Abstract
Older persons may particularly benefit from pharmacogenetic diagnostics, but there is little clinical evidence on that question. We quantitatively analyzed the effects of age and genotype in drugs with consensus on a therapeutically relevant impact of a genotype. Assuming additive effects of age and genotype, drugs may be classified in groups with different priorities to consider either age, or genotype, or both, in therapy. Particularly interesting were those studies specifically analyzing the age‐by‐genotype interaction.
BACKGROUND AND AIMS
Heritable genomic biomarkers are increasingly used for an individually optimized drug therapy. Many of these polymorphisms modulate activity in drug membrane transport and drug metabolism. Numerous studies have shown that genomic variation can have significant effects on the concentrations of drugs at their target sites. However, pharmacogenetic knowledge comes mainly from studies in young and middle‐aged individuals. The proportion of older people is growing worldwide and older patients are exceptionally vulnerable to adverse drug effects and may therefore particularly benefit from individualized therapy.
Physiological mechanisms like the mean age‐related decline in renal function, the decline in volume of distribution for hydrophilic drugs, and the relative increase in volume of distribution for lipophilic drugs are generally accepted facts.1, 2, 3 According to earlier studies in homogenates from liver biopsies, expression and in vitro activity of many drug‐metabolizing enzymes does not significantly decline in old age.4 However, age‐related decline in liver mass and in hepatic blood flow is well established5 and may explain lower total clearance in older people. Lower clearance in older people is more consistently found with high‐clearance drugs than with low‐clearance drugs. That may be explained by the fact that elimination of low‐clearance drugs is more likely to depend on plasma protein binding. Considering the free drug concentrations might even more consistently show a lower hepatic clearance in older people.6 But there are not enough experimental data on free drug concentrations in older people to allow an analysis of that in the present review. Age‐related microstructural changes impeding transfer of drugs to the hepatocytes7 may further be relevant for reduced hepatic drug clearance in old age.
In this review we do not want to answer why pharmacokinetics can change in old age, but we want to compare how much it is changing by using robust pharmacokinetic parameters relevant for drug dosing. In general, and irrespective of the route of elimination, the systemic exposure to drugs is assumed to increase in old age. Here we focused on systemic exposure measured as area under the curve (AUC) or alternatively trough blood concentration, which are parameters having the advantage of being proportional to the eventual dose adjustments in the case of relevant age‐ and/or genotype‐related differences.8 There are only a few substances with therapeutically relevant nonlinear pharmacokinetics (e.g., saturable transport or metabolism) for which this approach may not be valid.
It is reasonable to assume that in older people the variation in pharmacokinetics (PK) and pharmacodynamics (PD) will become greater due to acquired factors. Thus, the contribution of inherited variation to the interindividual variation may become less relevant.9 But there are only limited empirical data on that, and one may also find arguments that consideration of genomic variation in drug therapy may be even more important in old age than in young age. Therefore, we reviewed existing data to quantitatively summarize the effects of age and genotype on PK in older people.
We focused on those drugs for which the regulatory authorities (in particular the US Food and Drug Administration (FDA)) consider genetic polymorphisms important enough to include data in the information for prescribers.10 In particular, we included those drugs for which already actionable recommendations exist on how to consider the genotype in drug dosing.10, 11, 12, 13 In addition, some other drugs affected by genetic polymorphisms in a similar manner and some prototypic probe drugs of drug membrane transporters and drug‐metabolizing enzymes were included.
We quantitatively compared the mean effects exerted by defined genetic polymorphisms with the mean effects exerted by old age. We also present the boundary conditions of this research, like sample size and age range. Furthermore, we compared the interindividual variation in PK between young and old people. Finally, we analyzed the studies in which both genotypes and age (and the genotype–age interaction) have been studied. A main hypothesis stimulating the present meta‐analysis was that specific transporters and specific drug‐metabolizing enzymes are changing differently in their activity during life. Therefore, we analyzed differences in the systemic exposure parameters between drug groups defined by the transporter or enzyme presumed to be most relevant for PK.
For medical practice, this analysis might give physicians arguments on whether and how to consider both age and genotype in older patients. Although still not universally applied, pharmacogenetic diagnostics is increasingly used in medical practice nowadays and physicians taking care of older patients may ask whether or not to consider pharmacogenetic diagnostics.
SYSTEMIC EXPOSURE TO DRUGS IN OLD AGE
To compare different drugs we used the ratios of the PK parameter for systemic exposure (AUC or trough concentrations) in older vs. younger people, hereafter referred to as the age PK ratio. The age PK ratio was adjusted for dose if doses were different between the younger and older sample (details in the Supplementary text and Table S1). Only for 84 of the 108 drugs quantitative data on age‐related changes were found. Summarizing all studied drugs, the mean (min–max) age PK ratio was 1.55 (0.57–7.83), which means that the older people had on average a 1.55‐fold higher systemic exposure compared to the young. In 87% of the drugs a nominal increase in the exposure was found. Thus, for many drugs the age‐related increase in systemic exposure was only minor to moderate (Table 1 ; Table S1). However, the age PK ratio was 2‐fold or more for a few drugs, including digoxin, zolpidem, desmethyldiazepam, clomipramine, paroxetine, and fluvoxamine. With these drugs dose adjustment in older persons may be particularly justified. Some exceptional age‐related changes may not be reproducible. For instance, a big age effect with capecitabine (Figure 1) was found in only one study. Other age studies on capecitabine showed no or only minor age effects (Table S1) but did not provide quantitative data and thus could not be included in the calculations here.
Table 1.
Summary of age and genotype effects for drugs considered to be affected in a relevant manner by genomic variation
| Drug | Enzyme/transporter | Age PK ratio (systemic exposure in elderly/young) | Genotype PK ratio (slowest metabolizer/ normal metabolizer) | Ultrarapid genotype PK ratio (fastest metabolizer/normal metabolizer) |
|---|---|---|---|---|
| Acenocoumarol, (R) enantiomer | CYP2C9 | 0.99 | ||
| Amitriptyline | CYP2C19 | 1.16 | 1.51 | 0.75 |
| Amitriptyline | CYP2D6 | 1.16 | 1.69 | 0.82 |
| Aripiprazole | CYP2D6 | 1.25 | 1.75 | 0.83 |
| Atazanavir | CYP3A5 | 0.88 | 1.03 | |
| Atazanavir | UGT1A1 | 0.88 | a1.03 | |
| Atomoxetine | CYP2D6 | 9.07 | ||
| Atorvastatin | OATP1B1 | 1.34 | 1.82 | |
| Azathioprine | TPMT | 17.01 | ||
| Brexpiprazole | CYP2D6 | 1.05 | 4.28 | |
| Brivartaracetam | CYP2C19 | 1.15 | 1.42 | |
| Capecitabine | DPD | 3.48 | b10.00 | |
| Celecoxib | CYP2C9 | 1.49 | 3.34 | |
| Ciclosporin | CYP3A4 | 1.32 | 1.56 | |
| Citalopram | CYP2C19 | 1.71 | 1.82 | 0.83 |
| Clobazam | CYP2C19 | 1.47 | 7.39 | |
| Clomipramine | CYP2D6 | 2.56 | 2.17 | 0.79 |
| Clomipramine | CYP2C19 | 2.56 | 1.54 | 0.85 |
| Clopidogrel | CYP2C19 | 1.44 | 0.73 | |
| Clozapine | CYP2C19 | 0.92 | 1.69 | 0.88 |
| Codeine | CYP2D6 | d2.59 | d0.05 | d1.45 |
| Darifenacin | CYP2D6 | 1.23 | 1.70 | |
| Desipramine | CYP2D6 | 1.32 | 5.41 | 0.65 |
| Desmethyldiazepam | CYP2C19 | 2.63 | 2.37 | |
| Dexlansoprazole | CYP2C19 | 1.35 | 4.61 | |
| Dextromethorphan | CYP2D6 | 1.49 | ||
| Diazepam | CYP2C19 | 1.12 | 1.48 | |
| Dihydrocodeine | CYP2D6 | 1.85 | 1.12 | |
| Dolasetron | CYP2D6 | 1.31 | 2.00 | |
| Dolutegravir | UGT1A1 | 0.86 | 1.46 | |
| Doxepin | CYP2D6 | 1.91 | 3.87 | 0.65 |
| Dronabinol | CYP2C9 | 3.09 | ||
| Duloxetine | CYP2D6 | 1.24 | 1.65 | 0.84 |
| Efavirenz | CYP2B6 | 1.05 | 3.11 | |
| Eliglustat | CYP2D6 | 7.00 | ||
| Endoxifen | CYP2D6 | 1.23 | 0.40 | |
| Escitalopram | CYP2C19 | 1.23 | 1.74 | 0.68 |
| Esomeprazole | CYP2C19 | 1.58 | ||
| Etoposide | UGT1A1 | 1.28 | 1.06 | |
| Ezetemibe | UGT1A1 | 2.11 | 1.77 | |
| Fesoterodine | CYP2D6 | 1.00 | 2.31 | |
| Flecainide | CYP2D6 | 1.29 | 1.28 | |
| Flibanserin | CYP2C9 | 0.64 | 1.30 | |
| Fluorouracil | DPD | 1.12 | 10.00 | |
| Fluoxetine | CYP2D6 | 0.88 | 3.61 | |
| Fluoxetine, (R) enantiomer | CYP2C9 | 1.86 | ||
| Fluoxetine, (S) enantiomer | CYP2D6 | 1.37 | 0.74 | |
| Flurbiprofen | CYP2C9 | 1.21 | 1.63 | |
| Fluvastatin | CYP2C9 | 1.11 | 2.13 | |
| Fluvoxamine | CYP2D6 | 2.91 | 1.73 | 0.8 |
| Galantamine | CYP2D6 | 1.38 | 1.33 | |
| Glibenclamide | CYP2C9 | 0.74 | 1.69 | |
| Glimepiride | CYP2C9 | 1.48 | ||
| Glipizide | CYP2C9 | 0.90 | 5.45 | |
| Haloperidol | CYP2D6 | 1.23 | 1.86 | 1.1 |
| Hydralazine | NAT2 | 1.22 | 2.25 | |
| Ibuprofen | CYP2C9 | 0.99 | 1.81 | |
| Ibuprofen, (S) enantiomer | CYP2C9 | 1.26 | ||
| Imipramine | CYP2D6 | 1.86 | 4.70 | 0.72 |
| Irinotecan | UGT1A1 | 1.31 | c1.61 | |
| Isoniazid | NAT2 | 1.55 | 2.49 | |
| Lansoprazole | CYP2C19 | 1.50 | 4.91 | |
| Lesinurad | CYP2C9 | 1.96 | ||
| Lorazepam | UGT2B15 | 1.29 | 1.72 | |
| Losartan | CYP2C9 | 1.35 | ||
| Mercaptopurine | TPMT | 2.09 | ||
| Methadone, (R) enantiomer | CYP2B6 | 1.83 | ||
| Metoprolol | CYP2D6 | 1.17 | 2.28 | 0.43 |
| Morphine | OCT1 | 1.57 | 1.70 | |
| Nateglinide | CYP2C9 | 1.35 | ||
| Nilotinib | UGT1A1 | a | ||
| Nortriptyline | CYP2D6 | 1.63 | 3.32 | 0.66 |
| Olanzapine | CYP2D6 | 1.32 | 2.00 | |
| Omeprazole | CYP2C19 | 1.99 | 8.83 | |
| Ondansetron | CYP2D6 | 1.42 | 1.04 | |
| Ondansetron | OCT1 | 1.42 | 3.00 | |
| Oxazepam | UGT2B15 | 1.48 | 2.08 | |
| Oxycodone | CYP2D6 | 1.55 | ||
| Pantoprazole | CYP2C19 | 1.39 | 5.06 | 0.67 |
| Paroxetine | CYP2D6 | 3.82 | 2.45 | 0.74 |
| Pazopanib | UGT1A1 | a | ||
| Perphenazine | CYP2D6 | 2.58 | 0.71 | |
| Phenytoin | CYP2C9 | 1.34 | 1.55 | |
| Pimozide | CYP2D6 | 2.00 | 0.4 | |
| Piroxicam | CYP2C9 | 0.98 | 1.67 | |
| Pitavastatin | OATP1B1 | 2.62 | ||
| Pravastatin | OATP1B1 | 1.48 | 1.92 | |
| Propafenone | CYP2D6 | 3.77 | ||
| Propafenone, (R) enantiomer | CYP2D6 | |||
| Rabeprazole | CYP2C19 | 1.88 | 3.60 | |
| Risperidone | CYP2D6 | 2.08 | 6.85 | 0.47 |
| Rosuvastatin | OATP1B1 | 0.87 | 1.72 | |
| Sertraline | CYP2C19 | 1.06 | 1.92 | 0.92 |
| Simvastatin | OATP1B1 | 1.45 | 2.62 | |
| Sirolimus | CYP3A4 | 2.49 | ||
| Sumatriptan | OCT1 | 2.15 | ||
| Tacrolimus | CYP3A5 | 1.64 | 2.10 | |
| Tenoxicam | CYP2C9 | 1.02 | 1.59 | |
| Theophylline | CYP1A2 | 1.31 | ||
| Thioridazine | CYP2D6 | 2.59 | 2.98 | 0.81 |
| Timolol | CYP2D6 | 1.67 | 0.66 | |
| Tolbutamide | CYP2C9 | 4.77 | ||
| Tolterodine | CYP2D6 | 1.29 | 4.77 | |
| Torsemide | OATP1B1 | 1.50 | ||
| Tramadol, (+) enantiomer | CYP2D6 | 1.09 | 3.02 | 0.85 |
| Trimipramine | CYP2D6 | 1.99 | 0.8 | |
| Tropisetron | OCT1 | 2.00 | ||
| Tropisetron | CYP2D6 | 2.89 | 0.84 | |
| Venlafaxine | CYP2D6 | 2.31 | 1.41 | 0.63 |
| Voriconazole | CYP2C19 | 1.61 | 2.78 | 0.81 |
| Vorinostat | UGT2B17 | 1.25 | ||
| Vortioxetine | CYP2D6 | 1.32 | ||
| Warfarin | CYP2C9 | 1.34 | 1.94 | |
| Warfarin, (S) enantiomer | CYP2C9 | 1.35 | 3.60 | |
| Zolpidem | CYP3A4 | 2.18 | ||
| Zopiclone | CYP3A4 | 1.68 | ||
| Zopiclone, (S) enantiomer | CYP3A4 | 1.57 | ||
| Zuclopenthixol | CYP2D6 | 2.05 | 0.8 |
Age PK ratios reflect the effect of old age on drug exposure (calculated as AUC or trough concentration in elderly over the same parameter in the young). Genotype PK ratios reflect the increase in systemic drug exposure in poor metabolizers relative to normal metabolizers. The ultrarapid genotype PK ratio is the ratio of drug exposure in ultrarapid metabolizers over normal metabolizers; ultrarapid genotypes are only known for some drug metabolizing enzymes.
No known PK difference but drug induced hyperbilirubinemia dependent on UGT1A1 genotype.
Estimated for 5‐fluorouracil.
Genotype data for active metabolite SN38.
In age studies measurement of codeine, in genotype studies measurement of morphine.
Figure 1.
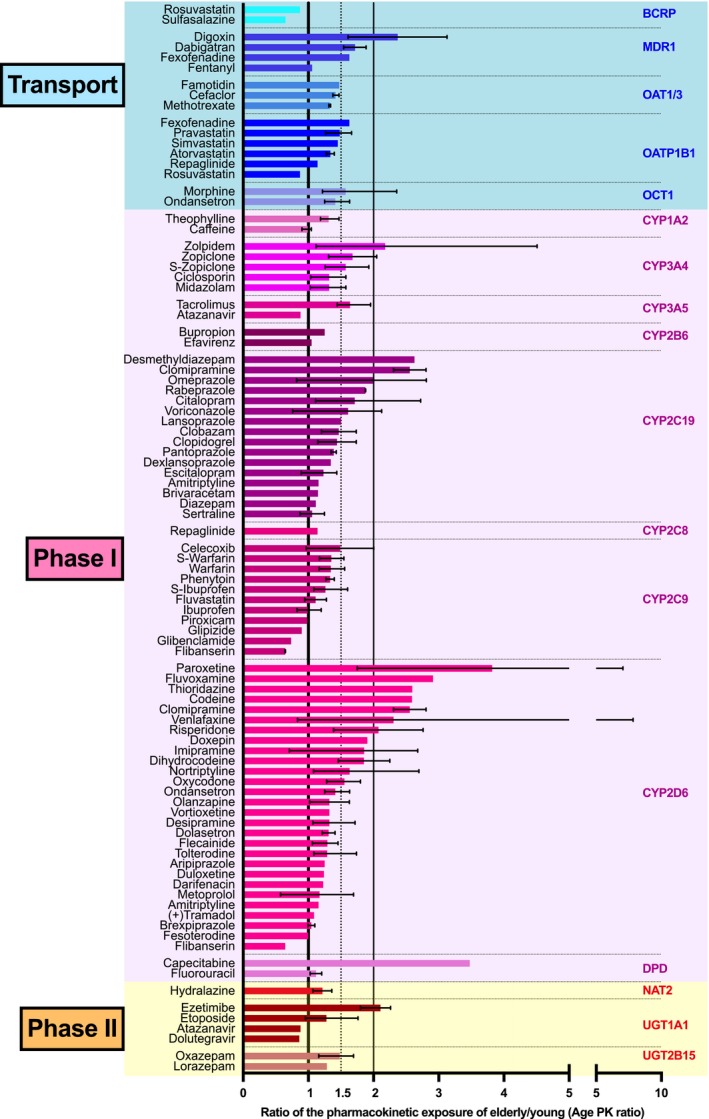
Age‐related increase in the systemic exposure to drugs. A PK ratio of 1 indicates no difference between younger and older people having received the same drug dose. Ratios indicating 1.5‐fold and 2‐fold increase in the systemic exposure in the elderly are marked by the dotted and the right solid line, respectively. If more than one study was available for a drug‐enzyme/transporter combination, we calculated means of ratios and in these instances; error bars indicate minimum and maximum of mean estimates of the multiple studies. The big age effect for capecitabine was not reproduced in other studies. As seen, a significant increase in the systemic exposure in old age was seen particularly for some CYP2C19 and CYP2D6 substrates, while for the majority of drugs only a moderate age‐dependent increase between 1‐ and 1.5‐fold was found.
Only in 11 drugs there was a nominal decrease in age‐related systemic exposure. Most of these differences were statistically not significant and so small that it challenges medical relevance (Figure 1). One extreme was flibanserin, where the exposure in older women was only 0.57‐fold the exposure in the young group. But even in this example, for safety reasons one would not recommend dose adjustments. Even less so since one explanation for that apparently higher clearance may be dependence of total clearance from plasma protein content.6 This phenomenon is also known from several drugs in moderate impairment of liver function. If the age‐related increase in clearance of a drug is indeed due to a higher free drug concentration, then increasing the dose might even be dangerous.
When starting this quantitative review, we hypothesized that age effects differ depending on the specific membrane transporters or metabolizing enzymes dominating the clearance. Gene expression data may indicate differential changes in older age14 and, for instance, cytochrome P450 enzymes are more susceptible to hypoxia compared with phase II enzymes. Furthermore, known age‐dependent changes in endogenous regulators like steroid hormones could lead to differential downregulation or upregulation. However, as illustrated in Figure 1, none of the drug groups ordered according to the presumed clearance‐relevant transporters or enzymes appeared exceptionally sensitive to age‐related alterations. The mean age PK ratio varied between 1.1 for substrates of BCRP and 1.8 for the substrates of MDR1. One‐way analysis of variance (ANOVA), however, showed no significant group differences in the age PK ratio across all enzyme transporters (P = 0.34, mean effects shown in Table 2). Nevertheless, this lack of difference between the effects of age on polymorphic transporters and drug‐metabolizing enzymes may be due to limited statistical power in combination with the heterogeneity of the various substrates of each transporter or enzyme. As an overall conclusion from these data, the average drug dose in older persons might only be reduced to about 60% to 75%, but with drug‐specific differences (Figure 1 , Table 1) and ignoring possible differences in PD and susceptibility to adverse effects.
Table 2.
Summary of age and genotype effects over all drugs classified according to the 16 polymorphic genes in drug transport and biotransformation
| Gene | Gene effect | Age effect | ||||
|---|---|---|---|---|---|---|
| Mean | Min | Max | Mean | Min | Max | |
| BCRP | 2.82 | 2.64 | 3 | 0.95 | 0.65 | 1.32 |
| CYP1A2 | 0.65 | 0.65 | 0.65 | 1.15 | 0.99 | 1.31 |
| CYP2B6 | 2.13 | 1.46 | 3.11 | 1.15 | 1.05 | 1.25 |
| CYP2C19 | 3.05 | 0.73 | 8.83 | 1.54 | 0.92 | 2.63 |
| CYP2C8 | 1.14 | 1.14 | 1.14 | |||
| CYP2C9 | 2.25 | 0.99 | 5.45 | 1.11 | 0.64 | 1.49 |
| CYP2D6 | 2.90 | 0.05 | 10 | 1.62 | 0.64 | 3.82 |
| CYP3A4 | 1.50 | 1.43 | 1.56 | 1.61 | 1.32 | 2.18 |
| CYP3A5 | 2.30 | 2.10 | 2.49 | 1.26 | 0.88 | 1.64 |
| DPD | 10 | 10 | 10 | 2.30 | 1.12 | 3.48 |
| MDR1 | 1.24 | 1.10 | 1.38 | 1.69 | 1.06 | 2.37 |
| NAT2 | 2.37 | 2.25 | 2.49 | 1.38 | 1.22 | 1.55 |
| OAT1, OAT3 | 1.41 | 1.33 | 1.47 | |||
| OATP1B1 | 2.16 | 1.5 | 2.90 | 1.32 | 0.87 | 1.63 |
| OCT1 | 2.21 | 1.70 | 3 | 1.49 | 1.42 | 1.57 |
| TPMT | 9.55 | 2.09 | 17 | |||
| UGT1A1 | 1.39 | 1.03 | 1.77 | 1.29 | 0.86 | 2.11 |
| UGT2B15 | 1.90 | 1.72 | 2.08 | 1.38 | 1.29 | 1.48 |
DESIGN ISSUES AND GENERAL TRENDS IN THE AGING AND PHARMACOGENETIC STUDIES
The age distributions in the studies reviewed here (Figure 2 a) confirmed the impression of investigators in the field that pharmacogenetic clinical research was preferentially performed in younger adults. Overall, the mean age in the pharmacogenetic studies was 34 years and only very few studies had a mean age above 60 years. The modus, the most commonly found mean age, was even 5 years younger in the pharmacogenetic studies than in the younger comparison group in the studies on age effects. Drugs typically used by younger and older people like ibuprofen were often studied in samples with younger age, but many drugs typically used in the elderly such as pravastatin, repaglinide, acenocoumarol, losartan, and metoprolol were also studied in young adults. A few of the pharmacogenetic studies indeed have included subjects with a mean age of 60 and above, including clopidogrel, propafenone, tramadol, or oxycodone apparently being studied in their target population. In studies on age effects, the mean age of the young samples was 34 (range 20–64 years) and the mean age of the elderly sample was 72 (range 56–88 years).
Figure 2.
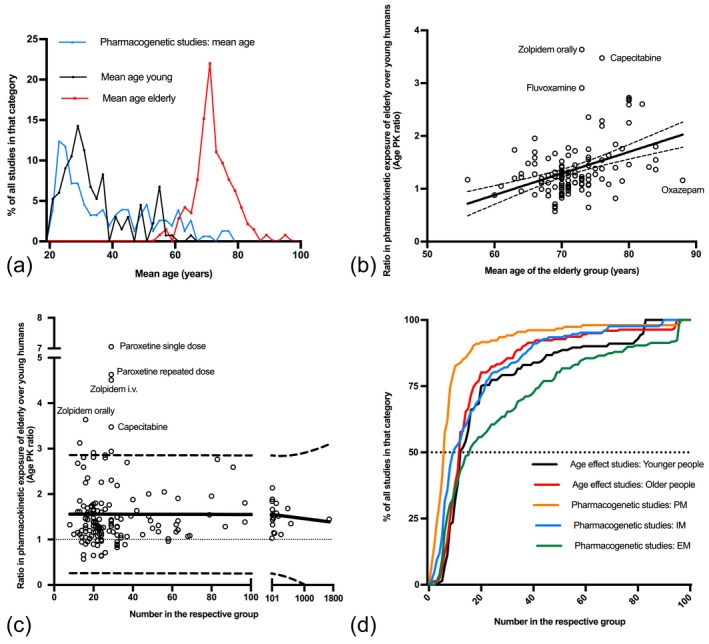
(a) Mean age of the study samples. The black and red lines show the mean age of the young and elderly groups in the studies on age effect on pharmacokinetics. The blue line shows the mean age in the pharmacogenetic studies. Apparently, in the pharmacogenetic studies mean age tended to be even lower than in the young group of the age studies. The modal values were 23, 30, and 70 years in the pharmacogenetic, young age group, and old age samples, respectively. Several of the studies included in the present meta‐analysis did not report the mean age and were thus not included. (b) Age effect on drug exposure (expressed as ratio of exposure in elderly over drug exposure in young) in relation to the mean age in the elderly group. The solid line shows the linear regression line with 90% confidence intervals shown. Of course, optimally this quantitative relationship putting together all drugs for which the data were available (Table S1) should be established for each drug separately. (c) Age‐related increase in drug exposure in the elderly in relationship to the sample size of the respective study (sum of the sample sizes in the young and the elderly group). As illustrated by the linear regression line with 90% confidence intervals, there was no significant dependency, arguing against major publication bias. (d) Cumulative frequency distributions of the samples size in the relevant subgroups in the age studies and pharmacogenetic studies. Black and red lines show the sample size in the young and elderly groups, respectively, for age studies. The orange, blue and green lines show the distributions of samples size in pharmacogenetic studies for the subgroups of the poor (PM), intermediate (IM), and extensive (EM) metabolizers or transporters, respectively. The dotted line crosses the cumulative distribution curves at their medians.
Age‐related differences should be bigger if the age difference between the two groups becomes bigger. Indeed, Figure 2 b illustrates a minor positive correlation between the mean age difference of the respective study and the age‐related increase in drug exposure (expressed as age PK ratio). The respective correlation coefficient was 0.399 (P < 0.0001) and according to the regression equation the age PK ratio was 0.041 × age – 1.57 (P for slope < 0.0001). Correspondingly, the overall mean PK ratios for age 60, 70, and 80 were 0.9, 1.3, and 1.7, respectively. However, there was a large scatter in that regard, and with several drugs age‐related changes per decade of life were much smaller (Figure 2 b).
To check for publication bias or other types of systematic error (e.g., prematurely terminating studies when large effects are seen), we analyzed the correlation between the sample size and the magnitude of the age‐related difference in PK (age PK ratio). As illustrated in Figure 2 c, the sample size of the five studies with the strongest age‐related effect in PK was indeed relatively small (less then 35). However, there were also numerous small studies with small age effects. Statistical analysis of the quantitative data available showed no significant correlation between effect and sample size, arguing against a relevant publication bias (slope = 0.0001; P = 0.81).
The frequency distribution of the sample size of all studies included in this meta‐analysis is depicted in Figure 2 d. The mean sample size for the young group was 28 (min–max: 4–266) and 29 for the group of elderly (min–max: 1–1,425). In general, sample sizes were small, and the relatively large means are due to a few drugs that were studied extensively. Indeed, quite small median sample sizes for the groups of young and elderly are apparent from Figure 2 d. This clearly shows that much of the age‐related data reviewed here is subject to considerable uncertainty. Sample size in the pharmacogenetic studies largely depends on the study design and genotype frequencies. Except for a very few genes with relevant genotype frequencies of around 50%, like NAT2, the functionally relevant genotypes are relatively rare (around 10% or below). Thus, unless extensive efforts for enrichment of these rare genotypes are made, the sample size of extensive metabolizers is significantly larger than those of intermediate and poor metabolizers. This is confirmed by the data as illustrated in Figure 2 d.
IS INTERINDIVIDUAL VARIATION INCREASING WITH AGE?
A widespread opinion is that the between‐subject variation increases with age. We tested this view based on the standard deviation (SD). Similar as for the effects, we used the ratio of the SD in the elderly over the SD in the young (age SD ratio). This analysis (illustrated in Figure 3 ; single study data in Table S2) showed that the SD in the elderly was even smaller than in the young group in as many as 37% of the datasets (n = 145), but in 63% it was as great as or even greater than that in the young group. However, only in 25 (17%) of the datasets did the SD in the older group exceed that in the younger group by more than 2‐fold. These findings did not change in any relevant manner by adjusting the SDs for the age range covered in the young and elderly group, with then 35% percent of the studies showing lower SDs in the elderly. Studies in the same subjects with different doses of the same drug are also depicted and demonstrate that the age SD ratios are rather stable, if not for a certain drug in general than at least for repeated administration of a certain drug to the same sample.
Figure 3.

This figure illustrates our analysis of whether and how much pharmacokinetic variation increases with old age. The age SD ratio was calculated as the ratio of the SD reported in the older participant sample over the younger participant sample from each study available. An SD ratio of 1 indicates the same SD in the young and old sample; SD ratios above 1 indicate higher SD in the elderly compared with the young samples. If more than one study was found, we calculated means of ratios; error bars indicate minimum and maximum. As shown, only for a few drugs there was a significant increase in variation in old age and in several drugs there was even lower variation in pharmacokinetics in old age compared with young age.
In conclusion, variation increased with age in more studies than it decreased. But as demonstrated by all data available in the present context, an increase in PK variation is not a general feature of old age (Figure 3). This is in contrast to the general conception of an increasing variation with increasing age15; however, one has to be aware of a possible selection bias. Groups like older people, frail elderly patients, and patients on multiple medications causing variation by drug–drug interactions are often excluded from PK studies such as those reviewed here.
HOW BIG ARE THE EFFECTS OF GENOMIC VARIATION ON SYSTEMIC EXPOSURE?
We quantified the impact of pharmacokinetically relevant genotypes on systemic exposure (hereafter termed genotype PK ratio) with the ratio of drug exposure in the poor metabolizers over drug exposure in the extensive metabolizers. If there were no poor metabolizers (e.g., because in most traits such complete deficiency is very rare), we used intermediate metabolizers for the calculation instead. Since a complete meta‐analysis of all pharmacogenomic data on the 107 drugs was beyond the scope of this review, we utilized one indicator study for each drug–genotype combination (Figure 4; Table S4; selection criteria for the indicator studies according to the Methods sections). Among these representative indicator studies the highest ratio of carriers of the impaired over carriers of the normal genotypes was 17‐fold and the overall mean effect of the genotypes was 2.78. Concerning individualized drug dosing, this would mean that an appropriate dose reduction in an average carrier of the impaired genotype may be by that factor (e.g., considering the mean ratio of 2.78 this would correspond to 36 mg instead of 100 mg). Of course, in medical practice not the mean but only the factors specific for each gene–drug combination are meaningful and relevant; these clinically relevant factors are given in Table 1 . In some individual studies comparing the frequent normally active genotype with the most functionally impaired genotype, even genotype PK ratios up to 36‐fold were found (Table S4). Thus, apparently the effect of the pharmacogenetics genotypes was significantly larger then the effect of age. However, this conclusion is only valid for drugs in which a monogenetic trait has a significant effect on PK.
Figure 4.
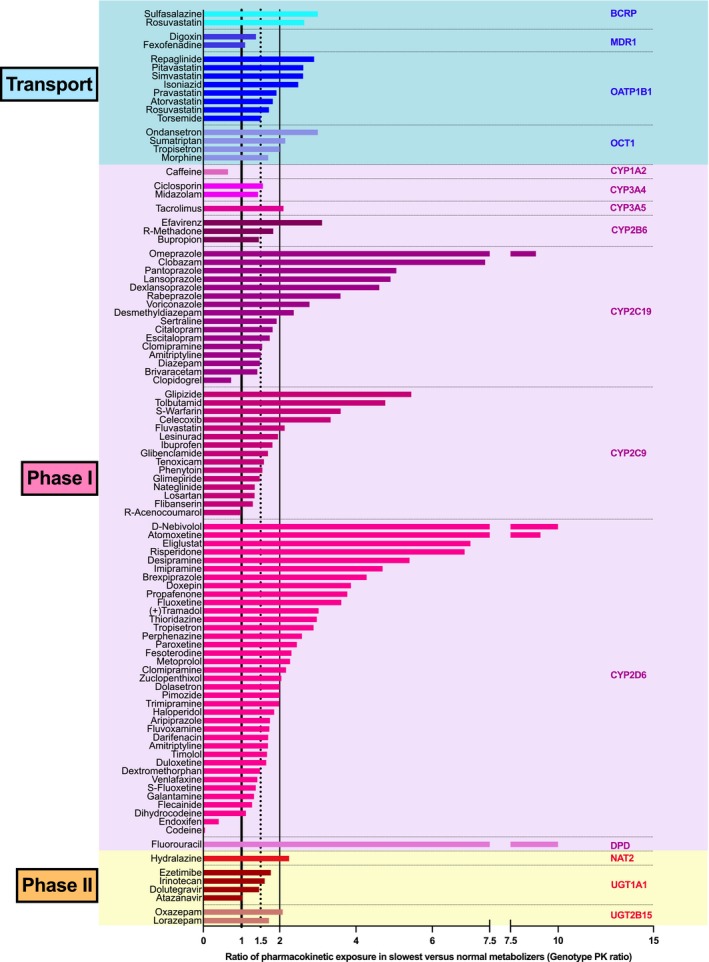
To allow comparison of age effects with genotype effects, we illustrate typical genotype effects. Relative drug exposure in carriers of the lowest activity in drug transport or metabolism is shown relative to the carriers of the genotypes coding for the normal function. A ratio of 1 indicates no difference and a ratio of 2 corresponding to a 2‐fold genotype‐dependent increase of systemic exposure. For the prodrugs clopidogrel (metabolite not specified), tamoxifen (endoxifen), and codeine (morphine) the data for the therapeutically relevant metabolite are given, thereby explaining why the ratio is below unity. As can be seen, the systemic exposure in carriers of the low activity genotypes was twice as high or even higher in about 50% if the drugs included in the present selection.
The genotype effect in each drug is gene‐ and drug‐specific. When grouping all drugs according to the relevant gene, as expected, some of the strongest effects were seen in studies on drugs affected by genes with a null‐function genotype (e.g., CYP2C19, CYP2D6, OCT1); but also in drugs affected by enzymes or transporters with reduced‐function genotypes (e.g., CYP2B6, CYP2C9), quite strong differences were found (Figure 4).
COMPARING AGE AND GENOTYPE EFFECTS
For identification of those drugs deserving particular attention due to both age and genotype, we correlated both factors (Figure 5). There is no reason to expect an overall correlation between the strength of the genotype effect and the strength of the age effects, but that correlation plot is nevertheless interesting, since it defines groups with possible particular relevance of age and/or genotype. Separating the space defined by age and genotype effects (Figure 5) at a factor of 2 in both directions may be reasonable because halving our doubling of drug dose should typically result in relevant differences in efficacy or tolerability. With drugs in which both age and genotype effects are below a factor of 2 (gray quadrant in Figure 5), often no particular consideration of age or genotype may be necessary. The therapeutic index is not considered in this figure and for drugs with a narrow therapeutic index, the four quadrants may have to be defined by factors of 1.5 in the genotype and age directions. Drugs in the red quadrant II are strongly affected by age but only moderately by genotype, whereas in all drugs in the yellow quadrant III, both genotype‐ and age‐related changes apparently may have to be considered. Drugs in the blue quadrant IV are mostly affected by genotype.
Figure 5.
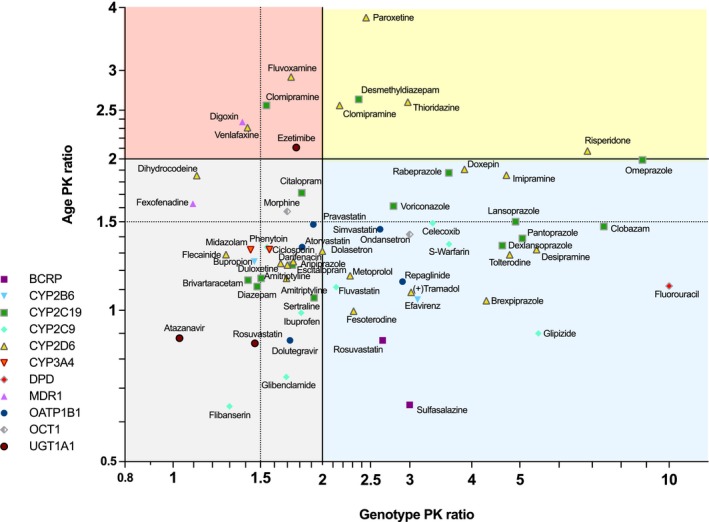
Combined presentation of age and genotype effects. The abscissa represents the age PK ratio representing the ratio of the systemic exposure in elderly over drug exposure in young given the same dose. The ordinate shows the genotype PK ratio representing the ratio of systemic drug exposure in the slowest metabolizer group (poor metabolizers if existing or intermediate metabolizer) over drug exposure in the normal metabolizer group. Four quadrants are highlighted illustrating the particular need to particularly consider age, genotype, or both parameters in therapy with the given drugs. The lines of 2 for age or genotype effect were highlighted because appropriate consideration of the respective factor in dosing would mean administering 50% of the standard dose only. Drugs in which both genotype and age ratios are above 2 may be of particular concern, but drugs above the 1.5 lines may also be of concern, depending on the therapeutic index of the respective drug.
While there is no clear‐cut overall pattern suggesting that some enzymes or transporters are producing large age effects, there appears indeed to be a tendency that drugs predominantly metabolized by CYP2C19 are affected by both, by relevant age effects as well as by relevant genotype effects. Also, some CYP2D6 substrates were strongly affected by both age and genotype effects (Figure 5).
The combined genotype and age effects illustrated in Figure 5 were not derived from the same study. Therefore, it was particularly not possible to identify more than additive (higher genotype effect in old age) or less than additive (lower genotype effect in old age) genotype–age interactions. Only for a very few drugs the statistical interaction of genotype and age has been studied specifically, but may result with some drugs in an unexpectedly high drug exposure in the elderly with genetic impairment in drug metabolism (Figure 6). Despite extensive research, we found PK data on the interaction between age and genotype for 10 drugs only. Probably the first study showing such a more than additive genotype–age interaction was in isoniazide, showing a slightly more than an additive interaction between old age and the slow acetylation phenotype due to the NAT2 polymorphism.16 However, in 1980 NAT2 genotyping was not yet widely used and the antimode between slow and rapid acetylators used in that study may also depend on age. That type of unsolvability of the problem is nowadays solved by genotyping. Still, such combined effects of genotype and age should optimally be studied using the latin square study design in which both the relevant genotype groups and age groups are studied in a factorial fashion. Data of 10 drugs studied in that way were found and are presented in Figure 6. For flecainide and omeprazole, two additional studies of the same groups of investigators are available but showed essentially the same effects. Therefore, we only present one study per drug.17, 18, 19, 20 Interestingly, with omeprazole there is a tendency towards smaller genotype effects in the elderly. Nevertheless, the CYP2C19 genotype remains relevant also in old age, but differences between the genotypes were smaller than in the young.19, 20 For the largest group of drugs (including tacrolimus, escitalopram, flecainide, haloperidol, and celecoxib) there seems to be no significant age‐by‐genotype interaction.17, 18, 21, 22, 23, 24, 25 This means that the genotype was equally important among the younger and the older people. In a third group, including warfarin, metoprolol, and especially venlafaxine and tolterodine, an age‐by‐genotype interaction was seen with an increasing effect size caused by drug exposure by the poor metabolizer status in older people.21, 25, 26, 27 According to independent studies on PK changes in the elderly, there is no age effect for the administration of a single dose of venlafaxine and only a 24% increase of exposure in the elderly in steady state, and according to meta‐analytical data there is only a 40% increase of exposure due to poor metabolizer status in CYP2D6.28, 29 Nonetheless, the interaction study by Waade et al. reported a 15.7‐fold increase of serum concentration per dose between extensive metabolizer patients under the age of 40 and poor metabolizer patients above the age of 65,21 this being about a 9.5 times higher effect than to be expected based on a purely additive model of age and genotype effect. For tolterodine, the FDA review reports a 23.2‐fold increase of AUC in elderly poor metabolizers compared to young extensive metabolizers, whereas studies on age effects from the same review suggest an 8–16% increase of exposure with age and a 4.8‐fold increase resulting from poor metabolizer status. Hence, the interaction exceeds the expected additive effect of genotype and age of 4.6‐fold. Although only a few studies on the genotype–age interaction were found, the data clearly indicates that such studies may reveal unexpected and potentially medically very important combined effects. These effects illustrated in Figure 6 could not be deduced from those studies which have analyzed only one of the factors, either age or genotype, per study.
Figure 6.
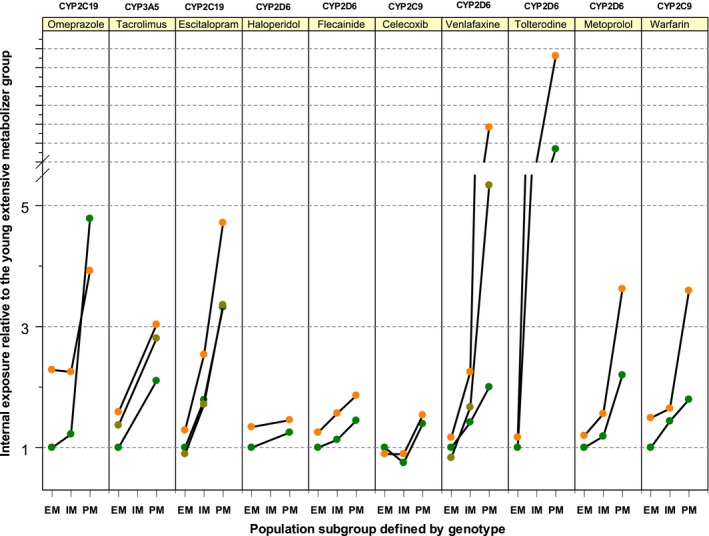
Results from genotype–age interaction studies. The relevant enzyme–drug combinations are given on top. The ordinate illustrates the drug exposure relative to the young extensive metabolizers. The abscissa marks the metabolizer status with EM, IM, and PM corresponding to extensive, intermediate, and poor metabolism. The colored dots stand for separate groups within a study, with orange being the elderly, olive being middle age, and green being the young group. Only data from studies on both factors, age and genotype, could be included in this analysis. The left example (omeprazole) represents an interaction with less than additive effects (still existing but lower impact of genotype in old age), while the examples from tacrolimus to celecoxib represent no interactions, but just additive effects of age and genotype. The right examples (venlafaxine, tolterodine, metoprolol, and warfarin) represent more‐than‐additive interactions between age and genotype, with the genotype becoming even more important in old age.
Discussion
Considering excessive systemic drug exposure due to old age and genotypes
The ratios of systemic drug exposure of older vs. younger people (age PK ratios) and the ratios of systemic drug exposure of carriers of impaired genotypes over carriers with the normal genotypes (genotype‐PK‐ratios) may be directly transformed into therapeutic action. For instance, an age PK ratio of 2 would mean that one would have to administer only 50% of the standard dose in older patients to achieve the same systemic exposure as in the younger population. Or correspondingly a genotype PK ratio of 3 would mean that one would have to administer 33% of the standard dose to carriers of the respective deficient genotype to achieve the same systemic exposure as in the normal metabolizers group. Concerning genotypes, this type of calculation means that the dose recommended in the information to prescribers is the optimal dose for the normal metabolizers. Alternatively, a slightly different but more complex mode of dose adjustment has been described earlier.30 Concerning age, in older patients only dose reductions may have to be considered for drugs with an age PK ratio of 1.5 or above (Figures 1 , 5 ; Table 1), but of course possible alterations in PD and tolerability will have to be considered in addition.
The mean increase of systemic exposure due to genotype was on average twice as high as the increase of systemic exposure due to age (Figures 1 , 4 , 5). One might object that, nonetheless, genotype effects could be dominated by other age‐related changes. But at least as long as we can trust in our main clinical PK endpoint—AUC or trough concentrations—neither the specifically designed genotype–age interaction studies nor the general pattern of PK variation in the elderly nor the subgroup of pharmacogenetic studies performed in older patients support the assumption that genetic variation becomes generally less important or even irrelevant in old age. The presented meta‐analysis of PK variation showed a trend towards higher variation in the elderly, but that was not ubiquitous and not so excessive that it could eliminate the genotype effects in old age.
As illustrated in Figure 2 a, most pharmacogenetic studies were performed in younger persons, but when restricting our analysis on the genotype effects found in study samples with an average age of 50 and older (mean 60.4 with minimum mean age of 50.9 and maximum mean age of 77.6), the mean genotype PK ratio was 3.99 with a maximum of 36.38. Interestingly, this genotype effect measured in people above a mean of 50 years even exceeded the overall effect of genotype among all studies. This analysis was based on studies on 24 different drugs only, because in many drugs the genotype effect has been studied in younger samples. These data strongly support a medically relevant role of the PK genotypes in the elderly; however, one cannot finally conclude a general increase of genotype effect with age since there are exceptions to that rule (e.g., omeprazole) and with most drugs the appropriate genotype–age interaction studies have not yet been performed.
Both types of interactions (overproportionally increasing impact of genotype with old age and overproportionally decreasing impact) were found. Thus, both genotype and age may affect the medically relevant systemic exposure to the drug in an unexpected fashion. This indicates that in the future the effects of age and genotype should more often be studied jointly. For the studies on venlafaxine and tolterodine, the systemic exposure was 10‐times, respectively 5‐times, higher than to be expected from studies separately dealing with age and genotype and assuming an additive effect.10, 21 This might lead to serious adverse drug reactions. For venlafaxine these risks especially include cardiovascular adverse effects, but less commonly also acute angle closure glaucoma31 or even the serotonergic syndrome. With tolterodine the risk corresponding to the superadditive interactions (Figure 6) may include all the anticholinergic effects known to be bad for older people and even the anticholinergic syndrome or hyponatremia caused by the syndrome of inappropriate antidiuretic hormone secretion (SIADH).32 In these drugs, like venlafaxine or tolterodine, the genotype may be particularly important for the elderly.
How to explain excessive systemic exposure in elderly poor metabolizers
As briefly outlined in the introductory paragraphs of this review, several mechanisms relevant in healthy older people as well as disease‐related alterations may contribute to differences in PK between younger and elderly individuals. Whereas these physiological changes in the elderly may explain some age‐related differences in PK, because of the complexity of the PK processes they may fall short in predicting an interaction effect between age and genotype. But of course, this is an interesting topic for future physiologically based PK/PD modeling. There are several possible explanations for the interesting age‐by‐genotype interactions (more than additive or less than additive combined effects). A simple analogy for the more than additive effects may be the picture of the rope that always breaks at the thinnest point. Age‐dependent physiological and morphological changes may lead to a reduced clearance based on another (possibly nonpolymorphic) pathway with the consequence of a polymorphic enzyme becoming even more involved. This hypothesis needs further experimental confirmation.
The second scenario is the opposite: physiological and morphological changes lead to an attenuated role of a polymorphic enzyme. This was impressively shown with two studies on omeprazole where the genotype‐related difference in old age was smaller than in young age. The mechanisms behind this may also need further scrutiny, but a simple and plausible explanation is that the deficient metabolizer having zero CYP2C19 activity cannot get less than zero CYP2C19 activity, whereas the activity in the extensive metabolizer can decline. In this respect, it may also be medically relevant what will happen with the genetically determined ultrarapid metabolic activity of CYP2D6 and CYP2C19 (see Figure S1) in older people. However, we did not find clinical PK data on that.
All such scenarios could also be caused by epigenetic mechanisms; for instance, an age‐dependent increase in DNA methylation.33 Epigenetic effects in poor and extensive metabolizers may even change differentially with age, thereby directly causing an interaction. Unfortunately, currently there are only scarce empirical data on that hypothesis.
Defining old age and frailty
New drug approval requires studies in the geriatric population. But who is an older person and what defines medically relevant old age? Although there are some recommendations and guidelines on that, the wide range of mean group age in the studies reviewed here (Figure 2 a) may indicate a need for further standardization. Chronological age is not the best criterion here, and additional parameters reflecting biological age should be recorded. One important aspect of very old age and frailty is unspecific systemic inflammation, partially due to dysregulation of the immune system. Systemic inflammation may result in downregulation of cytochrome P450 enzymes, increasing the risk of individual overdose in older people,34 and similar processes may be relevant in drug membrane transport. The wide scatter of age effects (Figures 1 , 3 ; Table S1) may in part be due to differences in the biological age of the samples. A more comprehensive definition using markers of biological age could lead to improved comparability between studies. Such markers of old age and frailty may include liver volume,5, 35, 36 renal clearance, and clearance of probe drugs,37, 38, 39, 40 as well as markers of systemic inflammation like C‐reactive protein, interleukin 6, and p16‐INK4a.41 DNA methylation patterns may correlate surprisingly closely with age,42 but there are currently no data on the predictive power of these age‐specific methylation patterns for age‐related changes in PK. Also, clinical frailty markers have been discussed with regard to drug exposure,41 renal clearance,43 and minimizing inappropriate medication.44, 45 Although currently there appears to be no validated and consensual aging biomarker, use of such markers in drug research and further research on that should be encouraged.
Limitations and Future Perspectives
The data summarized here may be medically relevant for the largest proportion of people older than 65 years of age. But the data may not optimally reflect the dose requirements of the oldest subgroup of older people and particularly for frail older people, because these were typically not included in the studies reviewed here. Selection bias is also a problem in this context. For example, in anticancer drugs only those older patients expected to be able to tolerate the drugs will be included. Although it may be reasonable to assume that the trends found here comparing younger and older people will intensify in very old age and in frailty, more clinical research in these subgroups is needed.
Here we focused on the changes in PK, since the right drug concentration at the target site is undeniably an essential prerequisite for any beneficial effects. But PK is not the only factor. For instance, we found no or only minor age‐related changes in the PK of most nonsteroidal antiinflammatory drugs in the elderly (Figure 1 , Table 1), even though there is at least a 2‐fold increase in risk from upper intestinal bleeding from these drugs in old age.46 Psychotropic drugs are known to be associated with a significantly higher risk in old age,47 but older people have a much increased systemic exposure only with a few of the psychotropic drugs (Figure 1).
Many studies on PK in older people were relatively small, with a median sample size below 10 in more than 50% of the studies (Figure 2 d); the mean age in the studies varied over a wide range and indicators of biological age have not been recorded systematically in many studies. Therefore, the age‐related changes summarized in this quantitative review cannot be very precise for many of the drugs. However, at least it is reassuring that data from replication studies were mostly in the same direction and the same range, if there were replication studies (Figures 1 , 3).
Many small studies on age effects did not report quantitative data, particularly if no significant age‐related differences were found. According to our inclusion criteria, and because we could not simply set the age effect as zero in this situation, the age effect illustrated and quantified in the present review may in reality even be slightly smaller with several drugs.
Conclusion
In drugs with PK significantly modulated by genetic polymorphisms, old age caused on average a moderate about 1.5‐fold increase in systemic exposure, but in a few drugs systemic exposure in the elderly was 2‐fold or more. In dosing of these drugs in the elderly particular attention should be paid to the adverse effects. On average, the genetic polymorphisms caused a larger effect on systemic exposure than age, but both old age and genetic polymorphisms should be considered in dosing of most of the drugs reviewed here.
Classification of the drugs according to the enzymes and transporters dominating the respective PK revealed that none of the transporters or drug‐metabolizing enzymes were associated with particularly strong age‐related changes. The combined effects of age and metabolic genotype have been specifically analyzed only in a few clinical studies. But some surprising supraadditive PK effects of age and genotype were found. This indicates that such age–genotype interactions may be a problem underestimated thus far, and supports future testing of such interactions using factorial designs or appropriately powered population PK studies. In the future, especially for those drugs with large genotype effects, large age effects and/or with a narrow therapeutic range, the combined effect of age and genotype should be specifically analyzed and considered in therapy. The age effects found in the different drugs and in the different studies varied over a wide range. This confirms that an age‐related increase in systemic exposure is drug‐specific, but due to limitations of several studies it was not always clear whether these are drug‐related differences or differences due to the different selection criteria and other design issues of the studies. The review should stimulate more extensive use of pharmacogenetic diagnostics in older patients. In research it should stimulate further efforts towards even more standardization of the clinical studies, further research on, and utilization of, biomarkers of biological age and further studies on the medically relevant environmental, physiological, and epigenetic factors modulating drug disposition in older people.
FUNDING
Both authors did this work as members of the Georg August University Göttingen, Germany. No external funding was received for this review.
Conflict of Interest
The authors declare no competing interests for this work.
Supporting information
References
- 1. Klotz, U. Pharmacokinetics and drug metabolism in the elderly. Drug Metab. Rev. 41, 67–76 (2009). [DOI] [PubMed] [Google Scholar]
- 2. McLean, A.J. & Le Couteur, D.G. Aging biology and geriatric clinical pharmacology. Pharmacol. Rev. 56, 163–184 (2004). [DOI] [PubMed] [Google Scholar]
- 3. Schlender, J.F. et al Development of a whole‐body physiologically based pharmacokinetic approach to assess the pharmacokinetics of drugs in elderly individuals. Clin. Pharmacokinet. 55, 1573–1589 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang, X. et al Systematic genetic and genomic analysis of cytochrome P450 enzyme activities in human liver. Genome Res. 20, 1020–1036 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wynne, H.A. , Cope, L.H. , Mutch, E. , Rawlins, M.D. , Woodhouse, K.W. & James, O.F. The effect of age upon liver volume and apparent liver blood flow in healthy man. Hepatology 9, 297–301 (1989). [DOI] [PubMed] [Google Scholar]
- 6. Butler, J.M. & Begg, E.J. Free drug metabolic clearance in elderly people. Clin. Pharmacokinet. 47, 297–321 (2008). [DOI] [PubMed] [Google Scholar]
- 7. Le Couteur, D.G. et al Old age and the hepatic sinusoid. Anat. Record 291, 672–683 (2008). [DOI] [PubMed] [Google Scholar]
- 8. Kirchheiner, J. et al Pharmacogenetics of antidepressants and antipsychotics: the contribution of allelic variations to the phenotype of drug response. Mol. Psychiatry 9, 442–473 (2004). [DOI] [PubMed] [Google Scholar]
- 9. McLachlan, A.J. , Hilmer, S.N. & Le Couteur, D.G. Variability in response to medicines in older people: phenotypic and genotypic factors. Clin. Pharmacol. Ther. 85, 431–433 (2009). [DOI] [PubMed] [Google Scholar]
- 10. US Food and Drug Administration . Table of pharmacogenomic biomarkers in drug labeling. 2017; Available from: <https://www.fda.gov/Drugs/ScienceResearch/ucm572698.htm>
- 11. Genes‐Drugs . 2017; Available from: <https://cpicpgx.org/genes-drugs/>
- 12. List Dosing Guidelines . 2017; Available from: <https://www.pharmgkb.org/view/dosing-guidelines.do>
- 13. Canadian Pharmacogenomics Network for Drug Safety . 2017; Available from: <http://cpnds.ubc.ca/>
- 14. Glass, D. et al Gene expression changes with age in skin, adipose tissue, blood and brain. Genome Biol. 14, R75 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schmucker, D.L. Liver function and phase I drug metabolism in the elderly: a paradox. Drugs Aging 18, 837–851 (2001). [DOI] [PubMed] [Google Scholar]
- 16. Advenier, C. , Saint‐Aubin, A. , Gobert, C. , Houin, G. , Albengres, E. & Tillement, J.P. Pharmacokinetics of isoniazid in the elderly. Br. J. Clin. Pharmacol. 10, 167–169 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Doki, K. , Homma, M. , Kuga, K. , Aonuma, K. & Kohda, Y. Effects of CYP2D6 genotypes on age‐related change of flecainide metabolism: involvement of CYP1A2‐mediated metabolism. Br. J. Clin. Pharmacol. 68, 89–96 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Doki, K. , Homma, M. , Kuga, K. , Aonuma, K. & Kohda, Y. CYP2D6 genotype affects age‐related decline in flecainide clearance: a population pharmacokinetic analysis. Pharmacogenet. Genomics 22, 777–783 (2012). [DOI] [PubMed] [Google Scholar]
- 19. Ishizawa, Y. , Yasui‐Furukori, N. , Takahata, T. , Sasaki, M. & Tateishi, T. The effect of aging on the relationship between the cytochrome P450 2C19 genotype and omeprazole pharmacokinetics. Clin. Pharmacokinet. 44, 1179–1189 (2005). [DOI] [PubMed] [Google Scholar]
- 20. Niioka, T. , Uno, T. , Sugimoto, K. , Sugawara, K. , Hayakari, M. & Tateishi, T. Estimation of CYP2C19 activity by the omeprazole hydroxylation index at a single point in time after intravenous and oral administration. Eur. J. Clin. Pharmacol. 63, 1031–1038 (2007). [DOI] [PubMed] [Google Scholar]
- 21. Waade, R.B. , Hermann, M. , Moe, H.L. & Molden, E. Impact of age on serum concentrations of venlafaxine and escitalopram in different CYP2D6 and CYP2C19 genotype subgroups. Eur. J. Clin. Pharmacol. 70, 933–940 (2014). [DOI] [PubMed] [Google Scholar]
- 22. Jacobson, P.A. et al Lower calcineurin inhibitor doses in older compared to younger kidney transplant recipients yield similar troughs. Am. J. Transplant. 12, 3326–3336 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ohara, K. , Tanabu, S. , Ishibashi, K. , Ikemoto, K. , Yoshida, K. & Shibuya, H. Effects of age and the CYP2D6*10 allele on the plasma haloperidol concentration/dose ratio. Prog. Neuropsychopharmacol. Biol. Psychiatry 27, 347–350 (2003). [DOI] [PubMed] [Google Scholar]
- 24. Brenner, S.S. et al Influence of age and cytochrome P450 2C9 genotype on the steady‐state disposition of diclofenac and celecoxib. Clin. Pharmacokinet. 42, 283–292 (2003). [DOI] [PubMed] [Google Scholar]
- 25. Bae, J.W. , Choi, C.I. , Lee, J.H. , Jang, C.G. , Chung, M.W. & Lee, S.Y. Effects of UDP‐glucuronosyltransferase polymorphisms on the pharmacokinetics of ezetimibe in healthy subjects. Eur. J. Clin. Pharmacol. 67, 39–45 (2011). [DOI] [PubMed] [Google Scholar]
- 26. Taguchi, M. et al Effect of CYP2D6*10 on pharmacokinetic variability of routinely administered metoprolol in middle‐aged and elderly Japanese patients. Eur. J. Clin. Pharmacol. 59, 385–388 (2003). [DOI] [PubMed] [Google Scholar]
- 27. Loebstein, R. et al Interindividual variability in sensitivity to warfarin—nature or nurture? Clin. Pharmacol. Ther. 70, 159–164 (2001). [DOI] [PubMed] [Google Scholar]
- 28. Klamerus, K.J. , Parker, V.D. , Rudolph, R.L. , Derivan, A.T. & Chiang, S.T. Effects of age and gender on venlafaxine and O‐desmethylvenlafaxine pharmacokinetics. Pharmacotherapy 16, 915–923 (1996). [PubMed] [Google Scholar]
- 29. Stingl, J.C. , Brockmöller, J. & Viviani, R. Genetic variability of drug‐metabolizing enzymes: the dual impact on psychiatric therapy and regulation of brain function. Mol. Psychiatry 18, 273–287 (2013). [DOI] [PubMed] [Google Scholar]
- 30. Kirchheiner, J. et al CYP2D6 and CYP2C19 genotype‐based dose recommendations for antidepressants: a first step towards subpopulation‐specific dosages. Acta Psychiatr. Scand. 104, 173–192 (2001). [DOI] [PubMed] [Google Scholar]
- 31. Ezra, D.G. , Storoni, M. & Whitefield, L.A. Simultaneous bilateral acute angle closure glaucoma following venlafaxine treatment. Eye 20, 128–129 (2006). [DOI] [PubMed] [Google Scholar]
- 32. Bryan, M.K. , Nguyen, M.T. & Hilas, O. Syndrome of inappropriate antidiuretic hormone associated with tolterodine therapy. Consult. Pharm. 25, 320–322 (2010). [DOI] [PubMed] [Google Scholar]
- 33. Seripa, D. , Panza, F. , Daragjati, J. , Paroni, G. & Pilotto, A. Measuring pharmacogenetics in special groups: geriatrics. Expert Opin. Drug Metab. Toxicol. 11, 1073–1088 (2015). [DOI] [PubMed] [Google Scholar]
- 34. Morgan, E.T. Impact of infectious and inflammatory disease on cytochrome P450‐mediated drug metabolism and pharmacokinetics. Clin. Pharmacol. Ther. 85, 434–438 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wynne, H.A. , Cope, L.H. , Herd, B. , Rawlins, M.D. , James, O.F. & Woodhouse, K.W. The association of age and frailty with paracetamol conjugation in man. Age Ageing 19, 419–424 (1990). [DOI] [PubMed] [Google Scholar]
- 36. Wynne, H.A. , Yelland, C. , Cope, L.H. , Boddy, A. , Woodhouse, K.W. & Bateman, D.N. The association of age and frailty with the pharmacokinetics and pharmacodynamics of metoclopramide. Age Ageing 22, 354–359 (1993). [DOI] [PubMed] [Google Scholar]
- 37. Opdam, F.L. et al CYP2D6 metabolism in frail elderly compared to non‐frail elderly: a pilot feasibility study. Drugs Aging 32, 1019–1027 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schwartz, J.B. Erythromycin breath test results in elderly, very elderly, and frail elderly persons. Clin. Pharmacol. Ther. 79, 440–448 (2006). [DOI] [PubMed] [Google Scholar]
- 39. Zeeh, J. et al Influence of age, frailty and liver function on the pharmacokinetics of brofaromine. Eur. J. Clin. Pharmacol. 49, 387–391 (1996). [DOI] [PubMed] [Google Scholar]
- 40. Hilmer, S.N. et al Gentamicin pharmacokinetics in old age and frailty. Br. J. Clin. Pharmacol. 71, 224–231 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dumond, J.B. et al Population pharmacokinetics modeling of unbound efavirenz, atazanavir, and ritonavir in HIV‐infected subjects with aging biomarkers. CPT Pharmacometrics Syst. Pharmacol. 6, 128–135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 14, R115 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Johnston, C. , Hilmer, S.N. , McLachlan, A.J. , Matthews, S.T. , Carroll, P.R. & Kirkpatrick, C.M. The impact of frailty on pharmacokinetics in older people: using gentamicin population pharmacokinetic modeling to investigate changes in renal drug clearance by glomerular filtration. Eur. J. Clin. Pharmacol. 70, 549–555 (2014). [DOI] [PubMed] [Google Scholar]
- 44. Fried, L.P. et al Frailty in older adults: evidence for a phenotype. J. Gerontol. A Biol. Sci. Med. Sci. 56, M146–156 (2001). [DOI] [PubMed] [Google Scholar]
- 45. Poudel, A. , Hubbard, R.E. , Nissen, L. & Mitchell, C. Frailty: A key indicator to minimize inappropriate medication in older people. QJM 106, 969–975 (2013). [DOI] [PubMed] [Google Scholar]
- 46. Bateman, D.N. & Kennedy, J.G. Non‐steroidal anti‐inflammatory drugs and elderly patients. BMJ 310, 817–818 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Just, K.S. , Schneider, K.L. , Schurig, M. , Stingl, J.C. & Brockmöller, J. Falls: the adverse drug reaction of the elderly and the impact of pharmacogenetics. Pharmacogenomics 18, 1281–1297 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


