimize the counterselection conditions prior to starting a new mutation.
As Fig. 5 shows, the srtA knockout can be generated at reasonable rates even without counterselection. There was considerable variability from one experimental replicate to the next, but the mean recovery rate of knockouts in the sucrose-negative conditions was 29 to 57%, regardless of whether the mutagenesis plasmid was pHY304 or pMBsacB. In contrast, the mean recovery rate in the pMBsacB sucrose counterselection condition was 99%, with low variability.
In the case of low-fitness mutations, however, rates of recovery without counterselection can be much lower (38). The last step in the mutagenesis workflow—autoexcision and curing of the plasmid—essentially establishes a competition assay between the single-cross strain and the intended mutant (12). If the mutant has a survival defect, the odds of randomly selecting a mutant colony from among the single-cross population is very low. By shrinking the single-cross background, the pMBsacB counterselection system increases the odds of isolating the desired mutant. Particularly when the goal is the mutation of genes with a high contribution to the fitness of the organism, we have found that it is important to confirm sucrose sensitivity of the transformant and single-cross intermediate since spontaneous mutations in the sacB gene could lead to escape of unmodified genes with a high contribution to fitness once sucrose counterselection is applied.
In the case of unmarked mutations, where no new antibiotic resistance is introduced into the chromosome, the bacterial population after counterselection is expected to be a mixture of the desired mutant and wild-type bacteria that reverted following plasmid autoexcision. This occurred in our cylE knockout experiment, where roughly equal fractions of the post-counterselection population showed knockout and wild-type β-hemolysin/cytolysin pigmentation phenotypes.
This means that for mutations that do not cause an easily assayable phenotype (such as pigment expression), sequence-based confirmation will be necessary. There are several possible ways to perform this confirmation. Here, we used PCR followed by Sanger sequencing, but probe- or quantitative PCR (qPCR)-based SNP assays are alternative strategies. Confirmation by some means is important, given that plasmid autoexcision can leave unintended deletions, as was the case in several colonies in our cylE experiment.
In summary, we have presented a new, straightforward counterselection-based approach to generating flexible mutations in GBS. Our future plans for this work involve modifications to coding and noncoding sequences, including promoter alterations, addition of fluorescent and affinity tags to natively expressed genes, and generation of multiple knockout strains.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
GBS strains CNCTC 10/84 (serotype V, sequence type 26) and A909 (serotype Ia, sequence type 7) and B. subtilis strain 168 were maintained as frozen glycerol stocks and were grown on tryptic soy (TS; product number DF0370-17-3; Fisher Scientific) agar plates or stationary in TS broth at 37°C or 28°C. Chemically competent E. coli DH5α was purchased from New England Biolabs (product number C2987H) and was stored and transformed according to the manufacturer’s instructions. E. coli growth was at 37°C unless transformed with a temperature-sensitive plasmid, in which case the growth temperature was 28°C (Table 1). Antibiotic concentrations were as follows: erythromycin, 5 μg/ml (GBS) or 300 μg/ml (E. coli), and chloramphenicol, 1 μg/ml (GBS) or 10 μg/ml (E. coli). The 0.5 M and 0.75 M sucrose-containing broth and solid medium were prepared by diluting a filter-sterilized 2 M sucrose stock solution in an appropriate medium and adding any necessary antibiotics to the final mixture.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference, source, or product no. |
|---|---|---|
| Strains | ||
| E. coli DH5α | Chemically competent cloning strain | New England Biolabs item no. C2987H |
| B. subtilis 168 | Legacy strain derived from B. subtilis Marburg | 39 |
| GBS CNCTC 10/84 | Serotype V, sequence type 26 | 40 |
| GBS A909 | Serotype Ia, sequence type 7 | 41 |
| GBS AR1598 | Serotype V, sequence type 26 10/84 mutant with barcoded premature stop codon in cylE gene | This study |
| Plasmids | ||
| pDC123 | Shuttle vector with the chloramphenicol acetyltransferase gene used as PCR template | 42 |
| pHY304 | Broad-host-range, temperature-sensitive mutagenesis plasmid; pWV01 derivative; erythromycin resistant | 12 |
| pMBsacB | pHY304 with p23-sacB cassette cloned into AvaII site, for sucrose-counterselectable GBS mutagenesis | This study |
| pORI23 | Shuttle vector with oriColE1 and Lactococcus-derived Gram-positive origin of replication; erythromycin resistant; p23 promoter adjacent to BamHI restriction site | 43 |
| pSacB23 | pORI23 with sacB cloned into BamHI site for expression off p23 promoter | This study |
Cloning technique.
The construction of all shuttle vectors and derivatives used in this study was obtained in E. coli DH5α, from which plasmid DNA for downstream applications was purified using the Qiagen QIAprep miniprep kit (product number 27104) according to the manufacturer’s instructions.
Isolation of genomic DNA.
Genomic DNA from GBS and B. subtilis was isolated using the Applied Biosystems MagMAX Core kit (product number A32700) with a KingFisher magnetic bead processing system according to the manufacturer’s instructions, with the following minor modifications. Overnight liquid culture volumes were 1 to 10 ml. After pelleting by centrifugation at 3,200 × g, the bacteria were lysed in a solution containing 100 μl manufacturer-supplied proteinase K and PK buffer, 50 μl lysozyme (100 mg/ml in water), and 5 μl mutanolysin (10 kU in 2 ml 0.1 M potassium phosphate buffer, pH 6.2). Lysis was performed at 37°C for 30 min, then 55°C for 30 min, followed by a 2-min centrifugation at 3,200 × g. The rest of the extraction followed manufacturer instructions, using the MagMAX Core Flex KingFisher protocol file.
Construction of pSacB23 and pMBsacB.
The sacB coding sequence was amplified from B. subtilis 168 genomic DNA using primers sacB_pORI23_F and sacB_pORI23_R, which contain Gibson assembly overhang sequences compatible with pORI23 digested with BamHI (Table 2). Successful clones were identified by Sanger sequencing (data not shown), after which the p23 promoter-sacB cassette was amplified with p23sacB_cassette_F and p23sacB_cassette_R and subcloned into pHY304 digested with AvaII (see Fig. 1). Outgrowth of the pMBsacB cloning reaction was performed at 28°C, and successful clones were identified with Sanger sequencing (data not shown).
TABLE 2.
Primers used in this study
| Primer | Sequence (5′ to 3′) | Description |
|---|---|---|
| cat_F | ATGGAGAAAAAAATCACTGGATATACCACC | Amplifies cat gene for allelic exchange knockout screening and for overlap extension PCR to make ΔsrtA mutagenesis cassette (from pDC123 template) |
| cat_R | CCCGCCCTGCCACTCATCGC | |
| cylE_farout_F | TACACGCGAGATCGGTTAGC | Binds GBS chromosome outside of the cylEmut mutagenesis cassette region, for confirming single- and double-crossover events at the cylE locus |
| cylE_farout_R | CTGGTGTTCCTGAAGCGAGT | |
| cylEmut_GA_F | ATTGGGTACCGGGCCCCCCCAGATGCTATAAAAGCAGC | Amplifies cylEmut mutagenesis cassette (from AR1958 template) for Gibson assembly into pMBsacB NotI/XhoI digest |
| cylEmut_GA_R | TGGAGCTCCACCGCGGTGGCCCTGTTTACTTGTTCCGATAAAAAG | |
| cylEmut_Conf_F | CCAACGAAGCCACTGTCTCT | Sequencing primer to confirm barcoded stop codon in the cylE gene |
| ΔsrtA_DS_F | GCGATGAGTGGCAGGGCGGGGCGTAAAAGGTAGTTAGAATTATGAAATTAAAGGCTGTTC | Amplifies downstream homology arm for ΔsrtA mutagenesis cassette; overlap extension PCR compatible |
| ΔsrtA_DS_R | GCGCGCGCCTCGAGGCGTTTTAACTTTCGTGTCTCTAGATTCATCATAATTCAGAAGT | |
| ΔsrtA_GA_F | GCTTCCAAGGAGCTAAAGAGGAATTCGAAAAGCCCTGAC | Amplifies ΔsrtA mutagenesis cassette for Gibson assembly into pMBsacB NotI/XhoI digest |
| ΔsrtA_GA_R | ATTCACTACTTTTAGTTAAGTTATTTGTTAACTGTTAATTGTCC | |
| ΔsrtA_US_F | GCGCGCGCGGATCCAATTTAGGGCGTGTGTATCGTTTGAAAGCATAT | Amplifies upstream homology arm for ΔsrtA mutagenesis cassette; overlap extension PCR compatible |
| ΔsrtA_US_R | GGTGGTATATCCAGTGATTTTTTTCTCCATGAAACCATGTGATTTTTTTTTATT | |
| p23sacB_cassette_F | GCTTCCAAGGAGCTAAAGAGGTCCTTAACTAAAAGTAGTGAATTTTTGATTTTTG | Amplifies p23 promoter-sacB gene from pSacB23 for Gibson assembly into pHY304 AvaII digest |
| p23sacB_cassette_R | AAATTCCCCGTAGGCGCTAGGCATGCGTTATTTGTTAACTGTTAATTGTCC | |
| pMBsacB_MCS_F | CAATACGCAAACCGCCTCTC | Binds pMBsacB, for use with “farout” primers for confirmation of single-cross insertion |
| sacB_pORI23_F | TATGAATGACAATGATGTTGATGAACATCAAAAAGTTTGC | Amplifies sacB from B. subtilis 168 for Gibson assembly into pORI23 BamHI digest |
| sacB_pORI23_R | AGCTTGGCTGCAGGTCGACGTTATTTGTTAACTGTTAATTGTCC | |
| srtA_farout_F | AGAGCACAAAAACGTGGAGG | Binds GBS chromosome outside of the ΔsrtA mutagenesis cassette region, for confirming single- and double-crossover events at the srtA locus |
| srtA_farout_R | ACTGCTAAGGTATCGTTAGACCC |
Transformation of GBS with pSacB23 and pMBsacB.
Electrocompetent GBS suitable for transformation with sacB-containing plasmids was prepared following methods outlined by Holo and Nes (31) and Framson et al. (30), modified to prevent toxicity from sucrose osmoprotectant.
Single 10/84 or A909 colonies from TS agar plates were used to seed 5 ml M17 (product number 218561; BD Difco) liquid cultures supplemented with 0.5% glucose, which were grown at 37°C to stationary phase. A total of 500 μl from these cultures was then used to seed filter-sterilized 50 ml M17 plus 0.5% glucose, 2.5% (A909) or 0.6% (10/84) glycine, and 25% (mass/mass) PEG 6000.
Following overnight growth at 37°C, this culture was diluted in 130 ml of the same prewarmed medium and allowed to grow for 1 h. The entire volume was pelleted at 3,200 × g, then washed twice in ice cold 25% PEG 6000 plus 10% glycerol. Following these washes, the samples were resuspended in 1 ml of wash solution and either used immediately for transformation or stored in aliquots at −80°C.
Electroporation and transformation of competent GBS were performed as described by Holo and Nes (31), except that sucrose in the outgrowth medium was replaced with 25% PEG 6000, and—in the case of pMBsacB—outgrowth was performed at 28°C instead of 37°C.
Sucrose killing assays.
Wild-type GBS or transformants were grown overnight in appropriate antibiotic selection without supplemental sucrose. For killing assays on agar plates, overnight cultures were serially diluted and plated directly on TS agar with appropriate antibiotic selection, with or without supplemental sucrose. CFU quantification was performed after 1 to 2 days of growth. For killing assays during planktonic growth, the overnight cultures were diluted 1:50 in broth without sucrose, grown to log phase, and then exposed to sucrose supplementation (or control outgrowth with only sterile water added to the broth) for two hours, at which time the cultures were diluted and plated on appropriate solid medium for CFU quantification after 1 to 2 days of growth.
Transmission electron microscopy.
Strain 10/84 transformed with pMBsacB or pHY304 was grown to mid-log phase in selective broth. That culture was used to seed a new culture with or without supplemental sucrose. After outgrowth to early mid-log phase, the bacteria were fixed with 2.5% glutaraldehyde and 2% paraformaldehyde and washed with cacodylate buffer (50 mM, pH 7.2), then postfixed with 2% osmium tetroxide. Bacteria were embedded in 2% agar, then cut and stained in the dark with 0.5% (wt/vol) uranyl acetate. Samples were dehydrated with alcohol, transferred to propylene oxide/Epon mixtures and finally embedded in EMbed 812 (Electron Microscopy Sciences, Hatfield, PA). Thin sections were cut, adsorbed on electron microscope grids, and stained with uranyl acetate and lead citrate. Stained grids were then imaged in a Philips CM12 electron microscope (FEI, Eindhoven, The Netherlands) and photographed with a Gatan (4k × 2.7k) digital camera (Gatan, Inc., Pleasanton, CA).
Construction of pMBsacB-ΔsrtA and pMBsacB-cylEmut.
The ΔsrtA mutagenesis cassette consisted of the cat gene with 822-bp upstream and 795-bp downstream homology arms that matched the 10/84 chromosomal regions flanking the srtA gene. The three fragments were PCR amplified from template DNA (10/84 genomic DNA or pDC123, a shuttle vector containing cat). The primers used to generate the upstream, coding, and downstream cassette regions of the mutagenesis cassette were ΔsrtA_US_F/R, cat_F/R, and ΔsrtA_DS_F/R, respectively.
Next, overlap extension PCR was used to join the three regions (34). The upstream homology arm was first joined to cat, after which the downstream homology arm was attached. The two-fragment intermediate and the final cassette were gel extracted and used for TOPO cloning into pCR2.1-TOPO, followed by Sanger sequence confirmation (data not shown). After successful construction, the complete cassette was digested out of pCR2.1-TOPO with NotI and KpnI and then ligated with T4 ligase overnight at 16°C into pHY304 double-digested with the same restriction enzymes. For insertion of the ΔsrtA cassette into pMBsacB, the construct was amplified from pHY304-ΔsrtA using primers ΔsrtA_GA_F and ΔsrtA_GA_R and then cloned into pMBsacB at the NotI and XhoI sites using Gibson assembly.
The 1,500-bp cylEmut cassette was amplified from GBS strain AR1598, a derivative of 10/84 in which the cylE gene has a barcoded premature stop codon (Fig. 6B), using primers cylEmut_GA_F and cylEmut_GA_R. This amplicon was similarly cloned into the NotI and XhoI sites of pMBsacB using Gibson assembly.
Allelic exchange using sucrose counterselection against pMBsacB.
After transformation of GBS, using the method described above, with pMBsacB (or pHY304 control) bearing a mutagenesis cassette (ΔsrtA or cylEmut), successful transformants were grown in TS broth with appropriate antibiotic selection (erythromycin with or without chloramphenicol) at 28°C. Sucrose sensitivity of the transformants was confirmed by plating serial dilutions on TS agar with 0.5 M (for A909) or 0.75 M (for 10/84) sucrose and appropriate antibiotic selection at 28°C.
To generate single-cross intermediates, after overnight growth, transformants were serially passaged three times at 28°C at a 1:100 dilution with erythromycin selection. The third passage was then used to seed (with a 1:100 dilution) another culture at 37°C with erythromycin selection, which was grown overnight. Serial dilutions of the final culture were plated on TS agar with erythromycin at 37°C. Individual colonies were grown and tested for sucrose sensitivity. Genomic DNA was also extracted and tested by PCR for proper vector insertion using either cylE_farout_F or srtA_farout_F, which match chromosomal sites outside of the homology arms, and pMBsacB_MCS_F, which binds pMBsacB and pHY304 upstream of the cloning sites used in this study (data not shown).
Single-cross intermediate strains with the correct sucrose sensitivity phenotype and PCR-confirmed genotype were then grown in TS broth without antibiotics at 28°C and passaged after overnight growth three times at a 1:100 dilution in order to enrich for spontaneous double-cross events. To counterselect against pMBsacB, the third passage was used to seed TS broth with sucrose at 37°C. In srtA knockout experiments, chloramphenicol was added to the sucrose-containing broth. The sucrose culture was similarly passaged three times at 37°C, and then serial dilutions were plated on TS agar with or without chloramphenicol. Simultaneous plating on erythromycin-containing TS agar (with or without chloramphenicol) was used to quantify the effectiveness of counterselection against pMBsacB.
Knockout candidates from the non-erythromycin plate were confirmed to be erythromycin sensitive by patching to a new plate. Genomic DNA extraction followed by PCR and Sanger sequencing confirmed plasmid excision and the correct knockout DNA sequence. For the srtA knockout, the region was amplified using primers srtA_farout_F/R, and these amplicons were sequenced using srtA_farout_F and cat_F primers. For the cylE knockout, the region was amplified using cylE_farout_F/R, and the barcoded mutation was confirmed by sequencing with the cylEmut_conf_F primer.
The pHY304 knockout controls were subjected to the same steps, including sucrose exposure, unless otherwise noted. Control conditions were identical except that the three final passages of the single-cross strain at 37°C were performed in the absence of sucrose.
ACKNOWLEDGMENTS
We thank the NYU Langone Health DART Microscopy Laboratory for consultation and assistance with transmission electron microscopy work.
This work was supported by NIH/NIAID grant K08AI132555 to T.A.H. and grant R56 AI136499 to A.J.R.
A.J.R. has served as a consultant to Pfizer. The other authors have no financial or other conflicts of interest to disclose.
REFERENCES
- 1.Baker CJ. 2013. The spectrum of perinatal group B streptococcal disease. Vaccine 31:D3–D6. doi: 10.1016/j.vaccine.2013.02.030. [DOI] [PubMed] [Google Scholar]
- 2.Stoll BJ, Hansen NI, Sánchez PJ, Faix RG, Poindexter BB, Van Meurs KP, Bizzarro MJ, Goldberg RN, Frantz ID, Hale EC, Shankaran S, Kennedy K, Carlo WA, Watterberg KL, Bell EF, Walsh MC, Schibler K, Laptook AR, Shane AL, Schrag SJ, Das A, Higgins RD, Eunice Kennedy Shriver National Institute Of Child Health Human Development Neonatal Research Network. 2011. Early onset neonatal sepsis: the burden of group B streptococcal and E. coli disease continues. Pediatrics 127:817–826. doi: 10.1542/peds.2010-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schrag SJ, Farley MM, Petit S, Reingold A, Weston EJ, Pondo T, Hudson Jain J, Lynfield R. 2016. Epidemiology of invasive early-onset neonatal sepsis, 2005 to 2014. Pediatrics 138:e20162013. doi: 10.1542/peds.2016-2013. [DOI] [PubMed] [Google Scholar]
- 4.Muñoz P, Llancaqueo A, Rodríguez-Créixems M, Peláez T, Martin L, Bouza E. 1997. Group B streptococcus bacteremia in nonpregnant adults. Arch Intern Med 157:213–216. [DOI] [PubMed] [Google Scholar]
- 5.Reingold A, Watt JP. 2015. Group B streptococcus infections of soft tissue and bone in California adults, 1995–2012. Epidemiol Infect 143:3343–3350. doi: 10.1017/S0950268815000606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kayansamruaj P, Pirarat N, Hirono I, Rodkhum C. 2014. Increasing of temperature induces pathogenicity of Streptococcus agalactiae and the up-regulation of inflammatory related genes in infected Nile tilapia (Oreochromis niloticus). Vet Microbiol 172:265–271. doi: 10.1016/j.vetmic.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 7.Botelho ACN, Ferreira AFM, Fracalanzza SEL, Teixeira LM, Pinto TCA. 2018. A perspective on the potential zoonotic role of Streptococcus agalactiae: searching for a missing link in alternative transmission routes. Front Microbiol 9:608. doi: 10.3389/fmicb.2018.00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalimuddin S, Chen SL, Lim CTK, Koh TH, Tan TY, Kam M, Wong CW, Mehershahi KS, Chau ML, Ng LC, Tang WY, Badaruddin H, Teo J, Apisarnthanarak A, Suwantarat N, Ip M, Holden MTG, Hsu LY, Barkham T, Singapore Group B Streptococcus Consortium. 2017. 2015 epidemic of severe Streptococcus agalactiae sequence type 283 infections in Singapore associated with the consumption of raw freshwater fish: a detailed analysis of clinical, epidemiological, and bacterial sequencing data. Clin Infect Dis 64:S145–S152. doi: 10.1093/cid/cix021. [DOI] [PubMed] [Google Scholar]
- 9.Tan S, Lin Y, Foo K, Koh HF, Tow C, Zhang Y, Ang LW, Cui L, Badaruddin H, Ooi PL, Lin RTP, Cutter J. 2016. Group B Streptococcus serotype III sequence type 283 bacteremia associated with consumption of raw fish, Singapore. Emerg Infect Dis 22:1970–1973. doi: 10.3201/eid2211.160210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Håvarstein LS, Hakenbeck R, Gaustad P. 1997. Natural competence in the genus Streptococcus: evidence that streptococci can change pherotype by interspecies recombinational exchanges. J Bacteriol 179:6589–6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brochet M, Rusniok C, Couve E, Dramsi S, Poyart C, Trieu-Cuot P, Kunst F, Glaser P. 2008. Shaping a bacterial genome by large chromosomal replacements, the evolutionary history of Streptococcus agalactiae. Proc Natl Acad Sci U S A 105:15961–15966. doi: 10.1073/pnas.0803654105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yim HH, Rubens CE. 1998. Site-specific homologous recombination mutagenesis in group B streptococci. Methods Cell Sci 20:13–20. doi: 10.1023/A:1009810002276. [DOI] [Google Scholar]
- 13.Sheen TR, Jimenez A, Wang N-Y, Banerjee A, van Sorge NM, Doran KS. 2011. Serine-rich repeat proteins and pili promote Streptococcus agalactiae colonization of the vaginal tract. J Bacteriol 193:6834–6842. doi: 10.1128/JB.00094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quach D, van Sorge NM, Kristian SA, Bryan JD, Shelver DW, Doran KS. 2009. The CiaR response regulator in group B Streptococcus promotes intracellular survival and resistance to innate immune defenses. J Bacteriol 191:2023–2032. doi: 10.1128/JB.01216-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura GS, Bratt DS, Yim HH, Nittayajarn A. 2005. Use of glnQ as a counterselectable marker for creation of allelic exchange mutations in group B streptococci. Appl Environ Microbiol 71:587–590. doi: 10.1128/AEM.71.1.587-590.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meng G, Fütterer K. 2003. Structural framework of fructosyl transfer in Bacillus subtilis levansucrase. Nat Struct Biol 10:935–941. doi: 10.1038/nsb974. [DOI] [PubMed] [Google Scholar]
- 17.Marvasi M, Visscher PT, Casillas Martinez L. 2010. Exopolymeric substances (EPS) from Bacillus subtilis: polymers and genes encoding their synthesis. FEMS Microbiol Lett 313:1–9. doi: 10.1111/j.1574-6968.2010.02085.x. [DOI] [PubMed] [Google Scholar]
- 18.Porras-Domínguez JR, Ávila-Fernández Á, Miranda-Molina A, Rodríguez-Alegría ME, Munguía AL. 2015. Bacillus subtilis 168 levansucrase (SacB) activity affects average levan molecular weight. Carbohydr Polym 132:338–344. doi: 10.1016/j.carbpol.2015.06.056. [DOI] [PubMed] [Google Scholar]
- 19.Ortiz-Soto ME, Rudiño-Piñera E, Rodriguez-Alegria ME, Munguia AL. 2009. Evaluation of cross-linked aggregates from purified Bacillus subtilis levansucrase mutants for transfructosylation reactions. BMC Biotechnol 9:68. doi: 10.1186/1472-6750-9-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abdel-Fattah AF, Mahmoud DAR, Esawy MAT. 2005. Production of levansucrase from Bacillus subtilis NRC 33a and enzymic synthesis of levan and fructo-oligosaccharides. Curr Microbiol 51:402–407. doi: 10.1007/s00284-005-0111-1. [DOI] [PubMed] [Google Scholar]
- 21.Ortiz-Soto ME, Rivera M, Rudiño-Piñera E, Olvera C, López-Munguía A. 2008. Selected mutations in Bacillus subtilis levansucrase semi-conserved regions affecting its biochemical properties. Protein Eng Des Sel 21:589–595. doi: 10.1093/protein/gzn036. [DOI] [PubMed] [Google Scholar]
- 22.Marx CJ. 2008. Development of a broad-host-range sacB-based vector for unmarked allelic exchange. BMC Res Notes 1:1. doi: 10.1186/1756-0500-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Thompson CM, Lipsitch M. 2014. A modified Janus cassette (Sweet Janus) to improve allelic replacement efficiency by high-stringency negative selection in Streptococcus pneumoniae. PLoS One 9:e100510. doi: 10.1371/journal.pone.0100510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gay P, Le Coq D, Steinmetz M, Berkelman T, Kado CI. 1985. Positive selection procedure for entrapment of insertion sequence elements in Gram-negative bacteria. J Bacteriol 164:918–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamoun S, Tola E, Kamdar H, Kado CI. 1992. Rapid generation of directed and unmarked deletions in Xanthomonas. Mol Microbiol 6:809–816. [DOI] [PubMed] [Google Scholar]
- 26.Blomfield IC, Vaughn V, Rest RF, Eisenstein BI. 1991. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature‐sensitive pSC101 replicon. Mol Microbiol 5:1447–1457. doi: 10.1111/j.1365-2958.1991.tb00791.x. [DOI] [PubMed] [Google Scholar]
- 27.Pelicic V, Reyrat JM, Gicquel B. 1996. Expression of the Bacillus subtilis sacB gene confers sucrose sensitivity on mycobacteria. J Bacteriol 178:1197–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jäger W, Schäfer A, Pühler A, Labes G, Wohlleben W. 1992. Expression of the Bacillus subtilis sacB gene leads to sucrose sensitivity in the gram-positive bacterium Corynebacterium glutamicum but not in Streptomyces lividans. J Bacteriol 174:5462–5465. doi: 10.1128/jb.174.16.5462-5465.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun X, Yang D, Wang Y, Geng H, He X, Liu H. 2013. Development of a markerless gene deletion system for Streptococcus zooepidemicus: functional characterization of hyaluronan synthase gene. Appl Microbiol Biotechnol 97:8629–8636. doi: 10.1007/s00253-013-5058-8. [DOI] [PubMed] [Google Scholar]
- 30.Framson PE, Nittayajarn A, Merry J, Youngman P, Rubens CE. 1997. New genetic techniques for group B streptococci: high-efficiency transformation, maintenance of temperature-sensitive pWV01 plasmids, and mutagenesis with Tn917. Appl Environ Microbiol 63:3539–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holo H, Nes IF. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol 55:3119–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Canedo M, Jimenez-Estrada M, Cassani J, López-Munguía A. 1999. Production of maltosylfructose (erlose) with levansucrase from Bacillus Subtilis. Biocatal Biotransformation 16:475–485. doi: 10.3109/10242429909015223. [DOI] [Google Scholar]
- 33.Seibel J, Moraru R, Götze S, Buchholz K, Na'amnieh S, Pawlowski A, Hecht H-J. 2006. Synthesis of sucrose analogues and the mechanism of action of Bacillus subtilis fructosyltransferase (levansucrase). Carbohydr Res 341:2335–2349. doi: 10.1016/j.carres.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 34.Lalioui L, Pellegrini E, Dramsi S, Baptista M, Bourgeois N, Doucet-Populaire F, Rusniok C, Zouine M, Glaser P, Kunst F, Poyart C, Trieu-Cuot P. 2005. The SrtA sortase of Streptococcus agalactiae is required for cell wall anchoring of proteins containing the LPXTG motif, for adhesion to epithelial cells, and for colonization of the mouse intestine. Infect Immun 73:3342–3350. doi: 10.1128/IAI.73.6.3342-3350.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whidbey C, Harrell MI, Burnside K, Ngo L, Becraft AK, Iyer LM, Aravind L, Hitti J, Waldorf KMA, Rajagopal L. 2013. A hemolytic pigment of group B Streptococcus allows bacterial penetration of human placenta. J Exp Med 210:1265–1281. doi: 10.1084/jem.20122753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosa-Fraile M, Dramsi S, Spellerberg B. 2014. Group B streptococcal haemolysin and pigment, a tale of twins. FEMS Microbiol Rev 38:932–946. doi: 10.1111/1574-6976.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Randis TM, Gelber SE, Hooven TA, Abellar RG, Akabas LH, Lewis EL, Walker LB, Byland LM, Nizet V, Ratner AJ. 2014. Group B Streptococcus β-hemolysin/cytolysin breaches maternal-fetal barriers to cause preterm birth and intrauterine fetal demise in vivo. J Infect Dis 210:265–273. doi: 10.1093/infdis/jiu067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Link AJ, Jeong KJ, Georgiou G. 2007. Beyond toothpicks: new methods for isolating mutant bacteria. Nat Rev Microbiol 5:680–688. doi: 10.1038/nrmicro1715. [DOI] [PubMed] [Google Scholar]
- 39.Zeigler DR, Prágai Z, Rodriguez S, Chevreux B, Muffler A, Albert T, Bai R, Wyss M, Perkins JB. 2008. The origins of 168, W23, and other Bacillus subtilis legacy strains. J Bacteriol 190:6983–6995. doi: 10.1128/JB.00722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hooven TA, Randis TM, Daugherty SC, Narechania A, Planet PJ, Tettelin H, Ratner AJ. 2014. Complete genome sequence of Streptococcus agalactiae CNCTC 10/84, a hypervirulent sequence type 26 strain. Genome Announc 2:e01338-14. doi: 10.1128/genomeA.01338-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lancefield RC, McCarty M, Everly WN. 1975. Multiple mouse-protective antibodies directed against group B streptococci. Special reference to antibodies effective against protein antigens. J Exp Med 142:165–179. doi: 10.1084/jem.142.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaffin DO, Rubens CE. 1998. Blue/white screening of recombinant plasmids in Gram-positive bacteria by interruption of alkaline phosphatase gene (phoZ) expression. Gene 219:91–99. [DOI] [PubMed] [Google Scholar]
- 43.Que YA, Haefliger JA, Francioli P, Moreillon P. 2000. Expression of Staphylococcus aureus clumping factor A in Lactococcus lactis subsp. cremoris using a new shuttle vector. Infect Immun 68:3516–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]