Summary
During the last centuries, humans have transformed global ecosystems. With their temporal dimension, herbaria provide the otherwise scarce long‐term data crucial for tracking ecological and evolutionary changes over this period of intense global change. The sheer size of herbaria, together with their increasing digitization and the possibility of sequencing DNA from the preserved plant material, makes them invaluable resources for understanding ecological and evolutionary species’ responses to global environmental change. Following the chronology of global change, we highlight how herbaria can inform about long‐term effects on plants of at least four of the main drivers of global change: pollution, habitat change, climate change and invasive species. We summarize how herbarium specimens so far have been used in global change research, discuss future opportunities and challenges posed by the nature of these data, and advocate for an intensified use of these ‘windows into the past’ for global change research and beyond.
Keywords: ancient DNA, biological invasions, climate change, habitat change, herbarium, phenology, pollution
Introduction
Global environmental change is one of the major challenges of the 20th and 21st centuries. It has been evident since the age of industrialization in the late 18th century – sometimes also referred to as the advent of the anthropocene – and has continuously gained momentum (Fig. 1a; Steffen et al., 2011; Hamilton, 2016). Biologists study global change for its broad ecological impact, and its negative effects on biodiversity. Also, as it represents an unplanned, long‐term and large‐scale experiment, studying global change can promote understanding of fundamental processes such as rapid adaptation. Experimental approaches to study these topics are usually locally focused, and limited to a duration of a few decades (Leuzinger et al., 2011). Although observational methods are often more large‐scale and long‐term, they are with few exceptions still restricted to a time frame of 50–80 yr (Fig. 1a; Fitter & Fitter, 2002; Thomas et al., 2004). To understand both the extent of global change as a long‐term process, and its full ecological and evolutionary impact, global data that go back to the onset of industrialization are crucial.
Figure 1.
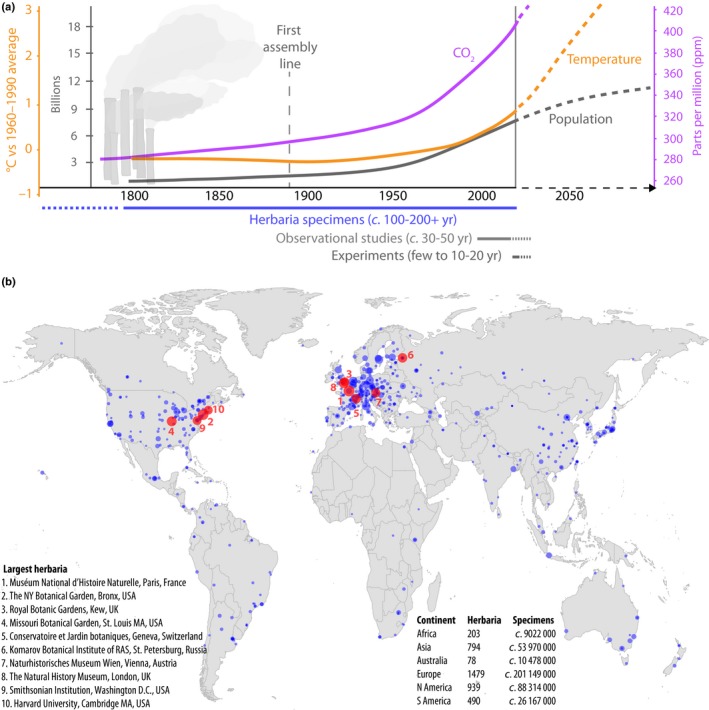
Herbaria as global change witnesses. (a) Timeline of global change, with lines tracking changes in world population, air temperature and atmospheric CO 2 during the last c. 200 years. Dashed line ends indicate future projections. Bars below plot indicate the typical temporal extent of herbarium samples vs observational studies and experiments. (Population growth: United Nations, Department of Economic and Social Affairs, Population Division (2017); World Population Prospects: The 2017 Revision. http://esa.un.org/unpd/wpp/; temperature: representative concentration pathway 8.5, Intergovernmental Panel on Climate Change, www.ipcc.ch; (Marcott et al., 2013); CO 2: (Neftel et al., 1994)). (b) Map with global distribution of herbaria (for visual clarity displaying only herbaria of > 100 000 specimens), names of the largest 10 herbaria, and number of herbaria and herbarium specimens curated per continent (reflecting places of storage of specimens, not their origins; Herbarium data from Index Herbariorum, http://sweetgum.nybg.org/science/api/v1/institutions/. Accessed in April 2018).
In this context, natural history collections are an underused treasure of temporally and geographically broad samples that we have just begun to dust off (Holmes et al., 2016). Especially rich is the botany section of this vault: plants collected, pressed and preserved, in most cases together with meta‐information on species, collection site, date and collector (Fig. 2): In terms of extent, there are > 350 million specimens in almost 3000 herbaria world‐wide (Fig. 1b; Thiers, 2017; http://sweetgum.nybg.org/science/ih/), sampled from the 16th century up to today (Sprague & Nelmes, 1931), and the collections’ potential uses range from classical taxonomy and systematics, to archaeobotany, archaeoecology and climate change research (Funk, 2003). Because plants are sessile, they are particularly exposed to environmental change. The time courses of many of their responses to environmental change are preserved in herbarium specimens, which therefore provide unique spatiotemporal data for studying global change (Primack & Miller‐Rushing, 2009; Lavoie, 2013; Vellend et al., 2013; Meineke et al., 2018).
Figure 2.
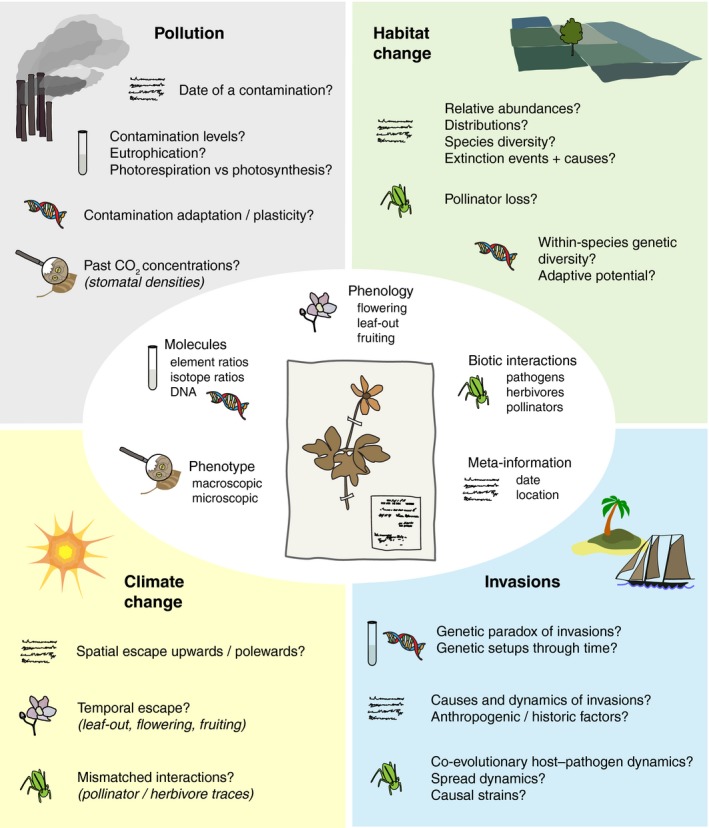
Diversity of herbarium data and their applications. Herbarium sheet in the centre surrounded by types of data that can be obtained from a specimen, with the questions that these data can help to answer around, ordered by respective global change driver. Symbols indicate the type of data used to address each question.
Recent studies have emphasized the scientific value of herbaria for a broad range of global change‐related topics (Fig. 2; e.g. Zschau et al., 2003; Miller‐Rushing et al., 2006; Feeley & Silman, 2011; Willis et al., 2017). Dense time‐series of herbarium specimens even permit studying long‐term processes such as recent invasions and their genetic population history (Exposito‐Alonso et al., 2018a).
Even though herbaria were used as early as in the 1960s to study global change (e.g. Ruhling & Tyler, 1968, 1969), and are in the process of being made available online via digitization (> 46 700 000 specimens in the Integrated Digitized Biocollections portal alone; as of 18 July 2018 https://www.idigbio.org/portal/ (search terms: type of record – PreservedSpecimen, kingdom – Plantae)), the community has not fully adopted herbaria as valuable ‘time machines’ to the past (Lavoie, 2013; Meineke et al., 2018). Especially with the advent of high‐throughput methods and recent technical developments in image analysis, the value of these collections is now more apparent than ever (Munson & Long, 2017).
Simultaneously, next generation sequencing (NGS) techniques now allow for in‐depth genetic analysis of century‐old specimens up to whole genome sequencing of plants and even of their equally preserved pathogens (e.g. Martin et al., 2013; Yoshida et al., 2013; Durvasula et al., 2017; Exposito‐Alonso et al., 2018a). This extends the spectrum of available long‐term data far beyond morphology or phenology. For instance, dense sampling of such full genetic information across time – and geography – enables population genetics studies, to follow speciation processes through time, or to quantify changes in genetic diversity in historical contexts. Working with these small samples of degraded DNA – so‐called ancient DNA (aDNA) – retrieved from historic collections is technically challenging and has recently boomed in the animal field (e.g. Shapiro & Hofreiter, 2014; Orlando et al., 2015; Marciniak & Perry, 2017), yet in the plant field it is still rarely used (Gutaker & Burbano, 2017).
Here, we present an overview of the different types of herbaria analyses possible in global change research (Fig. 2). Following a timeline from industrialization onwards, we divide herbarium‐related approaches into four main areas related to four main drivers of global change: industrialization causing increased pollution, which coincides with increasing loss of habitat and changes in land use as well as climate change, and finally global trade and transport resulting in an increasing number of invasive species world‐wide. In addition, in excursions dedicated to molecular methods (Box 1), collection biases (Box 2) and the digitization challenge (Box 3), we provide insight into three key methodological issues that herbaria research is currently dealing with, and hopefully inspire with ideas for extended utilization of botanical collections. Our aim is to advocate broader use of herbaria as ‘witnesses’ of global change. We believe that they have the potential to fast‐forward our understanding of the impacts of this unplanned biological experiment, to substantiate our predictions of its long‐term outcomes, and to inform conservation measures.
Box 1. Molecular analyses and degradation.
The age of herbarium specimens is both their strength and their weakness, as aging is a corrosive process. For most chemicals, the extent, rate and end‐results of this process are not defined in herbarium samples. Still, it is clear that age, but also preservation practices or storage conditions can alter tissue chemical contents. This is evident, for example, when N concentrations measured in stored tissues diverge from the results of previous methods and studies – in this case likely due to post‐collection contamination (Nielsen et al., 2017). Hence, in‐depth analyses of correlations between the age and chemical compound quantities in old samples are necessary in order to make claims about historical absolute abundance values (Nielsen et al., 2017).
For DNA from historical samples – aDNA – age‐related degradation dynamics are fairly well‐characterized (Allentoft et al., 2012; Weiß et al., 2016). Due to chemical modifications, DNA in dead tissue gets increasingly fragmented over time (Fig. B1a), and particularly in fragment ends, aDNA‐characteristic deamination drives nucleotide‐substitutions of cytosine with thymine ((Weiß et al., 2016); Fig. B1b). This per se does not lessen the potential of aDNA‐studies (Gutaker & Burbano, 2017): specialized protocols even allow extraction of ultra‐short fragments of < 50 bp (Gutaker et al., 2017), and the correlation of nucleotide misincorporations with time enables its use as authenticity criterion of ancient DNA (Sawyer et al., 2012; Weiß et al., 2016). Still, these particular characteristics call for categorical rules for herbarium genetics to minimize contamination risks, verify authenticity and maximize the information gained from precious old plants: samples have to be processed in clean room facilities to avoid contaminations with fresh DNA, and sequenced to a certain depth to yield useful information. Pure PCR analyses on the contrary are inappropriate for aDNA studies, as they do not allow the necessary authenticity verification and, due to the fragmentation of aDNA, are unlikely to yield consistent results.
Such quality requirements are particularly important due to the limitation of available material. Unlike traditional approaches that rely on metadata or morphology of historical samples, molecular analyses require tissue probes and hence destructive sampling of specimens. Therefore, it is the duty of any molecular herbarium scientist to optimize their methods, minimize the amount of sample needed, and employ state‐of‐the‐art analyses to retrieve maximum information from their samples. In the same vein, molecular herbarium scientists and curators should aim to maximize the detail of meta‐information that can be gathered from samples. Knowledge, for example, about temporary field collection in alcohol, or post‐collection specimen treatments with heavy metals (as insecticides or fungicides) is indispensable to assess the suitability of specimens for molecular approaches. Furthermore, both curators and researchers need to assess specimen‐label and specimen‐sample pairs for their correctness, and remain cautious particularly regarding the interpretation of trends in (molecular) data observed only in few or single samples.
Figure B1.
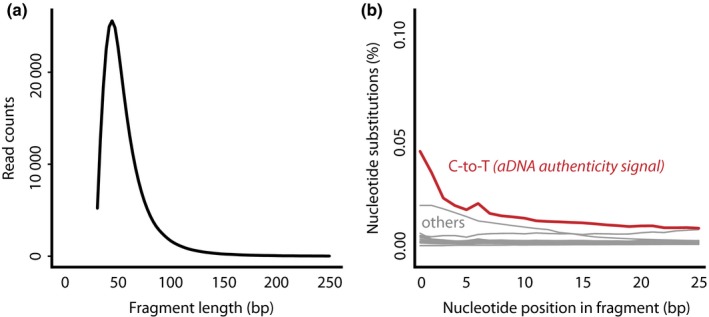
Typical molecular characteristics of herbarium DNA. (a) Fragment size distribution and (b) damage pattern found in ancient DNA (sample data from Weiß et al. (2016), publicly available at ENA ID ERR964451).
Box 2. Collection biases.
Imbalanced sampling is a well‐acknowledged issue for the use of herbaria, for example, to map species distributions or assess diversity (e.g. Meyer et al., 2016; Daru et al., 2018). Temporal biases are caused by intense collection periods, and seasonal preferences (Holmes et al., 2016). Also, collections often concentrate on easily accessible or much‐frequented sites (geographic bias; e.g. Sofaer & Jarnevich, 2017), and on common or particularly interesting species which – depending on the collectors – can change over time (taxonomic bias; e.g. Feeley, 2012). When working with herbarium data, it is necessary to explicitly test for these biases, for example to avoid a few dominant species generating trends in a dataset (Jácome et al., 2007). Depending on the type of question or analysis, biases may need to be corrected for by different means: normalizing collection efforts with different types of reference sets (e.g. Hedenäs et al., 2002; Law & Salick, 2005; Case et al., 2007), measuring invader distributions in relation to native species (Delisle et al., 2003), or verifying trends with additional, nonherbarium datasets (e.g. Lienert et al., 2002; Kouwenberg et al., 2003; or even those from citizen science, Spellman & Mulder, 2016). In particular when models are based on historical records, comparisons with modern data can support extrapolations or generalizations, but only if biases have been dealt with: models, for example, in the context of invader dynamics and spread, have to take species persistence into account, because historic occurrence does not equal contemporary presence and may cause overestimation of plants’ distribution and abundance (Pergl et al., 2012). This is particularly the case for species targeted by eradication measures, such as the human health hazard Heracleum mantegazzianum, where herbarium specimens can indicate suitable habitats, but not current occurrence or general invasion dynamics (Pergl et al., 2012). Furthermore, there are often no data on early invasion stages, because herbarium records indicate only the presence of a species, whereas its absence is not reliably documented by a lack of records. Conclusions based on modeling and statistical analysis, particularly of early invasion stages, should hence be used as indications rather than be over‐relied upon (Hyndman et al., 2015). Finally, the currently rising bias of low collection effort is a well‐known problem for tropical areas (Feeley & Silman, 2011), yet is threatening to become global, via overall declining collections (Prather et al., 2004). Although this particularly jeopardizes studies of new or recent invasions (Lavoie et al., 2012), it strongly affects all herbarium‐based research.
Box 3. Digitization challenge.
Large‐scale digitization is crucial to make biodiversity data more accessible, balance the unequal distribution of collections world‐wide (Drew et al., 2017; see also locations of all herbaria with > 100 000 specimens world‐wide, Fig. 1b), increase the use of herbaria in general, the number of specimens included per study specifically (Lavoie, 2013), and fuel novel research (see Soltis, 2017; Soltis et al., 2018). Various online databases already offer access to vast amounts of data (e.g. https://www.idigbio.org/, www.gbif.org, http://vh.gbif.de/vh/or http://avh.chah.org.au/), but the digitization task is enormous – with over 350 million specimens to process – and expensive. To optimize and speed up the process, various larger and smaller institutions have developed affordable digitization workflows (Haston et al., 2012; Nelson et al., 2015; Thiers et al., 2016; Harris & Marsico, 2017). Depending on data needs, digitization could be done in a prioritized way. In conservation biology, for instance, a fraction of available specimens appears to be enough to reliably detect threatened species and trigger conservation efforts (Rivers et al., 2011). How and towards which end such prioritization is carried out, and how large‐scale digitization projects would be funded, is a question that needs to be addressed.
Apart from cost and speed, the transcription of meta‐information, and particularly georeferencing information, is another digitization bottleneck.
Optical character recognition may help sorting entries by collector or country (Drinkwater et al., 2014), as might the development of semi‐automated imaging pipelines (Tegelberg et al., 2014). Other projects use citizen science approaches to transcribe specimen labels ((Hill et al., 2012); https://www.notesfromnature.org/active-expeditions/Herbarium), and computer vision or machine learning (re‐)classify specimens that are unidentified, or whose identification was based on an old taxonomy (Unger et al., 2016; Carranza‐Rojas et al., 2017; Gehan & Kellogg, 2017). Still, imprecise or wrong georeferencing is common in herbarium data (Yesson et al., 2007), an issue that is particularly problematic in conservation, for species distribution assessments, or prediction approaches (Feeley & Silman, 2010). Although care with location data from herbaria is, hence, necessary, digital field notebook apps such as ColectoR may at least help guarantee complete and correct meta‐information for novel collections (Maya‐Lastra, 2016). Finally, in light of concerns about misidentification of up to 50% of tropical specimens world‐wide (Goodwin et al., 2015) and the continuously evolving taxonomy, such notebooks, together with the aforementioned computerized identification approaches and even molecular methods, as well as rigorous and continuous manual verification of specimen identities, are crucial to ensure the value of herbaria and herbaria databases.
Pollution
Technological developments and the mechanization of work in the second half of the 18th century, known as industrialization, changed the landscape world‐wide. Key contributors were improved efficiency of steam engines, the replacement of biofuels with coal and the emergence of a chemical industry. A larger average income, increasing population sizes and accelerated urbanization led to the production of previously unseen quantities of waste and exhausts (Fig. 1a). Herbarium specimens can be used to track historical pollution levels, to serve as a baseline for pre‐pollution conditions, and to connect waste production with species’ reactions – even at the genetic level in the context of local adaptation, or to study long‐term effects of singular events such as the Chernobyl nuclear disaster (Heinrich et al. 1994).
Heavy metals
Metals from the atmosphere, soils and groundwater are deposited on or taken up by plants, and remain present in herbarium specimens, so the latter can be used as indicators of pollution, and due to their meta‐information facilitate the dating of contamination (Lee & Tallis, 1973; Shotbolt et al., 2007; Rudin et al., 2017). Depending on species, their morphology, physiology and proximity to a pollution source, plants are exposed to and take up more or less pollutants (Lawrey & Hale, 1981; Rudin et al., 2017). Studying lead pollution levels, for example, the isotopic lead composition in moss or lichen samples collected at roadsides reflects fluctuations in local motor vehicle traffic, efforts to reduce lead emissions and changes in petrol origin or composition over time (Farmer et al., 2002). In addition to lead, herbarium samples also track concentrations of other metals such as cadmium, copper and zinc to follow their temporal and spatial trends in relation to anthropogenic activities (Zschau et al., 2003; Shotbolt et al., 2007; Rudin et al., 2017). Combining pollution records and genetic information from historical and contemporary samples from contaminated sites can even enable studies of plants’ adaptation to pollution at the genetic, heritable level, for example by studying the association between pollution levels and specific alleles, and thus give indications about long‐term adaptation to changing conditions. Such approaches are already well‐established for contemporary data alone (Kawecki & Ebert, 2004; Turner et al., 2010; Arnold et al., 2016).
Anthropogenic nitrogen
Similarly, herbaria document human influences on global nitrogen (N) cycling, that started with the rise of the chemical industry and the production of fertilizers, and has peaked since c. 1960 (Millennium Ecosystem Assessment, 2005). Moss leaf N‐contents (as well as concentrations of phosphate and sulfur) determined from stable isotope ratios enable inferences about realized N sources and further cycling processes (Peñuelas & Filella, 2001). Such analyses show a retention of additional, anthropogenic N within terrestrial ecosystems (Peñuelas & Filella, 2001). Improved knowledge of these nutrient dynamics within different ecosystems helps us to understand eutrophication. Additional detail on the biotic effects of N fluctuations could be retrieved via shotgun‐sequencing of historical plant roots, given that bona fide microbiomes could be recovered, as it has been shown that the bacterial species composition of roots (and soils) is heavily influenced by overabundance of N (Dynarski & Houlton, 2018).
Increased carbon dioxide
Pollutants such as N or carbon dioxide (CO2) can influence overall organismal morphology, making their effects partially measurable without destructive sampling. Increased fossil fuel combustion and the concurrent increase in CO2 concentrations since the industrial revolution, for example, correlate with a reduction of stomatal densities on the leaves of herbarium specimens. This trend was already observed in 1987 in a 200‐yr spanning study of woody angiosperm herbaria samples. Further analyses under controlled experimental conditions (Woodward, 1987; Peñuelas & Matamala, 1990) confirm historic samples as proxies to reconstruct past CO2 concentrations.
In addition to morphological studies, herbarium specimens enable complementary measurement of global change effects on plant carbon metabolism. Using mass spectrometry to estimate the relative abundances of different carbon isotopes, studies indicate increased water‐use efficiency – the ratio of photosynthesis to water loss – with rising CO2 concentrations (Peñuelas & Azcón‐Bieto, 1992; Pedicino et al., 2002). With time‐series of genetic variation from herbaria, it is now further possible to determine what part long‐term adaptive changes or phenotypic plasticity play in such physiological or chemical responses.
There is, however, one caveat for measurements of any type of chemical compounds in long‐term stored historical samples: Do chemicals suffer degradation processes similar to hydrolytic damages occurring in DNA over time (see Box 1)? If so, to which extent and at what rate do compounds degrade, and what influence do factors like species, specimen mounting or general storage conditions have on such a decay? Systematic studies of chemical degradation through time will permit the assessment of whether absolute or relative values should be used in historical specimens‐based long‐term comparisons.
Habitat loss and land‐use changes
Apart from pollution, increasing human population densities, urbanization and, in particular, modern agriculture have caused extensive losses, fragmentation or changes of natural habitats. This affects plants’ geographic distribution and densities, for example causing range reductions to more pristine environments (Hallingbäck, 1992). Information about such habitat alterations in response to global change are documented in herbaria. Herbarium sheets normally contain information about the presented species and sometimes other, associated species (referred to in accompanying meta‐information, or co‐sampled with the focal species, e.g. pathogens). Importantly, herbarium sheets also state the time and place of collection. Hence, comparison between past and present localities serves to infer a species’ distribution through time (Hallingbäck, 1992).
Distribution changes
Many factors have contributed to converting the landscape into a patchwork of agricultural fields, interspersed with cities and roads: industrialization‐associated population growth, urbanization, increasing agricultural acreages due to mechanization of work, or expansion of railroads and other transport systems. Overall, species abundances tend to decrease with habitat and land‐use changes, as is the case, for example, for American ginseng (Panax quinquefolius), both as a result of deforestation and of heavy harvesting of wild populations (Case et al., 2007). In light of an area's geography, such data also can inform species’ conservation and future trends (Case et al., 2007). However, retrospective studies of species’ abundance in a certain location based on historical collections are sensitive both to the quality of available georeferencing data, and to fluctuating collection efforts and other biases (see Box 2). A reference set of specimens picked from the herbarium randomly and independent of species identity can be used to establish a general ‘expected collecting frequency’, which can balance these biases (e.g. Hedenäs et al., 2002).
When herbarium records are used to relocate historical populations, current data complement herbarium‐inferred distributions and abundances (Lienert et al., 2002; Stehlik et al., 2007). Herbaria may in some cases be the only documentation of (likely) extinct species (Chomicki & Renner, 2015). Revisiting surveys can detect such local extinction events, and, in correlation with current land‐use practices or site protection status, be used to study their causes (Lienert et al., 2002). They can further document changes in overall plant diversity, which, too, is affected by habitat fragmentation (Stehlik et al., 2007). Such approaches are particularly useful to evaluate changes in the local flora and motivate biodiversity monitoring campaigns, and can inform large‐scale diversity surveys, as well as modeling‐based inferences or predictions.
Indirect effects of habitat fragmentation
Similar to farming‐related landscape changes, urbanization is a prominent driver of biotic interaction changes. One of the most crucial, commercially important types of plant–animal interaction jeopardized, among others, by urbanization and diversity loss, is pollination. Depending on a plant's anatomy, herbaria also house documentation of such interactions, and can illustrate pollinator species decrease or loss. Presence or absence of pollinaria in herbarium specimens of the orchid Pterygodium catholicum, for example, reflects the historical pollination rate that depends strictly on a specific bee (Rediviva peringueyi) (Pauw & Hawkins, 2011). The bee's decrease following urbanization is consistent with a shift in local orchid communities towards selfing species (Pauw & Hawkins, 2011). Impairment of interactions between plants and their pollinators, caused for instance by such abundance decreases or temporal mismatches, likely also leaves genetic signatures. Given that affected biotic interactions could be identified using historical plant and insect collections, these signatures could be traced through time and inform the potential of other species‐pairs to overcome future mismatches.
Besides the apparent decrease of species diversity, losses of within‐species genetic diversity are a less conspicuous consequence of habitat loss, and are a result of shrinking and increasingly isolated populations (Ellstrand & Elam, 1993; Young et al., 1996). Improved high‐throughput sequencing techniques and novel molecular approaches have recently made within‐species genetic diversity – as preserved in herbaria – accessible (see Box 1). This ancient genetic information extends the information on habitat loss and decreasing relative abundances to the genetic level (Cozzolino et al., 2007; Martin et al., 2014b), with already few specimens giving insights into a population's genetic background. This is crucial knowledge for conservation measures, as genetic diversity, especially in times of increasingly fluctuating environmental conditions, is an indispensable resource for heritable phenotypic variation and rapid adaptation (Huenneke, 1991; Exposito‐Alonso et al., 2018b). Reduction of genetic diversity via abrupt decimation of a population, referred to as a bottleneck, can hamper the population's persistence, as selection is less efficient in small populations, where there is more stochasticity and less standing variation to act upon (Ellstrand & Elam, 1993; Young et al., 1996; Hartl & Clark, 2007). Comparison of contemporary vs historical genetic diversity can serve to prioritize the conservation of specific populations over others, and to identify genetically diverse source populations for potential reintroductions to balance bottlenecks (Cozzolino et al., 2007).
Climate change
Some factors on the rise since the start of industrialization, and potentially even before that, have less direct, but long‐term effects on ecosystems: the so‐called greenhouse gases such as methane (CH4) and CO2 (Fig. 1). Their atmospheric increase – for CO2 a result of enhanced fossil fuel burning in factories, power plants and for transportation – causes global warming and as a result climate change (Millennium Ecosystem Assessment, 2005). Thus, in addition to the earlier mentioned direct effects of the pollutant CO2 on plant morphology and physiology (see the ‘Pollution’ section), progressive CO2‐related global warming influences plant life cycles, as is observed for instance already in shifts of plant life cycles, as is observed for instance already in shifts of plant phenology (timing of life cycle events such as flowering and fruiting) to earlier dates. However, herbaria not only directly track these climate‐related plant responses, but also give insights into their ripple‐effects on pollinators, herbivores and even nutrient cycling.
Range shifts as spatial escape
One possible response of plants to global warming can be distributional shifts when plants escape from unfavorable conditions, which is traceable using herbarium time‐series. Comparison of field with herbarium data verifies predictions that with progressive global warming, species will move both upslope and poleward, following their original climatic niches. For instance, historic time‐series have monitored movements and consecutive diversity shifts in California, Costa Rica and South America as a whole (Feeley, 2012; Feeley et al., 2013; Wolf et al., 2016), and hence can differentiate successfully moving species from those that may not persist under continuously changing conditions (Feeley et al., 2013).
Phenology timing
Instead of spatial movements, plants also can escape global warming ‘in time’ by shifting phenological events like flowering or fruiting towards more favorable conditions. To track such changes in the past, flowering timing, for example, can be approximated from collection dates of flowering herbarium specimens. Using a combination of contemporary flowering time observations with a herbarium specimen series across > 100 yr and 37 genera, Primack and colleagues (Primack et al., 2004) were the first to connect meteorological data with earlier flowering, which was to a great part explained by increasing spring temperatures. This trend has been confirmed by multiple analogous studies (e.g. Davis et al., 2015) and also broader approaches that integrated herbarium data with phenology records obtained from field notes and photographs to cover recent years of herbarium record scarcity (Panchen et al., 2012).
Spatial scale and statistical power are important factors for these types of studies. Because phenology also depends on latitude, altitude and other environmental factors, broad sampling is necessary to separate climate change effects from other influences. Moreover, as phenology is partly species‐ or plant functional type‐specific, it is useful to study contrasting flowering seasons, native status, pollination syndromes or growth forms (Calinger et al., 2013). All of this is facilitated by large‐scale digitization and hence improved accessibility of specimens world‐wide (Lavoie, 2013; Box 3). Such studies, for example, showed that annual plants are generally more responsive to climate change than perennials (Calinger et al., 2013; Munson & Long, 2017). Compilation of large cross‐species datasets furthermore allows the search for phylogenetic signals and thus to identify evolutionary processes involved in shaping the observed responses (Rafferty & Nabity, 2017). Apart from interspecies or ‐family variation, plant responses also vary across geographic regions. Combination of world‐wide herbaria allows to capture such responses, enabling to include remote localities across the globe into analyses (Hart et al., 2014; Panchen & Gorelick, 2017).
Flowering is not the only phenological event heavily influenced by climate change that can be tracked from herbarium specimens. Depending on a plant's reproductive structures, seed dispersal timing also can be evaluated. At least for the Arctic, dispersal timing, too, seems to advance with increasing temperatures, in correspondence with associated flowering data (Panchen & Gorelick, 2017). Contrariwise, it was also estimated from collection meta‐information (Kauserud et al., 2008) that autumnal mushroom fruiting, especially of early fruiting species, is delayed in Norway, possibly reflecting a prolonged growth period due to warm autumn and winter temperatures.
Another parameter that affects entire communities and ecosystem processes is the leaf‐out timing of deciduous trees, as it impacts trophic interactions as well as nutrient and water cycling (Polgar & Primack, 2011). Such data collected from herbarium records track long‐term leaf‐out trends (Zohner & Renner, 2014) and, for example, confirm large‐scale patterns of earlier leaf‐out inferred with satellite data (Everill et al., 2014).
Mismatching biotic interactions
Naturally, these climate change‐related phenomena also affect biotic relationships beyond plants, and hence cannot be seen only as isolated processes. Changes of their timing are likely to affect evolutionarily synchronized relationships, and even their breaking‐up over time is, together with flowering change, partially recorded in herbaria. Combined with entomological museum specimens, herbaria for example document disruption of the plant–pollinator relationship between the bee Andrena nigroaenea and the orchid Ophrys sphegodes (Robbirt et al., 2014). In herbivory relationships, herbarium specimens can actually directly reflect insect reactions to warming. For example, increased traces left by the scale insect Melanaspis tenebricosa on maple tree leaves collected in warmer years evidence a higher insect density, perfectly in accordance with observations in the field (Youngsteadt et al., 2015). Herbaria can thus help overcome the lack of historical insect abundance records and facilitate evaluation of climate change effects beyond plants alone.
The greatest challenge of most aforementioned approaches investigating species’ responses to pollution, and habitat and climate change, is their inability to distinguish between plastic responses and evolutionary adaptation (Leger, 2013; Munson & Long, 2017), and thus whether observed differences among herbaria specimens reflect genetic changes or just environmentally induced phenotypic changes caused, for instance, by physiological processes (Bradshaw, 1965; Nicotra et al., 2010). Quantitative genetics methods using herbarium time‐series could help in disentangling these two alternative hypotheses (Gienapp et al., 2008; Tiffin & Ross‐Ibarra, 2014). Once the genetic basis of phenotypic differences is identified, local adaptation can be further tested using traditional approaches such as common garden experiments and reciprocal transplant studies (Savolainen et al., 2013).
Biological invasions
Natural long‐distance dispersal of plants is rare (Nathan & Muller‐Landau, 2000), but as a side effect of global change, plants increasingly move long distances (van Kleunen et al., 2015a). This movement massively increased with human migration waves towards the New World in the 16th century, and further accelerated with growing trade and faster transportation – coinciding with the core time range of herbarium collections. Today, jet‐setting plant stowaways establish as ‘neophytes’, ‘aliens’ or ‘invaders’ wherever conditions are favorable enough. With this growing alien species richness, the global species distribution is getting more homogenous (Winter et al., 2009). Local plants lose habitats and thus genetic diversity to the invaders, which are therefore considered a threat to biodiversity (Millennium Ecosystem Assessment, 2005).
Understanding invasion dynamics
Understanding the causes and spatiotemporal dynamics of invasions is indispensable to prevent further damage, preserve natural ecosystems and prioritize management actions (Vilà et al., 2011; van Kleunen et al., 2015b). Although contemporary surveys depict the current status of invasive species, herbaria track invasions from the first recorded colonizer onwards – which can serve as a proxy, even if it is not the actual first colonizer. In conjunction with contemporary collections and literature surveys, herbaria are crucial to establish inventories of introduced species that monitor their status of naturalization – or invasion – and inform management strategies (Magona et al., 2018). With native plants as baseline for collection efforts and abundance, herbaria illustrate geographical and temporal spreads (Crawford & Hoagland, 2009) that may – in search for invasion causes – be connected with historic events. For instance, a map of Chilean alien expansions uncovers two spread peaks, one connected to the spread of agriculture, the other to its increased mechanization (Fuentes et al., 2008). Understanding such causalities can feed early preventive measures: retrospectively mapped invasions identify geographic invasion hotspots, and the environmental and anthropogenic factors crucial for their creation. In this way, herbaria can contribute to understanding the general invasibility of particular habitats (Aikio et al., 2012; Dawson et al., 2017). Furthermore, combined with contemporary data, they can help to identify characteristics of successful invaders, and to quantitatively connect and established naturalization risk with external factors, and rank potential new invaders (Dodd et al., 2016).
Herbaria also provide a means of assessing the continued success of invasive species after establishment in a new environment. Previous studies have used them both to predict and to verify predictions of the climatic niche that plants can potentially occupy. For example, the size of the native range of an invasive species has been found to be highly correlated with its abundance in the new range, as documented for many highly invasive Eurasian species around Québec (Lavoie et al., 2013). Herbaria also can enable estimation of a weediness index – or how much a plant associates with human‐caused disturbance – which often also overlaps with plant invasiveness (Robin Hart, 1976). Such estimates hold well in comparison with field surveys (Hanan‐A et al., 2015). More precise forecasts of a species’ spread can further include its native climate range, again extrapolated from herbarium records, thereby roughly visualizing occupation of a possible climatic niche (Bradley et al., 2015). Much as surveying and modeling the dynamics and spread of invaders is crucial to inform containment measures, it is very sensitive to biases and errors in historical collections – one crucial and common error being misidentification and misnaming (Jacobs et al., 2017) – and increasingly at risk from decreasing collection efforts (see Box 2).
Genetic changes of invaders
Irrespective of whether invasive species stay within their native climatic range or move beyond, they face challenges when establishing in new environments. Successful invasive species often adjust to the novel conditions, and it is therefore important to understand such changes in the invasive range.
Adjustment of morphological traits to novel environments is often well‐captured in herbaria, as demonstrated with Australian invasives where 70% of surveyed species showed at least one phenotypic trait changing over time (Buswell et al., 2011). With NGS, it is now possible to define whether this trait variation is associated with genomic changes – caused either by drift or potentially adaptive – or more likely the result of phenotypic plasticity. In addition, these methods can potentially solve the ‘genetic paradox of invasion’: the surprising success and spread of colonizers in spite of their reduced genetic diversity (Estoup et al., 2016): Do these species adapt based on their (reduced) standing genetic variation, do they borrow pre‐adapted standing variation from native species (adaptive introgression; Keller & Taylor, 2010; Arnold et al., 2016), or do they rely on de novo mutations and hence novel variation (Exposito‐Alonso et al., 2018a)?
Comparison of historic native and invasive populations with contemporary genetic diversity can also point to diversification or hybridization events before species expansion. A recent herbarium genetics study, for example, has shown strong divergences of flowering time genes particularly during the establishment phase of the invader Sisymbrium austriacum ssp. chrysanthum, possibly enabling a subsequent spread (Vandepitte et al., 2014). Such patterns change over the course of invasion. In the Eurasian Alliaria petiolata invading North America, invasive success declines along with population age and reduced phytotoxin production in late stages of invasion (Lankau et al., 2009). Contrary to that, chemical analyses of herbarium specimens of the phototoxic Pastinaca sativa, a European weed also invading North America, displays increased concentrations of phytochemicals over time since invasion, which coincide with the emergence of the herbivore Depressaria pastinacella (Zangerl & Berenbaum, 2005). Studies using ancient DNA also have pointed to anthropogenic landscape disturbances causing genetic admixture in Ambrosia artemisiifolia's native populations before its introduction to new habitats, potentially a prerequisite for later invasive success (Martin et al., 2014b). In this sense, herbarium material allows us to compare genetic composition through time, and to identify so‐called ‘cryptic’ (i.e. hidden) invasions, where native genotypes are dispelled by phenotypically indistinguishable but more successful and aggressively spreading non‐native relatives (Saltonstall, 2002).
Hitchhiking invaders: pathogens and herbivores
Moving beyond plant invasions, herbaria even harbor information about hitchhikers traveling with the original plant stowaways, pathogens, purposely or unknowingly sampled together with their hosts (Yoshida et al., 2014). Thereby, they track the invasion (success) stories of plant pathogens such as Phytophthora infestans, the microbe at the root of potato late blight and the Irish potato famine (Martin et al., 2013, 2014a; Yoshida et al., 2013). Other preserved pathogens of particular interest for agriculture include rust fungi and downy‐mildew‐causing oomycetes. Herbaria allow identification of causal strains, their genetic characteristics and their tracking to contemporary pathogen diversity. Coupled with host plant analyses, they provide a (genetic) timeline of host–pathogen dynamics to study and illustrate co‐evolutionary principles such as the arms race between hosts and their pathogens. Genetic analysis of such systems can hence provide crucial insight into spread dynamics of pathogens that could have devastating consequences on crop monocultures world‐wide.
Even for invasive herbivores, historic samples may contain a genetic record. The horse chestnut leaf‐mining moth Cameraria ohridella, for example, is preserved pressed and dried in leaves of its host plant (Lees et al., 2011). Genetics can backtrack the moth's spread from its native Balkan region, and in conjunction with host plant analyses may identify resistant cultivars and biocontrol agents for the invasive pest (Lees et al., 2011).
Conclusions and outlook
Plants preserved in herbaria offer unique perspectives on global change and its consequences, as they are directly affected victims (Fig. 2). Thus, they represent an invaluable temporal, geographical and taxonomic extension of currently available data employed to understand global environmental change, predict its course and inform conservation measures. To fully take advantage of this potential, and to increase and sustain the value of herbaria for the future, three core areas demand particular attention: the maintenance and curation of herbaria including continued collection efforts, the digitization of collections, and herbarium genomics (see also Boxes 1, 2, 3).
Even though many herbaria are already investing in digitization, only a fraction of the c. 350 million specimens world‐wide have been digitized so far. Large‐scale digitization would both encourage the use of herbaria for research, and strengthen projects (e.g. Munson & Long, 2017), as studies including digitized material are able to use large sample sizes (Lavoie, 2013). Fast processing of specimens at consistently high data quality is crucial for making digital herbaria truly useful (Yesson et al., 2007), as is substantial funding to enable this task and secure databases’ continuity. Yet, even with increased digitization, the actual power of herbaria – for climate change study amongst other types of research – lies in their continuity through time. Despite growing recognition of the value of herbaria, this characteristic is currently threatened by declining collection efforts (i.e. Prather et al., 2004; Meyer et al., 2016) and a frequent lack of support for herbaria world‐wide. Consequences of reduced data for modeling and other analyses can already be seen in the tropics, where collections are generally sparse (Feeley & Silman, 2011). To maintain herbaria as the treasure they are today, continued and consistent collection world‐wide is essential, especially because they have recently revealed themselves as a browsable repository of genetic variation and diversity. This drastically increases the value of herbaria for climate change research, and for understanding principles of adaptation and evolution in this context. To date, herbaria are still underused in this aspect (Lavoie, 2013), and in particular, high‐quality sequencing data are scarce. With firm guidelines for protocols and quality standards, pointing also to the necessity of DNA preservation‐informed sequencing efforts, this neglect is likely to change in the coming years.
Hence, being aware of the answers herbaria can give if we use the right methods to ask, it is up to us to keep herbaria alive and well, define what we need to know, and start the questioning.
Author contributions
O.B., H.A.B., F.M.W., P.L.M.L. and J.F.S. developed the ideas for this review; F.M.W. and P.L.M.L. undertook the literature research and P.L.M.L. designed the figures and wrote the paper with input from all authors.
Acknowledgements
We thank Moises Exposito‐Alonso, Clemens Weiß and other members of the Research Group for Ancient Genomics and Evolution for support and suggestions. We also thank the three anonymous referees for their helpful comments, and apologize to colleagues whose work could not be cited owing to space constraints. This work was supported by the German Research Foundation (DFG; projects BO 3241/7‐1 and BU 3422/1‐1) and by the Presidential Innovation Fund of the Max Planck Society. The authors declare no competing or financial interests.
Contributor Information
Hernán A. Burbano, Email: hernan.burbano@tuebingen.mpg.de.
Oliver Bossdorf, Email: oliver.bossdorf@uni-tuebingen.de.
References
- Aikio S, Duncan RP, Hulme PE. 2012. The vulnerability of habitats to plant invasion: disentangling the roles of propagule pressure, time and sampling effort. Global Ecology and Biogeography 21: 778–786. [Google Scholar]
- Allentoft ME, Collins M, Harker D, Haile J, Oskam CL, Hale ML, Campos PF, Samaniego JA, Gilbert MTP, Willerslev E et al 2012. The half‐life of DNA in bone: measuring decay kinetics in 158 dated fossils. Proceedings of the Royal Society B 279: 4724–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold BJ, Lahner B, DaCosta JM, Weisman CM, Hollister JD, Salt DE, Bomblies K, Yant L. 2016. Borrowed alleles and convergence in serpentine adaptation. Proceedings of the National Academy of Sciences, USA 113: 8320–8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley BA, Early R, Sorte CJB. 2015. Space to invade? Comparative range infilling and potential range of invasive and native plants. Global Ecology and Biogeography 24: 348–359. [Google Scholar]
- Bradshaw AD. 1965. Evolutionary significance of phenotypic plasticity in plants In: Caspari EW, Thoday JM, eds. Advances in genetics. Amsterdam, the Netherlands: Academic Press, 115–155. [Google Scholar]
- Buswell JM, Moles AT, Hartley S. 2011. Is rapid evolution common in introduced plant species? Journal of Ecology 99: 214–224. [Google Scholar]
- Calinger KM, Queenborough S, Curtis PS. 2013. Herbarium specimens reveal the footprint of climate change on flowering trends across north–central North America. Ecology Letters 16: 1037–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carranza‐Rojas J, Goeau H, Bonnet P, Mata‐Montero E, Joly A. 2017. Going deeper in the automated identification of Herbarium specimens. BMC Evolutionary Biology 17: 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case MA, Flinn KM, Jancaitis J, Alley A, Paxton A. 2007. Declining abundance of American ginseng (Panax quinquefolius L.) documented by herbarium specimens. Biological Conservation 134: 22–30. [Google Scholar]
- Chomicki G, Renner SS. 2015. Watermelon origin solved with molecular phylogenetics including Linnaean material: another example of museomics. New Phytologist 205: 526–532. [DOI] [PubMed] [Google Scholar]
- Cozzolino S, Cafasso D, Pellegrino G, Musacchio A, Widmer A. 2007. Genetic variation in time and space: the use of herbarium specimens to reconstruct patterns of genetic variation in the endangered orchid Anacamptis palustris . Conservation Genetics 8: 629–639. [Google Scholar]
- Crawford PHC, Hoagland BW. 2009. Can herbarium records be used to map alien species invasion and native species expansion over the past 100 years? Journal of Biogeography 36: 651–661. [Google Scholar]
- Daru BH, Park DS, Primack RB, Willis CG, Barrington DS, Whitfeld TJS, Seidler TG, Sweeney PW, Foster DR, Ellison AM et al 2018. Widespread sampling biases in herbaria revealed from large‐scale digitization. New Phytologist 217: 939–955. [DOI] [PubMed] [Google Scholar]
- Davis CC, Willis CG, Connolly B, Kelly C, Ellison AM. 2015. Herbarium records are reliable sources of phenological change driven by climate and provide novel insights into species’ phenological cueing mechanisms. American Journal of Botany 102: 1599–1609. [DOI] [PubMed] [Google Scholar]
- Dawson W, Moser D, van Kleunen M, Kreft H, Pergl J, Pyšek P, Weigelt P, Winter M, Lenzner B, Blackburn TM et al 2017. Global hotspots and correlates of alien species richness across taxonomic groups. Nature Ecology & Evolution 1: 0186. [Google Scholar]
- Delisle F, Lavoie C, Jean M, Lachance D. 2003. Reconstructing the spread of invasive plants: taking into account biases associated with herbarium specimens. Journal of Biogeography 30: 1033–1042. [Google Scholar]
- Dodd AJ, McCarthy MA, Ainsworth N, Burgman MA. 2016. Identifying hotspots of alien plant naturalisation in Australia: approaches and predictions. Biological Invasions 18: 631–645. [Google Scholar]
- Drew JA, Moreau CS, Stiassny MLJ. 2017. Digitization of museum collections holds the potential to enhance researcher diversity. Nature Ecology & Evolution 1: 1789–1790. [DOI] [PubMed] [Google Scholar]
- Drinkwater RE, Cubey RWN, Haston EM. 2014. The use of Optical Character Recognition (OCR) in the digitisation of herbarium specimen labels. PhytoKeys 38: 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durvasula A, Fulgione A, Gutaker RM, Alacakaptan SI, Flood PJ, Neto C, Tsuchimatsu T, Burbano HA, Picó FX, Alonso‐Blanco C et al 2017. African genomes illuminate the early history and transition to selfing in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 114: 5213–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynarski KA, Houlton BZ. 2018. Nutrient limitation of terrestrial free‐living nitrogen fixation. New Phytologist 217: 1050–1061. [DOI] [PubMed] [Google Scholar]
- Ellstrand NC, Elam DR. 1993. Population genetic consequences of small population size: implications for plant conservation. Annual Review of Ecology and Systematics 24: 217–242. [Google Scholar]
- Estoup A, Ravigné V, Hufbauer R, Vitalis R, Gautier M, Facon B. 2016. Is there a genetic paradox of biological invasion? Annual Review of Ecology, Evolution, and Systematics 47: 51–72. [Google Scholar]
- Everill PH, Primack RB, Ellwood ER, Melaas EK. 2014. Determining past leaf‐out times of New England's deciduous forests from herbarium specimens. American Journal of Botany 101: 1293–1300. [DOI] [PubMed] [Google Scholar]
- Exposito‐Alonso M, Becker C, Schuenemann VJ, Reiter E, Setzer C, Slovak R, Brachi B, Hagmann J, Grimm DG, Chen J et al 2018a. The rate and potential relevance of new mutations in a colonizing plant lineage. PLoS Genetics 14: e1007155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exposito‐Alonso M, Vasseur F, Ding W, Wang G, Burbano HA, Weigel D. 2018b. Genomic basis and evolutionary potential for extreme drought adaptation in Arabidopsis thaliana . Nature Ecology & Evolution 2: 352–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer JG, Eades LJ, Atkins H, Chamberlain DF. 2002. Historical trends in the lead isotopic composition of archival Sphagnum mosses from Scotland (1838−2000). Environmental Science & Technology 36: 152–157. [DOI] [PubMed] [Google Scholar]
- Feeley KJ. 2012. Distributional migrations, expansions, and contractions of tropical plant species as revealed in dated herbarium records. Global Change Biology 18: 1335–1341. [Google Scholar]
- Feeley KJ, Hurtado J, Saatchi S, Silman MR, Clark DB. 2013. Compositional shifts in Costa Rican forests due to climate‐driven species migrations. Global Change Biology 19: 3472–3480. [DOI] [PubMed] [Google Scholar]
- Feeley KJ, Silman MR. 2010. Modelling the responses of Andean and Amazonian plant species to climate change: the effects of georeferencing errors and the importance of data filtering. Journal of Biogeography 37: 733–740. [Google Scholar]
- Feeley KJ, Silman MR. 2011. The data void in modeling current and future distributions of tropical species. Global Change Biology 17: 626–630. [Google Scholar]
- Fitter AH, Fitter RSR. 2002. Rapid changes in flowering time in British plants. Science 296: 1689–1691. [DOI] [PubMed] [Google Scholar]
- Fuentes N, Ugarte E, Kühn I, Klotz S. 2008. Alien plants in Chile: inferring invasion periods from herbarium records. Biological Invasions 10: 649–657. [Google Scholar]
- Funk V. 2003. 100 uses for an herbarium (well at least 72). American Society of Plant Taxonomists Newsletter 17: 17–19. [Google Scholar]
- Gehan MA, Kellogg EA. 2017. High‐throughput phenotyping. American Journal of Botany 104: 505–508. [DOI] [PubMed] [Google Scholar]
- Gienapp P, Teplitsky C, Alho JS, Mills JA, Merilä J. 2008. Climate change and evolution: disentangling environmental and genetic responses. Molecular Ecology 17: 167–178. [DOI] [PubMed] [Google Scholar]
- Goodwin ZA, Harris DJ, Filer D, Wood JRI, Scotland RW. 2015. Widespread mistaken identity in tropical plant collections. Current Biology 25: R1066–R1067. [DOI] [PubMed] [Google Scholar]
- Gutaker RM, Burbano HA. 2017. Reinforcing plant evolutionary genomics using ancient DNA. Current Opinion in Plant Biology 36: 38–45. [DOI] [PubMed] [Google Scholar]
- Gutaker RM, Reiter E, Furtwängler A, Schuenemann VJ, Burbano HA. 2017. Extraction of ultrashort DNA molecules from herbarium specimens. BioTechniques 62: 76–79. [DOI] [PubMed] [Google Scholar]
- Hallingbäck T. 1992. The effect of air pollution on mosses in southern Sweden. Biological Conservation 59: 163–170. [Google Scholar]
- Hamilton C. 2016. Define the Anthropocene in terms of the whole Earth. Nature 536: 251. [DOI] [PubMed] [Google Scholar]
- Hanan‐A AM, Vibrans H, Cacho NI, Villaseñor JL, Ortiz E, Gómez‐G VA. 2015. Use of herbarium data to evaluate weediness in five congeners. AoB Plants 8: plv144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KM, Marsico TD. 2017. Digitizing specimens in a small herbarium: a viable workflow for collections working with limited resources. Applications in Plant Sciences 5: 1600125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart R. 1976. An index for comparing weediness in plants. Taxon 25: 245–247. [Google Scholar]
- Hart R, Salick J, Ranjitkar S, Xu J. 2014. Herbarium specimens show contrasting phenological responses to Himalayan climate. Proceedings of the National Academy of Sciences, USA 111: 10 615–10 619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL, Clark AG. 2007. Principles of population genetics. Sunderland, MA, USA: Sinauer Associates. [Google Scholar]
- Haston E, Cubey R, Pullan M, Atkins H, Harris DJ. 2012. Developing integrated workflows for the digitisation of herbarium specimens using a modular and scalable approach. ZooKeys 209: 93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedenäs L, Bisang I, Tehler A, Hamnede M, Jaederfelt K, Odelvik G. 2002. A herbarium‐based method for estimates of temporal frequency changes: mosses in Sweden. Biological Conservation 105: 321–331. [Google Scholar]
- Heinrich VG, Oswald K, Müller H. 1994. Zur Kontamination von Flechten in der Steiermark vor und nach dem Reaktorunglück von Chernobyl. Mitteilungen des Naturwissenschaftlichen Vereins für Steiermark 124: 173–189. [Google Scholar]
- Hill A, Guralnick R, Smith A, Sallans A, Gillespie R, Denslow M, Gross J, Murrell Z, Conyers Tim, Oboyski P et al 2012. The notes from nature tool for unlocking biodiversity records from museum records through citizen science. ZooKeys 209: 219–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes MW, Hammond TT, Wogan GOU, Walsh RE, LaBarbera K, Wommack EA, Martins FM, Crawford JC, Mack KL, Bloch LM et al 2016. Natural history collections as windows on evolutionary processes. Molecular Ecology 25: 864–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huenneke LF. 1991. Ecological implications of genetic variation in plant populations In: Falk DAI, Holsinger KE, eds. Genetics and conservation of rare plants. New York, NY, USA: Oxford University Press, 31–44. [Google Scholar]
- Hyndman RJ, Mesgaran MB, Cousens RD. 2015. Statistical issues with using herbarium data for the estimation of invasion lag‐phases. Biological Invasions 17: 3371–3381. [Google Scholar]
- Jacobs LEO, Richardson DM, Lepschi BJ, Wilson JRU. 2017. Quantifying errors and omissions in alien species lists: the introduction status of Melaleuca species in South Africa as a case study. Neobiota 32: 89–105. [Google Scholar]
- Jácome J, Kessler M, Smith AR. 2007. A human‐induced downward‐skewed elevational abundance distribution of pteridophytes in the Bolivian Andes. Global Ecology and Biogeography: a Journal of Macroecology 16: 313–318. [Google Scholar]
- Kauserud H, Stige LC, Vik JO, Okland RH, Høiland K, Stenseth NC. 2008. Mushroom fruiting and climate change. Proceedings of the National Academy of Sciences, USA 105: 3811–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawecki TJ, Ebert D. 2004. Conceptual issues in local adaptation. Ecology Letters 7: 1225–1241. [Google Scholar]
- Keller SR, Taylor DR. 2010. Genomic admixture increases fitness during a biological invasion. Journal of Evolutionary Biology 23: 1720–1731. [DOI] [PubMed] [Google Scholar]
- van Kleunen M, Dawson W, Essl F, Pergl J, Winter M, Weber E, Kreft H, Weigelt P, Kartesz J, Nishino M et al 2015a. Global exchange and accumulation of non‐native plants. Nature 525: 100–103. [DOI] [PubMed] [Google Scholar]
- van Kleunen M, Dawson W, Maurel N. 2015b. Characteristics of successful alien plants. Molecular Ecology 24: 1954–1968. [DOI] [PubMed] [Google Scholar]
- Kouwenberg LLR, McElwain JC, Kürschner WM, Wagner F, Beerling DJ, Mayle FE, Visscher H. 2003. Stomatal frequency adjustment of four conifer species to historical changes in atmospheric CO2 . American Journal of Botany 90: 610–619. [DOI] [PubMed] [Google Scholar]
- Lankau RA, Nuzzo V, Spyreas G, Davis AS. 2009. Evolutionary limits ameliorate the negative impact of an invasive plant. Proceedings of the National Academy of Sciences, USA 106: 15 362–15 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie C. 2013. Biological collections in an ever changing world: herbaria as tools for biogeographical and environmental studies. Perspectives in Plant Ecology, Evolution and Systematics 15: 68–76. [Google Scholar]
- Lavoie C, Saint‐Louis A, Guay G, Groeneveld E, Villeneuve P. 2012. Naturalization of exotic plant species in north‐eastern North America: trends and detection capacity. Diversity and Distributions 18: 180–190. [Google Scholar]
- Lavoie C, Shah MA, Bergeron A, Villeneuve P. 2013. Explaining invasiveness from the extent of native range: new insights from plant atlases and herbarium specimens. Diversity and Distributions 19: 98–105. [Google Scholar]
- Law W, Salick J. 2005. Human‐induced dwarfing of Himalayan snow lotus, Saussurea laniceps (Asteraceae). Proceedings of the National Academy of Sciences,USA 102: 10218–10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrey JD, Hale ME. 1981. Retrospective study of lichen lead accumulation in the Northeastern United States. Bryologist 84: 449–456. [Google Scholar]
- Lee JA, Tallis JH. 1973. Regional and historical aspects of lead pollution in Britain. Nature 245: 216–218. [DOI] [PubMed] [Google Scholar]
- Lees DC, Lack HW, Rougerie R, Hernandez‐Lopez A, Raus T, Avtzis ND, Augustin S, Lopez‐Vaamonde C. 2011. Tracking origins of invasive herbivores through herbaria and archival DNA: the case of the horse‐chestnut leaf miner. Frontiers in Ecology and the Environment 9: 322–328. [Google Scholar]
- Leger EA. 2013. Annual plants change in size over a century of observations. Global Change Biology 19: 2229–2239. [DOI] [PubMed] [Google Scholar]
- Leuzinger S, Luo Y, Beier C, Dieleman W, Vicca S, Körner C. 2011. Do global change experiments overestimate impacts on terrestrial ecosystems? Trends in Ecology & Evolution 26: 236–241. [DOI] [PubMed] [Google Scholar]
- Lienert J, Fischer M, Diemer M. 2002. Local extinctions of the wetland specialist Swertia perennis L. (Gentianaceae) in Switzerland: a revisitation study based on herbarium records. Biological Conservation 103: 65–76. [Google Scholar]
- Magona N, Richardson DM, Le Roux JJ, Kritzinger‐Klopper S, Wilson JRU. 2018. Even well‐studied groups of alien species might be poorly inventoried: Australian Acacia species in South Africa as a case study. NeoBiota 39: 1. [Google Scholar]
- Marciniak S, Perry GH. 2017. Harnessing ancient genomes to study the history of human adaptation. Nature Reviews Genetics 18: 659–674. [DOI] [PubMed] [Google Scholar]
- Marcott SA, Shakun JD, Clark PU, Mix AC. 2013. A reconstruction of regional and global temperature for the past 11,300 years. Science 339: 1198–1201. [DOI] [PubMed] [Google Scholar]
- Martin MD, Cappellini E, Samaniego JA, Zepeda ML, Campos PF, Seguin‐Orlando A, Wales N, Orlando L, Ho SYW, Dietrich FS et al 2013. Reconstructing genome evolution in historic samples of the Irish potato famine pathogen. Nature Communications 4: 2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MD, Ho SYW, Wales N, Ristaino JB, Gilbert MTP. 2014a. Persistence of the mitochondrial lineage responsible for the Irish potato famine in extant new world Phytophthora infestans . Molecular Biology and Evolution 31: 1414–1420. [DOI] [PubMed] [Google Scholar]
- Martin MD, Zimmer EA, Olsen MT, Foote AD, Gilbert MTP, Brush GS. 2014b. Herbarium specimens reveal a historical shift in phylogeographic structure of common ragweed during native range disturbance. Molecular Ecology 23: 1701–1716. [DOI] [PubMed] [Google Scholar]
- Maya‐Lastra CA. 2016. ColectoR, a digital field notebook for voucher specimen collection for smartphones. Applications in Plant Sciences 4: 1600035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meineke EK, Davis CC, Davies TJ. 2018. The unrealized potential of herbaria for global change biology. Ecological Monographs. doi: 10.1002/ecm.1307. [DOI] [Google Scholar]
- Meyer C, Weigelt P, Kreft H. 2016. Multidimensional biases, gaps and uncertainties in global plant occurrence information. Ecology Letters 19: 992–1006. [DOI] [PubMed] [Google Scholar]
- Millennium Ecosystem Assessment . 2005. Ecosystems and human well‐being: synthesis. Washington, DC, USA: Island Press. [Google Scholar]
- Miller‐Rushing AJ, Primack RB, Primack D, Mukunda S. 2006. Photographs and herbarium specimens as tools to document phenological changes in response to global warming. American Journal of Botany 93: 1667–1674. [DOI] [PubMed] [Google Scholar]
- Munson SM, Long AL. 2017. Climate drives shifts in grass reproductive phenology across the western USA. New Phytologist 213: 1945–1955. [DOI] [PubMed] [Google Scholar]
- Nathan R, Muller‐Landau HC. 2000. Spatial patterns of seed dispersal, their determinants and consequences for recruitment. Trends in Ecology & Evolution 15: 278–285. [DOI] [PubMed] [Google Scholar]
- Neftel A, Friedli H, Moor E, Loetscher H, Oeschger H, Siegenthaler U, Stauffer B. 1994. Historical CO2 record from the Siple Station ice core. In trends: a compendium of data on global change. Oak Ridge, TN, USA: Carbon Dioxide Information Analysis Center, Oak Ridge National Laboratory, US Department of Energy. [Google Scholar]
- Nelson G, Sweeney P, Wallace LE, Rabeler RK, Allard D, Brown H, Carter JR, Denslow MW, Ellwood ER, Germain‐Aubrey CC et al 2015. Digitization workflows for flat sheets and packets of plants, algae, and fungi. Applications in Plant Sciences 3: 1500065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicotra AB, Atkin OK, Bonser SP, Davidson AM, Finnegan EJ, Mathesius U, Poot P, Purugganan MD, Richards CL, Valladares F et al 2010. Plant phenotypic plasticity in a changing climate. Trends in Plant Science 15: 684–692. [DOI] [PubMed] [Google Scholar]
- Nielsen TF, Larsen JR, Michelsen A, Bruun HH. 2017. Are herbarium mosses reliable indicators of historical nitrogen deposition? Environmental Pollution 231: 1201–1207. [DOI] [PubMed] [Google Scholar]
- Orlando L, Gilbert MTP, Willerslev E. 2015. Reconstructing ancient genomes and epigenomes. Nature Reviews Genetics 16: 395–408. [DOI] [PubMed] [Google Scholar]
- Panchen ZA, Gorelick R. 2017. Prediction of Arctic plant phenological sensitivity to climate change from historical records. Ecology and Evolution 7: 1325–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchen ZA, Primack RB, Anisko T, Lyons RE. 2012. Herbarium specimens, photographs, and field observations show Philadelphia area plants are responding to climate change. American Journal of Botany 99: 751–756. [DOI] [PubMed] [Google Scholar]
- Pauw A, Hawkins JA. 2011. Reconstruction of historical pollination rates reveals linked declines of pollinators and plants. Oikos 120: 344–349. [Google Scholar]
- Pedicino LC, Leavitt SW, Betancourt JL, Van de Water PK. 2002. Historical variations in δ13C LEAF of herbarium specimens in the Southwestern U.S. Western North American Naturalist/Brigham Young University 62: 348–359. [Google Scholar]
- Peñuelas J, Azcón‐Bieto J. 1992. Changes in leaf Δ13C of herbarium plant species during the last 3 centuries of CO2 increase. Plant, Cell & Environment 15: 485–489. [Google Scholar]
- Peñuelas J, Filella I. 2001. Herbaria century record of increasing eutrophication in Spanish terrestrial ecosystems. Global Change Biology 7: 427–433. [Google Scholar]
- Peñuelas J, Matamala R. 1990. Changes in N and S leaf content, stomatal density and specific leaf area of 14 plant species during the last three centuries of CO2 increase. Journal of Experimental Botany 41: 1119–1124. [Google Scholar]
- Pergl J, Pyšek P, Perglová I, Jarošík V, Procheş S. 2012. Low persistence of a monocarpic invasive plant in historical sites biases our perception of its actual distribution. Journal of Biogeography 39: 1293–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polgar CA, Primack RB. 2011. Leaf‐out phenology of temperate woody plants: from trees to ecosystems. New Phytologist 191: 926–941. [DOI] [PubMed] [Google Scholar]
- Prather LA, Alvarez‐Fuentes O, Mayfield MH, Ferguson CJ. 2004. The decline of plant collecting in the United States: a threat to the infrastructure of biodiversity studies. Systematic Botany 29: 15–28. [Google Scholar]
- Primack D, Imbres C, Primack RB, Miller‐Rushing AJ, Del Tredici P. 2004. Herbarium specimens demonstrate earlier flowering times in response to warming in Boston. American Journal of Botany 91: 1260–1264. [DOI] [PubMed] [Google Scholar]
- Primack RB, Miller‐Rushing AJ. 2009. The role of botanical gardens in climate change research. New Phytologist 182: 303–313. [DOI] [PubMed] [Google Scholar]
- Rafferty NE, Nabity PD. 2017. A global test for phylogenetic signal in shifts in flowering time under climate change. Journal of Ecology 105: 627–633. [Google Scholar]
- Rivers MC, Taylor L, Brummitt NA, Meagher TR, Roberts DL, Lughadha EN. 2011. How many herbarium specimens are needed to detect threatened species? Biological Conservation 144: 2541–2547. [Google Scholar]
- Robbirt KM, Roberts DL, Hutchings MJ, Davy AJ. 2014. Potential disruption of pollination in a sexually deceptive orchid by climatic change. Current Biology 24: 2845–2849. [DOI] [PubMed] [Google Scholar]
- Rudin SM, Murray DW, Whitfeld TJS. 2017. Retrospective analysis of heavy metal contamination in Rhode Island based on old and new herbarium specimens. Applications in Plant Sciences 5: 1600108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhling A, Tyler G. 1968. An ecological approach to the lead problem. Botaniska Notiser 121: 321–342. [Google Scholar]
- Ruhling A, Tyler G. 1969. Ecology of heavy metals – a regional and historical study. Botaniska Notiser 122: 248–259. [Google Scholar]
- Saltonstall K. 2002. Cryptic invasion by a non‐native genotype of the common reed, Phragmites australis, into North America. Proceedings of the National Academy of Sciences, USA 99: 2445–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen O, Lascoux M, Merilä J. 2013. Ecological genomics of local adaptation. Nature Reviews Genetics 14: 807–820. [DOI] [PubMed] [Google Scholar]
- Sawyer S, Krause J, Guschanski K, Savolainen V, Pääbo S. 2012. Temporal patterns of nucleotide misincorporations and DNA fragmentation in ancient DNA. PLoS ONE 7: e34131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro B, Hofreiter M. 2014. A paleogenomic perspective on evolution and gene function: new insights from ancient DNA. Science 343: 1236573. [DOI] [PubMed] [Google Scholar]
- Shotbolt L, Büker P, Ashmore MR. 2007. Reconstructing temporal trends in heavy metal deposition: assessing the value of herbarium moss samples. Environmental Pollution 147: 120–130. [DOI] [PubMed] [Google Scholar]
- Sofaer HR, Jarnevich CS. 2017. Accounting for sampling patterns reverses the relative importance of trade and climate for the global sharing of exotic plants. Global Ecology and Biogeography 26: 669–678. [Google Scholar]
- Soltis PS. 2017. Digitization of herbaria enables novel research. American Journal of Botany 104: 1281–1284. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Nelson G, James SA. 2018. Green digitization: online botanical collections data answering real‐world questions. Applications in Plant Sciences 6: e1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman KV, Mulder CPH. 2016. Validating herbarium‐based phenology models using citizen‐science data. BioScience 66: 897–906. [Google Scholar]
- Sprague TA, Nelmes E. 1931. The herbal of Leonhart Fuchs. Botanical Journal of the Linnean Society 48: 545–642. [Google Scholar]
- Steffen W, Grinevald J, Crutzen P, McNeill J. 2011. The Anthropocene: conceptual and historical perspectives. Philosophical Transactions of the Royal Society A 369: 842–867. [DOI] [PubMed] [Google Scholar]
- Stehlik I, Caspersen JP, Wirth L, Holderegger R. 2007. Floral free fall in the Swiss lowlands: environmental determinants of local plant extinction in a peri‐urban landscape. Journal of Ecology 95: 734–744. [Google Scholar]
- Tegelberg R, Mononen T, Saarenmaa H. 2014. High‐performance digitization of natural history collections: automated imaging lines for herbarium and insect specimens. Taxon 63: 1307–1313. [Google Scholar]
- Thiers BM. 2017. Index Herbariorum: a global directory of public herbaria and associated staff. New York Botanical Garden's Virtual Herbarium. [WWW document] URL http://sweetgum.nybg.org/science/ih/. (accessed December 2017).
- Thiers BM, Tulig MC, Watson KA. 2016. Digitization of the New York botanical garden herbarium. Brittonia 68: 324–333. [Google Scholar]
- Thomas JA, Telfer MG, Roy DB, Preston CD, Greenwood JJD, Asher J, Fox R, Clarke RT, Lawton JH. 2004. Comparative losses of British butterflies, birds, and plants and the global extinction crisis. Science 303: 1879–1881. [DOI] [PubMed] [Google Scholar]
- Tiffin P, Ross‐Ibarra J. 2014. Advances and limits of using population genetics to understand local adaptation. Trends in Ecology & Evolution 29: 673–680. [DOI] [PubMed] [Google Scholar]
- Turner TL, Bourne EC, Von Wettberg EJ, Hu TT, Nuzhdin SV. 2010. Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nature Genetics 42: 260–263. [DOI] [PubMed] [Google Scholar]
- Unger J, Merhof D, Renner S. 2016. Computer vision applied to herbarium specimens of German trees: testing the future utility of the millions of herbarium specimen images for automated identification. BMC Evolutionary Biology 16: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepitte K, de Meyer T, Helsen K, van Acker K, Roldán‐Ruiz I, Mergeay J, Honnay O. 2014. Rapid genetic adaptation precedes the spread of an exotic plant species. Molecular Ecology 23: 2157–2164. [DOI] [PubMed] [Google Scholar]
- Vellend M, Brown CD, Kharouba HM, McCune JL, Myers‐Smith IH. 2013. Historical ecology: using unconventional data sources to test for effects of global environmental change. American Journal of Botany 100: 1294–1305. [DOI] [PubMed] [Google Scholar]
- Vilà M, Espinar JL, Hejda M, Hulme PE, Jarošík V, Maron JL, Pergl J, Schaffner U, Sun Y, Pyšek P. 2011. Ecological impacts of invasive alien plants: a meta‐analysis of their effects on species, communities and ecosystems. Ecology Letters 14: 702–708. [DOI] [PubMed] [Google Scholar]
- Weiß CL, Schuenemann VJ, Devos J, Shirsekar G, Reiter E, Gould BA, Stinchcombe JR, Krause J, Burbano HA. 2016. Temporal patterns of damage and decay kinetics of DNA retrieved from plant herbarium specimens. Royal Society Open Science 3: 160239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis CG, Ellwood ER, Primack RB, Davis CC, Pearson KD, Gallinat AS, Yost JM, Nelson G, Mazer SJ, Rossington NL et al 2017. Old Plants, new tricks: phenological research using herbarium specimens. Trends in Ecology & Evolution 32: 531–546. [DOI] [PubMed] [Google Scholar]
- Winter M, Schweiger O, Klotz S, Nentwig W, Andriopoulos P, Arianoutsou M, Basnou C, Delipetrou P, Didziulis V, Hejda M et al 2009. Plant extinctions and introductions lead to phylogenetic and taxonomic homogenization of the European flora. Proceedings of the National Academy of Sciences, USA 106: 21 721–21 725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf A, Zimmerman NB, Anderegg WRL, Busby PE, Christensen J. 2016. Altitudinal shifts of the native and introduced flora of California in the context of 20th‐century warming. Global Ecology and Biogeography 25: 418–429. [Google Scholar]
- Woodward FI. 1987. Stomatal numbers are sensitive to increases in CO2 from pre‐industrial levels. Nature 327: 617–618. [Google Scholar]
- Yesson C, Brewer PW, Sutton T, Caithness N, Pahwa JS, Burgess M, Gray WA, White RJ, Jones AC, Bisby FA et al 2007. How global is the global biodiversity information facility? PLoS ONE 2: e1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Burbano HA, Krause J, Thines M, Weigel D, Kamoun S. 2014. Mining herbaria for plant pathogen genomes: back to the future. PLoS Pathogens 10: e1004028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Schuenemann VJ, Cano LM, Pais M, Mishra B, Sharma R, Lanz C, Martin FN, Kamoun S, Krause J et al 2013. The rise and fall of the Phytophthora infestans lineage that triggered the Irish potato famine. eLife 2: e00731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A, Boyle T, Brown T. 1996. The population genetic consequences of habitat fragmentation for plants. Trends in Ecology & Evolution 11: 413–418. [DOI] [PubMed] [Google Scholar]
- Youngsteadt E, Dale AG, Terando AJ, Dunn RR, Frank SD. 2015. Do cities simulate climate change? A comparison of herbivore response to urban and global warming. Global Change Biology 21: 97–105. [DOI] [PubMed] [Google Scholar]
- Zangerl AR, Berenbaum MR. 2005. Increase in toxicity of an invasive weed after reassociation with its coevolved herbivore. Proceedings of the National Academy of Sciences, USA 102: 15529–15532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohner CM, Renner SS. 2014. Common garden comparison of the leaf‐out phenology of woody species from different native climates, combined with herbarium records, forecasts long‐term change. Ecology Letters 17: 1016–1025. [DOI] [PubMed] [Google Scholar]
- Zschau T, Getty S, Gries C, Ameron Y, Zambrano A, Nash TH 3rd. 2003. Historical and current atmospheric deposition to the epilithic lichen Xanthoparmelia in Maricopa County, Arizona. Environmental Pollution 125: 21–30. [DOI] [PubMed] [Google Scholar]


