Species are the fundamental units of ecology and evolution, and speciation leads to the astounding diversity of life on Earth. Studying speciation is thus of great significance to understand, protect, and exploit biodiversity, but it is a challenge in the microbial world. In this study, using population genomics, we placed Streptomyces albidoflavus strains in a spectrum of speciation and showed that the genetic differences between phylogenomic clusters evolved mainly by environmental selection and gene-specific sweeps. These findings highlight the role of ecology in structuring recombining bacterial species, making a step toward a deeper understanding of microbial speciation. Our results also raise concerns of an underrated microbial diversity at the intraspecies level, which can be utilized for mining of ecologically relevant natural products.
KEYWORDS: adaptive evolution, ecological differentiation, population genomics, speciation, streptomycetes
ABSTRACT
Deciphering the genomic variation that represents microevolutionary processes toward species divergence is key to understanding microbial speciation, which has long been under debate. Streptomycetes are filamentous bacteria that are ubiquitous in nature and the richest source of antibiotics; however, their speciation processes remain unknown. To tackle this issue, we performed a comprehensive population genomics analysis on Streptomyces albidoflavus residing in different habitats and with a worldwide distribution and identified and characterized the foundational changes within the species. We detected three well-defined phylogenomic clades, of which clades I and III mainly contained free-living (soil/marine) and insect-associated strains, respectively, and clade II had a mixed origin. By performing genome-wide association studies (GWAS), we identified a number of genetic variants associated with free-living or entomic (denoting or relating to insects) habitats in both the accessory and core genomes. These variants contributed collectively to the population structure and had annotated or confirmed functions that likely facilitate differential adaptation of the species. In addition, we detected higher levels of homologous recombination within each clade and in the free-living group than within the whole species and in the entomic group. A subset of the insect-associated strains (clade III) showed a relatively independent evolutionary trajectory with more symbiosis-favorable genes but little genetic interchange with the other lineages. Our results demonstrate that ecological adaptation promotes genetic differentiation in S. albidoflavus, suggesting a model of ecological speciation with gene flow in streptomycetes.
IMPORTANCE Species are the fundamental units of ecology and evolution, and speciation leads to the astounding diversity of life on Earth. Studying speciation is thus of great significance to understand, protect, and exploit biodiversity, but it is a challenge in the microbial world. In this study, using population genomics, we placed Streptomyces albidoflavus strains in a spectrum of speciation and showed that the genetic differences between phylogenomic clusters evolved mainly by environmental selection and gene-specific sweeps. These findings highlight the role of ecology in structuring recombining bacterial species, making a step toward a deeper understanding of microbial speciation. Our results also raise concerns of an underrated microbial diversity at the intraspecies level, which can be utilized for mining of ecologically relevant natural products.
INTRODUCTION
Debate has been ongoing for decades about the meaning and driving forces of speciation in microbes, because of their enormous genetic diversity, promiscuous gene flow, and asexual reproduction (1, 2). The availability of multiple genome sequences from the same species have facilitated attempts to systematically address basic questions in microbial speciation. The process of microbial speciation can be defined as any stage of the dynamic microevolutionary process of ecological and genetic differentiation (3). To depict this process, two major parallel concepts based on ecological divergence and barriers to recombination and their integration have been proposed (4–7). Microevolution is attributed to four main mechanisms: gene flow, mutation, natural selection, and genetic drift (8). As one of the main forms of gene flow, horizontal gene transfer (HGT), mediated by either homologous recombination or nonhomologous recombination (9), has been widely considered one of the most important factors affecting microbial evolution (10–12). Moreover, HGT and habitat isolation have been further emphasized as the key points of ecological speciation in bacteria and archaea (12). Meanwhile, recent studies have demonstrated that ecological divergence and natural selection play pivotal roles in microbial speciation (9, 13, 14), although geographic isolation and genetic drift also affect the distribution patterns of microorganisms and jointly drive microbial biodiversity (15, 16). Based on these, Shapiro et al. (3, 9, 17) propose that early stages of the microbial speciation process are driven either by gene-specific selective sweeps in recombining populations or by genome-wide selective sweeps in clonal populations as microbes adapt to new niches, and more advanced stages involve barriers to gene flow and divergent natural selection. Yet it is still unclear which are the foundational genotypic and phenotypic changes during microbial speciation and how the microevolution mechanisms leave imprints across the genomes.
The genus Streptomyces is an important source of useful natural products and has become one of the most intensively genome-sequenced genera (18, 19). Streptomycetes inhabit various habitats, providing ideal materials for studies of microbial ecology and evolution (20, 21). It has been proved that streptomycetes have linear chromosomes and plastic genomes (22) with high levels of recombination (13, 23–25), yet genomic analyses that trace their microevolutionary processes are scarce. Streptomyces albidoflavus is one of the early described species of Streptomyces and also an important source of bioactive metabolites (26–28). S. albidoflavus strains have a minimized genome (29) but can survive and reproduce in a range of habitats worldwide (13), which raises questions about how they cope with various environments and what changes have occurred in their genomes during local adaptation.
In a recent study based on multilocus sequence analysis (MLSA), we described the population structure of S. albidoflavus in relation to three habitat sources, insects, sea, and soil, indicating the importance of ecological difference in shaping the structure (13). Here we report a comprehensive population genomics analysis on 30 S. albidoflavus strains, aimed at uncovering the foundational changes between populations and the microevolutionary mechanisms accounting for adaptive divergences. Our results reveal a series of habitat-associated genes and alleles that are conducive to better survival in related environments and to the formation of population structure, highlighting the role of ecology in the microevolution and structuring of recombining bacterial species. Insights gained expand our knowledge of the genetic basis of environmental adaptation and the mechanisms of evolution in bacteria.
RESULTS
Genomic features of S. albidoflavus.
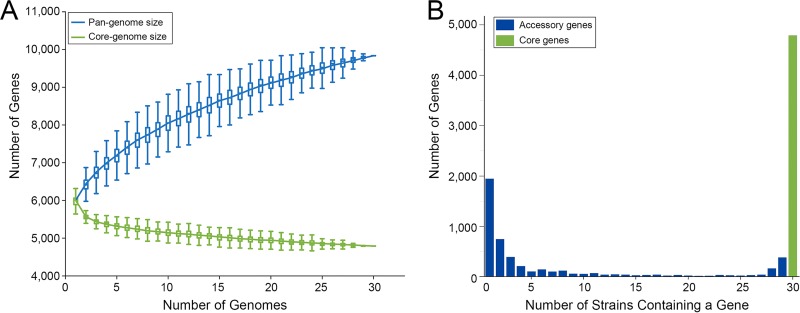
A summary of the general characteristics of the genomes of 30 S. albidoflavus strains and 3 closely related strains used in this study is given in Table 1. The genomes showed a highly conserved chromosome structure despite the fact that they were derived from strains inhabiting distinct habitats. The S. albidoflavus genomes showed >98.15% average nucleotide identity (ANI) values with one another (average ANI = 98.68%) but <96.15% ANI values with the genomes of the remaining three strains (PVA 94-07, GBA 94-10, and SCA2-2), which were defined as outgroups. Within S. albidoflavus, the genome size ranged from 6.84 to 7.61 Mb (mean, 7.09 Mb), which was smaller than most streptomycete genomes (18). The average number of filtered coding sequences (CDSs) was about 6,112 (range from 5,860 to 6,513), and 67.9% of these CDSs could be functionally categorized using the Clusters of Orthologous Groups (COG) database (30). Based on the gene content table obtained by OrthoMCL (31), the 30 S. albidoflavus strains had a pan-genome of 9,838 genes and a core genome of 4,791 genes, including 4,663 single-copy core genes (Fig. 1). The core genome represented 73.6% to 81.8% of the gene content of each strain and contained 20 core secondary metabolite biosynthetic gene clusters predicted by antiSMASH 3.0 (32) (Table S1). Although the S. albidoflavus strains were closely related and the core genome has approached an asymptote, the size of the pan-genome did not approach a constant as more genomes were sampled (α = 0.63, estimated by power-law regression), and most (3,300) accessory genes were unique or rare (existing in 1 to 4 strains) (Fig. 1). To define possible differences in the functions encoded by the core and accessory genomes of S. albidoflavus, a COG functional classification for each orthologous group (OG) was performed. The accessory genome was significantly enriched for genes of the prophages and transposons (COG X), replication, recombination and repair (COG L), defense mechanisms (COG V), and transcription (COG K) categories (P < 0.01; df = 1) (Fig. S1).
TABLE 1.
Genomic features and sources of strains used in this studya
| Strain | Genome size (Mb) | No. of contigs | Contig_N50 length (bp) | ANI (%) to J1074 | No. of filtered CDSs | Clade | Habitat (origin) | GenBank accession no. | Reference(s) |
|---|---|---|---|---|---|---|---|---|---|
| J1074 | 6.84 | 1 | — | — | 5,860 | I | Soil (—) | NC_020990 | 29 |
| FXJ2.339 | 7.12 | 49 | 377,764 | 99.02 | 6,147 | I | Soil (Kaifeng, Henan Province, China) | PKLK00000000 | 13 |
| NBRC 100770 | 7.16 | 41 | 334,066 | 98.93 | 6,126 | I | Soil (Caucasus) | PKLL00000000 | 13 |
| CGMCC 4.1677 | 6.98 | 66 | 364,125 | 98.97 | 6,018 | I | Muschelkalk (Werra, Germany) | PKLM00000000 | 13 |
| NBRC 12790 | 6.95 | 53 | 309,124 | 98.99 | 5,977 | I | River bank slime (Böhmischbruck, Upper Palatinate, Germany) | PKLN00000000 | 13 |
| DSM 40455T | 6.97 | 66 | 332,544 | 98.91 | 5,952 | I | Contaminated plate (Rome, Italy) | PKLO00000000 | 13 |
| NBRC 13083 | 7.04 | 56 | 278,969 | 98.95 | 6,118 | I | Potato scab (United Kingdom) | PKLP00000000 | 13 |
| “S. wadayamensis” A23 | 7.06 | 928 | 15,752 | 98.68 | 6,167 | I | XXXPlant tissue (Sao Paulo, Brazil) | JHDU00000000 | 89, 90 |
| DSM 40233 | 7.05 | 68 | 262,674 | 98.92 | 6,077 | I | Potato (Germany) | PKLQ00000000 | 13 |
| CGMCC 4.1681 | 7.23 | 54 | 319,881 | 98.88 | 6,283 | I | Contaminated fungus plate (Commercial Solvents Corporation, USA) | PKLR00000000 | 13 |
| D62 | 7.14 | 87 | 256,416 | 98.86 | 6,203 | I | Medicinal herb (Xishuangbanna, Yunnan Province, China) | PKLS00000000 | 13 |
| FXJ8.011 | 7.19 | 48 | 383,364 | 98.91 | 6,203 | I | Deep-sea water (southern Indian Ocean) | PKLT00000000 | 13 |
| FXJ8.008 | 7.17 | 147 | 97,051 | 98.89 | 6,245 | I | Deep-sea water (southern Indian Ocean) | PKLU00000000 | 13 |
| CR13 | 6.95 | 75 | 182,021 | 98.94 | 6,029 | I | Imperial moth (Guanacaste Conservation Area, Costa Rica) | PKLV00000000 | 13 |
| CGMCC 4.1615 | 6.96 | 39 | 447,068 | 98.73 | 5,966 | II | Soil (Champavathi River, Andhra Pradesh, India) | PKLW00000000 | 13 |
| CNY228 | 6.96 | 51 | 267,951 | 98.71 | 5,979 | II | Neritic sediment (coastal California, USA) | ARIN00000000 | — |
| FXJ8.031 | 6.96 | 39 | 494,739 | 98.73 | 5,967 | II | Deep-sea water (southwest Indian Ocean) | PKLX00000000 | 13 |
| FXJ6.189 | 7.36 | 42 | 368,474 | 98.72 | 6,465 | II | South China Sea sponge (Wanning port, Hainan Province, China) | PKLY00000000 | This study |
| CR46 | 7.05 | 74 | 247,280 | 98.69 | 6,071 | II | Owl butterfly (San José, Costa Rica) | PKNU00000000 | 13 |
| CR19 | 7.10 | 44 | 514,855 | 98.69 | 6,087 | II | Imperial moth (Guanacaste Conservation Area, Costa Rica) | PKNV00000000 | 13 |
| Ma24 | 6.96 | 56 | 317,189 | 98.74 | 5,981 | II | Imperial moth (Guanacaste Conservation Area, Costa Rica) | PKNW00000000 | 13 |
| LaPpAH-202 | 7.00 | 37 | 406,114 | 98.64 | 6,001 | II | Unknown insect (Cameroon) | ARDM00000000 | 91 |
| CR33 | 7.16 | 61 | 352,542 | 98.53 | 6,162 | III | Imperial moth (Guanacaste Conservation Area, Costa Rica) | PKNX00000000 | 13 |
| CR10 | 7.16 | 38 | 422,844 | 98.52 | 6,135 | III | Imperial moth (Guanacaste Conservation Area, Costa Rica) | PKNY00000000 | 13 |
| CR25 | 7.17 | 47 | 522,740 | 98.53 | 6,171 | III | Imperial moth (Guanacaste Conservation Area, Costa Rica) | PKNZ00000000 | 13 |
| Ma1 | 7.27 | 36 | 551,090 | 98.52 | 6,312 | III | Imperial moth (Guanacaste Conservation Area, Costa Rica) | PKOA00000000 | 13 |
| CR16 | 6.97 | 146 | 113,214 | 98.47 | 6,021 | III | Imperial moth (Guanacaste Conservation Area, Costa Rica) | PKOB00000000 | 13 |
| CR15 | 6.96 | 60 | 266,746 | 98.49 | 6,018 | III | Imperial moth (Guanacaste Conservation Area, Costa Rica) | PKOC00000000 | 13 |
| CR47 | 7.13 | 38 | 508,717 | 98.55 | 6,118 | III | Owl butterfly (San José, Costa Rica) | PKOD00000000 | 13 |
| S4 | 7.61 | 269 | 84,643 | 98.53 | 6,513 | III | Leaf-cutting ant (Trinidad) | CADY00000000 | 92 |
| PVA 94-07 | 7.01 | 34 | 527,991 | 96.15 | 5,916 | Outgroup | Shallow-water marine sponge (Trondheim fjord, Norway) | ASHE00000000 | 21 |
| GBA 94-10 | 7.03 | 20 | 559,062 | 96.13 | 5,965 | Outgroup | Shallow-water marine sponge (Trondheim fjord, Norway) | ASHF00000000 | 21 |
| SCA2-2 | 7.05 | 48 | 457,544 | 95.82 | 6,071 | Outgroup | Earthworm (Nanchang, Jiangxi Province, China) | PKMX00000000 | 13 |
—, information unavailable.
FIG 1.
The pan-genome, core genome, and accessory genome profiles of S. albidoflavus. (A) The sizes of core and pan-genomes in relation to numbers of genomes added into the gene pool. Box plots show the 25th and 75th percentiles, with medians shown as horizontal lines, and whiskers indicate the lowest and highest values within 1.5 times the interquartile range (IQR) from the first and third quartiles, respectively. The curve for the pan-genome is fitted by the power-law regression model (ypan = Apan xBpan + Cpan), with r2 = 0.999, Apan = 1488.57 ± 1.67, Bpan = 0.37, and Cpan = 4,502.37 ± 4.34. Bpan is equivalent to the parameter γ, and α (= 1 − γ) < 1 indicates that the pan-genome does not approach a constant as more genomes are sampled. The curve for the core genome is fitted by the exponential curve fit model (ycore = Acore eBcore.x + Ccore), with r2 = 0.938, Acore = 1,060.74 ± 21.92, Bcore = −0.13, and Ccore = 4,838.38 ± 5.57. (B) Distribution of genes across strains.
Phylogenomic relationship and population structure in S. albidoflavus related to habitat distribution.
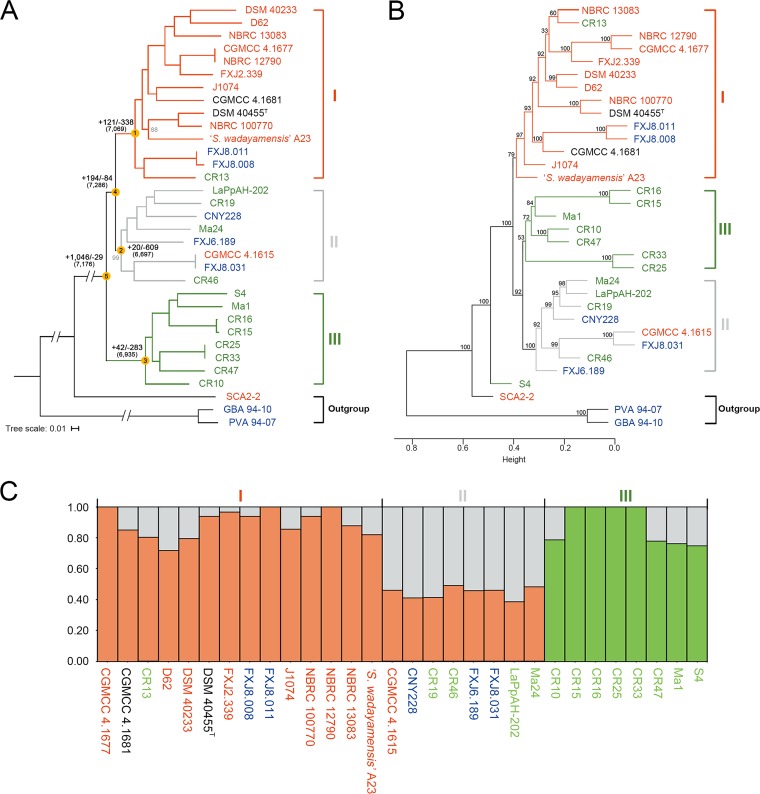
We constructed a phylogenomic maximum-likelihood (ML) tree (Fig. 2A) of the 33 strains based on concatenated single-copy core genes. Bootstrapping revealed that the tree topology was well-supported, showing three well-defined genetic clusters in S. albidoflavus, with clades I and II as sister groups. Both clades I and II contained strains isolated from different habitat types. However, most (11/14) strains in clade I could be assigned to soil (or plant-associated) habitats, and only one strain (CR13) in this clade was insect associated, while most (7/8) strains in clade II were isolated from insects and sea and only one in this clade was from soil. Clade III consisted of 8 definitely insect-associated strains isolated from imperial moth, owl butterfly, and leaf-cutting ant; this clade corresponds to clusters III and IV in our previous study (13). Ancestral reconstruction (Fig. 2A) revealed that the three clades experienced different levels of gene gain, with quite a number of the events occurring at the parental node of clades I and II (node 4) and then at the node of clade I (node 1). Substantial gene loss occurred at each clade node, notably at clade II (node 2).
FIG 2.
Phylogeny and population structure of S. albidoflavus. Strains collected from different habitats are represented in different colors (red, soil or plants; green, insects; blue, sea; black, uncertain). “I,” “II,” or “III” indicates genetic clusters or clades. (A) Maximum-likelihood phylogeny generated from concatenated 4,116 single-copy core genes of the 33 strains, including the three outgroup strains. Bootstrap values less than 100% are shown in gray at the nodes. The scale bar indicates 1% sequence divergence. Ancestral genome contents of S. albidoflavus were reconstructed using COUNT (81). The numbers of gene gain (+) and loss (−) events are indicated next to the deep nodes, and the total numbers of ancestral orthologous genes present at each of the nodes are shown in parentheses. (B) Hierarchical cluster analysis based on the presence or absence of dispensable genes in the 33 strains. Numbers above branches are bootstrap support values from 1,000 replicates. Height indicates the dissimilarity between genomes. (C) Structure plot based on concatenated 4,663 single-copy core genes of the 30 S. albidoflavus strains, showing the contribution to each S. albidoflavus strain from each of the three hypothetical ancestral populations.
A hierarchical clustering tree based on the content of dispensable genes showed three clades similar to those of the core genome tree, but with strain S4 located on the outskirts of the species and clade II gathered with clade III as sister groups (Fig. 2B). Structure (33) analysis of the S. albidoflavus strains also indicated three populations that were well in line with the three phylogenomic clades (Fig. 2C) and provided further evidence of shared genetic variants by different clades, which suggests gene flow. Clades I and II showed large amounts of genetic overlap with each other, and clade III shared only a few polymorphisms with the other clades.
Based on the isolation sources, the S. albidoflavus strains could be divided into two major ecological groups: a free-living (soil/marine) group (17 strains) and an insect-associated group (13 strains). It was obvious from the phylogeny and population structure that the strains were distributed differentially between the two ecologies, with clade I largely representing a free-living genetic cluster and clade III an obligate insect-associated cluster, while clade II was mixed.
Habitat-associated accessory OGs underlying differential adaptation.
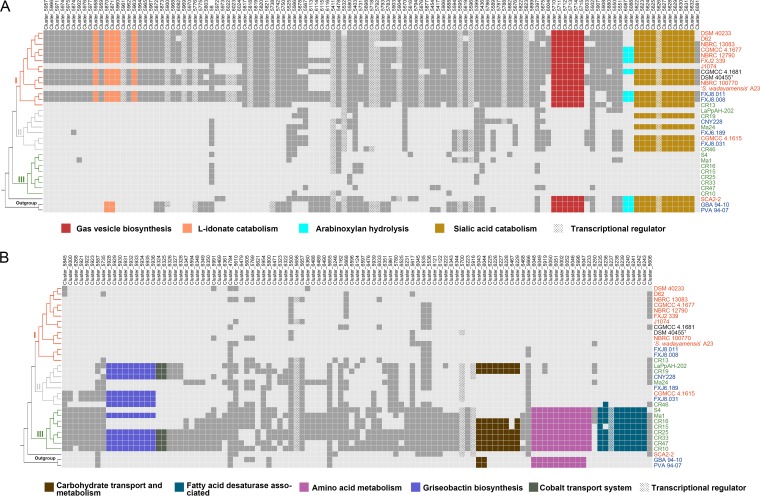
In order to record ecological variations in S. albidoflavus, we performed pan-genome-wide association studies (pan-GWAS) with Scoary (34) to identify OGs that could be associated with the free-living or entomic (denoting or relating to insects) habitat type in the accessory genome. As a result, we identified a total of 226 accessory OGs (Fig. 3; Data Set S1) showing significant habitat association (Benjamini-Hochberg P value < 0.05), of which 119 were correlated with free-living and 107 with entomic habitats. Within the S. albidoflavus phylogeny, 73% of the habitat-associated OGs were gained or lost at the nodes of the three clades (nodes 1, 2, and 3 in Fig. 2A; Data Set S1), indicating a correlation between these OGs and the population structure. Moreover, hierarchical cluster analysis based on these OGs also showed a three-clade structure consistent with that revealed by genome-based analyses, with strain S4 located in clade III, but the analysis based on the habitat-unassociated accessory OGs yielded a completely different and unstable tree topology (Fig. S2A). By mapping the habitat-associated OGs onto the phylogenomic tree, we noticed that 68 free-living habitat-associated OGs and 30 entomic habitat-associated OGs were specific for clades I and III, respectively, but none of the OGs were specific for clade II (Fig. S2B). In addition, some of the habitat-associated OGs were located adjacent to each other, forming continuous genomic regions which might perform certain complicated or special roles in extending the metabolic pathways or improving the energy utilization efficiency of the strains to handle different growth conditions.
FIG 3.
Distribution of the 226 accessory OGs significantly associated with free-living (soil/marine) (A) and entomic (denoting or relating to insects) (B) habitats. The OGs are ordered according to their relative locations in the genomes of strains NBRC 100770 (A) and CR33 (B). Dark gray and colored boxes indicate presence, with particularly described OGs in color (see key). Light gray boxes indicate absence. The tree on the left was derived from Fig. 2A; phylogenomic clades are indicated. Strains collected from different habitats are represented in different colors (red, soil or plants; green, insects; blue, sea; black, uncertain).
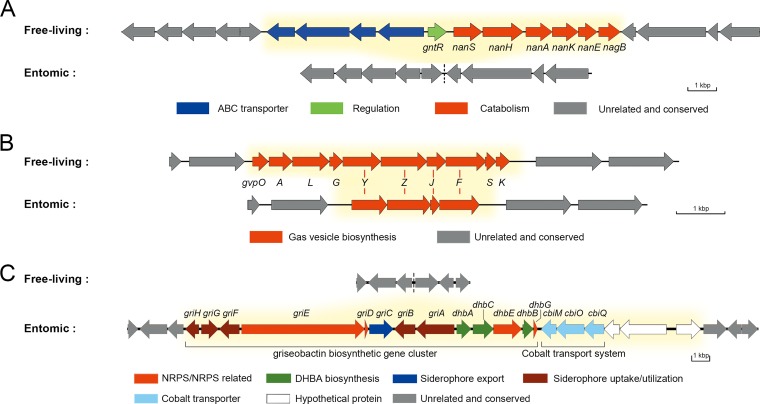
One of the free-living habitat-associated genomic regions was composed of a sialic acid catabolic gene cluster, nan (cluster_5622 to cluster_5632 [Fig. 3A and Fig. 4A]), located at ∼6.68 Mb of the chromosome. This cluster was widespread in clades I and II (17/22) and outgroup strains (3/3), enabling them to take up and utilize sialic acid as a carbon source from the surrounding environment (Fig. 5A), but was absolutely absent in clade III. Another region was the gvp gene cluster (cluster_5710 to cluster_5715) coding for gas vesicle proteins. All clade I strains had all 10 genes (gvpOALGYZJFSK) of the gvp gene cluster (35), but all clade II and III strains had only gvpYZJF (Fig. 3A and Fig. 4B). Three of the remaining gvp genes (gvpYZJ) were shorter (by codons corresponding to 35 amino acids, on average) than those in clade I strains, suggesting that the incompleteness of this gene cluster was due to gene deletion (Data Set S1). A third free-living habitat-associated region, present only in clade I stains (11/14) and located at the beginning (∼0.02 Mb) of the genome, contained five l-idonate catabolic genes (cluster_5870, -_5871, -_5958, -_5960, and -_5963 [Fig. 3A]), which probably enable the clade I strains to grow on l-idonate (36). Moreover, some free-living habitat-associated clusters could not be annotated to clear functions, several of which contained genes encoding transcriptional regulators such as TetR, MarR, and MerR (COG K [Fig. 3; Data Set S1]). In fact, a significantly greater proportion of transcriptional regulators were found in the free-living habitat-associated OGs than in the entomic habitat-associated OGs (P < 0.01; df = 1), which may give the free-living strains more possibilities facing various conditions (37).
FIG 4.
Genetic organizations of the regions containing habitat-associated gene clusters. Details of the habitat-associated accessory genes are shown in Data Set S1. Differential genes between the two ecological groups are marked with yellow background. (A) The nan gene cluster for sialic acid catabolism. (B) The gvp gene cluster for gas vesicle biosynthesis. Red lines between the two groups indicate corresponding homologous genes. (C) The gene clusters for griseobactin biosynthesis and the cobalt transport system. Genes griE, dhbA, dhbB, dhbG, and cbiO are habitat unassociated because some strains of clade I also contain corresponding homologous genes with low identity. DHBA, 2,3-dihydroxybenzoate; NRPS, nonribosomal peptide synthetase.
FIG 5.
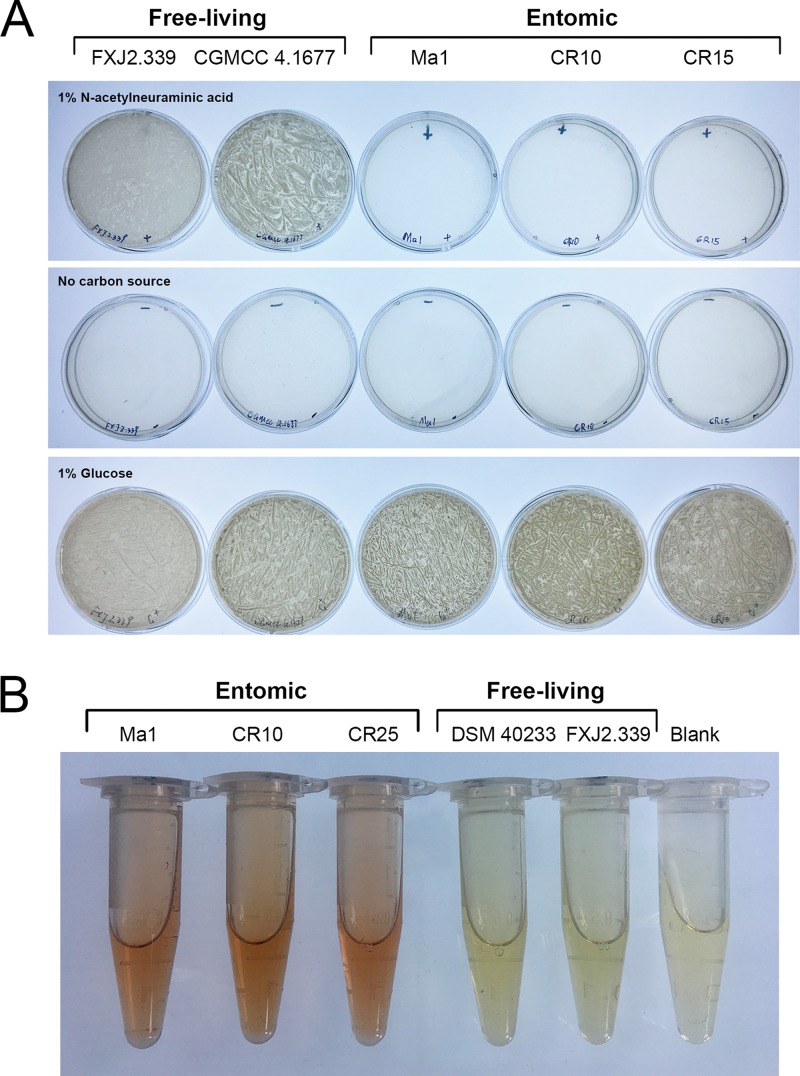
Functional verifications of sialic acid catabolic and griseobactin biosynthetic gene clusters. (A) The ability of representative free-living strains and insect-associated strains to utilize sialic acid as a sole carbon source. Strains were cultured for 5 days at 28°C on basal mineral salt medium agar plates supplemented with 1% N-acetylneuraminic acid or 1% glucose as a sole carbon source. (B) Arnow’s test of catechol-type siderophore production by representative insect-associated strains and free-living strains. The presence of a red color in the solution was recorded as a positive test for catechol.
Some entomic habitat-associated genomic regions were also found. A region located at ∼0.19 Mb of the genome encoded the biosynthetic pathway for griseobactin (cluster_5928 to cluster_5936) (38), a catechol-type siderophore, and a cobalt transport system (cbiMQO; cluster_6324 and -_6325) (39) (Fig. 3B and Fig. 4C). The griseobactin gene cluster was identified in 11 strains of clades II and III but not in clade I strains. Function of this cluster was confirmed by Arnow’s test (Fig. 5B) (40). The cbiMQO operon existed in 7 of the 11 strains and was adjacent to the griseobactin cluster. Another copy of this system was located at ∼5.46 Mb of the genome as core genes (with amino acid sequence identity of CbiMQO = 98.95% ± 0.29%) but shared a low identity (53.19% ± 0.24%) with the former. The observation that these genes shared low similarity with the other copies in the genome, together with the results of ancestral reconstruction (Data Set S1), provided evidence for HGT of this genomic region into the ancestor of the species followed by loss in clade I. Meanwhile, all strains in clade III contained a region putatively encoding amino acid metabolism (cluster_5919, -_5996, -_6002, -_6046 to -_6052, and -_6233), while strains from clades I and II only contained a truncated (47.8% shorter) major facilitator superfamily (MFS) transporter gene in the same region (Fig. 3B; Fig. S3). The remaining fragment in clades I and II showed an amino acid sequence identity of 78.9% with the MFS transporter gene of the region in clade III but shared no similarity with other MFS transporter genes in the genome, favoring gene loss in this region. In addition, a region (cluster_6043, -_6044, -_6225 to -_6229, and -_6467) comprising carbohydrate transport and metabolism-related genes was detected in 6 strains of clade III and 2 insect-associated strains of clade II but not in strains of clade I (Fig. 3B).
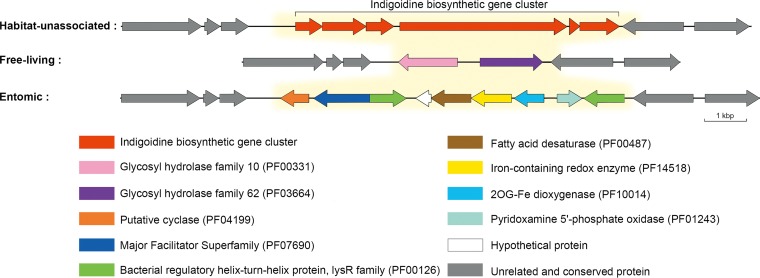
Furthermore, a clade III strain-specific region (cluster_6235 to -_6243) at the end of the genome showed a strong association with entomic habitats (Fig. 3B and Fig. 6). One particular gene in this region was annotated as fatty acid desaturase (IPR005804; cluster_6239), which plays an important role in the life history of some kinds of insects (41–43). Additional analysis proved that this is a versatile region with three genotypes in the species (Fig. 6). The second genotype existed in six free-living strains in clade I and was thus significantly associated with free-living habitats (Fig. 3A). This genotype comprised two carbohydrate-related genes (GH10 and GH62; cluster_6087 and -_6088), which synergistically act in extensive hydrolysis of arabinoxylan (44), which are commonly found in all major cereal grains (45). The last genotype embraced the essential part of the indigoidine biosynthetic gene cluster (46) but showed insignificant habitat association.
FIG 6.
Comparison of genetic organizations of the three genotypes in the versatile genomic region. The habitat-unassociated type exists in 8 strains (DSM 40233, D62, NBRC 13083, J1074, DSM 40455T, NBRC 100770, “S. wadayamensis” A23, and CR13) from clade I and all strains from clade II, the free-living type exists in the remaining 6 strains from clade I and all outgroup strains, and the entomic type exists in all strains from clade III. Differential genes between the genotypes are marked with yellow background.
Habitat-associated SNPs revealed by GWAS.
Among a total of 169,594 single nucleotide polymorphisms (SNPs) identified from the single-copy core genome alignment (4,663 OGs without outgroups), 6,511 (3.8%) showed significant association with free-living or/and entomic habitats (Benjamini-Hochberg P value < 0.05). The majority (65.8%) of the habitat-associated SNPs were located in the third codon position, while 19.0 and 15.2% of the SNPs were located in the first and second codon positions, respectively. A concatenation of these SNPs could also separate the three clades, notably, clade III from the other two (Fig. S4A). Moreover, among the habitat-associated SNPs, 93.7% showed a clear separation between the two habitat types (dimorphic-like SNPs with one allele significantly associated with free-living habitats and another to entomic habitats), of which 46.2% were distinct between clades I and III (with one allele in clade I and another in clade III). These results probably mean that the vast majority of the habitat-associated SNPs have become fixed and further shaped the population structure of S. albidoflavus.
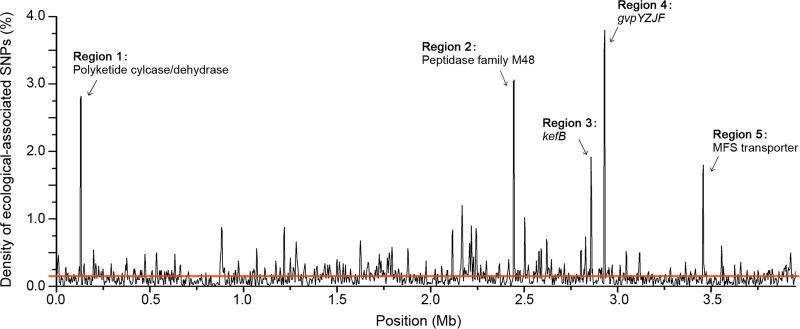
To evaluate the distribution of the habitat-associated SNPs across the genome, we calculated their density by establishing a sliding window of 5 kb. The resulting graph showed a nonuniform distribution of these SNPs through the core genome. Five regions embraced proportions of habitat-associated SNPs10 times higher (binomial test, P < 0.001) than the average (0.16%) and thus could be defined as highly divergent ecological-split genomic regions (Fig. 7; Table S2). Likewise, a concatenation of the OGs from these regions differentiates the three clades, especially clades I and III (Fig. S4B). The last two regions directly corresponded with the remaining genes, gvpYZJF (Fig. 4B) and the MFS transporter gene (Fig. S3), of the habitat-associated gene clusters resulting from gene loss events, where the shortened sequences most probably lost their original functions. However, neighboring structures of the other three ecological-split regions were relatively stable. One region corresponded to kefB (cluster_1226), a gene annotated as sodium/proton antiporter; and the remaining two regions contained OGs in cluster_2806 and cluster_4357, which encode a polyketide cyclase/dehydrase within the alteramide biosynthetic gene cluster and a peptidase of family M48, respectively.
FIG 7.
Density of habitat-associated SNPs along the core genome. The density of these SNPs was plotted along the core genome using a 5-kb sliding window (with an overlap of 2.5 kb between consecutive windows). The red line shows the average density of habitat-associated SNPs per 5 kb.
Homologous recombination within S. albidoflavus species.
We performed a series of analyses to evaluate the extent and role of recombination in S. albidoflavus. A NeighborNet network (47) based on the concatenated single-copy core genes (Fig. S5A) showed a reticulate structure, with the most complex reticulated interactions observed between strains of the same clades. Furthermore, the homoplasy index test (48) did find highly significant evidence for recombination within S. albidoflavus (P = 0.00). Using PhiPack (48), from the 4,116 single-copy core genes used to generate the phylogenomic tree (Fig. 2A), we detected 603 genes (14.7%) with evidence of genetic recombination among S. albidoflavus strains, which account for 28.2% of total SNPs of the core genome. After removing these recombinant OGs, we generated another concatenated phylogenomic tree, which showed incongruence with the former one especially for clades I and II (Fig. S5B). The intraclade relationships of clade I were obviously changed, and clade II was divided into two subclades, with subclade II-1 clustered with clade I and then with subclade II-2. Nevertheless, the demarcation of clades I and III was stable and clade III was kept distinct. In addition, based on the analysis of 40 randomly chosen consecutive genome blocks, we concluded that the average recombination rate (ρ)/mutation rate (θ) ratio of the whole species was 3.36 (median, 2.42), a value lower than that of the free-living group (mean, 5.67; median, 3.31) while similar to that of the entomic group (mean, 3.32; median, 2.61). The ρ/θ ratio was relatively high within each of the three clades, with mean values of 5.30, 4.41, and 5.71 (medians, 4.09, 3.52, and 3.71) for clades I, II, and III, respectively. These ρ/θ values gave a clear indication that recombination was more prevalent than point mutation in this species. Finally, the results of fastGEAR (49) showed that recombination was much more frequent between clades I and II than between clades III and either I or II (Fig. S5C), indicating relatively independent evolution of strains in clade III.
DISCUSSION
Correlation between phylogenomic clades and ecological habitats.
The differential distribution of S. albidoflavus strains between the free-living and entomic habitats (Fig. 2) suggests that some genomic variations have already occurred in this species to adapt to different habitat types, i.e., clade I to free-living habitats and clade III to entomic habitats. However, the mixed origin of clade II implies that it is very likely a transitional clade still undergoing dynamic evolution. This implication is supported by the changed interrelationship of clade II with the other two clades in the core- and accessory-genome trees (Fig. 2A and B). From the reconstruction of gene gain and loss events (Fig. 2A), it seems that the last common ancestor of the species (node 5), which had gained a large number of genes and possessed 7,176 orthologous gene families, was likely insect originated, because only a few genes were further gained in the formation of the insect-associated clade (node 3). In contrast, the two steps of considerable gene acquisition at nodes 4 and 1 might lead to a habitat expansion from insects into open environments. The second step of acquisition is almost negligible for clade II (node 2), with only 20 genes gained, suggesting that this clade is undergoing evolutionary transition from entomic to free-living habitats. The location of strain S4 on the outskirts of the species in the accessory genome tree (Fig. 2B) is probably because it has a larger genome size and more non-strain-specific accessory genes than the other S. albidoflavus strains, mainly due to the fact that it suffered the fewest number of gene losses (data not shown).
It seems that the S. albidoflavus lineages are at an advanced stage of the speciation process. Nevertheless, this process certainly has not finished, for the phylogenomic tree and population structure do not show clear ecological separation (Fig. 2), the neutral genes in the accessory genome are not separated either ecologically or genetically except for strain S4 (Fig. S2A), and the strains share high genome-wide ANI (>98.15%). These lineages are most likely on a trajectory toward speciation, providing an interesting sample with which to study streptomycete speciation.
Ecological adaptation strategies of S. albidoflavus and effects on the population structure.
It has been demonstrated that ecological niches affect the evolution of bacteria (9, 14). As the transition between insect-associated and free-living lifestyles is a dramatic ecological change for bacteria, adaptive evolutionary processes will occur, mediated by mechanisms such as gain of new functions through HGT, gene loss, and functional divergence of existing genes (50). By using GWAS, we identified a number of accessory genes that presumably contribute to adaptation of S. albidoflavus to free-living or entomic habitats. For example, it has been reported that host environments are deficient in iron (51) and the transport of cobalt is particularly important in host-associated bacteria (52); hence, the retained griseobactin gene cluster and cbiMQO likely endow the insect-associated strains with reinforced ability to acquire these precious elements within the host environment. We further identified two core hydroxamate-type siderophore biosynthetic gene clusters in the S. albidoflavus strains (Table S1). One is the well-studied desferrioxamine B gene cluster, which is very common and the product can be identified in our strains (Fig. S6), and the other is a putative aerobactin-like siderophore cluster. This finding is contrary to the function-related replacement in the model marine actinobacterium Salinispora, in which acquisition of one siderophore pathway co-occurs with the loss of another (53). On the other hand, the retention of capacities of sialic acid catabolism and gas vesicle biosynthesis may enable the free-living strains to survive complex conditions. Sialic acid and sialylation exist in certain larval stages during insect development (54, 55) and play a prominent role in the nervous system (56). Thus, the wide distribution of the nan gene cluster in the free-living and outgroup strains but absence in clade III strains (Fig. 3B) may illustrate that at the early stage of S. albidoflavus divergence, a subset of the insect-associated group most likely lost the sialic acid catabolic function by adaptive gene loss (57), so as to achieve harmonious coexistence with the hosts by avoiding competition for sialic acid. The association of the gvp gene cluster with free-living habitats is consistent with the previous observation that gvp is generally present in free-living actinomycetes with large genomes but absent in parasitic or pathogenic strains (35, 58). As actual gas vesicles have not been detected in streptomycetes until now, the gvp genes might have other functions (59), such as in response to temperature and osmotic upshifts (60, 61) and to stress caused by several kinds of antibiotics (62) in the open environment, whereas the loss of gvp genes may provide advantages in energy savings and spatial efficiency (57, 63) to the entomic strains. Furthermore, different genotypes in the same genomic region (Fig. 6) show different ecological adaptions, where fatty acid desaturase may give rise to mutualistic relationships with hosts (41–43) for the entomic strains and the ability of arabinoxylan hydrolysis may enable the free-living strains to metabolize cereal residues as additional carbon sources. Genome fluctuation, mediated by gene gain and loss, leads to the formation of genomic islands (GIs) and guarantees quick and economical genetic adaptation of corresponding S. albidoflavus strains to local environments.
Ecological divergence was also found in the core genome, with five regions highly enriched for habitat-associated SNPs. The formation of the habitat-associated SNPs partially connects with gene gain or loss events, as exemplified by the core regions 4 and 5 (Fig. 7), further emphasizing the important roles of HGT and gene loss in shaping the ecological differentiation processes. Moreover, the inhomogeneous distribution of the habitat-associated SNPs in the core genome indicates that the differentiation of the whole genome was initiated in several genome blocks rather than averagely along the chromosome. Such genetic changes probably enable the S. albidoflavus strains to specialize to different environments and then lead to their separation into different populations (Fig. S4). For example, the kefB gene may facilitate the insect-associated strains to survive the unnormal pH conditions that have been reported for many representatives of insect orders (64, 65), or it may participate in methylglyoxal detoxification in the free-living strains to protect their DNA during the exposure to toxic metabolites (66).
Meanwhile, it appears counterintuitive that the clustering patterns of the habitat-associated genes and alleles do not exactly follow habitat (Fig. S2A and S4). This may be due to the fact that the genes and alleles are habitat associated rather than strictly habitat specific (with one variant present in all free-living strains and a different variant in all insect-associated strains) (17). Actually, we did not detect any strictly habitat-specific genes or alleles in S. albidoflavus, largely because of the existence of clade II, which is likely still undergoing transition from entomic to free-living habitats. As the speciation process within S. albidoflavus has not finished, the habitat-specific variants are probably still in development, leading to the imperfect ecological distribution of the adaptive variants (Fig. 3). Intriguingly, the phylogenies based on the habitat-associated variants (Fig. S2A and S4) are similar to the phylogenomic tree (Fig. 2). This observation and the differential specificity of these variants among the three phylogenomic clades (Fig. S2) illustrate that ecological adaptations contribute to the population structure of S. albidoflavus, which is composed of two habitat-associated clades and a mixed-origin clade.
The role of recombination in shaping the evolutionary dynamics of S. albidoflavus.
Recombination, especially homologous recombination, is one of the main forces shaping bacterial evolution (6) and is widespread in streptomycetes, with high rates at the intraspecies level (13, 24, 25, 67). In this study, the results of four different methods provide compelling evidence for homologous recombination within S. albidoflavus, even though the recombination inferred may be underestimated by the traditional methods based on polymorphic segments (3). The relatively prevalent recombination detected in the whole species likely serves as a cohesive force to maintain S. albidoflavus, and the higher recombination rate within each phylogenomic clade may help to stabilize the genetic structure of the species. Recombination may also act as a powerful force in shaping the genetic diversity of the free-living strains, as this group has a similarly higher recombination rate. It is noticeable, however, that the ρ/θ ratio of all the entomic strains is lower than that of clade III, encompassing a major subset of the entomic group. This could be ascribed to the fact that an evident genetic difference, which is an important reason for recombination barriers (68), has already emerged between strains in clade III and the other entomic strains. It is perhaps the case that at one time, a portion of the ancestral entomic strains acquired and retained certain insect adaptation genes (such as cluster_6235 to -_6243, which contain a fatty acid desaturase gene) and/or lost certain useless genes (such as the nan gene cluster) (Data Set S1), causing differential distribution of the strains among habitats. Consequently, this portion of strains formed a genetically different subgroup that could escape from being homogenized with the other strains by recombination. Subsequently occurring adaptive mutations and genetic drift within the subgroup could spread in it by recombination in a gene-specific sweep manner but could hardly spread to other S. albidoflavus strains because the recombination was hindered by genetic distance (and habitat barriers as well in some cases). This process might lead to the formation of clade III and may finally give rise to a new species that is more suited to entomic habitats.
Potential contributions of geography to the formation of population structure and evolution of S. albidoflavus.
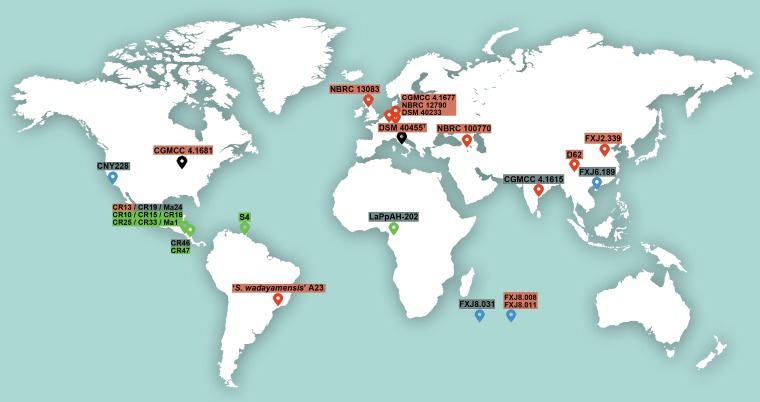
As the sample size of 30 S. albidoflavus strains is relatively small and the free-living and insect-associated strains are not sympatric, the fact that they largely cluster in different clades might also be due to geography and uneven sampling. However, although the free-living strains in clade I were collected from different areas all over the world (Fig. 8), they show a close evolutionary relationship to each other (Fig. 2). On the other hand, although strains “Streptomyces wadayamensis” A23, CGMCC 4.1681, and CNY228 were isolated from the American continent, they do not fall in clade III, which contains American insect-associated strains only. These observations suggest that the biogeographic pattern of S. albidoflavus strains cannot well explain the population structure, whereas environmental selection could be a considerable reason by making some strains in disparate geographic areas but similar habitats undergo similar variations. Hence, despite the fact that our strains were not sampled from sympatric niches, we can still perform a reverse ecology population genomic study (3) to obtain valuable information.
FIG 8.
Map depicting the sampling locations of strains used in this study. The location symbol colors indicate different habitats (red, soil or plants; green, insects; blue, sea; black, uncertain), and the text shadow colors indicate the different phylogenomic clades detected in the species (red, clade I; gray, clade II; green, clade III). The geographic origin of strain J1074 is unavailable. The map is derived from Rawpixel Ltd.
Yet the influence of geographic distribution should not be ignored. As shown in Fig. 8, all the entomic strains used in this study were collected from low-latitude areas, while the free-living strains came mainly from higher latitudes. It has been reported that the latitudinal diversity gradient observed for terrestrial streptomycetes can result from historical demographic range expansion, dispersal limitation, and genetic drift, though ecological and evolutionary processes cannot be excluded (16). Therefore, both ecology and geography may contribute to the formation of population structure and evolution of S. albidoflavus, and further efforts are needed to investigate larger sample sizes of both allopatric and sympatric populations in order to fully comprehend speciation in Streptomyces.
MATERIALS AND METHODS
Strains and genome sequencing.
We selected 30 S. albidoflavus strains that captured the previously characterized geographic distribution and genetic and habitat diversity of this species (13), plus 3 closely related strains as outgroups. Among them, 5 S. albidoflavus strains and 2 related strains have genome sequences available in NCBI and the remaining 26 strains were genome sequenced in this study (Table 1). Whole-genome sequencing was performed using the HiSeq 2500 sequencer (Illumina) by Novogene Bioinformatics Technology Co., Ltd. (Beijing, China). A PCR-free library was prepared for each sample to generate 125-bp paired-end reads. The library gave more than 140× coverage (sequencing depth) for each strain with >85% of bases having a quality score above Q20 (base-calling accuracy of 99%) and >80% of bases having a quality score above Q30 (base-calling accuracy of 99.9%). Genome sequences were then assembled using SOAPdenovo (69). Contigs were reordered against the complete genome of the S. albidoflavus reference strain J1074 using the Move Contig tool in Mauve 2.3.1 (70). Whole-genome ANI was calculated using the Jspecies package (71) based on MUMmer (ANIm) with default parameters.
Genome annotation and determination of orthologous groups.
All 33 genomes, including the previously published genomes, were analyzed under the same strategies, in order to reduce the system errors caused by different programs or parameters. De novo gene predictions were performed for CDSs with PRODIGAL v2.6.1 (72). Function annotation of proteins was performed by sequence comparison with the COG database (30), using BLASTP with an E value of <1e−5 and an identity of >40%. The significance of gene abundance differences in COG categories was examined using Fisher’s exact test as implemented in R. InterProScan (73) was used to identify Pfam and Interpro domains within the predicted protein sets. Secondary metabolite gene clusters were predicted by antiSMASH 3.0 (32).
All proteins longer than 10 amino acids were clustered using OrthoMCL (31) to identify OGs (BLASTP E value cutoff = 1e−5; inflation value = 1.5). Genome statistics were visualized using PanGP (74) with the distance guide sampling algorithm.
Phylogenomics and population structure analyses.
For phylogenomic analyses, only OGs containing exactly one gene copy for each of the 33 genomes were used. Codon-based alignments for each OGs were obtained by aligning the translated protein with MAFFT using the l-INS-i option (75) and back-translating with PAL2NAL (76). Poorly aligned regions of each codon alignment (mean, 7.92%; standard deviation, 19.79) were removed by Gblocks (77) with default parameters (except that −t = c was used). The aligned sequences were then concatenated as a single data set in an order of their locations in the completed genome of S. albidoflavus J1074. Based on SNPs of the alignment, a maximum-likelihood tree was built using FastTree 2, applying the generalized time-reversible model (GTR) (78). Phylogenomic analysis was also performed after removing the recombinant OGs identified in the following recombination estimates. In addition, according to OrthoMCL results, an absence/presence (0/1) matrix of dispensable genes was built and subjected to hierarchical clustering analysis with 1,000 bootstrap replicates, as implemented in the R package pvclust (79). The population structure of S. albidoflavus was analyzed based on the core genome SNPs using the model-based Bayesian method implemented in Structure 2.3.4 (33), in which the admixture model was used with a K value of 2 to 5. The most optimal value for K was generated using STRUCTURE HARVESTER (80). Reconstruction of gene gain and loss during the evolution of S. albidoflavus was performed using COUNT (81) with Dollo parsimony.
Detection of habitat-associated genes and SNPs.
Pan-GWAS was carried out using Scoary (34) to identify variants significantly associated with free-living or entomic habitat type. Two 0/1 matrixes were created, based on the presence/absence of “shell genes” (OGs exist in 16.7% to 86.7% of the S. albidoflavus strains, i.e., 5 to 26 strains) in the accessory genome and alleles of SNPs along the core genome, respectively. A binary matrix was also created using the habitat information of each strain, indicating free-living or insect associated. Traits with corrected P values (Benjamini-Hochberg) of association below 0.05 were considered significant. Genes and alleles of SNPs associated with free-living habitats were identified by an odds ratio of less than 1, and those associated with entomic habitats were identified by an odds ratio of greater than 1. The distribution of habitat-associated genes was illustrated by HemI (version 1.0.1) (82).
Test of sialic acid catabolism.
Strains possessing the nan gene cluster and strains negative for the presence of this gene cluster were parallelly tested for the ability to utilize sialic acid as a sole carbon source. Each strain was precultured on ISP (International Streptomyces Project) medium 2 (83) at 28°C for 5 days, and then spores on a 25-cm2 lawn of the culture were collected in 1.5 ml of distilled water. For the test, 60-μl quantities of the spore suspensions were inoculated, in triplicate, into basal mineral salt medium agar plates (84) supplemented with 1% (wt/vol) N-acetylneuraminic acid as the sole carbon source, and the plates were incubated at 28°C for 5 days. The growth of strains indicated their ability to catabolize sialic acid.
Detection of siderophore production and mass spectrometric measurement.
Strains were grown in liquid ISP medium 2 at 28°C for 10 days on a shaker at 150 rpm. The production of griseobactin was detected by the presence of catechol in the supernatant of culture broths with Arnow’s test (40). The presence of a red color in the solution was recorded as a positive test for catechol. To detect hydroxamate siderophores in culture broths, the FeCl3 test (85) was conducted. To further identify the hydroxamate siderophores, 100-ml quantities of the culture supernatant were treated by an adsorption process with 20 ml of macroporous resin HP-20 and eluted with 80 ml of alcohol. The eluate was concentrated in vacuo to evaporate the solvent and the residue was redissolved in 1.5 ml of methanol (MeOH) for analysis. After adding excessive FeCl3 solution, high-performance liquid chromatography (HPLC) analysis was performed on a Shimadzu Prominence HPLC system using a Waters Xbridge octyldecyl silane (ODS) column with a linear gradient of MeOH-H2O from 20:80 to 100:0 over 15 min. The effluent was monitored at 435 nm. The peak with a retention time of ca. 2.4 min in the HPLC analysis was collected and freeze-dried. The residue was resuspended in aqueous acetonitrile and the resulting solution was analyzed by high-resolution ultraperformance liquid chromatography-mass spectroscopy (UPLC-MS) on a Waters Acquity UPLC BEH C18 column (2.1 mm by 50 mm, 1.7 μm, and 45°C) connected to a Waters Acquity UPLC/Xevo G2 QTof MS system (Waters Corporation, Milford, MA), equipped with an electrospray source. The mass peak with m/z 614.2 [M+H]+ represented the iron complex of desferrioxamine B ([MH + Fe-3H]+ = ferrioxamine B) (86).
Recombination estimates.
SplitsTree version 4.13.1 (47) was employed to construct a network based on the concatenated single-copy core genes with the NeighborNet algorithm and to calculate the pairwise homoplasy index (48). In addition, we used PhiPack (48), which performs three different methods (neighbor similarity score, maximum chi, and phi), to identify genetic recombination occurred in each single-copy core gene. The potential recent recombination events were identified as having P values (computed from 1,000 permutations) lower than 0.05 in at least two of the three methods. The population-scaled mutation rate (θ, Watterson’s mutation parameter) and recombination rate (ρ) in 40 randomly chosen consecutive blocks (each with a length of 10 kb collected from the whole-core genome alignment) were estimated by LDHat (87) implemented in the RDP4 suite (88). The number of interlineage recombination events, for which donor-recipient relations could be inferred and Bayesian factors (BF) were >1, was estimated using fastGEAR (49).
Accession number(s).
The genome sequences reported in this paper have been deposited at GenBank under the accession numbers listed in Table 1. Sequencing reads have been deposited at the NCBI Sequence Read Archive (SRA) under accession numbers SRP127744 and SRP127754.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ning Liu (Institute of Microbiology, Chinese Academy of Sciences) for his assistance in siderophore identification and Xiao-Yang Zhi (Yunnan University) for his assistance in the initial analysis of the genomes. We are also grateful to the Ministry of Environment, Energy and Telecommunications and the Guanacaste Conservation Area for providing collection permits in Costa Rica (resolution no. ACG-PI-018-2012).
This work was supported by grants from the National Natural Science Foundation of China (31470140 and 31470142) and the External Cooperation Program of Chinese Academy of Sciences (GJHZ1108). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02555-18.
REFERENCES
- 1.Achtman M, Wagner M. 2008. Microbial diversity and the genetic nature of microbial species. Nat Rev Microbiol 6:431–440. doi: 10.1038/nrmicro1872. [DOI] [PubMed] [Google Scholar]
- 2.Doolittle WF, Papke RT. 2006. Genomics and the bacterial species problem. Genome Biol 7:116. doi: 10.1186/gb-2006-7-9-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shapiro BJ, Polz MF. 2014. Ordering microbial diversity into ecologically and genetically cohesive units. Trends Microbiol 22:235–247. doi: 10.1016/j.tim.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barraclough TG, Balbi KJ, Ellis RJ. 2012. Evolving concepts of bacterial species. Evol Biol 39:148–157. doi: 10.1007/s11692-012-9181-8. [DOI] [Google Scholar]
- 5.Cohan FM, Perry EB. 2007. A systematics for discovering the fundamental units of bacterial diversity. Curr Biol 17:R373–R386. doi: 10.1016/j.cub.2007.03.032. [DOI] [PubMed] [Google Scholar]
- 6.Fraser C, Hanage WP, Spratt BG. 2007. Recombination and the nature of bacterial speciation. Science 315:476–480. doi: 10.1126/science.1127573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraser C, Alm EJ, Polz MF, Spratt BG, Hanage WP. 2009. The bacterial species challenge: making sense of genetic and ecological diversity. Science 323:741–746. doi: 10.1126/science.1159388. [DOI] [PubMed] [Google Scholar]
- 8.Hufbauer RA, Roderick GK. 2005. Microevolution in biological control: mechanisms, patterns, and processes. Biol Control 35:227–239. doi: 10.1016/j.biocontrol.2005.04.004. [DOI] [Google Scholar]
- 9.Shapiro BJ, Leducq JB, Mallet J. 2016. What is speciation? PLoS Genet 12:e1005860. doi: 10.1371/journal.pgen.1005860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gogarten JP, Doolittle WF, Lawrence JG. 2002. Prokaryotic evolution in light of gene transfer. Mol Biol Evol 19:2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- 11.Ochman H, Lawrence JG, Groisman EA. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 12.Polz MF, Alm EJ, Hanage WP. 2013. Horizontal gene transfer and the evolution of bacterial and archaeal population structure. Trends Genet 29:170–175. doi: 10.1016/j.tig.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng K, Rong X, Pinto-Tomás AA, Fernández-Villalobos M, Murillo-Cruz C, Huang Y. 2015. Population genetic analysis of Streptomyces albidoflavus reveals habitat barriers to homologous recombination in the diversification of streptomycetes. Appl Environ Microbiol 81:966–975. doi: 10.1128/AEM.02925-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vos M. 2011. A species concept for bacteria based on adaptive divergence. Trends Microbiol 19:1–7. doi: 10.1016/j.tim.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Martiny JB, Bohannan BJ, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR, Morin PJ, Naeem S, Øvreås L, Reysenbach AL, Smith VH, Staley JT. 2006. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol 4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- 16.Andam CP, Doroghazi JR, Campbell AN, Kelly PJ, Choudoir MJ, Buckley DH. 2016. A latitudinal diversity gradient in terrestrial bacteria of the genus Streptomyces. mBio 7:e02200-15. doi: 10.1128/mBio.02200-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shapiro BJ, Friedman J, Cordero OX, Preheim SP, Timberlake SC, Szabó G, Polz MF, Alm EJ. 2012. Population genomics of early events in the ecological differentiation of bacteria. Science 336:48–51. doi: 10.1126/science.1218198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison J, Studholme DJ. 2014. Recently published Streptomyces genome sequences. Microb Biotechnol 7:373–380. doi: 10.1111/1751-7915.12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu G, Chater KF, Chandra G, Niu G, Tan H. 2013. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol Mol Biol Rev 77:112–143. doi: 10.1128/MMBR.00054-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian X, Zhang Z, Yang T, Chen M, Li J, Chen F, Yang J, Li W, Zhang B, Zhang Z, Wu J, Zhang C, Long L, Xiao J. 2016. Comparative genomics analysis of Streptomyces species reveals their adaptation to the marine environment and their diversity at the genomic level. Front Microbiol 7:998. doi: 10.3389/fmicb.2016.00998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ian E, Malko DB, Sekurova ON, Bredholt H, Rückert C, Borisova ME, Albersmeier A, Kalinowski J, Gelfand MS, Zotchev SB. 2014. Genomics of sponge-associated Streptomyces spp. closely related to Streptomyces albus J1074: insights into marine adaptation and secondary metabolite biosynthesis potential. PLoS One 9:e96719. doi: 10.1371/journal.pone.0096719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Z, Gu J, Li YQ, Wang Y. 2012. Genome plasticity and systems evolution in Streptomyces. BMC Bioinformatics 13:S8. doi: 10.1186/1471-2105-13-S10-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andam CP, Choudoir MJ, Vinh Nguyen A, Sol Park H, Buckley DH. 2016. Contributions of ancestral inter-species recombination to the genetic diversity of extant Streptomyces lineages. ISME J 10:1731–1741. doi: 10.1038/ismej.2015.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doroghazi JR, Buckley DH. 2010. Widespread homologous recombination within and between Streptomyces species. ISME J 4:1136–1143. doi: 10.1038/ismej.2010.45. [DOI] [PubMed] [Google Scholar]
- 25.Doroghazi JR, Buckley DH. 2014. Intraspecies comparison of Streptomyces pratensis genomes reveals high levels of recombination and gene conservation between strains of disparate geographic origin. BMC Genomics 15:970. doi: 10.1186/1471-2164-15-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seipke RF, Hutchings MI. 2013. The regulation and biosynthesis of antimycins. Beilstein J Org Chem 9:2556–2563. doi: 10.3762/bjoc.9.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seipke RF. 2015. Strain-level diversity of secondary metabolism in Streptomyces albus. PLoS One 10:e0116457. doi: 10.1371/journal.pone.0116457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skinner FA. 1953. Inhibition of Fusarium culmorum by Streptomyces albidoflavus. Nature 172:1191. doi: 10.1038/1721191a0. [DOI] [PubMed] [Google Scholar]
- 29.Zaburannyi N, Rabyk M, Ostash B, Fedorenko V, Luzhetskyy A. 2014. Insights into naturally minimised Streptomyces albus J1074 genome. BMC Genomics 15:97. doi: 10.1186/1471-2164-15-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, Kiryutin B, Galperin MY, Fedorova ND, Koonin EV. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res 29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L, Stoeckert CJ Jr, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Muller R, Wohlleben W, Breitling R, Takano E, Medema MH. 2015. antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res 43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Falush D, Stephens M, Pritchard JK. 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brynildsrud O, Bohlin J, Scheffer L, Eldholm V. 2016. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol 17:238. doi: 10.1186/s13059-016-1108-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Keulen G, Hopwood DA, Dijkhuizen L, Sawers RG. 2005. Gas vesicles in actinomycetes: old buoys in novel habitats? Trends Microbiol 13:350–354. doi: 10.1016/j.tim.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 36.Pitt D, Mosley MJ. 1985. Enzymes of gluconate metabolism and glycolysis in Penicillium notatum. Antonie Van Leeuwenhoek 51:353–364. doi: 10.1007/BF02275041. [DOI] [PubMed] [Google Scholar]
- 37.Cuthbertson L, Nodwell JR. 2013. The TetR family of regulators. Microbiol Mol Biol Rev 77:440–475. doi: 10.1128/MMBR.00018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patzer SI, Braun V. 2010. Gene cluster involved in the biosynthesis of griseobactin, a catechol-peptide siderophore of Streptomyces sp. ATCC 700974. J Bacteriol 192:426–435. doi: 10.1128/JB.01250-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siche S, Neubauer O, Hebbeln P, Eitinger T. 2010. A bipartite S unit of an ECF-type cobalt transporter. Res Microbiol 161:824–829. doi: 10.1016/j.resmic.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 40.Arnow LE. 1937. Colorimetric determination of the components of 3,4-dihydroxyphenylalanine-tyrosine mixtures. J Biol Chem 118:531–537. [Google Scholar]
- 41.Helmkampf M, Cash E, Gadau J. 2015. Evolution of the insect desaturase gene family with an emphasis on social Hymenoptera. Mol Biol Evol 32:456–471. doi: 10.1093/molbev/msu315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kocher SD, Li C, Yang W, Tan H, Yi SV, Yang X, Hoekstra HE, Zhang G, Pierce NE, Yu DW. 2013. The draft genome of a socially polymorphic halictid bee, Lasioglossum albipes. Genome Biol 14:R142. doi: 10.1186/gb-2013-14-12-r142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Ommen Kloeke AE, van Gestel CA, Styrishave B, Hansen M, Ellers J, Roelofs D. 2012. Molecular and life-history effects of a natural toxin on herbivorous and non-target soil arthropods. Ecotoxicology 21:1084–1093. doi: 10.1007/s10646-012-0861-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Christakopoulos P, Katapodis P, Hatzinikolaou DG, Kekos D, Macris BJ. 2000. Purification and characterization of an extracellular α-L-arabinofuranosidase from Fusarium oxysporum. Appl Biochem Biotechnol 87:127–133. doi: 10.1385/ABAB:87:2:127. [DOI] [PubMed] [Google Scholar]
- 45.Izydorczyk MS, Biliaderis CG. 1995. Cereal arabinoxylans: advances in structure and physicochemical properties. Carbohydr Polym 28:33–48. doi: 10.1016/0144-8617(95)00077-1. [DOI] [Google Scholar]
- 46.Olano C, García I, González A, Rodriguez M, Rozas D, Rubio J, Sánchez-Hidalgo M, Braña AF, Méndez C, Salas JA. 2014. Activation and identification of five clusters for secondary metabolites in Streptomyces albus J1074. Microb Biotechnol 7:242–256. doi: 10.1111/1751-7915.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 48.Bruen TC, Philippe H, Bryant D. 2006. A simple and robust statistical test for detecting the presence of recombination. Genetics 172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mostowy R, Croucher NJ, Andam CP, Corander J, Hanage WP, Marttinen P. 2017. Efficient inference of recent and ancestral recombination within bacterial populations. Mol Biol Evol 34:1167–1182. doi: 10.1093/molbev/msx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toft C, Andersson SG. 2010. Evolutionary microbial genomics: insights into bacterial host adaptation. Nat Rev Genet 11:465–475. doi: 10.1038/nrg2798. [DOI] [PubMed] [Google Scholar]
- 51.Schaible UE, Kaufmann SH. 2004. Iron and microbial infection. Nat Rev Microbiol 2:946–953. doi: 10.1038/nrmicro1046. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y, Rodionov DA, Gelfand MS, Gladyshev VN. 2009. Comparative genomic analyses of nickel, cobalt and vitamin B12 utilization. BMC Genomics 10:78. doi: 10.1186/1471-2164-10-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bruns H, Crüsemann M, Letzel A-C, Alanjary M, McInerney JO, Jensen PR, Schulz S, Moore BS, Ziemert N. 2018. Function-related replacement of bacterial siderophore pathways. ISME J 12:320–329. doi: 10.1038/ismej.2017.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roth J, Kempf A, Reuter G, Schauer R, Gehring WJ. 1992. Occurrence of sialic acids in Drosophila melanogaster. Science 256:673–675. doi: 10.1126/science.1585182. [DOI] [PubMed] [Google Scholar]
- 55.Vimr ER, Kalivoda KA, Deszo EL, Steenbergen SM. 2004. Diversity of microbial sialic acid metabolism. Microbiol Mol Biol Rev 68:132–153. doi: 10.1128/MMBR.68.1.132-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koles K, Repnikova E, Pavlova G, Korochkin LI, Panin VM. 2009. Sialylation in protostomes: a perspective from Drosophila genetics and biochemistry. Glycoconj J 26:313–324. doi: 10.1007/s10719-008-9154-4. [DOI] [PubMed] [Google Scholar]
- 57.Albalat R, Cañestro C. 2016. Evolution by gene loss. Nat Rev Genet 17:379–391. doi: 10.1038/nrg.2016.39. [DOI] [PubMed] [Google Scholar]
- 58.Walsby AE, Dunton PG. 2006. Gas vesicles in actinomycetes? Trends Microbiol 14:99–100. doi: 10.1016/j.tim.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 59.Pfeifer F. 2012. Distribution, formation and regulation of gas vesicles. Nat Rev Microbiol 10:705–715. doi: 10.1038/nrmicro2834. [DOI] [PubMed] [Google Scholar]
- 60.Karoonuthaisiri N, Weaver D, Huang J, Cohen SN, Kao CM. 2005. Regional organization of gene expression in Streptomyces coelicolor. Gene 353:53–66. doi: 10.1016/j.gene.2005.03.042. [DOI] [PubMed] [Google Scholar]
- 61.Lee EJ, Karoonuthaisiri N, Kim HS, Park JH, Cha CJ, Kao CM, Roe JH. 2005. A master regulator σB governs osmotic and oxidative response as well as differentiation via a network of sigma factors in Streptomyces coelicolor. Mol Microbiol 57:1252–1264. doi: 10.1111/j.1365-2958.2005.04761.x. [DOI] [PubMed] [Google Scholar]
- 62.Hesketh A, Hill C, Mokhtar J, Novotna G, Tran N, Bibb M, Hong HJ. 2011. Genome-wide dynamics of a bacterial response to antibiotics that target the cell envelope. BMC Genomics 12:226. doi: 10.1186/1471-2164-12-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morris JJ, Lenski RE, Zinser ER. 2012. The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. mBio 3:e00036-12. doi: 10.1128/mBio.00036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lemke T, Stingl U, Egert M, Friedrich MW, Brune A. 2003. Physicochemical conditions and microbial activities in the highly alkaline gut of the humus-feeding larva of Pachnoda ephippiata (Coleoptera: Scarabaeidae). Appl Environ Microbiol 69:6650–6658. doi: 10.1128/AEM.69.11.6650-6658.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harrison JF. 2001. Insect acid-base physiology. Annu Rev Entomol 46:221–250. doi: 10.1146/annurev.ento.46.1.221. [DOI] [PubMed] [Google Scholar]
- 66.Ferguson GP, Battista JR, Lee AT, Booth IR. 2000. Protection of the DNA during the exposure of Escherichia coli cells to a toxic metabolite: the role of the KefB and KefC potassium channels. Mol Microbiol 35:113–122. doi: 10.1046/j.1365-2958.2000.01682.x. [DOI] [PubMed] [Google Scholar]
- 67.Cheng K, Rong X, Huang Y. 2016. Widespread interspecies homologous recombination reveals reticulate evolution within the genus Streptomyces. Mol Phylogenet Evol 102:246–254. doi: 10.1016/j.ympev.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 68.Thomas CM, Nielsen KM. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 69.Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Shi Y, Liu Y, Yu C, Wang B, Lu Y, Han C, Cheung DW, Yiu SM, Peng S, Xiaoqian Z, Liu G, Liao X, Li Y, Yang H, Wang J, Lam TW, Wang J. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Richter M, Rosselló-Móra R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. 2005. InterProScan: protein domains identifier. Nucleic Acids Res 33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao Y, Jia X, Yang J, Ling Y, Zhang Z, Yu J, Wu J, Xiao J. 2014. PanGP: a tool for quickly analyzing bacterial pan-genome profile. Bioinformatics 30:1297–1299. doi: 10.1093/bioinformatics/btu017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Suyama M, Torrents D, Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 78.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Suzuki R, Shimodaira H. 2006. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 80.Earl D, Vonholdt BM. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- 81.Csűrös M. 2010. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics 26:1910–1912. doi: 10.1093/bioinformatics/btq315. [DOI] [PubMed] [Google Scholar]
- 82.Deng W, Wang Y, Liu Z, Cheng H, Xue Y. 2014. HemI: a toolkit for illustrating heatmaps. PLoS One 9:e111988. doi: 10.1371/journal.pone.0111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shirling EB, Gottlieb D. 1966. Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340. doi: 10.1099/00207713-16-3-313. [DOI] [Google Scholar]
- 84.Pridham TG, Gottlieb D. 1948. The utilization of carbon compounds by some Actinomycetales as an aid for species determination. J Bacteriol 56:107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Neilands JB. 1981. Microbial iron compounds. Annu Rev Biochem 50:715–731. doi: 10.1146/annurev.bi.50.070181.003435. [DOI] [PubMed] [Google Scholar]
- 86.Winkelmann G, Busch B, Hartmann A, Kirchhof G, Süssmuth R, Jung G. 1999. Degradation of desferrioxamines by Azospirillum irakense: assignment of metabolites by HPLC/electrospray mass spectrometry. Biometals 12:255–264. doi: 10.1023/A:1009242307134. [DOI] [PubMed] [Google Scholar]
- 87.McVean G, Awadalla P, Fearnhead P. 2002. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics 160:1231–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. 2015. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.de Oliveira LG, Tormet Gonzalez GD, Samborsky M, Marcon J, Araujo WL, de Azevedo JL. 2014. Genome sequence of Streptomyces wadayamensis strain A23, an endophytic actinobacterium from Citrus reticulata. Genome Announc 2:e00625-14. doi: 10.1128/genomeA.00625-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Araújo WL, Marcon J, Maccheroni W Jr, Van Elsas JD, Van Vuurde JW, Azevedo JL. 2002. Diversity of endophytic bacterial populations and their interaction with Xylella fastidiosa in citrus plants. Appl Environ Microbiol 68:4906–4914. doi: 10.1128/AEM.68.10.4906-4914.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hanshew AS, McDonald BR, Díaz Díaz C, Djiéto-Lordon C, Blatrix R, Currie CR. 2015. Characterization of actinobacteria associated with three ant-plant mutualisms. Microb Ecol 69:192–203. doi: 10.1007/s00248-014-0469-3. [DOI] [PubMed] [Google Scholar]
- 92.Barke J, Seipke RF, Grüschow S, Heavens D, Drou N, Bibb MJ, Goss RJ, Yu DW, Hutchings MI. 2010. A mixed community of actinomycetes produce multiple antibiotics for the fungus farming ant Acromyrmex octospinosus. BMC Biol 8:109. doi: 10.1186/1741-7007-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.