Abstract
Melatonin is a pleiotropic molecule that, after a short-term sleep deprivation, promotes the proliferation of neural stem cells in the adult hippocampus. However, this effect has not been observed in long-term sleep deprivation. The precise mechanism exerted by melatonin on the modulation of neural stem cells is not entirely elucidated, but evidence indicates that epigenetic regulators may be involved in this process. In this study, we investigated the effect of melatonin treatment during a 96-hour sleep deprivation and analyzed the expression of epigenetic modulators predicted by computational text mining and keyword clusterization. Our results showed that the administration of melatonin under sleep-deprived conditions increased the MECP2 expression and reduced the SIRT1 expression in the dentate gyrus. We observed that let-7b, mir-132, and mir-124 were highly expressed in the dentate gyrus after melatonin administration, but they were not modified by sleep deprivation. In addition, we found more Sox2+/5-bromo-2′-deoxyuridine (BrdU)+ cells in the subgranular zone of the sleep-deprived group treated with melatonin than in the untreated group. These findings may support the notion that melatonin modifies the expression of epigenetic mediators that, in turn, regulate the proliferation of neural progenitor cells in the adult dentate gyrus under long-term sleep-deprived conditions. All procedures performed in this study were approved by the Animal Ethics Committee of the University of Guadalajara, Mexico (approval No. CI-16610) on January 2, 2016.
Keywords: sleep-deprivation, melatonin, microRNA, neurogenesis, Sirtuin 1, SIRT1, methyl-CpG-binding protein 2, MECP2, epigenetic, text-mining, mir-9, let-7b
Chinese Library Classification No. R453; R364
Introduction
Sleep deprivation (SD) is a common disorder that frequently occurs as a consequence of insomnia, light pollution, sleep apnea, and stress (Hall et al., 2000; Raap et al., 2015). SD is associated with immunosuppression, production of reactive oxygen species and cell death (Rechtschaffen et al., 1989; Everson and Toth, 2000; Hu et al., 2003; Yang et al., 2008). Sleep deprivation of the rapid eye movement (REM) phase alters behavioral and cognitive functions (Gonzalez-Castañeda et al., 2016; Soto-Rodriguez et al., 2016) and produces long-lasting changes in neural plasticity and circadian rhythm (Massart et al., 2014; Nilsson et al., 2016), which can modify the profile expression of microRNAs in the hippocampus (Davis et al., 2007).
Melatonin (MEL) is a hormone derived from tryptophan that regulates nocturnal sleep phases and plays a role as a circadian zeitgeber. MEL is also a powerful antioxidant (Galano et al., 2011; Vishwas et al., 2013) and a modulator of cell proliferation (Jung-Hynes et al., 2011; Cheng et al., 2013) that seems to reduce some of the deleterious effects of SD (Mueller et al., 2008, 2011), promotes hippocampal neurogenesis (Moriya et al., 2007; Chern et al., 2012), and increases the expression of anti-apoptotic peptides (Bcl-xL and Bcl-2) (López-Armas et al., 2016). However, the mechanism underlying these biological effects is not completely elucidated, but recent evidence indicates that some epigenetic mechanisms are involved in these events (Sharma et al., 2008; Niles et al., 2013; Hardeland, 2014).
Epigenetic regulations are heritable changes in the function of genetic elements without changing the DNA sequence (Bird, 2007) and have multiple implications in cell proliferation, differentiation, cell fate, brain plasticity, and genomic imprinting (Reik and Walter, 2001; Lee et al., 2006; Nelson et al., 2008; Kim et al., 2010; Asuthkar et al., 2012). Epigenetic regulations are mediated via RNA interference (microRNA –mir-), histone modifications, and DNA methylation. MicroRNAs are small interfering strands of RNA that modify protein translation by binding to mRNA. MicroRNA 124 (mir-124) and microRNA 9 (mir-9) are associated with neural gene expression, which modulates neural-stem-cell fate and proliferation (Cheng et al., 2009; Coolen et al., 2013) by suppressing RE-1 silencing transcription factor (REST) (Ballas and Mandel, 2005). On the other hand, microRNA 132 modulates cell maturation, circadian rhythmicity, and synaptic plasticity (Alvarez-Saavedra et al., 2011; Luikart et al., 2011; Clovis et al., 2012).
Histone modification via deacetylation by Sirtuin 1 (SIRT1) is involved in cell longevity and differentiation via a modification of DNA-protein binding in the nucleosome (Saharan et al., 2013; Cai et al., 2016). SIRT1 is an NAD+ dependent histone and its activity depends on the energy and redox state of the cell (Bernstein et al., 2007; Iwahara et al., 2009; Caito et al., 2010; Yao and Jin, 2014). Under SD conditions, the oxidative state is increased, but changes in SIRT1 expression are unknown. DNA methylation can be recognized by a series of specific binding proteins, such as methyl-CpG-binding protein 2 (MECP2) (Lunyak et al., 2004; Ballas et al., 2005; Bogdanović and Veenstra, 2009). MECP2 and REST/NSFR, which constitute a neural-gene-repression complex (Ballas et al., 2005; Chadwick and Wade, 2007; Li et al., 2014; Masserdotti et al., 2015) that modifies brain-derived neurotrophic factor (BNDF) expression and regulates cell proliferation (Zhou et al., 2006; Zocchi and Sassone-Corsi, 2012). Altogether, this evidence suggests that microRNAs let-7b, mir-124, mir-9, and mir-132, and proteins SIRT1, MECP2, and REST can be modified either by SD or MEL treatment. To date, epigenetic effects of SD and MEL on neural cell proliferation have not been studied in vivo. Therefore, the aim of this study was to evaluate whether the MEL administration modifies the expression of epigenetic mediators involved in the neural-stem-cell proliferation of sleep-deprived mice.
Materials and Methods
Animal housing
We used 40 BALB/c male (post-natal day 60) mice for this study. The mice were kept under normal housing conditions and low noise exposure in polycarbonate cages (59 cm × 38.5 cm × 20 cm). The mice were randomly divided into four groups (n = 10 per group): control group (intact animals), melatonin-treated group (MEL), sleep-deprived plus melatonin group (MSD), and the sleep-deprived group (SD). Before the SD protocol, animals were kept in a 12/12-hour light/dark cycle (lights on at 08:00 a.m.), the room temperature was set at 25 ± 2°C and humidity of 50 ± 20% with access to food and water ad libitum. All procedures used in this work were approved by the Animal Ethics Committee of the University of Guadalajara (approval No. CI-16610) on January 2, 2016 and performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publication No. 85-23, revised 1996).
Sleep-deprivation protocol
Twenty mice in the MSD and SD groups were subjected to 96-hour SD using the multiple platform method (van Hulzen and Coenen, 1981; Lopez-Armas et al., 2016; Soto-Rodriguez et al., 2016). Briefly, animals were placed in a 180-cm water tank containing 20 platforms. The water temperature was maintained at 26 ± 2°C throughout the study. Each platform was 2.5 cm wide and 20 cm high. Water and food were available ad libitum on all the platforms. To avoid overcrowding, the platforms outnumbered the animals and mice could freely move around on them. All platforms were 2 cm above the water level; thus, when the animals reached the REM phase and lost their muscle tone, they fell into the water and had to climb up again to the platform. Control animals were housed in their cages, which were allocated in the same experimental room to be kept in the same environment and maintained a normal sleep cycle.
MEL and 5-bromo-2′-deoxyuridine (BrdU) treatment
The MEL and MSD groups received 10 mg/kg MEL (Sigma-Aldrich, St. Louis, MO, USA) intraperitoneal (i.p.) per day dissolved in 1% ethanol (Figure 1) until sacrifice. Additionally, the mice received a single i.p. injection of 300 mg/kg BrdU (Sigma-Aldrich) to label the cell progeny derived from primary hippocampal precursor cells (Cameron and Mckay, 2001).
Figure 1.
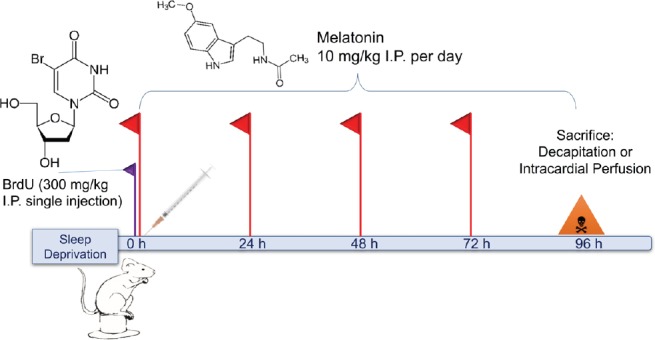
Experimental protocol scheme.
On the first day of the protocol, BrdU was injected (300 mg/kg, i.p) to all groups, and only the SD and MSD groups received SD. Melatonin was administered daily (10 mg/kg i.p.) at 8:00 a.m. After 96 hours (h) of SD, all groups are sacrificed for proper tissue processing. BrdU: 5-Bromo-2′-deoxyuridine; SD: sleep deprivation; MSD: melatonin; I.P. or i.p.: intraperitoneal.
Tissue processing
Immediately after the SD, five mice per group were sacrificed by decapitation, their brains were dissected out on ice, and the hippocampus was extracted and put promptly in cold (4°C) RNAlater solution (Ambion Chemicals, Carlsbad, CA, USA), and immediately stored at −80°C. The remaining animals (n = 5 per group) were used for immunoflourescence and sacrificed with 100 mg/kg pentobarbital followed by the transcardial perfusion. The mice were perfused with 0.1M phosphate buffered saline (PBS) solution at 37°C, followed by 4% paraformaldehyde in 0.1M phosphate buffer, and their brains were post-fixed overnight at 4°C in the same fixative. Later, the brains were cut at 35 µm using a VT1000E vibratome (Leica, Microsystems), starting from the rostral Bregma, −1.34 mm, to the caudal Bregma, −2.92 mm (Paxinos and Franklin, 2013). Tissue storage was done in PBS plus 0.3% sodium azide.
Text mining for detecting targets
To determine which microRNAs to study, we first performed text mining operations using the R studio (R Studio, Boston, MA, USA) and the Bioconductor and R package (https://www.bioconductor.org/) pubmed.mineR (Rani et al., 2015). PubMed abstracts were downloaded in text from January 01, 2000 to January 01, 2017. An abstract corpus was created using the following keywords “Epigenetic,” “Sleep,” “Neurogenesis,” and “Melatonin” (Additional Table 1). Then, the abstract corpus was subjected to word atomization to find the most recurring words. The list was consequently filtered to only include terms with the prefix mir- or the word microRNA. Terms that corresponded to MicroRNAs found were subjected to word clustering association using the apcluster package (Additional Table 2). After a brief review, we chose several key microRNAs after cluster analysis of a term matrix using keywords “neurogenesis”, “sleep,” and “melatonin.” An exhaustive literature review was performed to discard previously described pathways. We repeated this process one more time, and instead of filtering the list with prefixes “mir-” or the word “microRNA,” a gene filter was used. The most frequent terms were filtered based in the literature review, also circadian rhythm genes, basic metabolism enzymes, and globulin genes were excluded. These results were later stored and compared with mir-TAR-BASe. Positive targets for previously described microRNAs were found and a cluster analysis was run with the cluster analysis of a term matrix using keywords “neurogenesis,” “sleep,” and “melatonin.” Chosen targets were selected based on the cluster analysis and the literature review.
Additional Table 1.
Total abstracts analyzed and filtered with the appropriate keywords
| Word searched | Number of abstracts |
|---|---|
| Total of abstracts searched about “epigenetics” | 55178 |
| Abstracts containing “microRNA” | 3056 |
| Abstracts containing “neurogenesis” | 345 |
| Abstracts containing “sleep” | 188 |
| Abstracts containing “melatonin” | 74 |
| Abstracts containing wild cards of these words | 721 |
| Total containing all these keywords | 4384 |
Additional Table 2.
Number of gene incidences after keyword filtering
| Gene Symbol | Name | Frequency |
|---|---|---|
| EZH2 | enhancer of zeste homolog 2 (Drosophila) | 265 |
| T | T, brachyury homolog (mouse) | 261 |
| GC | group-specific component (vitamin D binding protein) | 161 |
| PTEN | phosphatase and tensin homolog | 131 |
| DNMT1 | DNA (cytosine-5-)-methyltransferase 1 | 129 |
| SIRT1 | sirtuin 1 | 77 |
| REST | RE1-silencing transcription factor | 67 |
| STAT3 | signal transducer and activator of transcription 3 (acute-phase response factor) | 67 |
| AR | androgen receptor | 62 |
| BDNF | brain-derived neurotrophic factor | 59 |
| DNMT3B | DNA (cytosine-5-)-methyltransferase 3 beta | 44 |
| MYC | v-myc myelocytomatosis viral oncogene homolog (avian) | 41 |
| VDR | vitamin D (1,25-dihydroxyvitamin D3) receptor | 41 |
| IGF2 | insulin-like growth factor 2 (somatomedin A) | 38 |
| YY1 | YY1 transcription factor | 37 |
| HDAC1 | histone deacetylase 1 | 34 |
| KRAS | v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | 32 |
| CTCF | CCCTC-binding factor (zinc finger protein) | 31 |
| HDAC3 | histone deacetylase 3 | 31 |
| TET1 | tet methylcytosine dioxygenase 1 | 31 |
| BRAF | v-raf murine sarcoma viral oncogene homolog B1 | 30 |
| EGFR | epidermal growth factor receptor | 29 |
| HDAC2 | histone deacetylase 2 | 28 |
| TP53 | tumor protein p53 | 28 |
| HDAC4 | histone deacetylase 4 | 27 |
| MECP2 | methyl CpG binding protein 2 (Rett syndrome) | 26 |
The frequency indicates the number of occurrences of a gene in the total abstracts analyzed and filtered
RNA and protein extraction
Brain samples were immersed and stored with the RNAlater (Ambion Chemicals, Carlsbad, CA, USA). MirVANA PARIS Kit (Life Technologies Burlington, Ontario, Canada) was used to extract both protein and RNA which employs a phenolic extraction of proteins and RNA followed by silica column purification (Boom et al., 1990; Kim et al., 2010). Tissue samples were processed following the kit instructions and employing the included reagents. Briefly, the extraction consisted of homogenizing the sample with the kit Cell Lysis buffer at 0°C from the MirVANA kit, and then, protein and RNA fractions were separated and stored in different vials. The RNA was purified via silica-cartridge adhesion and finally, eluted with RNase-free water also included in the kit. Both the protein and total RNA obtained from each sample were subsequently stored separately in vials at −80°C.
Quantitative PCR
We amplified and measured microRNA associated with neurogenesis. Since many microRNAs are expressed in different chromosomes, we evaluated the expression of primary microRNAs of two expressing alleles. First, the total RNA acquired via mirVANA was quantified using a BioTEK Synergy Htx (Biotek, Winooski, VT, USA) absorbance meter and Take3 (Biotek) 16-well microplate to determine the quality and quantity of RNA. Then, 2 µg of RNA was retrotranscribed using a RETROScript kit (Invitrogen, Vilnus, Lithuania) by utilizing an M-MuLV (Moloney Murine Leukemia Virus) reverse transcriptase and RETROScript Random Primers (Invitrogen, Vilnus, Lithuania). Retrotranscription consisted of 5 minutes of denaturalization at 65°C, 85 minutes of strand synthesis at 55°C plus 5 minutes of inactivation at 85°C. qPCR primers were ordered from IDT Technologies (San Diego, CA, USA) for Pri-Mir-124 (Ch 14), Pri-Mir-124-2 (Ch 3), Pri-Mir132 (Ch 11), Pri-Mir-9-1 (Ch3), Pri-Mir-9b (Ch13), and Pri-Let-7b (15), and we used SnoRNA234 and RNAU6 as our reference genes with the following sequences, which are provided in Table 1. The optimal melting temperature was determined with end-point PCR (Rychlik et al., 1990) and later confirmed via Real-time qPCR assay (Roche, Mannheim, Germany). It was near 62°C for both qPCR and end-point PCR. qPCR was processed using previously described primers with FastStart Green Essential Master Kit (Roche, Mannheim, Germany) in a Light Cycler 96 system (Roche Mannheim, Germany). This reaction consists of a premix solution with dNTPs, Polymerase, MgCl2, and SYBR Green Dye. This mix was poured into a vial of 0.2 mL filled with DNA and nuclease-free water by using the following standard conditions: (1) Preincubation at 94°C for 10 minutes, (2) 3 step amplification over 45 cycles at 95°C for 10 seconds, and then 65°C for 15 seconds, followed by 72°C for 14 seconds, and (3) a high-resolution melting from 65°C to 97°C in 10 minutes. All melting profiles were observed under these assay conditions demonstrating the amplification of a single unique product free of primer dimers or other anomalous products. Reaction efficiency was calculated by a slope using the following equation: Efficiency = (−1 + 10(−1/slope)) × 100. Average efficiency was 85%. The highest efficiency difference between transcripts was not larger than 0.3%. qPCR samples were run in duplicates, qPCR was evaluated by a fold change of expression 2ΔΔCt.
Table 1.
Primers used for detecting qPCR transcripts
| Sequence description | Number of bases | Sequence 5′–3′ |
|---|---|---|
| Let-7b | 20 | CCT CCT CCA GAA CAC GGA CA |
| Let-7b | 21 | CCA TTT AGC TTG CTG AGC GGG |
| mir-9-1 | 23 | AGC GAC TCG AGA CTA CGG AGG T |
| mir-9-1 | 24 | CTC GGG CTG AGC AAC CTT TGA AGG |
| mir-9-2 | 23 | AAG TAC CCC GGA GGA CTA CGC TT |
| mir-9-2 | 22 | TCT TTC CGG AAC GTT CCT CGG T |
| mir-124-1 | 20 | CCA TCC CCT CCC TTT CTT TC |
| mir-124-1 | 22 | ACC GCG TGC CTT AAT TGT ATG G |
| mir-124-2 | 22 | GGA GTA GGG ACT CCA AGC CTA |
| mir-124-2 | 20 | CTC CGC TCT TGG CAT TCA C |
| mir-132 | 20 | GTG CTG ACG TCA GCC TGC AA |
| mir-132 | 22 | TCC TCT TGC TCT GTA TCT GCC C |
| SnoRNA234 | 20 | GGG GTT AGG ATA GGA CCA AG |
| SnoRNA234 | 19 | GTC AGC CAG GGC TAT ACA G |
| Antisense RNAU6 | 21 | GAG AAA GAG GCA GGC CT |
| Sense RNAU6 | 17 | GGC TCT TCT GGC TTT CA |
Western blot assay
The total protein concentration was determined using the Lowry protein determination (Peterson, 1977) measured in a plate and read in BioTek Synergy Htx (Biotek) plate reader. Samples were loaded on 10% SDS- polyacrylamide gels, separated by electrophoresis, and then, transferred to PVDF membranes (Millipore, Darmstadt, Germany). Immunodetection was performed with Near InfraRed fluorescence detection. First, the membranes were blocked with 5% non-fat milk for 1 hour and incubated with primary antibodies overnight at the indicated dilutions: rabbit anti-REST (RE1-silencing factor) (1:200; Millipore, Billerica, MA, USA; Cat# 07-579), rabbit anti-MECP2 (Methyl-CpG-binding protein 2) (1:100; Boster, Pleasanton, CA, USA; Cat# CI1103), mouse anti-SIRT1 (1:100; Millipore, Billerica, MA, USA; Cat# 04-1557) and goat anti-β-actin (1:1000; Santa Cruz Biochemicals, Santa Cruz, CA, USA; Cat# SC1616), and with secondary antibodies goat (Cat # 925-32211, 925-32210 and 925-68071) and donkey (Cat# 925-32213, 925-68024 and 925-68073) at 1:20,000 (Li-Cor Lincoln, NE, USA) for 2 hours at room temperature. Membranes were read on an Odyssey Clx reader (Li-Cor, Lincoln, NE, USA) to normalize the signals of REST, MECP2, and SIRT1; the corresponding signals of β-actin were measured on the same blots. We analyzed the densities with ImageJ software (NIH, Madison, WI, USA).
Immunofluorescence analysis
For immunohistochemical processing, eight 35-µm-thick slices were randomly selected from each brain. Tissue samples were treated with 2 N HCl for 10 minutes at 37°C, followed by 0.1 M borate buffer at pH 8.5 for 10 minutes. Brain sections were rinsed four times in 0.1 M PBS and incubated in the blocking solution (PBS 0.1 M, Triton-X100 0.03% and 10% fetal bovine serum) for 50 minutes. Subsequently, the free-floating samples were incubated overnight with primary antibodies rat IgG anti-BrdU (Bromodeoxyuridine), a marker for cell proliferation (1:500; Bio-Rad, Kidlington, UK; Cat# OBT0030) and anti-Sox2, a marker for neural stem cells (1:500; Millipore, Billerica, MA, USA; Cat# AB5603) at 4ºC. Sections were then rinsed 4× with 0.1 M PBS and incubated with the same blocking solution containing the conjugated secondary antibodies (1:1000 Alexa Flour 488 anti-rat Cat# A-21208, and 1:1000 Alexa Flour 594 anti-rabbit Cat # R37117 Thermo-Fisher, Waltham, MA, USA) for 1 hour at room temperature. After rinsing (4× with 0.1 M PBS), nuclear staining was done with DAPI (Abcam Cambridge, MA, USA; Cat# ab104139). The sections were washed with 0.1 M PBS and mounted on glass slides and covered using Vectashield mounting media (Vector Laboratories, Burlingame, CA, USA; Cat# H-1000).
Cell counting
We assessed eight 35-µm-thick brain sections, 210 µm apart. The whole subgranular zone (SGZ) was quantified in all the collected slices, ranging from one field in rostral brain sections (−1.34 mm coordinates from Bregma) to three fields in dorsal brain sections (−2.92 mm coordinates from Bregma). We quantified manually the number of Sox2+/BrdU+ cells located in the SGZ of the dentate gyrus. Double labeling was always confirmed and quantified by matching cellular morphologies with clearly discernible nuclei (DAPI+ cells). This analysis was done in a Carl Zeiss AxioScope D1 instrument (Gottingen, Germany) with a 40× magnification objective (field area ≈ 0.15 mm2). Quantitative analysis was performed by a subject blinded to the group assignment.
Statistical methods
Data are expressed as the mean ± standard error. To determine the appropriate statistical test, we did a kurtosis analysis and according to these results, we used a parametric or a non-parametric test. Therefore, for multiple comparisons, we used the Kruskal-Wallis test and, for comparison between pairs, we used the Mann-Whitney U test. In all cases, the P < 0.05 value was chosen to establish statistically significant differences. The sample size used was validated by calculating the effect size of each experiment and the respective statistical power. All data were analyzed with Prism 6 (GraphPad Software, San Diego, CA, USA) and SPSS v23 (IBM, Armonk, NY, USA). For cell counting, we used Kruskar-Wallis test followed by a Mann-Whitney U test. For microRNA quantification ΔCt and Western Blot Mean Flourescence Ratio, we applied the Kruskal-Wallis test followed by the Mann-Whitney U test. In all cases, the significance level was set at P < 0.05.
Results
MEL administration maintains the number of Sox2+/BrdU+ neural progenitor cells in the dentate gyrus
To determine changes in Sox2 expression, we injected MEL for 4 days of REM SD and sacrificed animals on day 4 (n = 5 mice per group) (Figure 2). To label proliferative cells, we injected 300 mg/kg of BrdU on day 1. Our data indicated that the number of Sox2+ BrdU+ cells in the SD group (1.19 ± 0.18 cells /field) were significantly lesser as compared with the control group (2.13 ± 0.2 cells/field, P = 0.0023), the MSD group (2.66 ± 0.3 cells, P = 0. 003) and the MEL group (1.91 ± 0.28 cells/field, P = 0.041) (Figure 3). We did not find statistically significant differences between the control group and the MEL and MSD groups (P > 0.05). These data suggest that the MEL treatment helps preserve the proliferation of Sox2+ precursor cells in the dentate gyrus of sleep-deprived animals.
Figure 2.
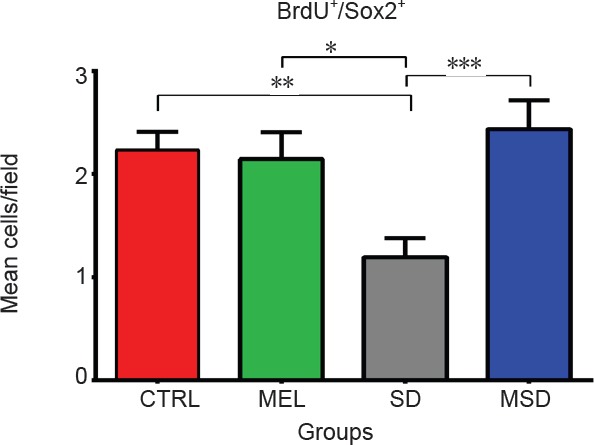
Quantitative analysis of Sox2+/BrdU+ cells in the subgranular zone of the dentate gyrus.
Melatonin-treated animals preserved the number of Sox2+/BrdU+ cells after the sleep deprivation exposure. Bars show the mean ± SEM (n = 5 mice per group). *P < 0.05, **P < 0.01, ***P < 0.01 (Mann-Whitney U test). CTRL: Control group; SD: sleep-deprived group; MEL: melatonin group; MSD: melatonin + sleep deprivation.
Figure 3.
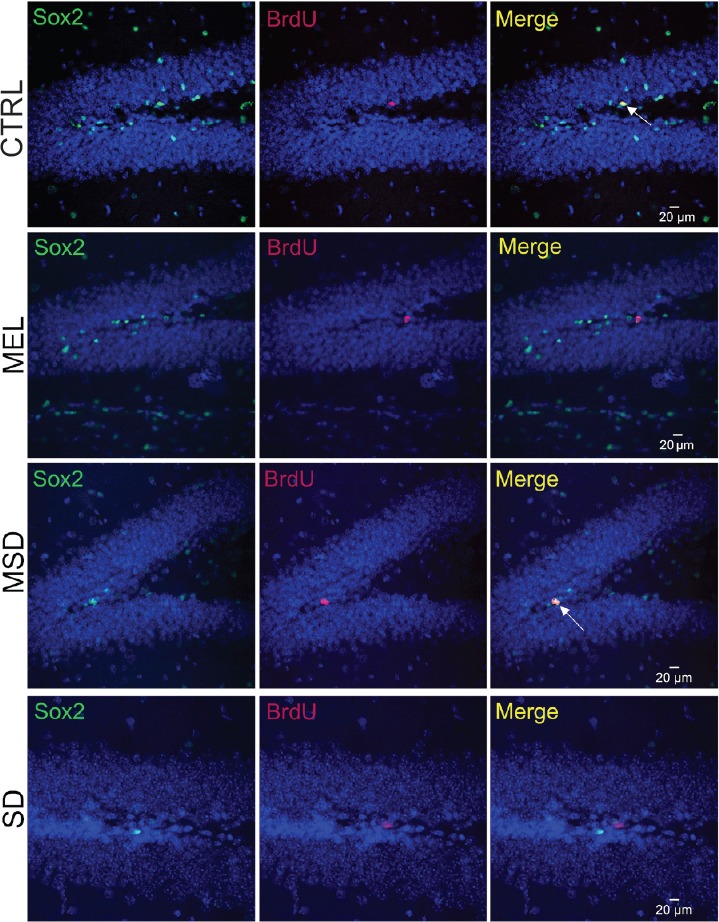
Sox2+/BrdU+ cells in the subgranular zone of the dentate gyrus.
Sox2+ cells are in green (the left panels) and BrdU+ cells are in red (central panels). Right panels show the co-expression patterns for each group. White arrows indicate double-positive cells. Scale bars: 20 μm. n = 5 per group. CTRL: Control group; SD: sleep-deprived group; MEL: melatonin group; MSD: melatonin + sleep deprivation; BrdU: 5-bromo-2′-deoxyuridine.
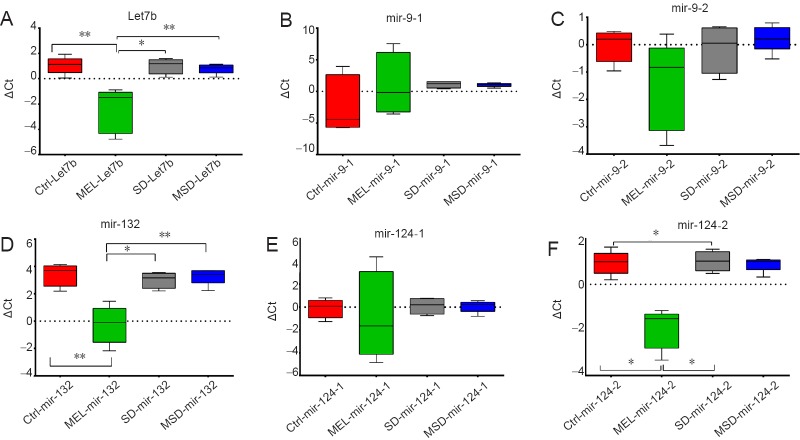
MEL increases microRNA transcription on specific loci
To identify whether the increased proliferation of Sox2+/BrdU+ cells in the hippocampus in sleep-deprived animals treated with MEL was due to a differential expression of epigenetic factors, we investigated several microRNAs involved in the cell proliferation of neural precursors. The regulation of mir-124, mir-132, let-7b and mir-9 transcripts was examined by qPCR analysis (n = 5 mice per group). For mir-124 and mir-9 that have more than one allele, we used two different transcripts for amplification, one for each locus, and was labeled with a dash and number, i.e. mir-124-2. We found that MEL per se increases the expression of some microRNAs when compared to the control group (Figure 4): Let-7b (MEL ΔCt −2.45 ± 0.78 vs. CTRL ΔCt 1.048 ± 0.30; P = 0.007), Mir 124-2 (MEL ΔCt −1.92 ± 0.39 vs. CTRL ΔCt 0.93 ± 0.23; P = 0.007) and mir-132 (MEL ΔCt −0.26 ± 0.61 vs. CTRL ΔCt 3.38 ± 0.36; P = 0.0079). This indicates that administration of MEL induces changes that alter the expression of Let7b, Mir 132 and Mir 124-2. However, the MEL-induced overexpression was reverted by SD effects as we did find statistically significant differences with the MSD group: Let-7b (MEL ΔCt −2.45 ± 0.78 vs. MSD ΔCt 0.79 ± 0.17, P = 0.007), Mir-124-2 (MEL ΔCt −1.92 ± 0.39 vs. MSD ΔCt 0.88 ± 0.14; P = 0.007) and mir-132 (MEL ΔCt −0.26 ± 0.61 vs. MSD ΔCt 3.24 ± 0.26; P = 0.0079). No statistically significant differences were found in the expression of mir-9-1 and mir 9-2 alleles among all groups.
Figure 4.
MicroRNA expression in the hippocampus.
In this case, the lower value indicates the greater expression after normalization with respect to the reference gene, i.e., SnoRNA234 and RNAU6. (A) Let-7b expression. (B) Mir-9-1 expression. (C) Mir-9-2 expression. (D) Mir-132 expression. (E) Mir-124-1 expression. (F) Mir-124-2 expression. Bars show the mean ± SEM (n = 5 mice per group). *P < 0.05, **P < 0.01 (Mann-Whitney U test). ΔCt: Cycle threshold difference; CTRL: control group; SD: sleep-deprived group; MEL: melatonin group; MSD: melatonin + sleep deprivation.
REM SD reduces SIRT1 expression and MEL increases MECP2 expression
We analyzed SIRT1, REST and MECP2 in the dentate gyrus on day 4 (n = 5 mice per group). Our results showed that SD significantly reduces the expression of SIRT1 (5.51 ± 0.61 fluorescence ratio) in comparison with the control group (163.9 ± 77.71 fluorescence ratio; P = 0.015) and this decrement was partially reversed by the effect of MEL as observed in the MSD group (12.65 ± 0.033 fluorescence ratio). We found that the administration of MEL during SD significantly increases MECP2 expression (4.1 ± 1 fluorescence ratio) when compared to the control group (1.582 ± 0.22 fluorescence ratio; P = 0.031) and the SD group (1.407 ± 0.032 fluorescence ratio; P = 0.016). We also did not find significant differences in MECP2 expression between the MEL group (4.325 ± 1.381 fluorescence ratio) and the MSD group (4.101 ± 1 fluorescence ratio; P = 0.9) (Figure 5). These data suggest that MEL treatment restores the MECP2 level under both physiological and SD conditions. In the analysis of REST, we did not find statistically significant differences in REST expression among all the groups analyzed: controls (3.949 ± 0.15 fluorescence ratio), MEL (4.206 ± 0.56 fluorescence ratio), SD (3.945 ± 0.25 fluorescence ratio) or MSD (4.51 ± 0.2 fluorescence ratio) with (P = 0.29). These findings indicate that the REST expression is not modified by the MEL treatment or sleep deprivation.
Figure 5.
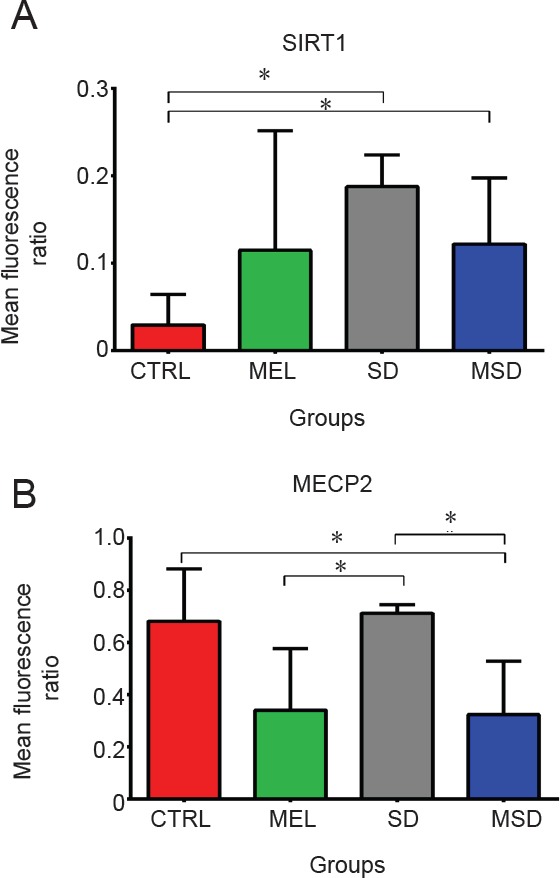
Expression of SIRT1 and MECP2 protein.
(A, B) Mean fluorescence intensity of SIRT1 and MECP2. Bars show the mean ± SEM. *P < 0.05 (Mann-Whitney U test, n = 5 mice per group). CTRL: Control group; SD: sleep-deprived group; MEL: melatonin group; MSD: melatonin + sleep deprivation.
Discussion
To the best of our knowledge, this is the first study that shows an epigenetic regulation in the adult hippocampus after the administration of MEL in vivo under SD conditions. Our results showed that the acute administration of MEL during the SD phase help recover the proliferation of neural precursor cells (Sox2+/BrdU+ cells). Sleep fragmentation and deprivation impair hippocampal proliferation and increase oxidative stress (Silva et al., 2004; Guzman-Marin et al., 2005; Mueller et al., 2008; Sportiche et al., 2010). However, it was unknown whether the administration of MEL during SD modifies cell proliferation of neural precursors in the dentate gyrus. Sox2 is an HMG-box transcription factor that is expressed by neural stem cells and is widely used to identify neural stem cells in the SGZ (Ellis et al., 2004; Suh et al., 2007; von Bohlen und Halbach, 2011). Previous findings have reported that MEL also increases the proliferation of nestin+ progenitor cells in the dentate gyrus (Lopez-Armas et al., 2016). This evidence suggests that MEL can efficiently modulate the hippocampal neurogenic niche. Several mechanisms could explain the increase in cell proliferation, and we propose that the cell expansion could be associated with changes in the expression of MECP2 and SIRT1 that, in turn, can modify the proliferation of neural progenitor cells. MEL may also promote changes in cell survival and proliferation via MT1 and MT2 receptors that activate the MAPK/ERK transduction pathway (Tocharus et al., 2014). This activation of the MAPK/ERK pathway triggers the transcription factor CREB that promotes cell survival by increasing the expression of the anti-apoptotic proteins Bcl-2, Bcl-xL and Bcl-1 (Luchetti et al., 2009). Interestingly, the administration of MEL in vivo increases the expression of Bcl-2 and Bcl-xL that may preserve the hippocampal neurogenesis affected by sleep deprivation (Lopez-Armas et al., 2016).
Our data indicate that MEL administration enhanced the expression of Let-7b, mir-132 and mir-124-2. The exogenous administration of mir-132 reduces the duration of NREM sleep, reduces the slow-wave activity and increases the duration of REM sleep (Davis et al., 2011), indicating that mir-132 could be involved in sleep regulation and phase change. Let-7 is a family of microRNAs that modulate neural maturation, cell fate, and apoptosis of neural stem cells (Shimizu et al., 2010; Zhao et al., 2010; Cimadamore et al., 2013). Under physiological conditions, mir-9, mir-132, mir-124, and let-7 are associated with epigenetic changes that regulate the proliferation and maturation of neural stem cells. Hence, this evidence suggests that MEL can be a strong regulator of these molecules in vivo.
Sleep deprivation modifies the expression of microRNAs in the hippocampus and the forebrain (Davis et al., 2007). We hypothesize that the microRNA changes that we found in the present study can be attributed to pleiotropic actions of MEL. In this study, we found that the MEL group showed a higher expression of mir-132 than the control group. MicroRNA 132 is part of a feedback loop involving BNDF and MECP2 (Klein et al., 2007). Mir-132 is a well-known circadian regulator that reduces the MECP2 and JARID1A expression (Alvarez-Saavedra et al., 2011). Therefore, we inferred that interactions between MEL, MECP2, and mir-132 are dependent on the circadian network regulation as suggested by other authors (Tsuchiya et al., 2015). We also found that the animals treated with MEL without REM SD exposure show an increase in the microRNA expression of mir-124-2 and Let-7b. These data indicate that REM SD may reduce the expression of these microRNA due to several factors, such as high ROS production (Mathangi et al., 2012) and changes in synaptic plasticity (Massart et al., 2014) and long-term potentiation (Romcy-Pereira and Pavlides, 2004). Yet, further miRNA characterization and long-term studies are required to support this hypothesis.
Our findings showed a partial increase in the expression of SIRT1 in the MSD group when compared to the SD group. The mild increase in SIRT1 levels induced by MEL treatment may be a compensatory mechanism against the oxidative stress that occurs during SD conditions. SIRT1 is a protein that modifies circadian clock, regulates cell longevity (Nakahata et al., 2009; Salminen and Kaarniranta, 2009) and maintains the metabolic homeostasis and the neuronal plasticity (Chang et al., 2009). SIRT1-mediated deacetylation contributes to maintaining the Sox2 expression that, in turn, helps preserve self-renewal and multipotency in mesenchymal stem cells (Yoon et al., 2014). Our data indicated that MEL treatment preserved the number of Sox2+ BrdU+ cells in the adult dentate gyrus. MECP2 and REST/NSFR are neural gene repressors that modify the nucleosome binding architecture or inhibit the transcription factor union (Ballas et al., 2005; Chadwick and Wade, 2007; Li et al., 2014; Masserdotti et al., 2015), which alter the BNDF expression and regulates cell proliferation (Zhou et al., 2006; Zocchi and Sassone-Corsi, 2012). Therefore, our findings suggest that the regulation of neural-progenitors induced by MEL in vivo requires SIRT1 (Brunet et al., 2004), MECP2 deacetylation, and BDNF expression (Zocchi and Sassone-Corsi, 2012). The phosphorylation of MECP2 induces the proliferation of neural stem cells and reduces the differentiation of newborn neurons (Kishi and Macklis, 2004; Zhong et al., 2018). Therefore, our data suggest that MEL regulates the pool of Sox2+ neural progenitor cells through SIRT1 and MECP2 modulation in the adult hippocampus.
We also analyzed the expression of REST, but we did not find statistically significant changes among groups. As previously mentioned, we found that the MEL administration increased the expression of mir-124-2, a well-known REST repressor (Visvanathan et al., 2007). REST/NSFR constitutes a neural-gene-repression complex that modifies the BNDF expression and regulates cell proliferation (Zhou et al., 2006; Zocchi and Sassone-Corsi, 2012). Hence, this evidence suggests that other pathways that do not involve REST may play a role in regulating the proliferation of neural progenitors of the adult brain.
Conclusion
MEL treatment administered during SD preserves the number of Sox2+/BrdU+ cells in the dentate gyrus, promotes the overexpression of Let-7b, mir-132, mir-124-2 transcripts, reduces the levels of SIRT1, and increases MECP2 expression. Taken together, this evidence indicates that, under SD conditions, MEL exerts a significant epigenetic modulation that, in turn, regulates the proliferation of neural precursor cells in the adult dentate gyrus of the adult hippocampus in vivo. Yet, the epigenetic role of MEL in the differentiation process of neural progenitor cells under sleep-deprived conditions remains to be elucidated.
Additional file 1: Open peer review reports 1 (106.3KB, pdf) and 2 (107.7KB, pdf) .
Footnotes
Conflicts of interest: The authors declare that there are no conflict of interests regarding the publication of this paper and concerning the funding that they have received.
Financial support: This work was supported by grants from Universidad de Guadalajara (PROSNI 2016, 2017-8) to REGC and partially by grants from Consejo Nacional de Ciencia y Tecnologia (CONACyT No. PN 2016-01-465 and INFR-280414) and PRODEP (213544) to OGP, and the CONACyT Fellowship grant (374823) to AHG.
Institutional review board statement: All procedures used in this work were approved by the Animal Ethics Committee of the University of Guadalajara, Mexico (approval No. CI-16610) on January 2, 2016.
Copyright license agreement: The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement: Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
Open peer reviewers: Lorenzo Di Cesare Mannelli, University of Florence, Italy; Willian Orlando Castillo, Universidade de Sao Paulo, Brazil.
Funding: This work was supported by grants from Universidad de Guadalajara (PROSNI 2016, 2017-8) to REGC and partially by grants from Consejo Nacional de Ciencia y Tecnologia (CONACyT No. PN 2016-01-465 and INFR-280414) and PRODEP (213544) to OGP, and the CONACyT Fellowship grant (374823) to AHG.
P-Reviewers: Di Cesare Mannelli L, Orlando Castillo W; C-Editor: Zhao M; S-Editor: Li CH; L-Editor: Song LP; T-Editor: Jia Y
References
- 1.Alvarez-Saavedra M, Antoun G, Yanagiya A, Oliva-Hernandez R, Cornejo-Palma D, Perez-Iratxeta C, Sonenberg N, Cheng HYMH-YM. miRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Hum Mol Genet. 2011;20:731–751. doi: 10.1093/hmg/ddq519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asuthkar S, Velpula KK, Chetty C, Gorantla B, Rao JS, Asuthkar S, Velpula KK, Chetty C, Gorantla B, Rao JS. Epigenetic regulation of miRNA-211 by MMP-9 governs glioma cell apoptosis, chemosensitivity and radiosensitivity. Oncotarget. 2012;3:1439–1454. doi: 10.18632/oncotarget.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 4.Ballas N, Mandel G. The many faces of REST oversee epigenetic programming of neuronal genes. Curr Opin Neurobiol. 2005;15:500–506. doi: 10.1016/j.conb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 6.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 7.Bogdanović O, Veenstra GJ. DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma. 2009;118:549–565. doi: 10.1007/s00412-009-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 10.Cai Y, Xu L, Xu H, Fan X. SIRT1 and neural cell fate determination. Mol Neurobiol. 2016;53:2815–2825. doi: 10.1007/s12035-015-9158-6. [DOI] [PubMed] [Google Scholar]
- 11.Caito S, Rajendrasozhan S, Cook S, Chung S, Yao H, Friedman AE, Brookes PS, Rahman I. SIRT1 is a redox-sensitive deacetylase that is post-translationally modified by oxidants and carbonyl stress. FASEB J. 2010;24:3145–3159. doi: 10.1096/fj.09-151308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cameron HA, Mckay RD. Adult neurogenesis produces a large pool of new granule cells in the dentate gyrus. J Comp Neurol. 2001;435:406–417. doi: 10.1002/cne.1040. [DOI] [PubMed] [Google Scholar]
- 13.Chadwick LH, Wade PA. MeCP2 in Rett syndrome: transcriptional repressor or chromatin architectural protein? Curr Opin Genet Dev. 2007;17:121–125. doi: 10.1016/j.gde.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Chang HM, Wu UI, Lan CT. Melatonin preserves longevity protein (sirtuin 1) expression in the hippocampus of total sleep-deprived rats. J Pineal Res. 2009;47:211–220. doi: 10.1111/j.1600-079X.2009.00704.x. [DOI] [PubMed] [Google Scholar]
- 15.Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng Y, Cai L, Jiang P, Wang J, Gao C, Feng H, Wang C, Pan H, Yang Y. SIRT1 inhibition by melatonin exerts antitumor activity in human osteosarcoma cells. Eur J Pharmacol. 2013;715:219–229. doi: 10.1016/j.ejphar.2013.05.017. [DOI] [PubMed] [Google Scholar]
- 17.Chern CM, Liao JF, Wang YH, Shen YC. Melatonin ameliorates neural function by promoting endogenous neurogenesis through the MT2 melatonin receptor in ischemic-stroke mice. Free Radic Biol Med. 2012;52:1634–1647. doi: 10.1016/j.freeradbiomed.2012.01.030. [DOI] [PubMed] [Google Scholar]
- 18.Clovis YM, Enard W, Marinaro F, Huttner WB, Tonelli DDP. Convergent repression of Foxp2 3′UTR by miR-9 and miR-132 in embryonic mouse neocortex: implications for radial migration of neurons. Development. 2012;139:3332–3342. doi: 10.1242/dev.078063. [DOI] [PubMed] [Google Scholar]
- 19.Coolen M, Katz S, Bally-Cuif L. miR-9: a versatile regulator of neurogenesis. Front Cell Neurosci. 2013;7:220. doi: 10.3389/fncel.2013.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis CJ, Bohnet SG, Meyerson JM, Krueger JM. Sleep loss changes microRNA levels in the brain: A possible mechanism for state-dependent translational regulation. Neurosci Lett. 2007;422:68–73. doi: 10.1016/j.neulet.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellis P, Fagan BM, Magness ST, Hutton S, Taranova O, Hayashi S, McMahon A, Rao M, Pevny L. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev Neurosci. 2004;26:148–165. doi: 10.1159/000082134. [DOI] [PubMed] [Google Scholar]
- 22.Everson CA, Toth LA. Systemic bacterial invasion induced by sleep deprivation. Am J Physiol Regul Integr Comp Physiol. 2000;278:R905–916. doi: 10.1152/ajpregu.2000.278.4.R905. [DOI] [PubMed] [Google Scholar]
- 23.Galano A, Tan DX, Reiter RJ. Melatonin as a natural ally against oxidative stress: A physicochemical examination. J Pineal Res. 2011;51:1–16. doi: 10.1111/j.1600-079X.2011.00916.x. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez-Castañeda RE, Galvez-Contreras AY, Martínez-Quezada CJ, Jauregui-Huerta F, Grcia-Estrada J, Ramos-Zuñiga R, Luquin S, Gonzalez-Perez O. Sex-related effects of sleep deprivation on depressive- and anxiety-like behaviors in mice. Exp Anim. 2016;65:97–107. doi: 10.1538/expanim.15-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guzman-Marin R, Suntsova N, Methippara M, Greiffenstein R, Szymusiak R, McGinty D. Sleep deprivation suppresses neurogenesis in the adult hippocampus of rats. Eur J Neurosci. 2005;22:2111–2116. doi: 10.1111/j.1460-9568.2005.04376.x. [DOI] [PubMed] [Google Scholar]
- 26.Hall M, Buysse DJ, Nowell PD, Nofzinger EA, Houck P, Reynolds CF, 3rd, Kupfer DJ. Symptoms of stress and depression as correlates of sleep in primary insomnia. Psychosom Med. 2000;62:227–230. doi: 10.1097/00006842-200003000-00014. [DOI] [PubMed] [Google Scholar]
- 27.Hardeland R. Melatonin, noncoding RNAs, messenger RNA stability and epigenetics--evidence, hints, gaps and perspectives. Int J Mol Sci. 2014;15:18221–18252. doi: 10.3390/ijms151018221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu J, Chen Z, Gorczynski CP, Gorczynski LY, Kai Y, Lee L, Manuel J, Gorczynski RM. Sleep-deprived mice show altered cytokine production manifest by perturbations in serum IL-1ra, TNFa, and IL-6 levels. Brain Behav Immun. 2003;17:498–504. doi: 10.1016/j.bbi.2003.03.001. [DOI] [PubMed] [Google Scholar]
- 29.Iwahara N, Hisahara S, Hayashi T, Horio Y. Transcriptional activation of NAD+-dependent protein deacetylase SIRT1 by nuclear receptor TLX. Biochem Biophys Res Commun. 2009;386:671–675. doi: 10.1016/j.bbrc.2009.06.103. [DOI] [PubMed] [Google Scholar]
- 30.Johnson R, Richter N, Jauch R, Gaughwin PM, Zuccato C, Cattaneo E, Stanton LW. Human accelerated region 1 noncoding RNA is repressed by REST in Huntington’s disease. Physiol Genomics. 2010;41:269–274. doi: 10.1152/physiolgenomics.00019.2010. [DOI] [PubMed] [Google Scholar]
- 31.Jung-Hynes B, Schmit TL, Reagan-Shaw SR, Siddiqui IA, Mukhtar H, Ahmad N. Melatonin, a novel Sirt1 inhibitor, imparts antiproliferative effects against prostate cancer in vitro in culture and in vivo in TRAMP model. J Pineal Res. 2011;50:140–149. doi: 10.1111/j.1600-079X.2010.00823.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kabra N, Li Z, Chen L, Li B, Zhang X, Wang C, Yeatman T, Coppola D, Chen J. SirT1 Is an Inhibitor of Proliferation and Tumor Formation in Colon Cancer. J Biol Chem. 2009;284:18210–18217. doi: 10.1074/jbc.M109.000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim AH, Reimers M, Maher B, Williamson V, McMichael O, McClay JL, van den Oord EJ, Riley BP, Kendler KS, Vladimirov VI. MicroRNA expression profiling in the prefrontal cortex of individuals affected with schizophrenia and bipolar disorders. Schizophr Res. 2010;124(1-3):183–191. doi: 10.1016/j.schres.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, Yabuuchi A, Takeuchi A, Cunniff KC, Hongguang H, McKinney-Freeman S, Naveiras O, Yoon TJ, Irizarry RA, Jung N, Seita J, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kishi N, Macklis JD. MECP2 is progressively expressed in post-migratory neurons and is involved in neuronal maturation rather than cell fate decisions. Mol Cell Neurosci. 2004;27:306–321. doi: 10.1016/j.mcn.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 36.Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10:1513–1514. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- 37.Lee ER, McCool KW, Murdoch FE, Fritsch MK. Dynamic changes in histone H3 phosphoacetylation during early embryonic stem cell differentiation are directly mediated by mitogen- and stress-activated protein kinase 1 via activation of MAPK pathways. J Biol Chem. 2006;281:21162–21172. doi: 10.1074/jbc.M602734200. [DOI] [PubMed] [Google Scholar]
- 38.Li H, Zhong X, Chau KF, Santistevan NJ, Guo W, Kong G, Li X, Kadakia M, Masliah J, Chi J, Jin P, Zhang J, Zhao X, Chang Q. Cell cycle-linked MeCP2 phosphorylation modulates adult neurogenesis involving the Notch signaling pathway. Nat Commun. 2014;5:5601. doi: 10.1038/ncomms6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.López-armas G, Flores-soto ME, Chaparro-huerta V, Jave-suarez LF, Soto-rodríguez S, Rusanova I, Acuña-castroviejo D, González-perez O, González-castañeda RE. Prophylactic role of oral melatonin administration on neurogenesis in adult Balb/C mice during REM sleep deprivation. Oxid Med Cell Longev. 2016;2016:35–45. doi: 10.1155/2016/2136902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luchetti F, Betti M, Canonico B, Arcangeletti M, Ferri P, Galli F, Papa S. ERK MAPK activation mediates the antiapoptotic signaling of melatonin in UVB-stressed U937 cells. Free Radic Biol Med. 2009;46:339–351. doi: 10.1016/j.freeradbiomed.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 41.Luikart BW, Bensen AL, Washburn EK, Perederiy JV, Su KG, Li Y, Kernie SG, Parada LF, Westbrook GL. MiR-132 mediates the integration of newborn neurons into the adult dentate gyrus. PLoS One. 2011;6:e19077. doi: 10.1371/journal.pone.0019077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lunyak VV, Prefontaine GG, Rosenfeld MG. REST and peace for the neuronal-specific transcriptional program. Ann N Y Acad Sci. 2004;1014:110–120. doi: 10.1196/annals.1294.011. [DOI] [PubMed] [Google Scholar]
- 43.Massart R, Freyburger M, Suderman M, Paquet J, El Helou J, Belanger-Nelson E, Rachalski A, Koumar OC, Carrier J, Szyf M, Mongrain V. The genome-wide landscape of DNA methylation and hydroxymethylation in response to sleep deprivation impacts on synaptic plasticity genes. Transl Psychiatry. 2014;4:e347. doi: 10.1038/tp.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masserdotti G, Gillotin S, Sutor B, Drechsel D, Irmler M, Jørgensen HF, Sass S, Theis FJ, Beckers J, Berninger B, Guillemot F, Götz M. Transcriptional mechanisms of proneural factors and REST in regulating neuronal reprogramming of astrocytes. Cell Stem Cell. 2015;17:74–88. doi: 10.1016/j.stem.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mathangi DC, Shyamala R, Subhashini AS. Effect of REM sleep deprivation on the antioxidant status in the brain of Wistar rats. Ann Neurosci. 2012;19:161–164. doi: 10.5214/ans.0972.7531.190405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moriya T, Horie N, Mitome M, Shinohara K. Melatonin influences the proliferative and differentiative activity of neural stem cells. J Pineal Res. 2007;42:411–418. doi: 10.1111/j.1600-079X.2007.00435.x. [DOI] [PubMed] [Google Scholar]
- 47.Mueller AD, Mear RJ, Mistlberger RE. Inhibition of hippocampal neurogenesis by sleep deprivation is independent of circadian disruption and melatonin suppression. Neuroscience. 2011;193:170–181. doi: 10.1016/j.neuroscience.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 48.Mueller AD, Pollock MS, Lieblich SE, Epp JR, Galea LA, Mistlberger RE. Sleep deprivation can inhibit adult hippocampal neurogenesis independent of adrenal stress hormones. Am J Physiol Regul Integr Comp Physiol. 2008;294:R1693–1703. doi: 10.1152/ajpregu.00858.2007. [DOI] [PubMed] [Google Scholar]
- 49.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nelson ED, Kavalali ET, Monteggia LM. Activity-dependent suppression of miniature neurotransmission through the regulation of DNA methylation. J Neurosci. 2008;28:395–406. doi: 10.1523/JNEUROSCI.3796-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niles LP, Pan Y, Kang S, Lacoul A. Melatonin induces histone hyperacetylation in the rat brain. Neurosci Lett. 2013;541:49–53. doi: 10.1016/j.neulet.2013.01.050. [DOI] [PubMed] [Google Scholar]
- 52.Nilsson EK, Boström AE, Mwinyi J, Schiöth HB. Epigenomics of Total Acute Sleep Deprivation in Relation to Genome-Wide DNA Methylation Profiles and RNA Expression. OMICS. 2016;20:334–342. doi: 10.1089/omi.2016.0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Niu W, Zang T, Smith DK, Vue TY, Zou Y, Bachoo R, Johnson JE, Zhang C-L SOX2 reprograms resident astrocytes into neural progenitors in the adult brain. Stem Cell Rep. 2015;4:780–794. doi: 10.1016/j.stemcr.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paxinos G, Franklin K. 4th ed. San Diego: Elsevier Academic Press; 2013. Paxinos and Franklin’s the mouse brain in stereotaxic coordinates. [Google Scholar]
- 55.Peterson GL. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal Biochem. 1977;83:346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- 56.Raap T, Pinxten R, Eens M. Light pollution disrupts sleep in free-living animals. Sci Rep. 2015;5:13557. doi: 10.1038/srep13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rani J, Shah AR, Ramachandran S. pubmed.mineR: An R package with text-mining algorithms to analyse PubMed abstracts. J Biosci. 2015;40:671–682. doi: 10.1007/s12038-015-9552-2. [DOI] [PubMed] [Google Scholar]
- 58.Rechtschaffen A, Bergmann BM, Everson CA, Kushida CA, Gilliland MA. Sleep deprivation in the rat: X. Integration and discussion of the findings. Sleep. 1989;12:68–87. [PubMed] [Google Scholar]
- 59.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 60.Romcy-Pereira R, Pavlides C. Distinct modulatory effects of sleep on the maintenance of hippocampal and medial prefrontal cortex LTP. Eur J Neurosci. 2004;20:3453–3462. doi: 10.1111/j.1460-9568.2004.03808.x. [DOI] [PubMed] [Google Scholar]
- 61.Rychlik W, Spencer WJ, Rhoads RE. Optimization of the annealing temperature for DNA amplification in vitro. Nucleic Acids Res. 1990;18:6409–6412. doi: 10.1093/nar/18.21.6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saharan S, Jhaveri DJ, Bartlett PF. SIRT1 regulates the neurogenic potential of neural precursors in the adult subventricular zone and hippocampus. J Neurosci Res. 2013;91:642–659. doi: 10.1002/jnr.23199. [DOI] [PubMed] [Google Scholar]
- 63.Salminen A, Kaarniranta K. SIRT1: Regulation of longevity via autophagy. Cell Signal. 2009;21:1356–1360. doi: 10.1016/j.cellsig.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 64.Seri B, García-Verdugo JM, Collado-Morente L, McEwen BS, Alvarez-Buylla A. Cell types, lineage, and architecture of the germinal zone in the adult dentate gyrus. J Comp Neurol. 2004;478:359–378. doi: 10.1002/cne.20288. [DOI] [PubMed] [Google Scholar]
- 65.Sharma R, Ottenhof T, Rzeczkowska PA, Niles LP. Epigenetic targets for melatonin: induction of histone H3 hyperacetylation and gene expression in C17.2 neural stem cells. J Pineal Res. 2008;45:277–284. doi: 10.1111/j.1600-079X.2008.00587.x. [DOI] [PubMed] [Google Scholar]
- 66.Silva RH, Abílio VC, Takatsu AL, Kameda SR, Grassl C, Chehin AB, Medrano WA, Calzavara MB, Registro S, Andersen ML, Machado RB, Carvalho RC, Ribeiro Rde A, Tufik S, Frussa-Filho R. Role of hippocampal oxidative stress in memory deficits induced by sleep deprivation in mice. Neuropharmacology. 2004;46:895–903. doi: 10.1016/j.neuropharm.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 67.Soto-Rodriguez S, Lopez-Armas G, Luquin S, Ramos-Zuñiga R, Jauregui-Huerta F, Gonzalez-Perez O, Gonzalez-Castañeda RE. Rapid eye movement sleep deprivation produces long-term detrimental effects in spatial memory and modifies the cellular composition of the subgranular zone. Front Cell Neurosci. 2016;10:1–13. doi: 10.3389/fncel.2016.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sportiche N, Suntsova N, Methippara M, Bashir T, Mitrani B, Szymusiak R, McGinty D. Sustained sleep fragmentation results in delayed changes in hippocampal-dependent cognitive function associated with reduced dentate gyrus neurogenesis. Neuroscience. 2010;170:247–258. doi: 10.1016/j.neuroscience.2010.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Suh H, Consiglio A, Ray J, Sawai T, D’Amour KA, Gage FH. In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell. 2007;1:515–528. doi: 10.1016/j.stem.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tocharus C, Puriboriboon Y, Junmanee T, Tocharus J, Ekthuwapranee K, Govitrapong P. Melatonin enhances adult rat hippocampal progenitor cell proliferation via ERK signaling pathway through melatonin receptor. Neuroscience. 2014;275:314–321. doi: 10.1016/j.neuroscience.2014.06.026. [DOI] [PubMed] [Google Scholar]
- 71.Tsuchiya Y, Minami Y, Umemura Y, Watanabe H, Ono D, Nakamura W, Takahashi T, Honma S, Kondoh G, Matsuishi T, Yagita K. Disruption of MeCP2 attenuates circadian rhythm in CRISPR/Cas9-based Rett syndrome model mouse. Genes Cells. 2015;20:992–1005. doi: 10.1111/gtc.12305. [DOI] [PubMed] [Google Scholar]
- 72.van Hulzen ZJ, Coenen AM. Paradoxical sleep deprivation and locomotor activity in rats. Physiol Behav. 1981;27:741–744. doi: 10.1016/0031-9384(81)90250-x. [DOI] [PubMed] [Google Scholar]
- 73.Vishwas DK, Mukherjee A, Haldar C, Dash D, Nayak MK. Improvement of oxidative stress and immunity by melatonin: An age dependent study in golden hamster. Exp Gerontol. 2013;48:168–182. doi: 10.1016/j.exger.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 74.Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.von Bohlen und Halbach O. Immunohistological markers for proliferative events, gliogenesis, and neurogenesis within the adult hippocampus. Cell Tissue Res. 2011;345:1–19. doi: 10.1007/s00441-011-1196-4. [DOI] [PubMed] [Google Scholar]
- 76.Yang RH, Hu SJ, Wang Y, Zhang WB, Luo WJ, Chen JY. Paradoxical sleep deprivation impairs spatial learning and affects membrane excitability and mitochondrial protein in the hippocampus. Brain Res. 2008;1230:224–232. doi: 10.1016/j.brainres.2008.07.033. [DOI] [PubMed] [Google Scholar]
- 77.Yao B, Jin P. Unlocking epigenetic codes in neurogenesis. Genes Dev. 2014;28:1253–1271. doi: 10.1101/gad.241547.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yoon DS, Choi Y, Jang Y, Lee M, Choi WJ, Kim SH, Lee JW. SIRT1 directly regulates SOX2 to maintain self-renewal and multipotency in bone marrow-derived mesenchymal stem cells: epigenetic regulation of SOX2 by SIRT1. Stem Cells. 2014;32:3219–3231. doi: 10.1002/stem.1811. [DOI] [PubMed] [Google Scholar]
- 79.Zhong X, Li H, Kim J, Chang Q. Regulation of neural differentiation, synaptic scaling and animal behavior by MeCP2 phophorylation. Neurobiol Learn Mem. 2018 doi: 10.1016/j.nlm.2018.04.014. doi: 10.1016/j.nlm.2018.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou Z, Hong EJ, Cohen S, Zhao W ning, Ho H yi H, Schmidt L, Chen WG, Lin Y, Savner E, Griffith EC, Hu L, Steen JAJ, Weitz CJ, Greenberg ME. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52:255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zocchi L, Sassone-Corsi P. SIRT1-mediated deacetylation of MeCP2 contributes to BDNF expression. Epigenetics. 2012;7:695–700. doi: 10.4161/epi.20733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.