The molecular diversity of the aquatic picocyanobacterial community cannot be accurately described using only the available universal 16S rRNA gene primers that target the whole bacterial and archaeal community. We show that the hypervariable regions V5, V6, and V7 of the 16S rRNA gene are better suited to study the diversity, community structure, and dynamics of picocyanobacterial communities at a fine scale using Illumina MiSeq sequencing. Due to its variability, it allows reconstructing phylogenies featuring topologies comparable to those generated when using the complete 16S rRNA gene sequence. Further, we successfully designed a new set of primers flanking the V5 to V7 region whose specificity for picocyanobacterial genera was tested in silico and validated in several freshwater and marine aquatic communities. This work represents a step forward for understanding the diversity and ecology of aquatic picocyanobacteria and sets the path for future studies on picocyanobacterial diversity.
KEYWORDS: 16S rRNA gene, Cyanobium, HTS, Prochlorococcus, Synechococcus, primer design
ABSTRACT
High-throughput sequencing (HTS) of the 16S rRNA gene has been used successfully to describe the structure and dynamics of microbial communities. Picocyanobacteria are important members of bacterioplankton communities, and, so far, they have predominantly been targeted using universal bacterial primers, providing a limited resolution of the picocyanobacterial community structure and dynamics. To increase such resolution, the study of a particular target group is best approached with the use of specific primers. Here, we aimed to design and evaluate specific primers for aquatic picocyanobacterial genera to be used with high-throughput sequencing. Since the various regions of the 16S rRNA gene have different degrees of conservation in different bacterial groups, we therefore first determined which hypervariable region of the 16S rRNA gene provides the highest taxonomic and phylogenetic resolution for the genera Synechococcus, Prochlorococcus, and Cyanobium. An in silico analysis showed that the V5, V6, and V7 hypervariable regions appear to be the most informative for this group. We then designed primers flanking these hypervariable regions and tested them in natural marine and freshwater communities. We successfully detected that most (97%) of the obtained reads could be assigned to picocyanobacterial genera. We defined operational taxonomic units as exact sequence variants (zero-radius operational taxonomic units [zOTUs]), which allowed us to detect higher genetic diversity and infer ecologically relevant information about picocyanobacterial community composition and dynamics in different aquatic systems. Our results open the door to future studies investigating picocyanobacterial diversity in aquatic systems.
IMPORTANCE The molecular diversity of the aquatic picocyanobacterial community cannot be accurately described using only the available universal 16S rRNA gene primers that target the whole bacterial and archaeal community. We show that the hypervariable regions V5, V6, and V7 of the 16S rRNA gene are better suited to study the diversity, community structure, and dynamics of picocyanobacterial communities at a fine scale using Illumina MiSeq sequencing. Due to its variability, it allows reconstructing phylogenies featuring topologies comparable to those generated when using the complete 16S rRNA gene sequence. Further, we successfully designed a new set of primers flanking the V5 to V7 region whose specificity for picocyanobacterial genera was tested in silico and validated in several freshwater and marine aquatic communities. This work represents a step forward for understanding the diversity and ecology of aquatic picocyanobacteria and sets the path for future studies on picocyanobacterial diversity.
INTRODUCTION
Picocyanobacteria are common components of plankton and, together with other groups of cyanobacteria, are the only prokaryotes capable of performing oxygenic photosynthesis. They have been present on Earth for 3.5 billon years (1), and during their history they have evolved ecological strategies to adapt to various environmental conditions from the poles to the tropics (2–7). Picocyanobacteria contribute substantially to the primary production of numerous aquatic systems and play a major role in aquatic ecosystem energy flow, as they are at the base of food webs (8–11). Phenotypically, they are single or colonial rod-shaped cells ranging from 0.2 to 3 µm and, based on their phylogeny, they belong to three genera, Synechococcus, Prochlorococcus, and Cyanobium (12). Prochlorococcus is the sister group of the marine Synechococcus and has been divided into distinct clades with ecologically relevant differences in light adaptation (high light [HL] or low light [LL] intensities) and nutrient utilization (13, 14). Synechococcus is a polyphyletic genus containing marine and freshwater taxa (1, 15). Phylogenetic reconstructions resolved three subclusters (named 5.1, 5.2, and 5.3) with marine representatives (16–18), and at least 13 clades of nonmarine picocyanobacteria (19). The 5.1 cluster encompasses most marine groups, and some freshwater strains have also been affiliated to clusters 5.2 and 5.3 (15, 19). Less information is available about the Cyanobium clade. It is a cosmopolitan group represented by strains from systems with contrasting limnological characteristics (i.e., from shallow hypertrophic to deep and oligotrophic lakes), and, based on 16S rRNA gene phylogenetic reconstruction, is closely related to some freshwater Synechococcus clusters (19, 20).
As it occurs with most prokaryotes, picocyanobacteria lack obvious morphological differences and, consequently, the use of molecular techniques is essential to discriminate the distinct taxa (13, 21, 22). During the last decade, new molecular and bioinformatic techniques have changed our perspective on microbial diversity. In particular, high-throughput sequencing (HTS) techniques applied to genetic markers, such as the 16S rRNA gene, have allowed a better understanding of the structure of bacterial communities and their responses to environmental changes (for examples, see references 23–27). To date, most researchers have used the 16S rRNA gene universal primers targeting the whole bacterial community to explore bacterial diversity. However, in these works, the number of reads recovered from the phylum Cyanobacteria hardly exceeds 20% of the total read abundance (25, 27–31). Given that natural environments are composed of a few dominant taxa, using universal primers may overlook the diversity and population genetic structure of less abundant organisms even when using deep sequencing. To increase such genetic resolution, the use of specific primers is the best approach to the study of a particular target group (32). This was the main goal of our work, in order to get a more realistic picture on the diversity and structure of picocyanobacteria in natural communities. Accurate information of picocyanobacterial genetic diversity has been obtained using high-resolution marker genes, such as petB, rbcL, rpoC, cpcBA, mpeBA, and mpeW, and the 16S-23S rRNA internal transcribed spacer (ITS) (4, 6, 15, 17, 18, 33–36). However, this information is strongly biased toward marine picocyanobacterial strains, leaving the freshwater species still undersampled in databases; thus, a comprehensive study of all aquatic cyanobacterial using such functional markers is not currently possible. As an alternative, using the 16S rRNA gene as a marker is still the best-suited approach.
In fact, 16S rRNA gene primer pairs specific for coccoid Cyanobacteria, CYA359F/CYA781(b)R (37), were developed and used in several denaturing gradient gel electrophoresis (DGGE) fingerprinting studies, in which the specificity was verified (38–41). In particular, Boutte et al. (42) observed that the primers designed by Nübel et al. (37) were the most specific for coccoid Cyanobacteria. These primers flank the V2 to V4 region of the 16S rRNA gene, encompassing ca. 500 bp, and could potentially be used in HTS. However, the 16S rRNA gene is a mosaic of conserved and variable regions with different degrees of conservation in different prokaryotic groups (43), and consequently the choice of the region for diversity studies requires careful consideration. The most used variable regions for the characterization of prokaryotic communities are the V4, V3 to V4, V4 to V5, and V6 regions (for examples, see references 26 and 44), but which hypervariable region is the most suitable for use in picocyanobacterium-specific diversity studies has never been evaluated.
With the aim of unveiling the hidden diversity within the picocyanobacteria and in order to establish a systematic approach based on 16S rRNA gene iTags that would facilitate evaluation of picocyanobacterial community structure, we (i) determined which hypervariable region of the 16S rRNA gene provides the most taxonomic and phylogenetic information for the genera Synechococcus, Prochlorococcus, and Cyanobium, (ii) designed specific 16S rRNA gene primers for these genera to be used in the Illumina MiSeq sequencing platform, and (iii) evaluated the specificity and performance of the newly designed primers in marine (Blanes Bay, NW Mediterranean) and freshwater (Chascomús, Carpincho, Bragado, and Monte lakes, the Pampa plain, Argentina) systems.
RESULTS
Defining the most adequate hypervariable region.
To evaluate which 16S rRNA gene region is the most informative in picocyanobacterial, we first created in silico libraries using public picocyanobacterial sequences. Two 16S rRNA gene libraries were constructed: full-length sequence (FLS) and short-length sequence (SLS) libraries; the first contained almost the complete 16S rRNA gene (ca. 1,400 bp). These FLS were then used as the seed to construct the SLS libraries. Two types of SLS libraries were generated, one with 16S rRNA gene fragments containing the hypervariable region V2 to V4 (SLSV2–V4) and another one with fragments containing the V5 to V7 regions (SLSV5–V7) (Table S1).
To test for different degrees of variability in the hypervariable regions for different clades, we organized the sequences into five phylogenetic groups. Three groups contained sequences of Synechococcus (designated Syn and listed with consecutive numbers), one contained sequences of Cyanobium (Cyab), and another one contained sequences of Prochlorococcus (Proch) (Table S1). Thus, a total of six FLS libraries were constructed, one containing all of the sequences (FLS-Pcy) and one for each phylogenetic group (Table S1). The 16S rRNA gene fragments of SLS were extracted from each of the 6 FLS libraries, resulting in a total of 12 SLS libraries (Table S1). Once the FLS and SLS libraries were ready, we constructed maximum likelihood (ML) phylogenetic trees and analyzed (i) the genetic variability and (ii) the phylogenetic information contained in each of the V2 to V4 and the V5 to V7 variable regions.
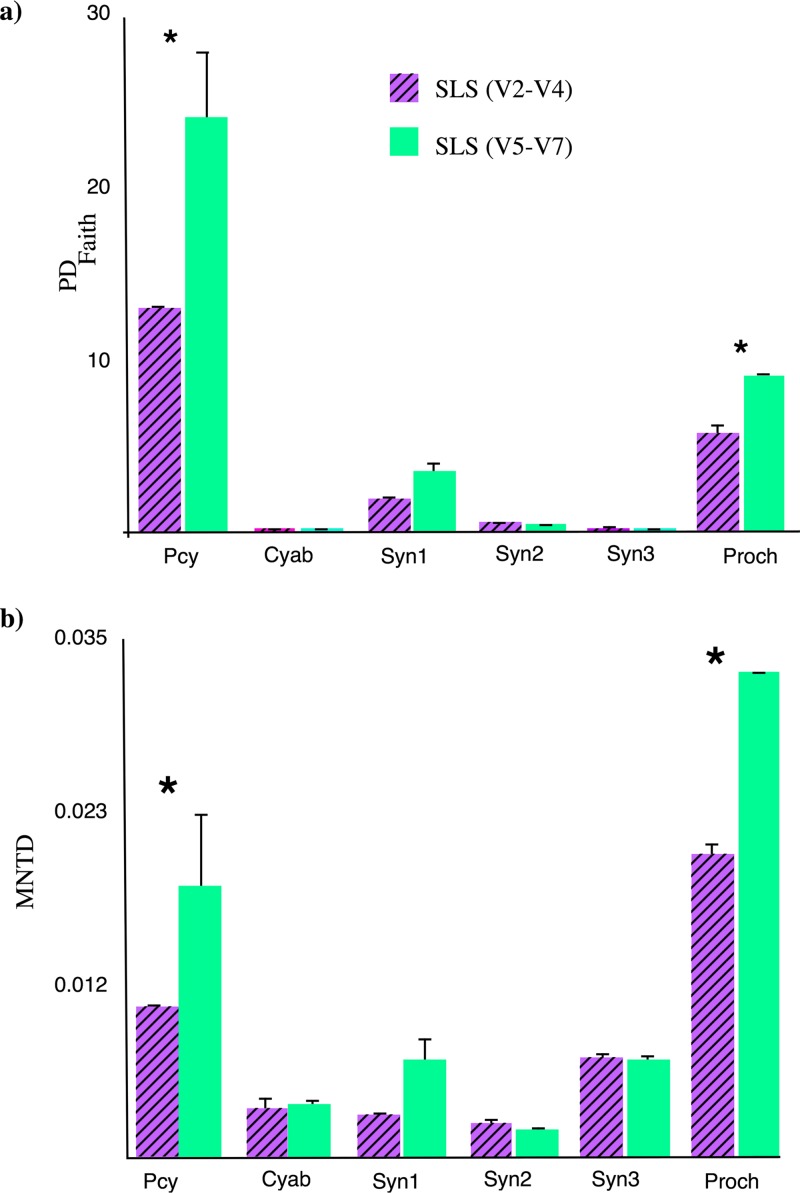
The genetic variability along the different 16S rRNA gene regions was explored through the Faith phylogenetic diversity index (PDFaith) (45) and the mean-nearest-taxon-distance (MNTD) (46). The PDFaith value is defined as the sum of the minimum lengths of all branches in a phylogenetic tree (45), while the MNTD value calculates the phylogenetic distance between each taxon within a community and its closest relative (46). Higher values of both metrics indicate greater phylogenetic diversity but MNTD is more sensitive to variations of closely related taxa than PDFaith (45). In this study, the higher values of PDFaith were observed in the trees constructed with the short-length sequence (SLS) libraries containing the V5 to V7 regions (i.e., SLSV5–V7) and were significantly higher than those from libraries containing the V2 to V4 regions (i.e., SLSV2–V4) (Fig. 1a), indicating larger genetic variability in the V5 to V7 16S rRNA gene hypervariable regions for picocyanobacterial taxa. The same pattern was observed when phylogenetic groups were analyzed separately (except for the Syn2 group) (Fig. 1a). The results obtained with the MNTD index were in agreement with those obtained with the PDFaith index (Fig. 1b). The highest values were observed in the SLSV5–V7 libraries, being significantly higher for SLS-Pcy (all picocyanobacterial sequences) and SLS-Proch (Prochlorococcus sequences). Thus, these results confirm that the fragment containing the V5 to V7 hypervariable region features more genetic variability than that of the V2 to V4 hypervariable region.
FIG 1.
Values of PDFaith (a) and MNTD (b) indices obtained from phylogenetic trees constructed from each short-length sequence library (SLS), consisting of one containing all the sequences (Pcy) and one for each phylogenetic group (Cyab, Cyanobium; Syn, Synechococcus; Proch, Prochlorococcus). Significant differences between the compared SLSs are indicated with an asterisk.
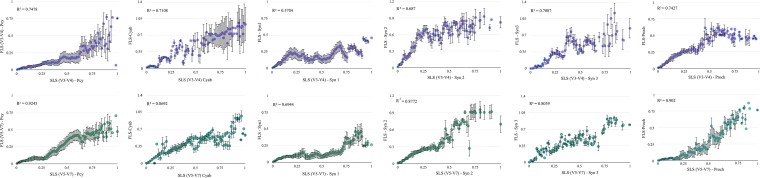
To evaluate the phylogenetic information contained in the different regions, we compared the topology of the phylogenetic trees constructed from the short- and full-length sequence libraries. First, we calculated the patristic distances (i.e., the sum of branch lengths that link two tips in the tree [PDist]) between all possible pairs of sequences in each constructed phylogeny and correlated each PDist of the SLS trees with those of the FLS trees (see methodology). The PDist correlations were always higher for FLS and SLS containing the V5 to V7 hypervariable region (SLSV5–V7) than for those in the SLSV2–V4. This occurred when using all the sequences and also when each phylogenetic group was studied separately (Table 1 and Fig. 2). We also calculated the Robinson-Foulds (RF) and weighted Robinson-Foulds (WRF) indices (47). Both metrics count the number of different bipartitions between two trees. The lower the values, the more similar the trees are with respect to their topology. The difference between the RF and WRF indices is that the latter takes into account the bootstrap support values of the bipartitions instead of looking only at their presence or absence, and thus bifurcations with lower support are less penalized. In all cases, the RF and WRF indices were significantly lower for the V5 to V7 fragment than for the V2 to V4 fragment (Table 1), indicating that the number of different bipartitions between the FLS and SLS trees occurred to a lesser degree in FLS versus SLS(V5–V7) than in FLS versus SLS(V2–V4) trees (Table 1). In addition, the WRF values were much lower than the values of their respective RFs, indicating that the different bipartitions occurred in nodes with low bootstrap values. Although this was observed for both regions, the lowest values were between FLS and SLS(V5–V7). To further strengthen our results, we evaluated the congruency between FLS and SLS tree topologies by the F measure, defined as the harmonic mean of precision and recall (see Materials and Methods). The F values range from zero for fully dissimilar trees to a maximum value for fully similar trees, value approaches one as the topologies of both trees become similar. For all compared trees, the SLS(V5–V7) libraries showed high F measure values compared to those of SLS(V2–V4) libraries, supporting the results described above (Table 1).
TABLE 1.
Summary of measures and indices used to compare the phylogenetic information in trees constructed using FLS and SLS libraries
| Library | Region | Replicate of SLS | Patristic PCa | RF distanceb | WRF distancec | F measured |
|---|---|---|---|---|---|---|
| FLS-Pcy | SLS-Pcy(V2–V4) | a | 0.546 | 2,460 | 46.777 | 0.072 |
| b | 0.544 | 2,462 | 47.606 | 0.077 | ||
| c | 0.548 | 2,433 | 46.880 | 0.080 | ||
| SLS-Pcy(V5–V7) | a | 0.612 | 2,402 | 44.327 | 0.096 | |
| b | 0.618 | 2,418 | 45.285 | 0.091 | ||
| c | 0.712 | 2,412 | 43.970 | 0.089 | ||
| FLS-Cyab | SLS-Cyab(V2–V4) | a | 0.442 | 132 | 1.500 | 0.083 |
| b | 0.542 | 133 | 1.580 | 0.082 | ||
| c | 0.426 | 134 | 1.600 | 0.069 | ||
| SLS-Cyab(V5–V7) | a | 0.541 | 120 | 1.470 | 0.142 | |
| b | 0.441 | 122 | 1.460 | 0.115 | ||
| c | 0.440 | 122 | 1.500 | 0.102 | ||
| FLS-Syn1 | SLS-Syn1(V2–V4) | a | 0.190 | 1,116 | 9.191 | 0.046 |
| b | 0.197 | 1,114 | 8.890 | 0.044 | ||
| c | 0.207 | 1,110 | 8.862 | 0.039 | ||
| SLS-Syn1(V5–V7) | a | 0.318 | 1,104 | 8.620 | 0.049 | |
| b | 0.311 | 1,107 | 8.624 | 0.053 | ||
| c | 0.323 | 1,102 | 8.590 | 0.054 | ||
| FLS-Syn2 | SLS-Syn2(V2–V4) | a | 0.712 | 418 | 6.967 | 0.087 |
| b | 0.6936 | 417 | 6.780 | 0.062 | ||
| c | 0.7121 | 420 | 6.770 | 0.054 | ||
| SLS-Syn2(V5–V7) | a | 0.763 | 402 | 6.480 | 0.122 | |
| b | 0.7631 | 408 | 6.210 | 0.105 | ||
| c | 0.8031 | 404 | 6.490 | 0.098 | ||
| FLS-Syn3 | SLS-Syn3(V2–V4) | a | 0.456 | 46 | 0.360 | 0.281 |
| b | 0.569 | 43 | 0.340 | 0.317 | ||
| c | 0.490 | 45 | 0.330 | 0.285 | ||
| SLS-Syn3(V5–V7) | a | 0.725 | 0 | 0.000 | 1.00 | |
| b | 0.689 | 2 | 0.090 | 0.968 | ||
| c | 0.698 | 1 | 0.087 | 0.984 | ||
| FLS-Proch | SLS-Proch(V2–V4) | a | 0.544 | 606 | 10.834 | 0.087 |
| b | 0.573 | 603 | 11.021 | 0.087 | ||
| c | 0.551 | 607 | 10.720 | 0.084 | ||
| SLS-Proch(V5–V7) | a | 0.737 | 576 | 10.272 | 0.132 | |
| b | 0.739 | 575 | 10.190 | 0.127 | ||
| c | 0.737 | 577 | 10.212 | 0.124 |
Patristic PC, Pearson correlation of patristic distances assessed by the Mantel test. In all cases, P < 0.05.
RF, Robinson-Fould.
WRF, weighted Robinson-Fould.
F measure = 2 × [(precision × recall)/(precision + recall)].
FIG 2.
Patristic distances of FLS and SLS(V2–V4) trees (upper panel) and FLS and SLS(V5–V7) trees (lower panel). The distances were normalized to a maximum value of one and ordered at intervals of 0.01according to the distance from the FLS tree. Each point represents the average of the FLS distance and corresponding averaged SLS distances in each interval. The standard deviations of the averaged SLS distances were used as error bars on the chart.
Furthermore, to test whether the topologies of the constructed SLS trees were a methodological artifact generated by sequence length, we correlated the PDist values of the three trees constructed from each SLS library. In all cases, the correlations between PDist values were close to one (Fig. S1), confirming that the differences in topology between the SLS and FLS trees described above are reliable.
Design of specific primers for picocyanobacteria.
Based on the previous results, and in order to cover Cyanobacteria of the genera Synechococcus, Prochlorococcus, and Cyanobium, we designed a set of primers flanking the hypervariable region V5 to V7 of the 16S rRNA gene (fragment size, 520 bp), Cya-771F (5′-AGGGGAGCGAAAGGGATTA-3′) and Cya-1294R (5′-GCCTACGATCTGAACTGAGC-3′). In silico analysis (Table 2) demonstrated a high specificity for the target sequences, i.e., more than 99% of the total sequences recovered by the primers Cya-771F/1294R belong to the three cyanobacterial picoplanktonic genera. Additionally, from 90.67% to 94.28% of total sequences within each genus (i.e., Prochlorococcus, Synechococcus, and Cyanobium) could be recovered with our primer set, indicating a high coverage rate for these specific picocyanobacterial genera (Table 2). We also compared our results with the coverage range and specificity obtained with the primers proposed by Nübel et al. (37) specific for coccoid Cyanobacteria. When perfect matches were considered (i.e., 0 mismatches), our set of primers displayed higher specificity and coverage for the sequences of the targeted organisms (Table 2), while a greater number of sequences for other Cyanobacteria genera (e.g., Leptolyngbyales, Nostocales, Oxyphotobacteria, and Phormidesmiales) were recovered using the Nübel et al. (37) primers. Furthermore, when 1 and 2 mismatches were allowed, both sets of tested primers increased their target group sequence coverage and decreased their specificity, as expected (Table S2 and Table S3). However, when considering 2 mismatches, Nübel et al. (37) primer specificity declined from 99.7% to 97.1%, while the change in specificity for our primers was lower (from 99.9% to 99.2%) (Table S3). Finally, we calculated the coverage and specificity of two sets of universal prokaryotic primers broadly used in microbial community studies, Bact_341F (5′-CCTACGGGNGGCWGCAG-3′) and Bact_805R (5′-GACTACHVGGGTATCTAATCC-3′), proposed by Herlemann et al. (48), and 515F-Y (5′-GTGYCAGCMGCCGCGGTAA-3′) and 926R (5′-GTGYCAGCMGCCGCGGTAA-3′) from Parada et al. (26). Regardless of the number of mismatches allowed (0,1, or 2), the coverage ranges for the targeted organisms were similar to those obtained with the specific primers; however, the specificity values obtained with universal primers were, in both cases, considerably lower (Tables S4 to S9 and Fig. S2).
TABLE 2.
In silico evaluation of specificity and coverage of the primer pairs designed in this work compared with those designed by Nübel et al. (37), with zero mismatches allowed
| Taxonomya | Coverage (%)b |
Matchc |
Eligibled |
Specificity (%)e |
Outgroup_matchedf |
Outgroup_matchableg |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| This work | Nübel et al. (1997) | This work | Nübel et al. (1997) | This work | Nübel et al. (1997) | This work | Nübel et al. (1997) | This work | Nübel et al. (1997) | This work | Nübel et al. (1997) | |
| Archaea | 0 | 0 | 0 | 0 | 15,292 | 23,948 | 99.74 | 99.49 | 1.650 | 3.313 | 645,343 | 647,636 |
| Bacteria | 0.29 | 0.58 | 1.650 | 3.314 | 572,584 | 575,263 | 100.00 | 100.00 | 0 | 0 | 88,051 | 96,341 |
| Bacteria; Actinobacteria | 0.00 | 0.00 | 1 | 0 | 59,305 | 59,486 | 99.73 | 0.54 | 1.649 | 3.313 | 601,330 | 612,098 |
| Bacteria; Proteobacteria | 0.00 | 0.00 | 1 | 1 | 228,791 | 229,791 | 99.62 | 99.25 | 1.649 | 3.312 | 431,844 | 441,793 |
| Bacteria; Cyanobacteria | 12.27 | 24.57 | 1.648 | 3.313 | 13,434 | 13,486 | 99.99 | 100.00 | 2 | 0 | 647,201 | 658,118 |
| Bacteria; Cyanobacteria; Oxyphotobacteria | 12.83 | 25.69 | 1.648 | 3.313 | 12,846 | 12,894 | 99.99 | 100.00 | 2 | 0 | 647,789 | 658,710 |
| Bacteria; Cyanobacteria; Oxyphotobacteria; Synechococcales | 90.15 | 89.73 | 1.638 | 1.652 | 1,817 | 1,841 | 99.99 | 99.75 | 12 | 1.661 | 658,818 | 669,763 |
| Bacteria; Cyanobacteria; Oxyphotobacteria; Synechococcales; Cyanobiaceae | 91.97 | 90.76 | 1.638 | 1.620 | 1,781 | 1,785 | 99.99 | 99.75 | 12 | 1.693 | 658,854 | 669,799 |
| Bacteria; Cyanobacteria; Oxyphotobacteria; Synechococcales; Cyanobiaceae; Prochlorococcus MIT9313 | 90.67 | 89.85 | 408 | 602 | 450 | 670 | 99.81 | 99.60 | 1.242 | 2.711 | 660,185 | 670,934 |
| Bacteria; Cyanobacteria; Oxyphotobacteria; Synechococcales; Cyanobiaceae; Synechococcus CC9902 | 90.88 | 89.14 | 578 | 402 | 636 | 451 | 99.84 | 99.57 | 1.072 | 2.911 | 659,999 | 671,133 |
| Bacteria; Cyanobacteria; Oxyphotobacteria; Synechococcales; Cyanobiaceae; Cyanobium PCC-6307 | 94.28 | 92.62 | 610 | 602 | 647 | 650 | 99.84 | 99.60 | 1.040 | 2.711 | 659,988 | 670,934 |
| Bacteria; Cyanobacteria; Oxyphotobacteria; Synechococcales; Cyanobiaceae; Synechococcus MBIC10613 | 100.00 | 91.04 | 5 | 579 | 5 | 636 | 99.75 | 99.59 | 1.645 | 2.734 | 660,630 | 670,948 |
| Bacteria; Cyanobacteria; Oxyphotobacteria; Synechococcales; Cyanobiaceae; spongiarum group | 85.71 | 76.19 | 36 | 32 | 42 | 42 | 99.76 | 99.51 | 1.614 | 3.281 | 660,593 | 671,542 |
| Bacteria; Cyanobacteria; Oxyphotobacteria; Synechococcales; Prochlorotrichaceae | 0.00 | 80.00 | 0 | 4 | 11 | 5 | 99.75 | 99.51 | 1.650 | 3.309 | 660,624 | 671,579 |
| Bacteria; Cyanobacteria; Oxyphotobacteria; Synechococcales; Synechococcaceae | 0.00 | 100.00 | 0 | 11 | 15 | 11 | 99.75 | 99.51 | 1.650 | 3.302 | 660,620 | 671,573 |
| Bacteria; Cyanobacteria; Oxyphotobacteria; Chloroplast | 0.00 | 0.12 | 0 | 7 | 6,031 | 6,040 | 99.75 | 99.50 | 1.650 | 3.306 | 654,604 | 665,544 |
| Bacteria; Cyanobacteria; Oxyphotobacteria; Eurycoccales | 0.00 | 22.22 | 0 | 10 | 45 | 45 | 99.75 | 99.51 | 1.650 | 3.303 | 660,590 | 671,539 |
| Bacteria; Cyanobacteria; Oxyphotobacteria; Leptolyngbyales | 3.06 | 89.34 | 6 | 176 | 196 | 197 | 99.75 | 99.53 | 1.644 | 3.137 | 660,439 | 671,387 |
| Bacteria; Cyanobacteria; Oxyphotobacteria; Limnotrichales | 5.88 | 82.35 | 1 | 14 | 17 | 17 | 99.75 | 99.51 | 1.649 | 3.299 | 660,618 | 671,567 |
| Bacteria; Cyanobacteria; Oxyphotobacteria; Neosynechococcales | 0.00 | 100.00 | 0 | 1 | 1 | 1 | 99.75 | 99.51 | 1.650 | 3.312 | 660,634 | 671,583 |
| Bacteria; Cyanobacteria; Oxyphotobacteria; Nostocales | 0.03 | 18.58 | 1 | 700 | 3,757 | 3,768 | 99.75 | 99.61 | 1.649 | 2.613 | 656,878 | 667,816 |
| Bacteria; Cyanobacteria; Oxyphotobacteria; Oxyphotobacteria incertae sedis | 0.41 | 85.45 | 2 | 423 | 493 | 495 | 99.75 | 99.57 | 1.648 | 2.890 | 660,142 | 671,089 |
| Bacteria; Cyanobacteria; Oxyphotobacteria; Phormidesmiales | 0.00 | 86.60 | 0 | 252 | 291 | 291 | 99.75 | 99.54 | 1.650 | 3.061 | 660,344 | 671,293 |
| Bacteria; Cyanobacteria; Oxyphotobacteria; Pseudanabaenales | 0.00 | 4.90 | 0 | 5 | 102 | 102 | 99.75 | 99.51 | 1.650 | 3.308 | 660,533 | 671,482 |
| Bacteria; Cyanobacteria; Oxyphotobacteria; RD011 | 0.00 | 87.50 | 0 | 7 | 8 | 8 | 99.75 | 99.51 | 1.650 | 3.306 | 660,627 | 671,576 |
| Bacteria; Cyanobacteria; Oxyphotobacteria; RD017 | 0.00 | 83.33 | 0 | 10 | 12 | 12 | 99.75 | 99.51 | 1.650 | 3.303 | 660,623 | 671,572 |
| Bacteria; Cyanobacteria; Oxyphotobacteria; SepB-3 | 0.00 | 87.50 | 0 | 7 | 8 | 8 | 99.75 | 99.51 | 1.650 | 3.306 | 660,627 | 671,576 |
SILVA taxonomy, SSU r132 database; Ref NR sequence collection.
Coverage % = (match/eligible) × 100.
Number of match hits in the taxonomic path.
Number of hits with sequence data at the primers’ position in the taxa path.
Specificity %= 100 − (outgroup_matches/outgroup_matchable) ×100.
Number of matched hits outside the taxonomic path.
Number of matchable hits outside the taxonomic path.
Testing the primers with environmental samples.
The specificity and usefulness of the designed primers were tested in both freshwater and marine samples using Illumina sequencing. We obtained totals of 21,477 and 56,610 high quality reads from the marine (Blanes Bay) and freshwater (Argentinian lakes) samples (see Materials and Methods), respectively. All sequences from Blanes Bay were assigned to either the genus Synechococcus or Prochlorococcus, and 96.3% of the reads retrieved from lakes belonged to the genera Synechococcus (32%), Cyanobium (12%), and environmental clones closely related to them (51%) (Table S10). The remaining percentage was assigned to filamentous Cyanobacteria (2.3%) of the Cylindrospermosis and Leptolynbya genera, as well as to heterotrophic bacteria (1.3%), particularly Planctomycetales and Verrucomicrobiales. Rarefaction curves showed an asymptotic tendency only in two out of the ten samples (in Chascomús lake during the spring of 2012 and in the summer sample from Blanes Bay), while in the rest of the samples, picocyanobacterial diversity did not reach saturation with the sequencing effort used in this work (Fig. S3). A total of 113 zero-radius operational taxonomic units (zOTUs) within the picocyanobacterial genera were defined, with 87 zOTUs being exclusive to fresh waters and 26 zOTUs being exclusive to the marine site (Blanes Bay).
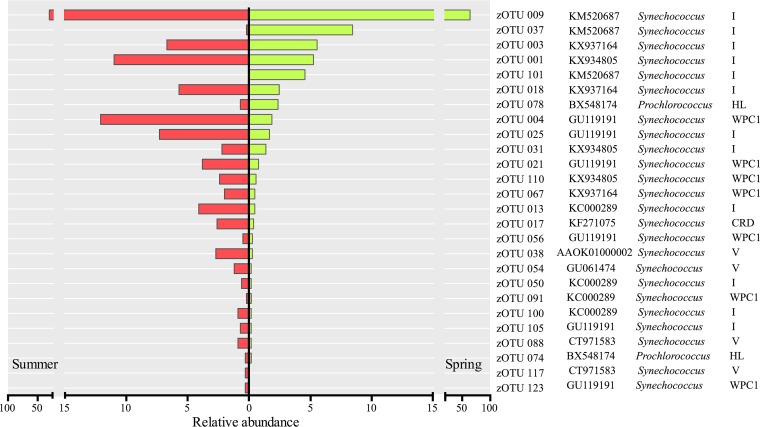
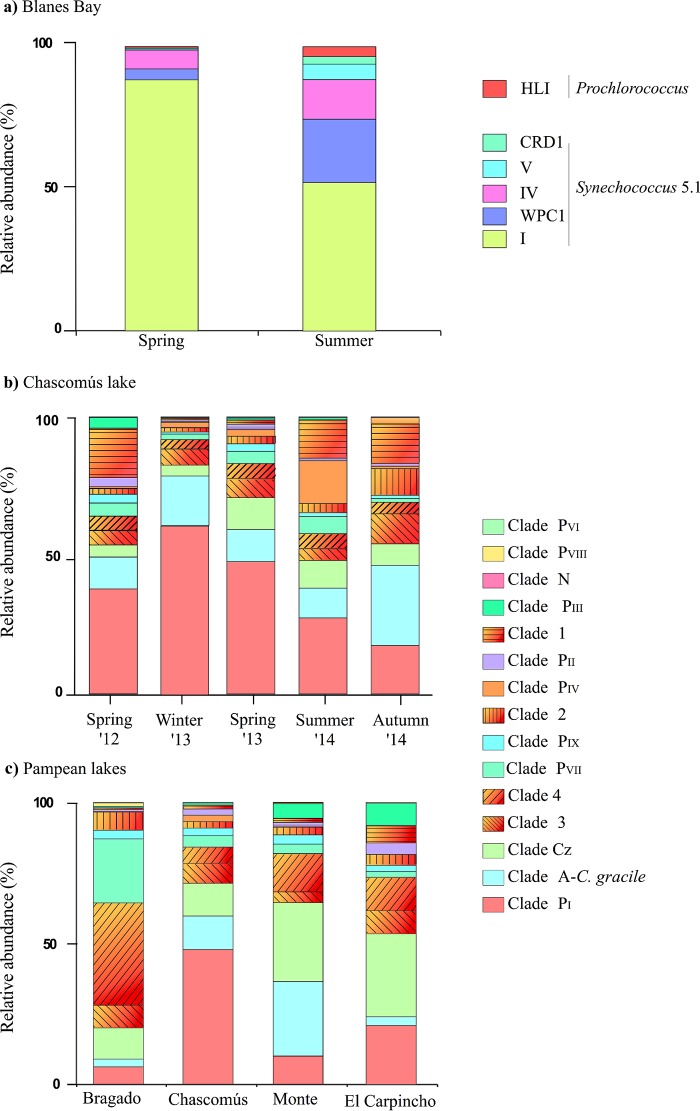
The samples from Blanes Bay correspond to two different periods of the year (spring and summer) when peaks of picocyanobacteria are frequently registered. Out of 26 zOTUs, 24 belonged to Synechococcus, and only 2 displayed high similarity with sequences of Prochlorococcus (Fig. 3), which contributed with less than 2% to the total abundance in each sample (Prochlorococcus spp. are more abundant in the fall-winter season in this site [49]). The taxonomic composition was very similar in both samples, including only three zOTUs exclusive to one season (zOTU-91 and zOTU-117 from summer, and zOTU-101 from spring), and the community in summer showed a higher diversity (Shannon index = 3.69; PDFaith = 1.8) than in spring (Shannon index = 3.44; PDFaith = 1.6) (Table 3). Despite community structure being significantly different between seasons (Student t test, P < 0.001), there was a single sequence (zOTU-9) that dominated in both seasons, contributing 63.4% of the total number of reads in spring and 30.6% in summer (Fig. 3). All Synechococcus zOTUs belonged to the marine subcluster 5.1, clustering into five phylogenetic clades defined in the reference tree (RT), I, IV, V, CRD1, and WPCI (Fig. 3 and Fig. S4). In both samples, the most abundant lineage was clade I, followed by clades WPC1 and IV, which exhibited a larger relative contribution in summer (Fig. 4a). Regarding the Prochlorococcus zOTUs, both of them belonged to clade HLI and were closely related to the MED4 strain retrieved from the Mediterranean Sea (Fig. 3 and Fig. S4).
FIG 3.
Relative abundance of zOTUs determined for the two samples from Blanes Bay (left. summer; right, spring). For each zOTU, the GenBank accession number and the taxonomic and phylogenetic clade assignments are presented.
TABLE 3.
Number and abundance of zOTUs per sample and values of the different indices used for the characterization of the communities
| Sample type | System | Season (sample date) | No. of zOTUs | No. of zOTUs >1%a | % of reads >1%b | Index value |
||
|---|---|---|---|---|---|---|---|---|
| Shannon | Chao1 | PDFaithc | ||||||
| Marine | Blanes Bay | Spring (April 13) | 24 | 10 | 96.5 | 3.44 | 21 | 1.6 |
| Blanes Bay | Summer (August 13) | 25 | 14 | 94.2 | 3.69 | 22 | 1.8 | |
| Freshwater (temporal) | Chascomús | Spring (October 12) | 66 | 24 | 86.5 | 3.32 | 71 | 0.28 |
| Chascomús | Winter (June 13) | 72 | 22 | 88.9 | 3.12 | 85 | 0.46 | |
| Chascomús | Spring (November 13) | 76 | 24 | 84.5 | 3.37 | 87 | 0.43 | |
| Chascomús | Summer (February 14) | 83 | 25 | 85.2 | 3.48 | 86 | 0.49 | |
| Chascomús | Autumn (May 14) | 84 | 25 | 80.2 | 3.59 | 86 | 0.49 | |
| Freshwater (spatial) | El Carpincho | Spring (December 09) | 59 | 30 | 87.0 | 2.79 | 73 | 0.26 |
| Bragado | Spring (December 09) | 78 | 13 | 84.2 | 3.37 | 80 | 0.44 | |
| Monte | Spring (December 09) | 68 | 24 | 91.3 | 3.39 | 71 | 0.45 | |
| Chascomús | Spring (November 13) | 65 | 23 | 89.4 | 3.12 | 69 | 0.36 | |
Number of zOTUs composing the “abundant fraction” in each sample (i.e., abundance of >1%).
Percentage contribution of zOTU abundant fraction to the number of total reads in each sample (i.e., abundance of >1%).
PDFaith, Faith phylogenetic diversity index.
FIG 4.
Bar graph showing the relative abundance of zOTUs per clades in (a) Blanes Bay, (b) Chascomús lake, and (c) Pampean lakes, based on the phylogenetic reconstruction shown in Fig. S4 to S6.
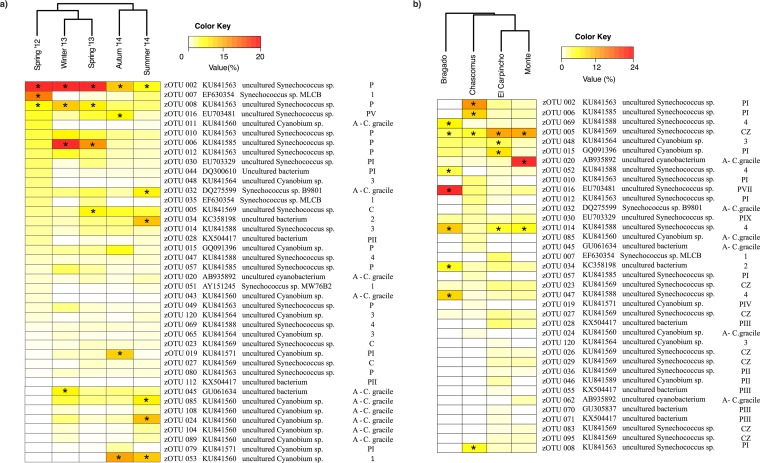
A total of 86 zOTUs were obtained from the five seasonal samples taken from Chascomús lake (CH). Richness varied between 66 (spring 2012) and 84 zOTUs (fall 2014), and the Shannon index varied little between samples (Table 3). However, the phylogenetic diversity (PDFaith) was notably lower in spring 2012 (Table 3). A cluster analysis revealed higher similarity in the picocyanobacterial community structure in samples within the same year, independent of the season (Fig. 5a). None of the environmental variables measured were significantly associated with community structure. In each sample, the abundant fraction of the communities was represented by 22 to 25 zOTUs (Table 3), accounting for more than 80% of total reads. Most zOTUs (67.8%) were found all year long. In particular, zOTU-2 was always present at >5% of relative abundance (Fig. 5a). Most zOTUs clustered into 11 clades previously defined in the reference tree (Fig. 5a and Fig. S5), while 4 new clades were formed exclusively by the newly obtained zOTUs (the new clades were named 1 to 4 in Fig. S5). zOTUs composing the core group (those that contributed with more than 5% of the reads) belonged to different phylogenetic clades, and their contribution changed markedly among samples (Fig. 4b and 5a). For instance, clade PI was prevalent in spring and winter and clade PIV in summer 2014, and clade A was more represented in 2014 (Fig. 4b).
FIG 5.
Cluster analysis of zOTU abundance from (a) Chascomús lake and (b) the four Pampean shallow lakes studied. The heat maps represent the relative abundances of the abundant zOTUs (i.e., abundance of ≥1%). The asterisks indicate the zOTUs with a relative abundance of >5%. For each zOTU, the GenBank accession number and the taxonomic and phylogenetic clade assignments are presented.
In the spatial comparison between the four Pampean lakes, zOTU richness varied between 78 in Chascomús and 59 in Bragado lakes. Bragado lake also had the lowest taxonomic and phylogenetic diversity (Table 3). Only 10 out of the total of 85 zOTUs were exclusive to any particular environment. The cluster analysis showed that the Monte and El Carpincho lakes presented a more similar community composition to each other than the other lakes (Fig. 5b), and the structure of the picocyanobacterial community was highly correlated with the quality of light (expressed as the relation between the vertical diffuse light attenuation coefficient for red [675 nm] and green [440 nm] [Kdred/Kdgreen]; Table S11) and total phosphorus concentration (Mantel test, r = 0.83, P < 0.05). The number of abundant zOTUs (i.e., relative abundances of >1%) varied between 13 (Bragado) and 30 (El Carpincho), and in all cases they contributed with more than 84% to the total reads of each lake (Table 3). The zOTUs were affiliated with the 13 clades previously described in the temporal study of Chascomús (Fig. S6). The dominant zOTUs of each environment belonged to different clades (Fig. 5b and Fig. S5). The most notable case was clade 4, which was almost exclusively represented by sequences from Bragado (Fig. 4c). Clade PVII was also more prevalent in this lake. Clade PI showed the highest relative abundance in Chascomús and El Carpincho lakes, and clade A was dominant in Monte Lake (Fig. 4c).
DISCUSSION
We found that the hypervariable region V5 to V7 of the 16S rRNA gene is the most appropriate for studying the diversity, structure, and dynamics of picocyanobacterial communities using high-throughput sequencing. Specifically, we found that (i) the V5 to V7 region features more variability in picocyanobacteria than other regions and (ii) that the phylogenies based on the V5 to V7 region mirror to a larger extent the phylogenies obtained with the complete 16S rRNA gene. Additionally, we successfully designed and used a set of primers flanking the V5 to V7 region, which target almost exclusively Synechococcus, Prochlorococcus, and Cyanobium, thus allowing the accurate characterization of natural picocyanobacterial communities.
We explored the taxonomic information contained in the hypervariable regions of the 16S rRNA gene using indices with different sensitivity to phylogenetic relatedness (i.e., PDFaith and MNTD). The results indicate that the V5 to V7 region allows for a better discrimination of both phylogenetically distant and closely related picocyanobacteria (46). Which region is suitable for studying prokaryotic diversity has mostly been explored for entire bacterial communities and other taxonomic groups (26, 50–52); however, to our knowledge, this is the first time that it has been evaluated for picocyanobacterial genera. Reviewing the information present in the literature (26, 50–52), we detected contrasting results about the suitability of 16S rRNA gene hypervariable regions for the study of prokaryotic communities, which alerted us about the importance of selecting a particular hypervariable region of 16S rRNA according to the taxonomic level/group at which the results need to be inferred (53).
The study of phylogenetic relationships (i.e., patristic distance, RF and WRF indices, and F value) indicated that the information present in fragments of the 16S rRNA gene containing the V5 to V7 region allows the inference of the phylogenetic relationships obtained from the full-length 16S rRNA gene for the picocyanobacterial groups. In previous studies, the V4 fragment was found to be the best for establishing phylogenetic relationships when the entire bacterial community was considered (26, 52, 54). However, this contrasts with our results, which suggest that the hypervariable region of the 16S rRNA gene that best reflects the full-length gene phylogeny depends on the taxonomic groups being studied. Additionally, we observed a high correlation between tree topologies constructed from the same SLS, which suggests that (i) the difference in the correlations of SLS(V2–V4) and SLS(V5–V7) with the FLS may be attributable to the phylogenetic information present in the fragments instead of the size of the sequences and (ii) the phylogenies obtained from partial sequences have a good reproducibility power.
The specificity of the new set of primers analyzed in silico was validated in natural communities. Nevertheless, some discrepancies in the recovered taxa were detected, indicating the importance performing primer tests on environmental samples. For example, some reads were assigned to genera that appeared in the in silico analyses allowing 1 or 2 mismatches (e.g., Planctomycetes and Verrucomicrobia), while some genera typical of aquatic systems (e.g., Pseudanabaenales, Actinobacteria, and Firmicutes) that were recovered in the in silico analysis did not appear in the zOTU table.
On the other hand, by defining operational taxonomic units as exact sequence variants (zOTUs) (55) we could achieve a fine-scale taxonomic resolution (55, 56), allowing us to detect greater genetic diversity among the picocyanobacteria, as well as specific patterns of community composition in different systems and seasons. In Blanes Bay, Synechococcus reads were much more abundant and diverse than those of Prochlorococcus, as was also observed in the cell abundances (Table S11). The number of operational taxonomic units defined here as zOTUs was greater than those established in previously studies, where bacterial primers 341F (5′-CCTACGGGNGGCWGCAG-3′) (48) and 806RB (5′-GGACTACNVGGGTWTCTAAT-3′) (44) were used to amplify partial 16S rRNA genes (i.e., 10 zOTUs of Synechococcus and 1 zOTU of Prochlorococcus; I. Ferrera and J. M. Gasol, personal communication). Picocyanobacterial community composition differed between spring and summer, yet there was a unique zOTU that dominated in both seasons. This supports observations from other sites that also revealed the presence of a single strain of Synechococcus throughout the year (57, 58), and points toward the existence of a single strain adapted to grow in different seasons characterized by quite different temperatures and nutrients (for example, see reference 49). When analyzing the phylogenetic structure of the communities, we observed that most Synechococcus zOTUs correspond to clades typical of coastal zones, clades I, IV, V, and WPC1 (15, 18, 59), which tend to cooccur (for examples, see references 4, 15, 29, 33, 59). However, zOTU-17 was assigned to clade CRD1, which has been described from the Costa Rica upwelling (60), as well as from the Pacific, Indian and Atlantic Oceans, although it is seemingly absent from the Mediterranean Sea (6). Regarding Prochlorococcus zOTUs, both belonged to the HLI clade, a dominant clade in surface waters with moderate stratification (17, 61–63).
In Chascomús lake there was a core of zOTUs (i.e., relative abundance of >5%) that changed their relative contribution between seasons. This pattern is usually observed in temperate lakes regardless of their limnological characteristics (22, 64) and suggests different ecological roles within genetically close strains. However, as in previous reports (22, 64), we failed to relate the distinct niches among the dominant zOTUs with environmental factors, such as nutrient concentrations, pH, underwater light, or temperature (Table S11). This may suggest that biotic feedback mechanisms, such as competition, viral infection, and prey-predator interactions are significant factors driving the temporal dynamics of the picocyanobacterial community, and further studies are needed to shed light on this issue. Regarding the spatial presence of the different zOTUs, each Pampean shallow lake was represented by a particular assemblage associated with local environmental factors. These results lend support to previous evidence about the importance of underwater light conditions and the availability of nutrients in governing freshwater picocyanobacterial distribution (35, 64–68).
Finally, the incorporation of zOTUs into the phylogenetic tree allowed the observation of a large number of coexisting clades with specific abundance dynamics changing over space and time. This has been well reported by other authors (20, 35, 69, 70) and seems to be a common feature in picocyanobacterial communities. What allows the coexistence of these very similar organisms? The great genetic microdiversity within picocyanobacterial genera may effectively be a good explanation. However, how environmental factors drive changes in clade abundances and whether cooccurring phylotypes within the same and different clades affect each other in beneficial or detrimental ways are questions that still need to be posed.
In summary, we determined that the V5 to V7 hypervariable region of the 16S rRNA gene provides more taxonomic and phylogenetic information for Synechococcus, Prochlorococcus, and Cyanobium than do other hypervariable regions, and we designed a set of specific primers for these genera. The results obtained from marine and freshwater environments showed the usefulness of the present approach and tools to explore the diversity and successional patterns in the picocyanobacterial assemblage via fine (i.e., zOTU) or coarse-grained (clade and subclade) taxonomic resolutions.
MATERIALS AND METHODS
Defining the most adequate hypervariable region.
To determine the most informative hypervariable 16S rRNA gene region, we followed the methodology proposed by Ghyselinck et al. (52). Genetic libraries were constructed using 1,334 sequences from representative picocyanobacterial clades selected from the public database SILVA 119 (https://www.arb-silva.de/download/arb-files/) (Table S1). The full-length sequence (FLS) libraries contained complete sequences with the information of all hypervariable regions (V1 through V9). To generated the short-length sequence (SLS) libraries, we searched for the V4 and V6 hypervariable regions from each FLS and trimmed out fragments of 500 bp in length containing either the V4 or the V6 region using V-Xtractor v 2.0 (71).
For each FLS and SLS library constructed, we aligned the sequences using Multiple Alignment using Fast Fourier Transform (MAFFT) v.7 with default options (72) and constructed maximum likelihood (ML) phylogenetic trees using FastTree v.2.1.5 (73) with default options using the Geneious v 9.1. interface (74). A single ML tree was constructed for each of the 6 FLS libraries, along with 3 trees for each of the 12 SLS libraries, using the general time reversible (GTR) model with a gamma-distributed rate of variation across sites. To estimate statistical support, 1,000 pseudoreplications were performed using the “rapid bootstrapping” algorithm. Three sequences of Gloeobacter sp. were used as an outgroup. In those analyses in which rootless trees were required, these outgroup sequences were discarded. The generated trees were used in downstream analyses.
Genetic variability was explored using the Faith phylogenetic diversity index (PDFaith) (45) and the mean nearest taxon distance (MNTD) (46). PDFaith is defined as the sum of the phylogenetic branch lengths connecting all taxa present in a community; higher values of PDFaith would indicate greater phylogenetic diversity and therefore greater genetic variability (45). The MNTD quantifies the phylogenetic distance between each taxon within a sample and its closest relative in the sample, returning an average (46). Thus, higher MNTD values indicate greater phylogenetic diversity among closely related taxa. To calculate PDFaith and MNTD values we used the package Picante (v.1.6) (75) within the R environment.
The phylogenetic information present in the different regions of the 16S rRNA gene was evaluated comparing the tree topology through the patristic distances (PDist), the Robinson-Foulds (RF) distances and the Robinson-Foulds weighted (WRF) distances (47). The PDist values were calculated between all possible pairs of sequences in each phylogeny constructed and were presented as distance matrices. Then, correlations between each SLS (in triplicate) and FLS distance matrix were tested using Mantel tests based on Pearson correlations. In order to visualize these data, the distances were normalized to a maximum value of one and ordered at intervals of 0.01 according to the distance from the FLS tree. For each interval, we calculated the average of the FLS distance and corresponding averaged SLS distances and then plotted them in a graph. The standard deviations of the averaged SLS distances were used as error bars in the chart (54).
Similarly, the RF distances allowed us to compare topologies of phylogenetic trees by counting the number of bipartitions that occur in one tree but not in the other, so a lower RF value indicates that a pair of trees is topologically more similar. We also calculated the WRF distance (47). This measure, unlike the RF distance, takes into account the bootstrap value of each bipartition instead of considering presence/absence only. For example, if a bipartition is present in one tree but not in another and has a bootstrap value of 0.3, then the WRF distance counts it as 0.3 instead of 1. Thus, by using the WRF distance, the bifurcations with low support are less penalized. When the WRF values are lower than the respective RF values, it means that the differences between trees occurred in clades with low bootstrap values. The patristic distances and RF/WRF metrics were calculated in the R environment using the stats and phangorn v.2.4 R packages (76).
Finally, to further strengthen our results, we compared the topologies between SLS and FLS trees using the precision and recall metrics. These measures were defined as follows: precision = TP/(TP + FP) and recall = TP/(TP + FN), where true positives (TP) are the set of bipartitions present in both the SLS tree and the FLS tree, false positives (FP) are the number of bipartitions present in the SLS tree but not in the FLS, tree and FN is the number of bipartitions present in the FLS tree but not in the SLS tree (77). These metrics were calculated in the R environment using the phangorn v.2.4 R package (76). The results were expressed as the F measure, which is defined as the harmonic mean of precision and recall and balances false positives and false negatives [F = 2 × (precision × recall)/(precision + recall)].
Primer design.
We designed a set of specific primers for each of the Synechococcus, Cyanobium, and Prochlorococcus genera. Using the FSL-Pcy library as a reference, we searched possible primers with Geneious v.9.1.5 (74), targeting a region that would start in the conserved area at the beginning of the V5 hypervariable region up to the end of the V7 hypervariable region, with an amplicon size of approximately 540 bp, suitable for Illumina MiSeq sequencing using 2 × 300 bp. Once the candidate primers were defined, we tested their specificity and coverage in silico against the SSU r132 RefNR database allowing 0, 1, and 2 mismatches using the Test Probe 1.0 feature from SILVA (http://www.arb-silva.de/?id=650).
Testing the primers with environmental samples: field sampling, PCR, and sequencing.
The specificity and usefulness of the designed primers were then tested in two sets of samples, as described below.
(i) Two marine samples (Table S11) were taken from the coastal Blanes Bay Microbial Observatory in the Mediterranean Sea (41°39.90′N, 2°48.03′E) (49). We selected one sample taken in spring and one in summer 2013 (April and August) based on previous knowledge that during these seasons the abundance of Synechococcus reaches maximum values (up to 8 × 104 cell per milliliter) in this coastal station (e.g., 49, 57, 58).
(ii) Eight freshwater samples were taken from four hypertrophic shallow lakes (Table S11) of the Salado River floodplain basin, located in the Pampa plain in Argentina (35°41′S, 59°35′W) (22, 68). The samples were selected in order to cover spatial and temporal variation. Three samples were taken during spring 2009 from the Bragado, El Carpincho, and Monte lakes. In addition, five samples from Chascomús lake were collected between October 2012 and May 2014 (Table S11). Site features and sampling details are provided in detail in Fermani et al. (68) and Huber et al. (22).
Sample collection and DNA extractions are described in Ferrera et al. (58) for the marine samples and in Huber et al. (22) for the freshwater samples. The volume of water processed is indicated in Table S11.
Sequencing was performed by the RTL Genomics division of Research and Testing Laboratories of the South Plains (Lubbock, TX) using Illumina MiSeq 2 ×300 paired-end reads (sequencing runs 1 and 2), with a sequencing depth of approximately 20,000 sequences per sample. The V5 to V7 region of the 16S rRNA gene was amplified using the primers designed in this work, Cya-771F and Cya-1294R (see above). The PCR conditions for these primers were as follows: 5 min preheating step at 94°C; 10 cycles of denaturation at 94°C for 1 min; an annealing touchdown procedure for 1 min in which the temperature decreased by 1°C every second cycle from 65 to 56°C; followed by 25 cycles of 1 min of denaturation at 94°C; and annealing at 55°C for 1 min and extension at 72°C for 3 min, with a final extension step of 10 min at 72°C.
Data analysis.
Raw sequences were processed using a modified version of the pipeline proposed by Logares (78) (https://github.com/ramalok). The reads were first analyzed for error correction using the algorithms based on Hamming graphs and Bayesian subclustering (BAYES HAMMER tool) (79) implemented in SPAdes v3.5.0 (80). Then, the forward and reverse sequences were assembled using the function fastq_mergepairs from USEARCH-v10 (81). The minimum overlap length was set to 20 bp and assemblage sequences with less than 100 nucleotides were discarded; the rest of the parameters were used as the default. Quality checking was performed using fastq_filter in USEARCH-v10 (81). Reads that passed the quality control were then analyzed using UNOISE2 (82) to define operational taxonomic units (OTUs) with no clustering (zero-radius OTUs [zOTUs] or amplicon sequence variants [ASVs]). These tool-defined zOTUs provide a higher accuracy than OTUs by achieving single-nucleotide resolution after correcting for Illumina sequencing errors and chimeras (55, 56). Finally, taxonomy assignment of zOTUs was done by BLAST (83), using the SILVA database (SSU Ref 132 NR 99) as a reference, and the zOTU table was created with the function otutab in USEARCH-v10 (81).
zOTUs with a relative abundance below 1% per sample were considered representative of the “rare biosphere,” while those whose abundances exceeded 1% were considered part of the “abundant fraction” (84). We constructed zOTU rarefaction curves for each environment, using vegan v.2.5 to evaluate whether we saturated richness. The taxonomic diversity and richness of the picocyanobacterial communities were estimated with the Shannon (85) and Chao1 (86) indices, respectively (Table 3). Bray-Curtis dissimilarity matrices were constructed to analyze the similarities in the composition of the assemblages. The abundance of each zOTU was previously transformed using Hellinger transformation, as recommended by Legendre and Gallagher (87). We also calculated phylogenetic diversity using the PDFaith index, based on maximum likelihood trees constructed using the Randomized Axelerated Maximum Likelihood (RAxML) v.8 program (88).
We also tested the environmental factors that determine community structure. For that purpose, we first selected possible drivers using the function BIOENV (89) within the R environment (90). After that, we constructed a Euclidian distance matrix with the selected variables and determined its correlation with the picocyanobacterial dissimilarity matrix using a Mantel test. The physical, chemical, and biological variables that were used in this analysis are summarized in Table S11.
To infer the phylogenetic positions of the zOTUs, we incorporated the amplicon short sequences into freshwater and marine phylogenetic reference trees (RT). First, we aligned the query sequences against the reference alignments using PaPaRa v.2.5 (91), and then we placed the queries onto the trees using the Evolutionary Placement Algorithm (EPA) (92) as implemented in RAxML (88). The RTs were constructed using sequences of the 16S rRNA gene (minimum length 700 bp) from the NCBI public database, ensuring that the main clades of picocyanobacteria were represented (6, 18–20, 22, 93) (Table S12). The marine RT included 94 sequences, 22 assigned to seven Prochlorococcus clades (HL and LL) and 72 to Synechococcus subclusters 5.2, 5.3, and 5.1 (with 11 clades assigned to 5.1). For the freshwater RT, we selected 181 published sequences and also incorporated 82 unpublished sequences corresponding to environmental clones of the complete 16S rRNA gene retrieved from two Pampean lakes (see Supplemental Material). Twenty-two clades were defined; 13 have been described in previous studies and 9 were novel, being constituted mostly of taxa retrieved from Pampean systems (designated by the letter P and listed with Roman numerals, i.e., PI to PIX); Synechococcus strain PCC 6301 was used as external group. The sequences were aligned using MAFFT v.7, and the RTs were constructed using the maximum likelihood method in RAxML v.8 (88) with the GTR+GAMMA model for freshwater taxa and the GTR+CAT+I model for marine taxa, considering 1,000 bootstraps in both cases. Trees were edited using the Interactive Tree Of Life (iTOL; http://itol.embl.de) (94).
Data availability.
All sequences generated in this study can be accessed through European Nucleotide Archive (ENA) under the study accession number PRJEB27291 and accession numbers ERR2639355 to ERR2639364.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Argentinean Network for the Assessment and Monitoring of Pampean Shallow Lakes (PAMPA2–CONICET), the ANPCyT (PICT-2014-1290 and PICT-2016-1079), and by MAGGY (grant CTM2017-87736-R) to S. G. Acinas and REMEI (grant CTM2015-70340-R) to J. M. Gasol from the Spanish Ministry of Economy and Competitiveness (MINECO).
We thank Roberto U. Escaray and Jose Bustingorry (INTECH) for technical support and M. Devercelli and M. L. Gelin for their advice on an early draft and for critical revision of the manuscript. We thank the members of the Laboratorio de Ecología y Fotobiología Acuática (IIB-INTECH) and the Departament de Biologia Marina i Oceanografia (ICM), and particularly the people involved in sampling the Blanes Bay Microbial Observatory, for their support.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02659-18.
REFERENCES
- 1.Dvořák P, Casamatta DA, Poulíčková A, Hašler P, Ondřej V, Sanges R. 2014. Synechococcus: 3 billion years of global dominance. Mol Ecol 23:5538–5551. doi: 10.1111/mec.12948. [DOI] [PubMed] [Google Scholar]
- 2.Weisse T. 1993. Dynamics of autotrophic picoplanckton in marine and freshwater systems. Adv Microb Ecol 13:327–370. doi: 10.1007/978-1-4615-2858-6_8. [DOI] [Google Scholar]
- 3.Stockner JG. 1991. Autotrophic picoplankton in freshwater ecosystems: the view from the summit. Int Revue Ges Hydrobiol Hydrogr 76:483–492. doi: 10.1002/iroh.19910760402. [DOI] [Google Scholar]
- 4.Sohm JA, Ahlgren NA, Thomson ZJ, Williams C, Moffett JW, Saito MA, Webb EA, Rocap G. 2016. Co-occurring Synechococcus ecotypes occupy four major oceanic regimes defined by temperature, macronutrients and iron. ISME J 10:333–345. doi: 10.1038/ismej.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wood SA, Maier MY, Puddick J, Pochon X, Zaiko A, Dietrich DR, Hamilton DP. 2017. Trophic state and geographic gradients influence planktonic cyanobacterial diversity and distribution in New Zealand lakes. FEMS Microbiol Ecol 93:fiw234. doi: 10.1093/femsec/fiw234. [DOI] [PubMed] [Google Scholar]
- 6.Farrant GK, Doré H, Cornejo-Castillo FM, Partensky F, Ratin M, Ostrowski M, Pitt FD, Wincker P, Scanlan DJ, Iudicone D, Acinas SG, Garczarek L. 2016. Delineating ecologically significant taxonomic units from global patterns of marine picocyanobacteria. Proc Natl Acad Sci U S A 113:E3365–E3374. doi: 10.1073/pnas.1524865113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Callieri C. 2008. Picophytoplankton in freshwater ecosystems: the importance of small-sized phototrophs. Freshw Rev 1:1–28. doi: 10.1608/FRJ-1.1.1. [DOI] [Google Scholar]
- 8.Stockner J, Callieri C, Cronberg G. 2000. Picoplankton and other non-bloom-forming cyanobacteria in lakes, p. 195–238. In Whitton BA, Potts M (ed), The ecology of cyanobacteria. Springer, Dordrecht, The Netherlands. [Google Scholar]
- 9.Scanlan DJ, West NJ. 2002. Molecular ecology of the marine cyanobacterial genera Prochlorococcus and Synechococcus. FEMS Microbiol Ecol 40:1–12. doi: 10.1111/j.1574-6941.2002.tb00930.x. [DOI] [PubMed] [Google Scholar]
- 10.Camacho A, Miracle MR, Vicente E. 2003. Which factors determine the abundance and distribution of picocyanobacteria in inland waters? A comparison among different types of lakes and ponds. Arch Hydrobiol 157:321–338. doi: 10.1127/0003-9136/2003/0157-0321. [DOI] [Google Scholar]
- 11.Andersson A, Tamminen T, Lehtinen S, Jürgens K. 2017. The pelagic food web, p. 281–332. In Snoeijs-Leijonmalm P, Schubert H, Radziejewska T (ed), Biological oceanography of the Baltic Sea, 1st ed Springer, Dordrecht, The Netherlands. [Google Scholar]
- 12.Sánchez-Baracaldo P, Hayes PK, Blank CE. 2005. Morphological and habitat evolution in the Cyanobacteria using a compartmentalization approach. Geobiology 3:145–165. doi: 10.1111/j.1472-4669.2005.00050.x. [DOI] [Google Scholar]
- 13.Rocap G, Distel DL, Waterbury JB, Chisholm SW. 2002. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl Environ Microbiol 68:1180–1191. doi: 10.1128/AEM.68.3.1180-1191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biller SJ, Berube PM, Berta-Thompson JW, Kelly L, Roggensack SE, Awad L, Roache-Johnson KH, Ding H, Giovannoni SJ, Rocap G, Moore LR, Chisholm SW. 2014. Genomes of diverse isolates of the marine cyanobacterium Prochlorococcus. Sci Data 1:140034. doi: 10.1038/sdata.2014.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahlgren NA, Rocap G. 2012. Diversity and distribution of marine Synechococcus: multiple gene phylogenies for consensus classification and development of qPCR assays for sensitive measurement of clades in the ocean. Front Microbiol 3:213. doi: 10.3389/fmicb.2012.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuller NJ, Marie D, Partensky F, Vaulot D, Post AF, Scanlan DJ. 2003. Clade-specific 16S ribosomal DNA oligonucleotides reveal the predominance of a single marine Synechococcus clade throughout a stratified water column in the Red Sea. Appl Environ Microbiol 69:2430–2443. doi: 10.1128/AEM.69.5.2430-2443.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR, Post a. F, Hagemann M, Paulsen I, Partensky F. 2009. Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev 73:249–299. doi: 10.1128/MMBR.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazard S, Ostrowski M, Partensky F, Scanlan DJ. 2012. Multi-locus sequence analysis, taxonomic resolution and biogeography of marine Synechococcus. Environ Microbiol 14:372–386. doi: 10.1111/j.1462-2920.2011.02514.x. [DOI] [PubMed] [Google Scholar]
- 19.Callieri C, Coci M, Corno G, Macek M, Modenutti B, Balseiro E, Bertoni R. 2013. Phylogenetic diversity of nonmarine picocyanobacteria. FEMS Microbiol Ecol 85:293–301. doi: 10.1111/1574-6941.12118. [DOI] [PubMed] [Google Scholar]
- 20.Crosbie ND, Pöckl M, Weisse T, Po M. 2003. Dispersal and phylogenetic diversity of nonmarine picocyanobacteria, inferred from 16S rRNA gene and cpcBA-intergenic spacer sequence analyses. Appl Environ Microbiol 69:5716–5721. doi: 10.1128/AEM.69.9.5716-5721.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann L, Komárek J, Kaštovský J. 2005. System of cyanoprokaryotes (cyanobacteria)—state in 2004. Algol Stud 117:95–115. doi: 10.1127/1864-1318/2005/0117-0095. [DOI] [Google Scholar]
- 22.Huber P, Diovisalvi N, Ferraro M, Metz S, Lagomarsino L, Llames ME, Royo-Llonch M, Bustingorry J, Escaray R, Acinas SG, Gasol JM, Unrein F. 2017. Phenotypic plasticity in freshwater picocyanobacteria. Environ Microbiol 19:1120–1133. doi: 10.1111/1462-2920.13638. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi S, Tomita J, Nishioka K, Hisada T, Nishijima M. 2014. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS One 9:e105592. doi: 10.1371/journal.pone.0105592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zinger L, Amaral-Zettler LA, Fuhrman JA, Horner-Devine MC, Huse SM, Welch DBM, Martiny JBH, Sogin M, Boetius A, Ramette A. 2011. Global patterns of bacterial beta-diversity in seafloor and seawater ecosystems. PLoS One 6:e24570. doi: 10.1371/journal.pone.0024570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruiz-González C, Niño-García JP, del Giorgio PA. 2015. Terrestrial origin of bacterial communities in complex boreal freshwater networks. Ecol Lett 18:1198–1206. doi: 10.1111/ele.12499. [DOI] [PubMed] [Google Scholar]
- 26.Parada AE, Needham DM, Fuhrman JA. 2016. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18:1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 27.Richa K, Balestra C, Piredda R, Benes V, Borra M, Passarelli A, Margiotta F, Saggiomo M, Biffali E, Sanges R, Scanlan DJ, Casotti R. 2017. Distribution, community composition, and potential metabolic activity of bacterioplankton in an urbanized Mediterranean Sea coastal zone. Appl Environ Microbiol 83:e00494-17. doi: 10.1128/AEM.00494-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mo Y, Zhang W, Yang J, Lin Y, Yu Z, Lin S. 2018. Biogeographic patterns of abundant and rare bacterioplankton in three subtropical bays resulting from selective and neutral processes. ISME J 12:2198–2210. doi: 10.1038/s41396-018-0153-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G, Djahanschiri B, Zeller G, Mende DR, Alberti A, Cornejo-Castillo FM, Costea PI, Cruaud C, D’Ovidio F, Engelen S, Ferrera I, Gasol JM, Guidi L, Hildebrand F, Kokoszka F, Lepoivre C, Lima-Mendez G, Poulain J, Poulos BT, Royo-Llonch M, Sarmento H, Vieira-Silva S, Dimier C, Picheral M, Searson S, Kandels-Lewis S, Boss E, Follows M, Karp-Boss L, Krzic U, Reynaud EG, Sardet C, Sieracki M, Velayoudon D, Bowler C, De Vargas C, Gorsky G, Grimsley N, Hingamp P, Iudicone D, Jaillon O, Not F, Ogata H, Pesant S, Speich S, Stemmann L, Sullivan MB, Weissenbach J, Wincker P, Karsenti E, Raes J, Acinas SG, Bork P. 2015. Structure and function of the global ocean microbiome. Science 348:1261359. doi: 10.1126/science.1261359. [DOI] [PubMed] [Google Scholar]
- 30.Guevara Campoverde NC, Hassenrück C, Buttigieg PL, Gärdes A. 2018. Characterization of bacterioplankton communities and quantification of organic carbon pools off the Galapagos Archipelago under contrasting environmental conditions. PeerJ 6:e5984. doi: 10.7717/peerj.5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doherty M, Yager PL, Moran MA, Coles VJ, Fortunato CS, Krusche AV, Medeiros PM, Payet JP, Richey JE, Satinsky BM, Sawakuchi HO, Ward ND, Crump BC. 2017. Bacterial biogeography across the Amazon River-ocean continuum. Front Microbiol 8:882. doi: 10.3389/fmicb.2017.00882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO. 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tai V, Palenik B. 2009. Temporal variation of Synechococcus clades at a coastal Pacific Ocean monitoring site. ISME J 3:903–915. doi: 10.1038/ismej.2009.35. [DOI] [PubMed] [Google Scholar]
- 34.Huang S, Wilhelm SW, Harvey HR, Taylor K, Jiao N, Chen F. 2012. Novel lineages of Prochlorococcus and Synechococcus in the global oceans. ISME J 6:285–297. doi: 10.1038/ismej.2011.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ernst A. 2003. Ecosystem-dependent adaptive radiations of picocyanobacteria inferred from 16S rRNA and ITS-1 sequence analysis. Microbiology 149:217–228. doi: 10.1099/mic.0.25475-0. [DOI] [PubMed] [Google Scholar]
- 36.Kashtan N, Roggensack SE, Rodrigue S, Thompson JW, Biller SJ, Coe A, Ding H, Marttinen P, Malmstrom RR, Stocker R, Follows MJ, Stepanauskas R, Chisholm SW. 2014. Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science 344:416–420. doi: 10.1126/science.1248575. [DOI] [PubMed] [Google Scholar]
- 37.Nübel U, Garcia-Pichel F, Muyzer G. 1997. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl Environ Microbiol 63:3327–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ban L, Pedro C, Casamayor EO, Scha H, Muyzer G, Schäfer H, Bañeras L, Pedrós-Alió C, Muyzer G. 2000. Identification of and spatio-temporal differences between microbial assemblages from two neighboring sulfurous lakes: comparison by microscopy and denaturing gradient gel electrophoresis. Appl Environ Microbiol 66:499–508. doi: 10.1128/AEM.66.2.499-508.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abed RMM, Safi NMD, Koster J, de Beer D, El-Nahhal Y, Rullkotter J, Garcia-Pichel F. 2002. Microbial diversity of a heavily polluted microbial mat and its community changes following degradation of petroleum compounds. Appl Environ Microbiol 68:1674–1683. doi: 10.1128/AEM.68.4.1674-1683.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geiß U, Selig U, Schumann R, Steinbruch R, Bastrop R, Hagemann M, Schoor A. 2004. Investigations on cyanobacterial diversity in a shallow estuary (Southern Baltic Sea) including genes relevant to salinity resistance and iron starvation acclimation. Environ Microbiol 6:377–387. doi: 10.1111/j.1462-2920.2004.00569.x. [DOI] [PubMed] [Google Scholar]
- 41.Zwart G, Kamst-van Agterveld MP, Van der Werff-Staverman I, Hagen F, Hoogveld HL, Gons HJ. 2005. Molecular characterization of cyanobacterial diversity in a shallow eutrophic lake. Environ Microbiol 7:365–377. doi: 10.1111/j.1462-2920.2005.00715.x. [DOI] [PubMed] [Google Scholar]
- 42.Boutte C, Grubisic S, Balthasart P, Wilmotte A. 2006. Testing of primers for the study of cyanobacterial molecular diversity by DGGE. J Microbiol Methods 65:542–550. doi: 10.1016/j.mimet.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 43.Hillis DM, Dixon MT. 1991. Ribosomal DNA: molecular evolution and phylogenetic inference. Q Rev Biol 66:411–453. doi: 10.1086/417338. [DOI] [PubMed] [Google Scholar]
- 44.Apprill A, McNally S, Parsons R, Weber L. 2015. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol 75:129–137. doi: 10.3354/ame01753. [DOI] [Google Scholar]
- 45.Faith DP. 1992. Conservation evaluation and phylogenetic diversity. Biol Conserv 61:1–10. doi: 10.1016/0006-3207(92)91201-3. [DOI] [Google Scholar]
- 46.Webb CO. 2000. Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. Am Nat 156:145–155. doi: 10.1086/303378. [DOI] [PubMed] [Google Scholar]
- 47.Robinson DF, Foulds LR. 1981. Comparison of phylogenetic trees. Math Biosci 53:131–147. doi: 10.1016/0025-5564(81)90043-2. [DOI] [Google Scholar]
- 48.Herlemann DPR, Labrenz M, Jürgens K, Bertilsson S, Waniek JJ, Andersson AF. 2011. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J 5:1571–1579. doi: 10.1038/ismej.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gasol JM, Cardelús C, G Morán XA, Balagué V, Forn I, Marrasé C, Massana R, Pedrós-Alió C, Montserrat Sala M, Simó R, Vaqué D, Estrada M. 2016. Seasonal patterns in phytoplankton photosynthetic parameters and primary production at a coastal NW Mediterranean site. Sci Mar 80:63–77. doi: 10.3989/scimar.04480.06E. [DOI] [Google Scholar]
- 50.Liu Z, Lozupone C, Hamady M, Bushman FD, Knight R. 2007. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res 35:e120. doi: 10.1093/nar/gkm541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, Sogin ML. 2008. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet 4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ghyselinck J, Pfeiffer S, Heylen K, Sessitsch A, De Vos P. 2013. The effect of primer choice and short read sequences on the outcome of 16S rRNA gene based diversity studies. PLoS One 8:e71360. doi: 10.1371/journal.pone.0071360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gloor GB, Hummelen R, Macklaim JM, Dickson RJ, Fernandes AD, MacPhee R, Reid G. 2010. Microbiome profiling by Illumina sequencing of combinatorial sequence-tagged PCR products. PLoS One 5:e15406. doi: 10.1371/journal.pone.0015406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jeraldo P, Chia N, Goldenfeld N. 2011. On the suitability of short reads of 16S rRNA for phylogeny-based analyses in environmental surveys. Environ Microbiol 13:3000–3009. doi: 10.1111/j.1462-2920.2011.02577.x. [DOI] [PubMed] [Google Scholar]
- 55.Edgar RC. 2018. Updating the 97% identity threshold for 16S ribosomal RNA OTUs. Bioinformatics 34:2371–2375. doi: 10.1093/bioinformatics/bty113. [DOI] [PubMed] [Google Scholar]
- 56.Callahan BJ, Sankaran K, Fukuyama JA, McMurdie PJ, Holmes SP. 2016. Bioconductor workflow for microbiome data analysis: from raw reads to community analyses. F1000Res 5:1492. doi: 10.12688/f1000research.8986.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruiz-González C, Simó R, Vila-Costa M, Sommaruga R, Gasol JM. 2012. Sunlight modulates the relative importance of heterotrophic bacteria and picophytoplankton in DMSP-sulphur uptake. ISME J 6:650–659. doi: 10.1038/ismej.2011.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ferrera I, Borrego CM, Salazar G, Gasol JM. 2014. Marked seasonality of aerobic anoxygenic phototrophic bacteria in the coastal NW Mediterranean Sea as revealed by cell abundance, pigment concentration and pyrosequencing of pufM gene. Environ Microbiol 16:2953–2965. doi: 10.1111/1462-2920.12278. [DOI] [PubMed] [Google Scholar]
- 59.Xia X, Vidyarathna NK, Palenik B, Lee P, Liu H. 2015. Comparison of the seasonal variations of Synechococcus assemblage structures in estuarine waters and coastal waters of Hong Kong. Appl Environ Microbiol 81:7644–7655. doi: 10.1128/AEM.01895-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saito MA, Rocap G, Moffett JW. 2005. Production of cobalt binding ligands in a Synechococcus feature at the Costa Rica upwelling dome. Limnol Oceanogr 50:279–290. doi: 10.4319/lo.2005.50.1.0279. [DOI] [Google Scholar]
- 61.Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EMS, Chisholm SW. 2006. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science 311:1737–1740. doi: 10.1126/science.1118052. [DOI] [PubMed] [Google Scholar]
- 62.Biller SJ, Berube PM, Lindell D, Chisholm SW. 2015. Prochlorococcus: the structure and function of collective diversity. Nat Rev Microbiol 13:13–27. doi: 10.1038/nrmicro3378. [DOI] [PubMed] [Google Scholar]
- 63.Chandler JW, Lin Y, Gainer PJ, Post AF, Johnson ZI, Zinser ER. 2016. Variable but persistent coexistence of Prochlorococcus ecotypes along temperature gradients in the ocean’s surface mixed layer. Environ Microbiol Rep 8:272–284. doi: 10.1111/1758-2229.12378. [DOI] [PubMed] [Google Scholar]
- 64.Ruber J, Geist J, Hartmann M, Millard A, Raeder U, Zubkov M, Zwirglmaier K. 2018. Spatio-temporal distribution pattern of the picocyanobacterium Synechococcus in lakes of different trophic states: a comparison of flow cytometry and sequencing approaches. Hydrobiologia 811:77–92. doi: 10.1007/s10750-017-3368-z. [DOI] [Google Scholar]
- 65.Postius C, Ernst A. 1999. Mechanisms of dominance: coexistence of picocyanobacterial genotypes in a freshwater ecosystem. Arch Microbiol 172:69–75. doi: 10.1007/s002030050742. [DOI] [PubMed] [Google Scholar]
- 66.Callieri C. 2010. Single cells and microcolonies of freshwater picocyanobacteria: a common ecology. J Limnol 69:257–277. doi: 10.4081/jlimnol.2010.257. [DOI] [Google Scholar]
- 67.Callieri C, Cronberg G, Stockner J. 2012. Freshwater picocyanobacteria: single cells, microcolonies and colonial forms, p. 229–269. In Whitton BA. (ed), Ecology of cyanobacteria II: their diversity in space and time. Springer, Dordrecht, The Netherlands. [Google Scholar]
- 68.Fermani P, Torremorell A, Lagomarsino L, Escaray R, Unrein F, Pérez G. 2015. Microbial abundance patterns along a transparency gradient suggest a weak coupling between heterotrophic bacteria and flagellates in eutrophic shallow Pampean lakes. Hydrobiologia 752:103–123. doi: 10.1007/s10750-014-2019-x. [DOI] [Google Scholar]
- 69.Sánchez-Baracaldo P, Handley B. a, Hayes PK. 2008. Picocyanobacterial community structure of freshwater lakes and the Baltic Sea revealed by phylogenetic analyses and clade-specific quantitative PCR. Microbiology 154:3347–3357. doi: 10.1099/mic.0.2008/019836-0. [DOI] [PubMed] [Google Scholar]
- 70.Becker S, Sánchez-Baracaldo P, Singh AK, Hayes PK. 2012. Spatio-temporal niche partitioning of closely related picocyanobacteria clades and phycocyanin pigment types in Lake Constance (Germany). FEMS Microbiol Ecol 80:488–500. doi: 10.1111/j.1574-6941.2012.01316.x. [DOI] [PubMed] [Google Scholar]
- 71.Hartmann M, Howes CG, Abarenkov K, Mohn WW, Nilsson RH. 2010. V-Xtractor: an open-source, high-throughput software tool to identify and extract hypervariable regions of small subunit (16S/18S) ribosomal RNA gene sequences. J Microbiol Methods 83:250–253. doi: 10.1016/j.mimet.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 72.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO. 2010. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26:1463–1464. doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- 76.Schliep KP. 2011. phangorn: phylogenetic analysis in R. Bioinformatics 27:592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kelly S, Maini PK. 2013. DendroBLAST: approximate phylogenetic trees in the absence of multiple sequence alignments. PLoS One 8:e58537. doi: 10.1371/journal.pone.0058537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Logares R. 2017. ramalok/amplicon_processing: workflow for analysing MiSeq amplicons based on Uparse v1.5. 10.5281/zenodo.259579. [DOI]
- 79.Nikolenko SI, Korobeynikov AI, Alekseyev MA. 2013. BayesHammer: Bayesian clustering for error correction in single-cell sequencing. BMC Genomics 14(Suppl 1):S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, Stepanauskas R, Clingenpeel SR, Woyke T, McLean JS, Lasken R, Tesler G, Alekseyev MA, Pevzner PA. 2013. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol 20:714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Edgar RC, Flyvbjerg H. 2015. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 31:3476–3482. doi: 10.1093/bioinformatics/btv401. [DOI] [PubMed] [Google Scholar]
- 82.Edgar RC. 2016. UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. bioRxiv 10.1101/081257. [DOI]
- 83.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 84.Pedrós-Alió C. 2012. The rare bacterial biosphere. Annu Rev Mar Sci 4:449–466. doi: 10.1146/annurev-marine-120710-100948. [DOI] [PubMed] [Google Scholar]
- 85.Shannon CE, Weaver W. 1949. The mathematical theory of communication. Math Theory Commun 27:117. [Google Scholar]
- 86.Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJM. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl Environ Microbiol 67:4399–4406. doi: 10.1128/AEM.67.10.4399-4406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Legendre P, Gallagher ED. 2001. Ecologically meaningful transformations for ordination of species data. Oecologia 129:271–280. doi: 10.1007/s004420100716. [DOI] [PubMed] [Google Scholar]
- 88.Stamatakis A. 2014. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Clarke KR, Somerfield PJ, Gorley RN. 2008. Testing of null hypotheses in exploratory community analyses: similarity profiles and biota-environment linkage. J Exp Mar Bio Ecol 366:56–69. doi: 10.1016/j.jembe.2008.07.009. [DOI] [Google Scholar]
- 90.R Development Core Team. 2011. R: a language and environment for statistical computing [2.12.2]. R Foundation for Statistical Computing, Vienna, Austria: http://www.r-project.org/. [Google Scholar]
- 91.Berger SA, Stamatakis A. 2011. Aligning short reads to reference alignments and trees. Bioinformatics 27:2068–2075. doi: 10.1093/bioinformatics/btr320. [DOI] [PubMed] [Google Scholar]
- 92.Berger SA, Krompass D, Stamatakis A. 2011. Performance, accuracy, and web server for evolutionary placement of short sequence reads under maximum likelihood. Syst Biol 60:291–302. doi: 10.1093/sysbio/syr010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jasser I, Królicka A, Karnkowska-Ishikawa A. 2011. A novel phylogenetic clade of picocyanobacteria from the Mazurian lakes (Poland) reflects the early ontogeny of glacial lakes. FEMS Microbiol Ecol 75:89–98. doi: 10.1111/j.1574-6941.2010.00990.x. [DOI] [PubMed] [Google Scholar]
- 94.Raes J, Letunic I, Yamada T, Jensen LJ, Bork P. 2011. Toward molecular trait-based ecology through integration of biogeochemical, geographical and metagenomic data. Mol Syst Biol 7:473. doi: 10.1038/msb.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequences generated in this study can be accessed through European Nucleotide Archive (ENA) under the study accession number PRJEB27291 and accession numbers ERR2639355 to ERR2639364.