Abstract
Circadian disruption has been linked to markers for poor health outcomes in humans and animal models. What is it about circadian disruption that is problematic? One hypothesis is that phase resetting of the circadian system, which occurs in response to changes in environmental timing cues, leads to internal desynchrony within the organism. Internal desynchrony is understood as acute changes in phase relationships between biological rhythms from different cell groups, tissues, or organs within the body. Do we have strong evidence for internal desynchrony associated with or caused by circadian clock resetting? Here we review the literature, highlighting several key studies from measures of gene expression in laboratory rodents. We conclude that current evidence offers strong support for the premise that some protocols for light-induced resetting are associated with internal desynchrony. It is important to continue research to test whether internal desynchrony is necessary and/or sufficient for negative health impact of circadian disruption.
Keywords: Circadian, entrainment, internal desynchrony, jetlag, suprachiasmatic
Introduction
Our physiology and behavior are shaped by our circadian rhythms, which are driven by a master biological pacemaker in the suprachiasmatic nuclei (SCN†) of the hypothalamus. Light exposure is the major source of circadian synchronization in the vast majority of studied organisms, and as such it can also drive circadian disruption when irregularly timed. The SCN is synchronized by the daily light-dark cycle, with lighting information transmitted to the SCN through pathways from the retina [1]. Both rod/cone and melanopsin photoreceptors play important roles in circadian responses [2]. The SCN then uses redundant mechanisms to relay this information to organs and tissues throughout the body, leading to temporal coordination of physiological functions and behaviors [3]. Molecular circadian rhythms are generated by a set of genes which comprise a regulatory feedback loop, and circadian expression is widespread throughout the body because of these intracellular molecular clocks. It is now evident that most peripheral organs and tissues can express circadian oscillations independently and respond differently to entraining signals, while still responding to inputs generated by the SCN in vivo. Recent evidence has shown that during circadian clock resetting the body may be in a state of internal desynchrony among the central and peripheral clocks, a form of circadian disruption that may be linked to poor health outcomes.
Circadian Disruption Has Been Linked to Markers of Poor Health
The literature on the negative health costs of circadian disruption is substantial, describing a variety of effects occurring at cell, tissue, and whole organism levels. The SCN coordinates endocrine and autonomic systems throughout the circadian cycle, such as fluctuations in insulin sensitivity throughout the day [4]. Environmental phase shifts can affect physiology in many ways, such as a reduction in leptin, an increase in glucose and blood pressure, or by reducing sleep efficacy [5]. These physiological effects result in metabolic disruptions, cardiovascular and immune dysfunctions, mood disorders, increased risks of cancer [6], and even an increase in mortality rate in aged mice when subjected to chronic phase shifts [7,8].
Because of the negative effects of disrupted circadian rhythms, it is important to consider carefully what is known about the state of circadian disruption. The term “circadian disruption” has been used to describe various scenarios of misalignment [9,10], such as misalignment between the internal circadian system and external environmental cues (e.g., light-dark cycle) or desynchrony among the rhythms within the organism. For example, studies in humans show the effect of short-term circadian disruption on alignment between physiological circadian rhythms and behavioral cycles. It has been shown that phase markers of SCN rhythms, such as melatonin and clock protein rhythms, were not greatly shifted by night shift work, but many circulating plasma metabolites became dissociated from the SCN rhythm and became aligned with the shifted behavioral cycles [9]. Evidence of desynchrony such as this might explain how shift work and metabolic diseases may be associated.
Overview of the Organization of this Review
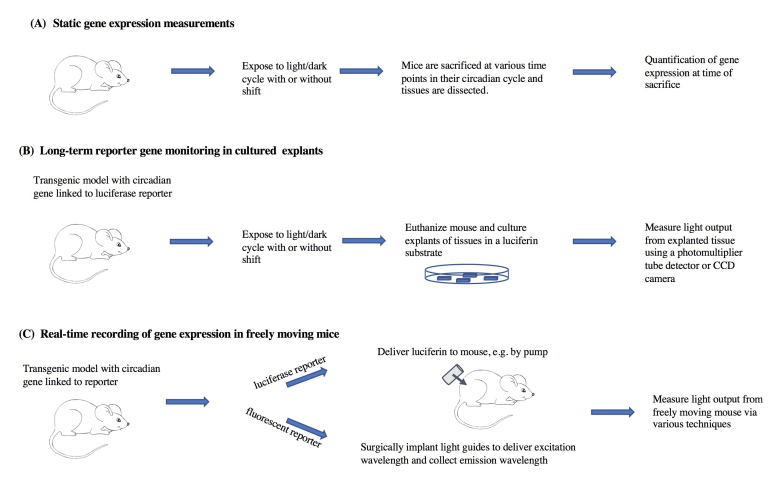
In this paper we will review measures of internal desynchronization that characterize circadian resetting in laboratory rodents. We will focus mostly on evidence arising from studies of gene expression rhythms during re-entrainment to phase shifts of the light-dark schedule. The review is organized by technique. We first highlight studies that measured clock gene expression at selected times of euthanasia (cross-sectional studies), and then describe studies using continuous measurements in transgenic reporter mouse models (longitudinal studies). We separately discuss in vitro methods using explanted tissues, and in vivo bioluminescence measures in free moving mice (Figure 1). We discuss pros and cons of the different techniques currently in use to determine genetic rhythms of circadian genes both ex vivo and in vivo, as well as critical assumptions of these techniques.
Figure 1.
Summary of some of the in vitro and in vivo methods for measurement of gene expression rhythms that will be discussed in this review.
What is Circadian Resetting and How Do We Study it in the Lab?
Under normal conditions, the rhythms controlled by the circadian system maintain what we assume is an optimized phase relationship with environmental cues and with each other. The SCN master pacemaker sends diverse signals throughout the organism to maintain this orchestration. When a sudden change in phase of an external environmental time cue occurs (if we fly from New York to Paris, or vice versa; or when our work shift changes), the system resets to the new phase over several days.
The process of resetting the circadian system can easily be reproduced in experimental settings using rodents and other animal models. Laboratory animals are typically housed under a 24 h cycle of light and darkness (e.g., 12 h of light followed by 12 h of darkness; LD). By convention, we define Zeitgeber Time 12 (ZT12) as the moment when the dark interval begins; in a symmetrical LD cycle, ZT0 is coincident with the lights-on time. A shift in the phase of the LD cycle (i.e., phase shift) occurs by shifting the onset and offset of darkness by a determined number of hours. Under these controlled environmental conditions, we can study the mechanisms by which circadian rhythms are reset to the new phase.
Experimental phase shifts of the LD cycle can vary in magnitude and direction (advanced or delayed relative to the original phase) and can be achieved through different protocols. Advances can be performed by shortening either the light or the dark intervals of the cycle by the desired number of hours. Likewise, delays can be performed by lengthening either interval. Additional considerations for chronic phase shifting schedules include the latency between subsequent shifts, and the pattern of phase shifts (e.g., repetitive chronic phase advances or alternating advances and delays). Experiments can also involve variations in the characteristics of the light stimulus (e.g., intensity, wavelength, duration). These features allow researchers to work with a large number of experimental scenarios, but also give rise to considerable inter-experimental variation when trying to compare and integrate findings from different studies.
In some studies, a researcher may choose to release studied animals into constant darkness (DD) after a number of cycles of the shifted LD in order to assess their internal phase independently from the continual effects of light exposure. For example, 4 days following an 8 h phase advance of a 12:12 LD cycle, wildtype C57BL/6 male mice released into DD showed no apparent phase shift in locomotor activity measured with an infrared sensor, and only a small shift in locomotor activity if they were housed with a running wheel (see supplementary data Figure S4 in [11]). Delays of the LD cycle shifted the locomotor activity circadian rhythm more effectively in mice, producing a ~5 h delay in circadian rhythms after 1-2 days of an 8 h delay in the LD [11].
Post-dissection Measurement of Gene Expression (Figure 1A): Evidence for Internal Phase Misalignment During Phase Resetting
The circadian system is now well understood at the molecular level. Transcriptional-translational loops between inter-related “canonical” clock genes generate daily cycles in both core clock genes (such as Per1, Per2, Cry1, Cry2, Bmal1, Clock) as well as tissue-specific clock-controlled output genes in the SCN and peripheral tissues. The sequence of this core loop involves the activation of Period (Per1, Per2, and Per3) and Cryptochrome (Cry1 and Cry2) genes transcriptions by a BMAL1/CLOCK heterodimer and, after a suitable delay built in by post-transcriptional mechanisms, the subsequent inhibition of this process by complexes containing PER and CRY proteins [12]. An additional loop involves the nuclear receptors Rev-erb and Ror, which can translocate into the nucleus to modulate Bmal1, Clock, and Npas2 transcription via opposing actions on a RORE sequence located in their promoter regions. These molecular feedback loops occur in cells throughout the body, and have a near-24 h cycle length.
What does jetlag look like from the vantage of the SCN as a whole? Circadian clock genes (Per1, Per2, Bmal1, and the “output” gene Dbp) measured in male mouse SCN tissue punches show days of misaligned clock gene expression within the SCN following a phase advance of the LD cycle. More specifically, after a 6 h advance of the LD cycle in mice, a dissociation occurs between Per and Cry, Dpb, Bmal1, and Rev-Erb genes, in which the Per genes show a fast adjustment to the new phase while the other circadian genes adjust more slowly [13,14]. However, following a 6 h delay of the LD cycle, a fast and coordinated resetting occurs in the Per and Cry genes with shifts complete after day 2 [13].
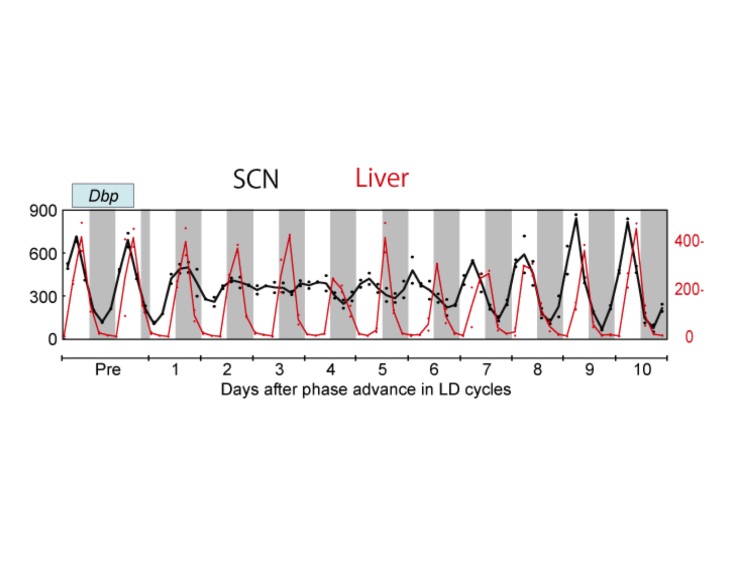
In agreement with this (but with many more time points), Yamaguchi et al. (2013) [11] found that, in the first few days after an 8 h advance of the light cycle, SCN clock gene expression profiles of Per1, Per2, Bmal1, and Dbp show no rhythm. The rhythm then slowly re-emerges near the new phase, albeit with a dampened amplitude. Recovery is seen gradually, with the Per genes appearing to recover their rhythms faster than Bmal1 and Dbp. Importantly, the authors highlight that the light-responsiveness of the Per genes makes this assessment uncertain (an important point to consider also when interpreting data from [13,14] discussed above). The peripheral organs examined (liver and kidney) showed slower shifting in response to the light cycle change, but with much less loss of rhythm amplitude (Figure 2). Several additional days were required for full re-entrainment of the peripheral tissues examined (liver and kidney). Because the phase of the SCN shifts more rapidly than that of the kidney and liver, the animals are in a state of internal desynchrony as well as external desynchrony for several days after the light cycle shift (Figure 2) [11].
Figure 2.
Gene expression rhythms after an 8 h advance jetlag. Using qPCR, expression of clock genes was measured every 4 h after the phase shift in laser-microdissected tissue samples. Dbp gene expression rhythm in the SCN (black; left side Y-axis) and in the liver (red; right side Y-axis) with quantification of gene expression normalized to 36b4 expression, units of mmol/mol 36b4 mRNA. Adapted from Yamaguchi et al. (2013) [11]; Courtesy of Drs. Okamura and Yamaguchi.
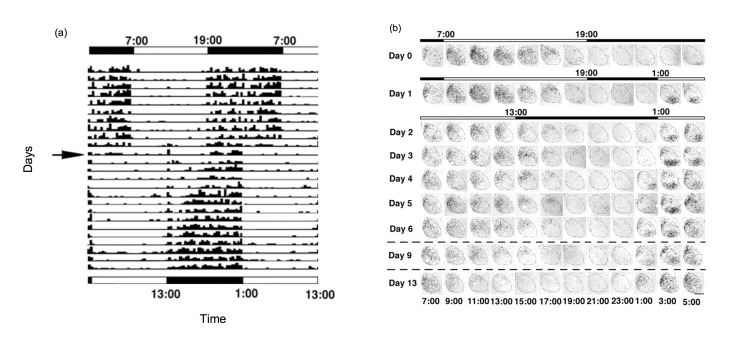
Other authors considered desynchrony at the cellular level. In response to a shift in the light cycle, cellular subpopulations within the SCN shift at different rates, leading to internal desynchrony within SCN. For example, following a 6 h phase advance shift of the LD cycle, rats demonstrate a 2-week period of behavioral resetting (Figure 3A) [15]. This gradual adjustment is accompanied by about a week of desynchrony in gene expression rhythms among regions of the SCN. Cells in the ventral SCN are light-responsive and show strong induction of the light-responsive gene Per1 (e.g., see samples from 3:00 on days 1 through 6 following the shifted LD cycle in Figure 3B). Cells in the dorsal portion of the SCN do not shift for multiple days and shift at a rate correlated with behavioral shifts (compare samples from 15:00 on days 2 through 13 in. Figure 3B). On the other hand, the ventral SCN rhythm in sensitivity to light-induced c-fos mRNA shifts phase slowly following a light cycle shift, readjusting at a rate similar to that of behavioral rhythms [16].
Figure 3.
(A) A double-plot actogram showing slow resetting of locomotor activity rhythms in male Wistar rats following a 6 h advance of the LD cycle. Every row illustrates the locomotor activity of the animal on a given day, and consecutive days are stacked below. The initial LD cycle is shown on top, with the shifted cycle shown on the bottom, and light and dark periods are indicated by the white and black bars respectively. The arrow points to the day of the shift. The activity of the animal is shown as black tick marks [15]. (B) The SCN shows internal disorder following a 6 h phase advance shift of the LD cycle. Coronal sections of SCN stained for Per1 mRNA from rats sacrificed every 2 h. The top bar indicates timing of the LD cycle and the lower bars indicate the new LD cycle. Time of day is indicated at the bottom of each column of images. Midline is to the left of each SCN image. On the baseline day (Day 0), ventrolateral SCN and dorsomedial SCN show similar timed Per1 expression. Following the LD advance, the two regions are desynchronized but eventually come together [15]. Note on Day 1, when the lights first interrupt the prior dark phase, Per1 mRNA label is observed for the first 2 samples (3:00 and 5:00) mainly in the ventrolateral SCN. On Day 2, we note that the signal in dorsomedial SCN is high in the sample taken at 11:00, a time that on the prior baseline day showed Per1 signal both in ventrolateral as well as dorsomedial SCN. As the days of the shifted cycle continue, the SCN readjusts to the new phase. Note that in the baseline samples there was little Per1 visible in the SCN in the beginning of the dark period. Following the shift, signal is apparent in the dorsomedial SCN at 15:00 even on Day 9, and is only no longer visible on Day 13 post-shift. The ventrolateral SCN appears shifted to the new LD cycle immediately, but may be reporting direct response to light exposure.
Internal desynchrony may be altered, depending upon cellular communication within the SCN. The speed of internal resetting across cell groups in the SCN, and thus the speed of resetting of downstream rhythms, can be increased by alterations in the expression of vasopressin receptors. Mice lacking the arginine vasopressin receptors showed faster phase resetting as measured in behavior, body temperature, and in SCN and peripheral tissue gene expression [11]. Both V1a and V1b receptors appear to play a role, and knocking out both receptors generates a phenotype of very rapid resetting in C57BL/6 mice. Experiments in which these receptors were pharmacologically blocked in wild type mice confirmed these results. This suggests that V1a- and V1b-mediated cell communication in the SCN plays a role in regulating the rate of re-entrainment. Other research shows that specifically timed treatment with vasoactive intestinal polypeptide (VIP) can speed resetting to an 8h advance of the LD cycle, perhaps attributable to the effect of this peptide treatment on cellular synchrony [17].
Tissue Explant Ex Vivo Approach (Figure 1B): Evidence for Internal Phase Misalignment During Phase Resetting
While in situ hybridization and immunohistochemistry methods can give information on the level of expression of a specific gene or protein, they can only provide a snapshot of a specific time of dissection. Transgenic techniques and the advent of genetically modified rodents, as well as virally expressed reporter genes that can be introduced into otherwise wild-type animals, have allowed the development of models carrying “reporter” genes that can be tracked continuously in real time. The gene for luciferase (Luc), the bioluminescent enzyme from the firefly, can be inserted into a mouse genome under the promoter of a circadian gene of interest, or even fused to the protein product generated from the rhythmically expressed gene. Tissues of interest can then be dissected and cultured in vitro in the presence of luciferase substrates, and emitted light can be recorded continuously to infer temporal patterns of expression of the gene of interest.
A seminal study presenting evidence of disruption in peripheral and central phase alignment during phase resetting was performed using a transgenic rat line in which luciferase was expressed under the control of the mouse Per1 (mPer1) promoter [18]. Tissue explants of SCN and various peripheral tissues (skeletal muscle, lung, and liver) were harvested, and the rhythmicity of the mPer1-controlled luciferase light emission was recorded. This study demonstrated that the SCN and peripheral tissue explants all showed rhythmic expression of the mPer1-driven reporter, with the peripheral tissue rhythms phase-lagging the SCN by approximately 7 to 11 hours. To investigate whether abrupt changes of the light cycle led to disorganization of circadian rhythms, rats had their 12:12 LD cycle either advanced or delayed by 6 h. The SCN explants shifted within two circadian cycles after shifts in either direction, while peripheral tissues shifted at different, slower rates, indicating a desynchronized circadian system.
Nakamura et al. (2005) [19] subjected Per1-luc rats to 6 h delays or advances of the LD cycle, and then dissected their SCN after 1, 3, or 6 days after the phase shift to determine their phase. Using this method, they were able to replicate the results of Nagano et al. (2003) [15] described in the previous section, showing with camera imaging that the ventral SCN re-entrains faster to the new phase than the dorsomedial region, and that this effect is more pronounced in the case of advances than delays. Similarly, when rat SCN slices were first cut to separate dorsal from ventral subregions, electrophysiological recordings show a differential rate of resetting after a phase shift [20].
Another important model using the luciferase reporter approach is the PER2::LUC transgenic mouse [21]. A knock-in technique was used to create a fusion protein reporter by fusing the Luc gene to the terminal exon of the endogenous Per2 locus. Their results showed that peripheral tissues phase-lagged the SCN by 2-6 h. In mice with SCN lesions, there were persistent circadian oscillations in PER2::LUC in peripheral tissues, although with a progressive loss in synchrony between the different tissues.
Mouse strains vary in their physiological/behavioral “jetlag.” Most studies of mice use highly inbred, ~99 percent genetically identical strains, with a majority of studies utilizing male C57BL/6 mice. The C57BL/6 strain, as reviewed above [9], typically shows relatively slow (5-10 day) shifts of locomotor rhythms following large (6-8 h) advances of the LD cycle. On the other hand, a different inbred strain, BALB/c, shows almost no evidence for jetlag in locomotor rhythms or in temporal sensitivity to light-induced Fos expression in the SCN, achieving large (up to 10 h) phase advances within 1-2 circadian cycles [22]. The BALB/c and C57BL/6 strains show similar circadian-day patterns of phase shifts to brief (1-3 h) pulses of light (“phase response curves”), and the BALB/c mice cannot entrain normally to short (< 3 h) photoperiods (e.g., 2L:22D) or skeleton photoperiods (light cycles defined by several short light pulses, e.g., at dawn and dusk, 1L:10D:1L:12D). These two lines of evidence support the hypothesis that the significantly larger magnitude phase shifts achievable by BALB/c mice are highly dependent on the duration of light exposure [23], unlike C57BL/6J and most other mouse models.
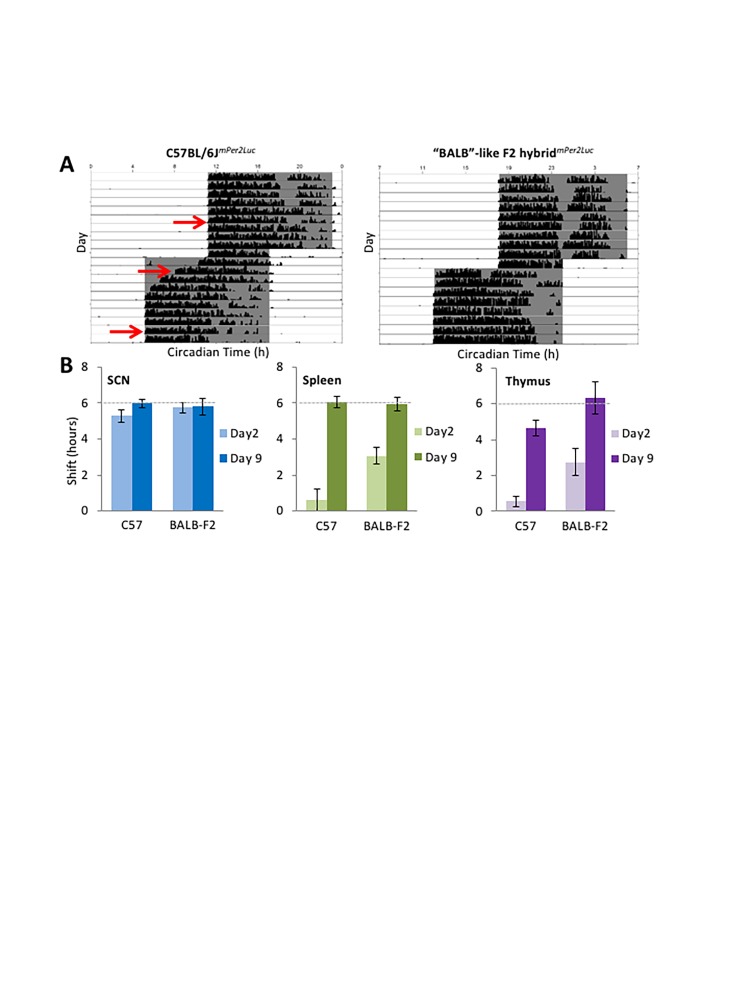
Some F2 hybrids of BALB/cJ and C57BL/6J mice display intermediate rates of re-entrainment of locomotor rhythms, while others show rapid or slow phenotypes typical of the parental strains, suggesting incomplete dominance of genetic contributions to entrainment in these two strains. A preliminary analysis of bioluminescence rhythms in tissue explants was performed on F2 hybrids of BALB/cJ and C57BL/6mPer2Luc mice. Just as in the parental C57BL/6JmPer2Luc controls, re-entrainment of PER2::LUC rhythms in SCN explants of F2 hybrids was completed within 2 days after a 6 h phase advance of the LD cycle (Figure 4; Weber and Harrington, unpublished data). Alternatively, phase shifts in spleen and thymus explants from F2 hybrids with the BALB/c-like rapid-shifting phenotype were significantly faster than those from C57BL/6JmPer2Luc controls, suggesting that rates of re-entrainment of molecular rhythms in peripheral tissues are being regulated downstream of, or independently of, the suprachiasmatic nucleus cells producing PER2::LUC bioluminescence rhythms.
Figure 4.
Molecular rhythms in peripheral tissues do not re-entrain at the same rate in different mouse strains. (A) Representative actograms show slow re-entrainment of wheel-running rhythms in C57BL/6J mice versus rapid re-entrainment in “BALB-like” F2 hybrids (similarly rapid as in BALB/cJ parental strains). Explants were taken 5 days prior, 2 days after, or 9 days after a 6 h phase advance of the LD cycle (red arrows). (B) SCN explants showed near-complete 6 h phase advances of mPer2Luc bioluminescence rhythms (dashed lines) by day 2 following the phase shift in both C57BL/6J and BALB-like F2 hybrids (left panel), while phase advances were significantly greater in spleen and thymus explants from BALB-like hybrids on day 2 following the phase shift (middle & right panels) (n=3-4/group). (Unpublished data, Weber and Harrington).
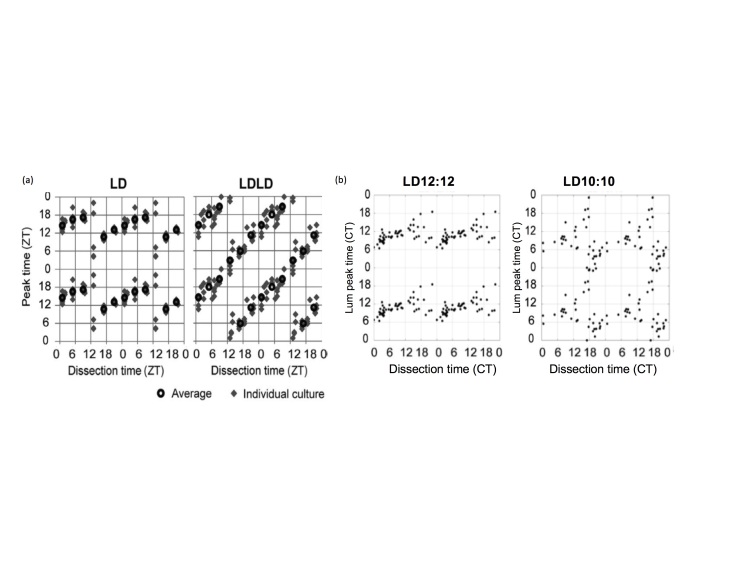
Recent evidence has suggested that under some specific light conditions, the assessment of the phase of PER2::LUC rhythms using this explant technique might not be reliable. Two studies have shown that exposure to disrupting light cycles alters SCN rhythms such that the peak time recorded from explant cultures is determined by the time of dissection. In one study, mice were maintained in a LD 14:10 cycle, and then transferred to a “bifurcated” light cycle (7L:5D:7L:5D) for at least 2 weeks. SCN from mice in LDLD showed rhythms which were strongly reset by the dissection process (Figure 5A) [24]. These results are supported by recent research in our lab [25]. Male mice were housed under a 20 h light cycle (LD10:10), to which C57BL/6J mice are unable to entrain, in order to induce disruption to the circadian system through light at night and phase resetting. Control animals were housed under LD12:12. The time of dissection strongly influenced circadian phase of PER2::LUC rhythms in the SCN from mice housed under LD10:10, while the effect on control mice was minimal (Figure 5B). Dissection time reset the phase of adipose tissue regardless of the lighting cycle, but it did not reset the phase of thymus explants. These studies indicate circadian disruption can disrupt the coordination of phase within the SCN and between the SCN and peripheral tissues, and may thereby alter network sensitivity to stimuli related to explantation procedures to which they are usually unresponsive. In light of these studies, results from ex vivo gene expression experiments involving prior disrupting or resetting conditions should be interpreted with considerable caution.
Figure 5.
(A) Relationship between peak time and dissection time of the SCN explants prepared from mice entrained to LD or LDLD bifurcated light periods, dissected at various time points across the day. Each gray diamond indicates a dissection time (ZT) and peak time of a single explant culture. Black circles indicate average peak times for each dissection time group. Data are quadruple plotted to aid visualization of the resetting pattern (from Noguchi et al., 2018). (B) Similar results found in our recent work using PER2::LUC mice, showing the effects of time of dissection on resetting of phase shifts. Each dot represents the time of dissection of the explanted SCN tissue and the peak time relative to Circadian Time (a timeframe based on the locomotor activity under constant dark conditions; CT) prior to dissection for that tissue. Adapted from Leise et al., 2018 [25].
In vivo Approach (Figure 1C): Evidence for Internal Phase Misalignment During Phase Resetting
Techniques have now been developed to measure circadian gene expression from the SCN as well as other tissues in the body in vivo in freely moving animals. These techniques allow us to record circadian rhythms and the effect of light-induced phase shifts in real-time. Although recordings of bioluminescence and fluorescence enable noninvasive and repeated measures of gene expression, they differ in several aspects. Bioluminescence imaging has the advantage of very low background noise but is influenced by substrate pharmacokinetics. On the other hand, fluorescent signals need to be distinguished from background autofluorescence, are prone to photobleaching, and require delivery of the excitatory light, raising concerns about phototoxicity. In terms of resolution, fluorescence has greater current capability to resolve single cells, especially for in vivo studies. Despite complications from animal movements, quantitative measurement of bioluminescence in vivo is possible if signals can be properly calibrated, as one group accomplished using light from scintillators positioned on the mouse and employing two CCD cameras to track the mouse in space [26].
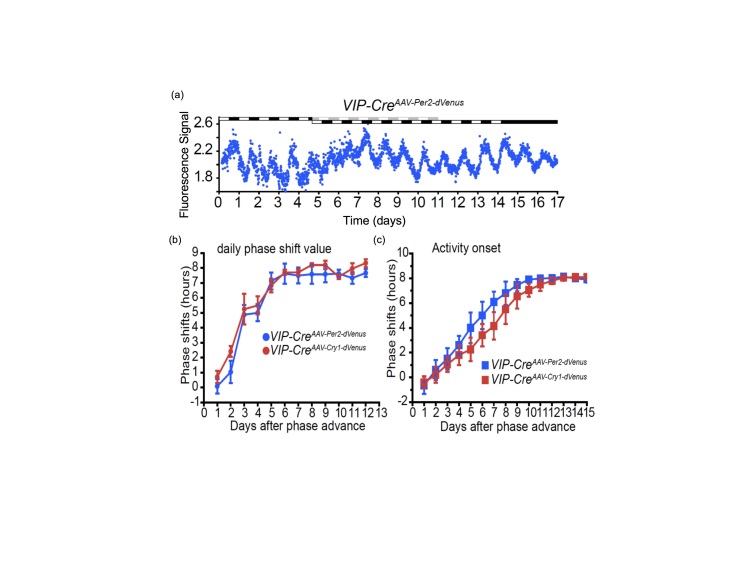
The potential of these techniques can be appreciated in a recent paper by Mei and collaborators that studied the impact of an LD phase shift on a subpopulation of SCN neurons through in vivo fluorescence recording [27]. These researchers transduced SCN neurons with a fluorescent reporter for Per2-Venus, which is expressed selectively in SCN neurons expressing vasoactive intestinal polypeptide (VIP). A fiber optic guide implanted directly above the SCN delivered the excitation light as well as collected the emitted light, which was then measured by a photomultiplier tube (PMT). In this way, they could record Per2 gene expression in the SCN of mice housed under a full LD cycle. (Unlike luciferase bioluminescence measurements, fluorescence measurements can be conducted without interference from environmental lighting). After an 8 h advance of the LD cycle, the Per2-Venus rhythms of the sub-population of VIP-expressing SCN neurons were suppressed and then phase shifted (Figure 6A), confirming the results of Yamaguchi et al. (Figure 2) with added cell-specificity. The study also confirmed that this cell population within the ventral SCN shifts more rapidly than behavioral rhythms (Figure 6C). Mei et al. determined that Per2 and Cry1 expression show similar rates of resetting (see Figure 6B), and a different group using a similar approach reported that Per1 and Bmal1 rhythms re-entrain at different speeds and are thus desynchronized during resetting [28].
Figure 6.
Initial SCN response to LD phase shift is loss of rhythmicity in VIP neurons, then a shift to the new phase precedes the shift of behavioral rhythms. (A) Recording clock gene expression in vivo from the SCN during an 8 h advance of the LD cycle. Light treatments are indicated by bars on top of the recordings. SCN VIP neurons are expressing a reporter for Per2 (example shown in blue) or Cry 1 (red lines) during 17 days. The phase shifts of SCN VIP neurons (B) or wheel-running behavior (C) are summarized. Running wheels were present in the experiments reported in (C) but not for the data in (B) [27].
These in vivo tools for recording gene expression provide new experimental possibilities. Saini et al. (2013) recorded the expression of clock genes in the liver of mice with SCN lesions and showed that hepatocytes synchronized more quickly to inverted feeding cycles than in control mice, suggesting that the SCN can “hold” the phase of the liver, resisting the signals of misaligned feeding rhythms [29]. Other groups have monitored rhythms of genes in regions of the brain other than the SCN. For example, in response to a single 8 h delaying light pulse, Per1-luc bioluminescence rhythms in the olfactory bulb shifted more rapidly than did the cerebral cortex and the skin [26]. Internal desynchrony during re-entrainment to a phase shift can also be observed at the behavioral level in laboratory rodents, as shown in Figure 3A. The resetting of rhythms in sleep stages of rats after a 6 h phase delay of the LD cycle was studied using electroencephalography (EEG) and electromyography (EMG) recordings [30]. The study determined that slow-wave sleep rhythms were immediately re-entrained to the new LD phase, while rapid-eye movement sleep rhythms took 3 to 5 days to return to their original phase. The main consequence of this transitory desynchronization of sleep stages is an abnormally high propensity for rapid-eye movement sleep during the early subjective day, which is normalized after full resetting of sleep stages rhythms. The authors also show that this desynchronization of sleep stages has its origin in the differences in the speed of re-entrainment between the ventrolateral and the dorsomedial SCN [30].
Conclusion and Outlook: Evidence for Internal Phase Misalignment During Phase Resetting
Our current understanding is that jetlag involves malaise that has two sources: during the period of re-entrainment, external lighting cues fall at biologically inappropriate parts of the circadian cycle (external desynchrony) and parts of the body shift their circadian rhythms at varied rates (internal desynchrony). Several studies stand out as providing strong evidence for the hypothesis that circadian phase resetting is accomplished after a transient internal desynchrony of many body clocks.
For example, we highlight the heroic study by Yamaguchi et al. (2013) [11], in which mice were euthanized every 4 h, before and up to 11 days after a shift of the LD cycle. After collecting 54 time points of gene expression measures, the authors showed that the SCN loses amplitude after a light cycle shift, then achieves the new phase several days prior to the liver or the kidney. The liver’s circadian Per2 expression across a population of mice takes over 10 days to shift in response to an 8 h LD phase advance, lagging the SCN which shifts faster [11]. Although the measures using this technique are necessarily static and population-based, the results provide strong support for this hypothesis.
We highlight an important complication in descriptions of SCN response. The ventral subpopulation of cells in the SCN shows rapid shifting, whereas the dorsal subpopulation lags [15]. Thus, the SCN is of “two minds” when it comes to adjusting to a new time zone. When a study analyzes tissue punches of SCN, we must recognize that results that do not support a strong rhythm may be signifying a diversity of responses within the SCN itself. New techniques are being developed that allow us to track the phase resetting dynamics of sub-populations of SCN cells [27].
Limitations
Techniques: Some of the strongest evidence we have highlighted here arises from studies using techniques that average responses across animals by collecting “snapshots” of circadian gene expression at set times (Figure 1A). Although this technique does not reveal the variable responses within a population of animals, when researchers collect frequent samples over long periods of time, results can provide detailed time series of resetting dynamics of multiple tissues within animals.
We have new concerns about veracity of past studies using ex vivo tissue explant techniques to measure circadian phase (Figure 1B). Recent research has shown that dissection itself may act as a stimulus that can cause phase resetting in tissues from mice previously exposed to circadian-disruptive conditions [24,25]. These results suggest that exposure to some forms of circadian disruption may desynchronize SCN neurons, thereby increasing network sensitivity. Tissues with weakened coupling, or with little to no coupling, as may be true of some peripheral tissues, will show larger resetting effects by the explantation procedure. Exposure to light at inconsistent circadian times on a recurring weekly basis disrupts locomotor activity rhythms and alters sensitivity of the SCN pacemaker to dissection time effects. Time of peak of Per1-luc rhythms recorded in vivo differed from those reported in ex vivo studies [26]. These results indicate we should be quite cautious in interpreting phase measurements from ex vivo circadian gene expression experiments. On the other hand, single-cell resolution is more feasible when working in vitro. An in vitro preparation of the entire Drosophila brain allowed visualization of detailed dynamics of desynchrony within neural components of circadian pacemakers in response to a single light pulse [31].
Newer in vivo techniques allow long-term recordings of gene expression of tissues, and should allow simultaneous recordings of behavior and physiology (Table 1). Currently it is most feasible to record from one tissue site, limiting the ability to track internal alignment of rhythms within one animal (but see [26]). One important consideration for future development of in vivo techniques is the ability to provide an environment optimal for animal welfare. In some studies reviewed here, mice are constrained to small recording chambers (e.g., 200 mm diameter in Hamada et al., 2016 [26]), are connected to pumps delivering substrates, or are tethered by probes entering the brain [18,27,32]. Ideal approaches to long-term monitoring of in vivo rhythms will minimize restraint, tethering, and surgical interventions.
Table 1. Some key papers using different in vivo approaches to measure internal desynchrony of circadian gene expression.
| In Vivo method | ||||
| Citation | Animal Model | Detection Method | ||
| Saini et al., 2013 [29] | Viral delivered Adv-Bmal1-luc or RevErba-luc; PER2::LUC mice | CCD camera, or PMT | ||
| Ono et al, 2017 [28] | Per1-luc, Bmal1-Eluc mice | Implanted optical fiber connected to a PMT | ||
| Hamada et al., 2016 [26] | Per1-luc, Bmal1-Eluc mice | 2 CCD cameras with scintillators used for quantification of signal from multiple sites | ||
| Mei et al., 2018 [27] | Injection of Per1 and Cry1 transcription reporter virus Venus fluorophore | Implanted optical fiber delivering excitation light and connected to a PMT | ||
PMT - Photomultiplier Tube
CCD - Charge-coupled device
Subject variables: As noted above, most studies use only a few common laboratory strains, such as the C57BL/6 mouse, and often researchers use only male mice. We described the dramatically different responses that can be observed when using BALB/c strain mice. Other subject variables may also be important. Few studies have been powered to compare males and females. Older animals show slower adjustment of phase in response to shifts [33]. It is important to consider a wider range of subject variables in order to better estimate the importance of our findings for the larger population of rodents, and extrapolation to other species, including humans, should be evidence-based.
Environmental factors: Timed food availability does not shift the SCN circadian rhythm, but can phase shift the liver circadian clock [34]. This shift can be accomplished more rapidly when the SCN is ablated [29] indicating that there are dynamic interactions between these components of the circadian system. Timed feeding can shift phase of other organs as well, such as heart, kidney, pancreas, and skin [34,35]. Interestingly, a somewhat extreme feeding protocol (24 h fast followed by restricting food to 2 h/day) at the time of new onset of darkness can accelerate the rate of re-entrainment to 6 h advance of the LD cycle in rats [36].
Social housing is a much less well-explored factor and offers an innovative future direction for our research. One study showed that housing mice in groups of 5, but not pairs, could influence circadian synchronization [37]. Housing mice in groups of 3 or 4 blocked effects of chronic circadian disruption on weight gain [38]. Impact of social factors highlights the importance of studies in ecologically relevant conditions [39-42]
We have shown that the use of running wheels can improve circadian phase resetting in older mice [33]. A running wheel alleviates the disruptive metabolic effects of a chronically advancing light schedule (6 h advance every 2 d) [38].
Future studies: We see several high priority directions for future research on this topic.
1. The need for technique development. The evidence presented in this review suggests that the field is at a point where we can utilize in vivo techniques for phase measures, to move forward in testing the impact of various environmental factors thought to alter circadian phase during resetting. To measure the phase misalignment of multiple internal circadian rhythms, we need to track multiple internally generated rhythms from an individual animal over several weeks. It is also possible that individual cells may show different resetting dynamics, and the rhythm from an individual tissue may show broadening of rhythm waveform, loss of amplitude, and lower signal:noise ratio during circadian phase resetting. These changes in waveform would be impossible to interpret clearly without in vivo techniques offering cellular resolution.
2. Health significance. It will be essential in the future to demonstrate whether internal desynchrony underlies the negative health impact of circadian disruption. If this can be established, we can aim to determine how to reset circadian phase while minimizing internal phase misalignment. We note that a shift of the LD cycle might also negatively impact health via effects of light during the subjective night [43], through changes in lipid metabolism [7], loss of sleep quantity or quality [44] or non-specific stress response. Researchers studying the impact of internal circadian phase misalignment will aim to manipulate just this aspect of the response to an LD shift (as, for example, accomplished through a genetic approach by van der Vinne et al., 2018 [41]). Such research is key to determine the health significance of internal desynchrony in gene expression rhythms as distinct from external desynchrony.
3. Understanding of SCN network responses. Our understanding of the neuronal network of the SCN suggests the ventral cells respond quickly to light but the dorsal cells are less responsive, taking longer to shift phase. Dorsal SCN cells appear to shift at the same rate as locomotor activity rhythms [15], but this should be verified with other techniques that allow measurements of both rhythms in the same animal. New papers suggest astrocytes play important roles in the SCN network [45-48], but their roles are as yet unclear in terms of light-mediated shifts and internal desynchrony.
Overall, we highlight here the past research studies of gene expression studies in lab rodents. Such highly controlled studies demonstrate the state of internal desynchrony that accompanies circadian clock phase resetting. As circadian clock resetting is associated with adverse health consequences in animal models as well as in epidemiological studies in humans, understanding the contribution of internal desynchrony to these effects is of experimental interest and public health importance.
Acknowledgments
We would like to thank Dr. Tanya Leise (Amherst College) and Dr. Dave Weaver (Univ. Mass. Medical School) for critical comments on a draft of this manuscript. We thank Drs. Okamura and Yamaguchi for helpful discussions and for preparation of Figure 2. MEH was supported by NIH grant 1R15GM126545-01.
Glossary
- CCD
charge-coupled device
- CT
Circadian Time
- DD
constant darkness
- EEG
electroencephalography
- EMG
electromyography
- LD
light:dark
- PMT
photomultiplier tube
- SCN
suprachiasmatic nuclei
- VIP
Vasoactive Intestinal Polypeptide
- ZT
Zeitgeber Time
Author Contributions
S. Nicholls prepared the first draft of the manuscript. All authors critically commented and wrote sections of the final manuscript.
References
- Morin LP. A Path to Sleep Is through the Eye. eNeuro. 2015. April;2(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirson SN, Brown LA, Pothecary CA, Benson LA, Fisk AS. Light and the laboratory mouse. J Neurosci Methods. 2018. April;300:26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35:445–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J, Man CD, Morris CJ, Cobelli C, Scheer FA. Differential effects of the circadian system and circadian misalignment on insulin sensitivity and insulin secretion in humans. Diabetes Obes Metab. 2018;20(10):2481–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plano SA, Casiraghi LP, García Moro P, Paladino N, Golombek DA, Chiesa JJ. Circadian and Metabolic Effects of Light: Implications in Weight Homeostasis and Health. Front Neurol. 2017;8:558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JA, Davidson AJ. Health consequences of circadian disruption in humans and animal models. Prog Mol Biol Transl Sci. 2013;119:283–323. [DOI] [PubMed] [Google Scholar]
- Boomgarden AC, Sagewalker GD, Shah AC, Haider SD, Patel P, Wheeler HE, et al. Chronic circadian misalignment results in reduced longevity and large-scale changes in gene expression in Drosophila. BMC Genomics. 2019. January;20(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AJ, Sellix MT, Daniel J, Yamazaki S, Menaker M, Block GD. Chronic jet-lag increases mortality in aged mice. Curr Biol. 2006. November;16(21):R914–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter C. Circadian disruption: what do we actually mean? Eur J Neurosci. 2018. November [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roenneberg T, Merrow M. The Circadian Clock and Human Health. Curr Biol. 2016. May;26(10):R432–43. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Suzuki T, Mizoro Y, Kori H, Okada K, Chen Y, et al. Mice Genetically Deficient in Vasopressin V1a and V1b Receptors Are Resistant to Jet Lag. Science. 2013. October;342(6154):85–90. [DOI] [PubMed] [Google Scholar]
- Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2017. March;18(3):164–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AB, Field MD, Maywood ES, Hastings MH. Differential Resynchronisation of Circadian Clock Gene Expression within the Suprachiasmatic Nuclei of Mice Subjected to Experimental Jet Lag. J Neurosci. 2002. September;22(17):7326–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiessling S, Eichele G, Oster H. Adrenal glucocorticoids have a key role in circadian resynchronization in a mouse model of jet lag. J Clin Invest. 2010. July;120(7):2600–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano M, Adachi A, Nakahama K, Nakamura T, Tamada M, Meyer-Bernstein E, et al. An Abrupt Shift in the Day/Night Cycle Causes Desynchrony in the Mammalian Circadian Center. J Neurosci. 2003. July;23(14):6141–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano M, Ikegami K, Minami Y, Kanazawa Y, Koinuma S, Sujino M, et al. Slow shift of dead zone after an abrupt shift of the light-dark cycle. Brain Res. 2019. February [DOI] [PubMed] [Google Scholar]
- An S, Harang R, Meeker K, Granados-Fuentes D, Tsai CA, Mazuski C, et al. A neuropeptide speeds circadian entrainment by reducing intercellular synchrony. Proc Natl Acad Sci USA. 2013. November;110(46):E4355–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, et al. Resetting Central and Peripheral Circadian Oscillators in Transgenic Rats. Science. 2000. April;288(5466):682–5. [DOI] [PubMed] [Google Scholar]
- Nakamura W, Yamazaki S, Takasu NN, Mishima K, Block GD. Differential Response of Period 1 Expression within the Suprachiasmatic Nucleus. J Neurosci. 2005. June;25(23):5481–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albus H, Vansteensel MJ, Michel S, Block GD, Meijer JH. A GABAergic mechanism is necessary for coupling dissociable ventral and dorsal regional oscillators within the circadian clock. Curr Biol. 2005. May;15(10):886–93. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, et al. PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004. April;101(15):5339–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeGates TA, Dunn D, Weber ET. Accelerated re-entrainment to advanced light cycles in BALB/cJ mice. Physiol Behav. 2009. October;98(4):427–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajtay TJ, St. Thomas JJ, Takacs TE, McGann EG, Weber ET. Duration and timing of daily light exposure influence the rapid shifting of BALB/cJ mouse circadian locomotor rhythms. Physiol Behav. 2017. October;179:200–7. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Harrison EM, Sun J, May D, Ng A, Welsh DK, et al. Circadian rhythm bifurcation induces flexible phase resetting by reducing circadian amplitude. Eur J Neurosci. 2018. July [DOI] [PubMed] [Google Scholar]
- Leise TL, Goldberg A, Michael J, Montoya G, Solow S, Molyneux P, et al. Recurring circadian disruption alters circadian clock sensitivity to resetting. Eur J Neurosci. 2018. September [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada T, Sutherland K, Ishikawa M, Miyamoto N, Honma S, Shirato H, Honma K-I. In vivo imaging of clock gene expression in multiple tissues of freely moving mice. Nat Commun. 2016;10(7):11705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei L, Fan Y, Lv X, Welsh DK, Zhan C, Zhang EE. Long-term in vivo recording of circadian rhythms in brains of freely moving mice. Proc Natl Acad Sci USA. 2018. April;115(16):4276–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono D, Honma S, Nakajima Y, Kuroda S, Enoki R, Honma K. Dissociation of Per1 and Bmal1 circadian rhythms in the suprachiasmatic nucleus in parallel with behavioral outputs. Proc Natl Acad Sci USA. 2017. May;114(18):E3699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini C, Liani A, Curie T, Gos P, Kreppel F, Emmenegger Y, et al. Real-time recording of circadian liver gene expression in freely moving mice reveals the phase-setting behavior of hepatocyte clocks. Genes Dev. 2013. July;27(13):1526–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ML, Swanson BE, de la Iglesia HO. Circadian timing of REM sleep is coupled to an oscillator within the dorsomedial suprachiasmatic nucleus. Curr Biol. 2009. May;19(10):848–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts L, Leise TL, Noguchi T, Galschiodt AM, Houl JH, Welsh DK, et al. Light evokes rapid circadian network oscillator desynchrony followed by gradual phase retuning of synchrony. Curr Biol. 2015. March;25(7):858–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono D, Honma K, Honma S. Circadian and ultradian rhythms of clock gene expression in the suprachiasmatic nucleus of freely moving mice. Sci Rep. 2015. July;5:12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leise TL, Harrington ME, Molyneux PC, Song I, Queenan H, Zimmerman E, et al. Voluntary exercise can strengthen the circadian system in aged mice. Age (Dordr). 2013. December;35(6):2137–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000. December;14(23):2950–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, van Spyk E, Liu Q, Geyfman M, Salmans M, Kumar V, et al. Time-restricted feeding shifts the skin circadian clock and alters UVB-induced DNA damage. Cell Rep. 2017. August;20(5):1061–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubaldo‐Reyes LM, Buijs RM, Escobar C, Ángeles‐Castellanos M. Scheduled meal accelerates entrainment to a 6-h phase advance by shifting central and peripheral oscillations in rats. Eur J Neurosci. 2017;46(3):1875–86. [DOI] [PubMed] [Google Scholar]
- Paul MJ, Indic P, Schwartz WJ. Social synchronization of circadian rhythmicity in female mice depends on the number of cohabiting animals. Biol Lett. 2015. June;11(6):20150204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casiraghi LP, Alzamendi A, Giovambattista A, Chiesa JJ, Golombek DA. Effects of chronic forced circadian desynchronization on body weight and metabolism in male mice. Physiol Rep. 2016. April;4(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daan S, Spoelstra K, Albrecht U, Schmutz I, Daan M, Daan B, et al. Lab Mice in the Field: Unorthodox Daily Activity and Effects of a Dysfunctional Circadian Clock Allele. J Biol Rhythms. 2011. April;26(2):118–29. [DOI] [PubMed] [Google Scholar]
- Fuchikawa T, Eban-Rothschild A, Nagari M, Shemesh Y, Bloch G. Potent social synchronization can override photic entrainment of circadian rhythms. Nat Commun. 2016;23(7):11662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vinne V, Swoap SJ, Vajtay TJ, Weaver DR. Desynchrony between brain and peripheral clocks caused by CK1δ/ε disruption in GABA neurons does not lead to adverse metabolic outcomes. Proc Natl Acad Sci USA. 2018. March;115(10):E2437–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanin S, Bhutani S, Montelli S, Menegazzi P, Green EW, Pegoraro M, et al. Unexpected features of Drosophila circadian behavioural rhythms under natural conditions. Nature. 2012. April;484(7394):371–5. [DOI] [PubMed] [Google Scholar]
- Emmer KM, Russart KL, Walker WH, 2nd, Nelson RJ, DeVries AC. Effects of light at night on laboratory animals and research outcomes. Behav Neurosci. 2018;132(4):302–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter C, Fischer D, Matera JL, Roenneberg T. Aligning Work and Circadian Time in Shift Workers Improves Sleep and Reduces Circadian Disruption. Curr Biol. 2015. March;25(7):907–11. [DOI] [PubMed] [Google Scholar]
- Barca-Mayo O, Pons-Espinal M, Follert P, Armirotti A, Berdondini L, De Pietri Tonelli D. Astrocyte deletion of Bmal1 alters daily locomotor activity and cognitive functions via GABA signalling. Nat Commun. 2017;10(8):14336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancaccio M, Patton AP, Chesham JE, Maywood ES, Hastings MH. Astrocytes Control Circadian Timekeeping in the Suprachiasmatic Nucleus via Glutamatergic Signaling. Neuron. 2017. March;93(6):1420–1435.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancaccio M, Edwards MD, Patton AP, Smyllie NJ, Chesham JE, Maywood ES, et al. Cell-autonomous clock of astrocytes drives circadian behavior in mammals. Science. 2019. January;363(6423):187–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tso CF, Simon T, Greenlaw AC, Puri T, Mieda M, Herzog ED. Astrocytes regulate daily rhythms in the suprachiasmatic nucleus and behavior. Curr Biol. 2017. April;27(7):1055–61. [DOI] [PMC free article] [PubMed] [Google Scholar]