Abstract
Pioneer transcription factors play a primary role in establishing competence for gene expression and initiating cellular programming and reprogramming, and their dysregulation causes severe effects on human health, such as promoting tumorigenesis. Although more than 200 transcription factors are expressed in each cell type, only a small number of transcription factors are necessary to elicit dramatic cell‐fate changes in embryonic development and cell‐fate conversion. Among these key transcription factors, a subset called “pioneer transcription factors” have a remarkable ability to target nucleosomal DNA, or closed chromatin, early in development, often leading to the local opening of chromatin, thereby establishing competence for gene expression. Although more key transcription factors have been identified as pioneer transcription factors, the molecular mechanisms behind their special properties are only beginning to be revealed. Understanding the pioneering mechanisms will enhance our ability to precisely control cell fate at will for research and therapeutic purposes.
This article is categorized under:
Biological Mechanisms > Cell Fates
Biological Mechanisms > Regulatory Biology
Developmental Biology > Lineages
Keywords: chromatin, epigenetic, pioneer transcription factor
1. INTRODUCTION
Gene regulatory mechanisms have evolved along with ways to package genomic DNA. In prokaryotes, genomic DNA is loosely packaged by histone‐like proteins, which are no obstacle to RNA polymerase gaining access to target DNA. Therefore, the ground state for prokaryotic transcription is nonrestrictive (Struhl, 1999). By contrast, in eukaryotes, genomic DNA is tightly packaged by an octamer of the four core histone proteins (H2A, H2B, H3, and H4) to make an array of nucleosomes, where linker histones facilitate further compaction to create a higher order chromatin structure. Thus, chromatin structure becomes a general obstacle for eukaryotic transcription by blocking the access of the basic RNA polymerase machinery, as well as most transcription factors. Interestingly, the DNA sequences at promoters, enhancers, and transcription factor‐binding sites generally have high affinity for histone octamers, which results in higher nucleosome occupancy (Tillo et al., 2010). Therefore, transcription factors and transcriptional machinery need to overcome this nucleosome barrier to activate their target genes.
Cell‐type‐specific gene regulation relies on transcription factors, which can bind directly to genomic DNA with specific sequences through their DNA‐binding domains. Most strikingly, only four transcription factors (Oct3/4, Sox2, Klf4, and c‐Myc) are needed to reprogram fibroblasts to induced pluripotent stem cells (iPSCs) (Takahashi & Yamanaka, 2006). Among these key transcription factors, a subset called “pioneer transcription factors” or “pioneer factors” have an intrinsic ability to target nucleosomal DNA sites in closed chromatin, often leading to the local opening of chromatin, thereby creating a permissive state for gene activation (Cirillo et al., 2002). In the context of embryonic development, pioneer factors start being expressed in the early stages and play a role in establishing competence for specific cell fates (Iwafuchi‐Doi & Zaret, 2014; Zaret & Mango, 2016). Not surprisingly, the aberrant activation or inhibition of pioneer factors can compromise large‐scale chromatin structure and ultimately human health. In many forms of cancer, pioneer factors are misregulated, mutated, or amplified in their genomic locus; alternatively, the binding sites of pioneer factors are mutated (Jozwik & Carroll, 2012). Because I and others have already overviewed the pioneering phenomena in several biological contexts, this review mainly focuses on the molecular mechanisms by which pioneer factors bind to nucleosomal DNA in closed chromatin and subsequently open their target sites. We are beginning to understand the molecular mechanisms and more detailed analyses will greatly enhance our ability to manipulate cell fate for diverse research and therapeutic purposes.
2. THE MOLECULAR MECHANISMS UNDERLYING PIONEERING ACTIVITY
Although pioneer factors belong to diverse structural classes of transcription factors, such as FoxA, Gata, Oct3/4, Sox2, Klf4, Pax7, Ascl1, and p53, they have common features that include binding their target sites in closed chromatin and subsequent chromatin opening. Here, I review the mechanistic insight into pioneering activities (Figure 1), focusing on well‐characterized pioneer factors.
Figure 1.
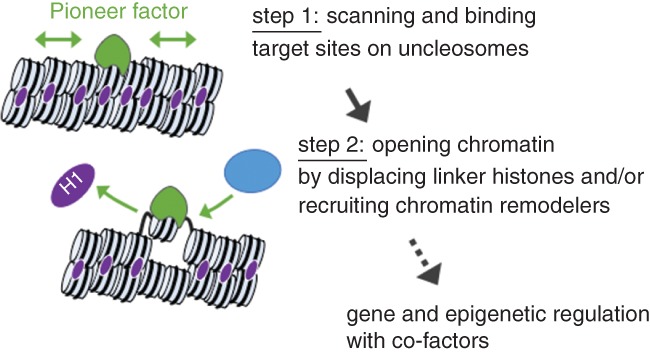
Steps of chromatin regulation by pioneer factors: binding, opening, and beyond
2.1. Step 1: Binding target sites on nucleosomes in closed chromatin
The FoxA family of transcription factors has been most extensively studied in mechanistic depth by biochemical, genetic, and genomic studies. The first evidence for the pioneering activity of FoxA came from attempts to model its binding with in vitro reconstituted nucleosomal arrays, which were compacted by the binding of linker histones. Remarkably, purified FoxA protein, and to a lesser extent Gata4, but not NF‐1 and C/EBP, can bind to their target sites on compacted nucleosomal DNA and open a local domain of compacted chromatin in vitro without ATP or ATP‐dependent chromatin remodelers (Cirillo et al., 2002). This nucleosome binding hierarchy of transcription factors is reflected in their mobility in nuclei. Fluorescence recovery after photobleaching assays revealed that FoxA moves much more slowly in nuclei than the other transcription factors tested. This slower nuclear mobility correlates with nonspecific chromatin binding, which is possibly describing chromatin scanning mechanisms of pioneer factors (Sekiya, Muthurajan, Luger, Tulin, & Zaret, 2009; Zaret, Lerner, & Iwafuchi‐Doi, 2016). Even during mitosis, FoxA and Gata1 remain bound in condensed chromatin and “bookmark” genes for rapid reactivation, as cells progress through the cell cycle (Caravaca et al., 2013; Kadauke et al., 2012). Notably, the DNA‐binding domain of FoxA possesses a “winged helix” structure, which binds to one side of a DNA helix and leaves the other side of DNA to bind a nucleosome core particle (Cirillo et al., 1998; Clark, Halay, Lai, & Burley, 1993; Ramakrishnan, Finch, Graziano, Lee, & Sweet, 1993). Thus, FoxA is structurally suited to recognize its DNA binding sites on a nucleosome.
The most dramatic example of cell‐fate conversion is the reprogramming of fibroblasts into iPSCs by only four transcription factors: Sox2 (HMG box DNA binding domain), Oct3/4 (Pou‐specific and Pou‐homeo‐domain), Klf4 (zinc finger DNA binding domain), and c‐Myc (bHLH DNA binding domain) (Takahashi & Yamanaka, 2006). c‐Myc alone prefers to engage open chromatin sites but can bind closed chromatin sites in conjunction with the other factors. In contrast, Sox2, Oct3/4, and Klf4 act as pioneer factors in that they can access closed chromatin whether they bind together or alone (Soufi, Donahue, & Zaret, 2012). It has also been shown that SOX2, OCT3/4, and KLF4 bookmark enhancers of stem cell‐related genes during mitosis in pluripotent stem cells, which play a role in maintaining pluripotency through cell division (Y. Liu et al., 2017). In contrast to the full DNA motif recognition of FoxA, the reprogramming factors (Sox2, Oct3/4, and Klf4) recognize one face of DNA on nucleosome by targeting a partial DNA sequence of their canonical binding motifs exposed on nucleosomes (Soufi et al., 2015). These findings introduce a predictive principle for pioneering activity and explain the nucleosome binding ability of a diverse set of transcription factors containing different structural classes of DNA binding domains.
Although pioneer factors can target closed chromatin, they are significantly enriched at only a fraction of their potential binding sites and exhibit cell‐type‐specific binding patterns (Hurtado, Holmes, Ross‐Innes, Schmidt, & Carroll, 2011; Lupien et al., 2008; Tsankov et al., 2015). However, a recent study showed that after decreasing the threshold for mapping its binding events, FOXA2 and GATA4 weakly occupied most target sites that were strongly occupied by them in other cell lines, and additional cofactors increase their occupancy (Donaghey et al., 2018). These new findings suggest that FOXA and GATA4 independently occupy diverse sites in common across different cells and allow cell‐type‐specific cofactors to access their target sites, which in turn stabilize the pioneer factors' occupancy. The binding of Oct3/4, Sox2, and Klf4 has also been shown to be facilitated by cofactors (Chen et al., 2014; Soufi et al., 2012; Z. Liu & Kraus, 2017).
The transcription factor p53 functions as a tumor suppressor that responds to a variety of stress‐inducing signals and is the most frequently mutated in all human tumors. As a consequence, p53 is one of the most extensively studied transcription factors in cancer research (Kastenhuber & Lowe, 2017). The first evidence of its pioneering activity was shown by a p53‐dependent in vitro transcription system using chromatin‐assembled p21 gene and its regulatory regions. In this system, p53 exhibited a higher affinity to its response site in the context of chromatin than in naked DNA and did not require chromatin remodeling or modifying complexes to interact with chromatin (Espinosa & Emerson, 2001). An in vivo study also showed that p53 was prebound to p21 regulatory sequences on nucleosomal DNA and its stable nucleosome binding required the binding sites positioned close to the edge, but not the center, of nucleosomal DNA (Laptenko, Beckerman, Freulich, & Prives, 2011). This is an interesting distinction from FoxA, which binds preferentially near the center of nucleosomes (Iwafuchi‐Doi et al., 2016). On the other hand, the other lines of evidence showed that p53 binding to nucleosomal DNA depended more on the rotational positioning of DNA binding sites; the strongest p53 binding occurred when the binding sites were exposed on the nucleosome surface and were bent in the same directions as observed for the p53‐DNA complex (Cui & Zhurkin, 2014; Sahu et al., 2010). Notably, strong p53‐bound sites reside preferentially within genomic regions whose DNA sequences encode high intrinsic nucleosome occupancy (Lidor Nili et al., 2010). Genome‐wide correlation between p53 binding and the dynamics of the local chromatin environment indicates that p53 could bind to the sites within closed chromatin lacking other chromatin features of active enhancers (Sammons, Zhu, Drake, & Berger, 2015). Another p53 family member, p63, plays an essential role in epidermal development, and its malfunction is involved in severe developmental disorders (Soares & Zhou, 2018). p63 has also been reported to function as a pioneer factor; its DNA binding domain shares high sequence and structural homology with p53 and can bind to nucleosomes in closed chromatin (Bao et al., 2015). It would be important to address how the mutant forms of p53 and p63 alter their pioneering activity in human tumors and epidermal defects, which will provide further insight into the molecular mechanisms for developing therapeutic options.
2.2. Step 2: Opening chromatin
As the second step of pioneering activity, pioneer factors locally open chromatin structure and create a permissive state for gene regulation. In contrast to the chromatin binding step, the molecular mechanisms underlying the chromatin opening step remain to be determined for most pioneer factors.
The chromatin opening that follows FoxA binding does not require ATP or ATP‐dependent chromatin remodelers (Cirillo et al., 2002), but we cannot rule out the possibility that chromatin remodelers are recruited to FoxA pioneering sites for further chromatin remodeling. A FoxA2 ChIP‐exo‐seq (chromatin immunoprecipitation with lambda exonuclease digestion followed by sequencing) experiment in adult mouse livers identified FoxA2 binding sites in single nucleotide resolution throughout the genome and revealed that FoxA2 binding events were enriched near the dyad axis (center) of the nucleosome (Iwafuchi‐Doi et al., 2016). Interestingly, the “winged helix” DNA‐binding domain of FoxA structurally resembles the globular, nucleosomal binding domain of linker histones (Clark et al., 1993; Ramakrishnan et al., 1993), and linker histones also bind near the dyad axis of the nucleosome (Goytisolo et al., 1996). in vitro biochemical and in vivo knockout studies indicate that FoxA binding locally displaces linker histones from the closed chromatin and keeps the nucleosome accessible (Cirillo et al., 1998; Iwafuchi‐Doi et al., 2016), providing a simple explanation for the underlying nucleosomes becoming more accessible. Furthermore, the C‐terminal domain of FoxA interacts directly with core histone proteins and is required for opening chromatin (Cirillo et al., 2002). The interaction with core histones could stabilize FoxA binding and enhance the displacement of linker histones. Similarly, a C‐terminal domain of EBF1, a key transcription factor for B cell programming, is required for chromatin opening at the majority of EBF1‐inducible DNase‐hypersensitive sites (Boller et al., 2016). It remains to be determined whether the C‐terminal domain of EBF1 directly interacts with chromatin or coregulatory factors for chromatin opening. These studies demonstrate the role of non‐DNA‐binding domains in pioneering activity.
As many transcription factors function by recruiting transcriptional coregulatory complexes, including chromatin remodelers, some pioneer factors can recruit chromatin remodelers and open chromatin structure. Large‐scale proteomic studies indicate that the pioneer factor OCT3/4 interacts with major chromatin remodeling factors (Ding, Xu, Faiola, Ma'ayan, & Wang, 2012; Pardo et al., 2010; van den Berg et al., 2010). A genetics and genomics study shows that OCT3/4 recruits the chromatin remodeler BRG1 (SMARCA4) and establishes accessible chromatin structure in pluripotent stem cells (King & Klose, 2017). The linker between the two Oct3/4 DNA‐binding domains (Pou‐specific and Pou‐homeodomain) functions as a protein–protein interaction interface with BRG1 and is required for the reprogramming activity of Oct3/4 (Esch et al., 2013). More pioneer factors, such as Sox2, p63, Gata, and AP‐1, have also been reported to recruit chromatin remodelers to their target sites (Bao et al., 2015; Engelen et al., 2011; Hu et al., 2011; Takaku et al., 2016; Vierbuchen et al., 2017). Further studies are required to determine the extent to which chromatin opening activity relies on the support of chromatin remodelers and whether the pioneer factors have the intrinsic ability to open chromatin by themselves as FoxA does.
2.3. Beyond the pioneering activity—epigenetic regulator?
The term “epigenetics” describes how a cell retains memory of past states and environmental influences. Pioneer factors actually have some of the required properties of an epigenetic regulator but are usually underappreciated in the epigenetics phenomena (Henikoff & Greally, 2016; Ptashne, 2013).
Mammalian DNA methylation is crucial for gene silencing, X‐inactivation, and cell‐fate determination. It has been demonstrated that pioneer factors FOXA and Pax7 are involved in the DNA demethylation process. After FOXA binding events, loss of DNA methylation was observed at some FOXA binding sites in a DNA replication‐dependent manner, suggesting passive mechanisms to block the maintenance of DNA methylation (Donaghey et al., 2018). Genome‐wide DNA methylomes and biochemical studies demonstrate that FOXA is associated with DNA repair complexes and regulates DNA demethylation at its genomic targets in a lineage‐specific fashion (Zhang et al., 2016). Furthermore, the pioneering action of Pax7 leads to loss of DNA methylation and ensures long‐term chromatin accessibility even after Pax7 withdrawal, at least for a time frame of 12 cell divisions (Mayran et al., 2018).
Other epigenetic changes, which are either acquired de novo or increased histone H3K4 methylation, follow or are concomitant with FoxA induction (Donaghey et al., 2018; Sérandour et al., 2011; Wang et al., 2015). A mass spectrometry using endogenous proteins in breast cancer cells identified histone‐lysine N‐methyltransferase (MLL3) as a top FOXA1‐interacting protein (Jozwik, Chernukhin, Serandour, Nagarajan, & Carroll, 2016). Genome‐wide occupancy of FOXA1 and MLL3 and their knockdown studies suggest that FOXA1 can recruit MLL3 to deposit H3K4 methylation on FOXA pioneering sites (Jozwik et al., 2016). Another example is that the p53 activation domain recruits histone acetyltransferase p300, establishes histone acetylation, and increases the ability to activate target gene expression (Espinosa & Emerson, 2001).
A recent report demonstrated that cell‐type‐restricted enhancers are “premarked” by binding of pluripotency transcription factors, including pioneer factors OCT3/4 and SOX2, in embryonic stem (ES) cells (Kim et al., 2018). This premarking is required for the robust future enhancer activation in differentiated cells. Interestingly, these enhancers are in an open configuration but do not exhibit canonical enhancer histone modifications (H3K27ac, H3K4me1/2) in ES cells. The persistent engagement of pioneer factors or handing over pioneering sites to other pioneer factors could be the key mechanism to transmit cellular memory without canonical histone modifications, namely “epigenetics” regulation by pioneer factors.
3. CONCLUSION
As seen from the above discussion, we are beginning to understand the molecular mechanisms underlying pioneering activities. A recent study showed that the pioneer factor Pax7 binds to its target sites rapidly, but subsequent chromatin opening and gene activation events occur slowly (Mayran et al., 2018). These findings suggest that the binding of pioneer factors is mechanistically separate from subsequent events. Given that misregulation of pioneer factors can compromise human health, a more detailed and comprehensive mechanistic understanding may enhance our ability to manipulate cell identity for therapeutic purposes.
CONFLICT OF INTEREST
The author has declared no conflicts of interest for this article.
RELATED WIREs ARTICLES
Genome‐wide maps of transcription regulatory elements
Dynamic enhancer function in the chromatin context
Cell‐specific integration of nuclear receptor function at the genome
Estrogen receptor‐positive breast cancer: a multidisciplinary challenge
A systems view of epigenetic networks regulating pancreas development and ß‐cell function
ACKNOWLEDGMENTS
I thank Ken Zaret and Marissa Granitto for comments on the manuscript.
Iwafuchi‐Doi M. The mechanistic basis for chromatin regulation by pioneer transcription factors. WIREs Syst Biol Med. 2019;11:e1427. 10.1002/wsbm.1427
REFERENCES
- Bao, X. , Rubin, A. J. , Qu, K. , Zhang, J. , Giresi, P. G. , Chang, H. Y. , & Khavari, P. A. (2015). A novel ATAC‐seq approach reveals lineage‐specific reinforcement of the open chromatin landscape via cooperation between BAF and p63. Genome Biology, 16(1), 1–17. 10.1186/s13059-015-0840-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller, S. , Ramamoorthy, S. , Akbas, D. , Nechanitzky, R. , Burger, L. , Murr, R. , … Grosschedl, R. (2016). Pioneering activity of the C‐terminal domain of EBF1 shapes the chromatin landscape for B cell programming. Immunity, 44(3), 527–541. 10.1016/j.immuni.2016.02.021 [DOI] [PubMed] [Google Scholar]
- Caravaca, J. M. , Donahue, G. , Becker, J. S. , He, X. , Vinson, C. , & Zaret, K. S. (2013). Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes. Genes & Development, 27(3), 251–260. 10.1101/gad.206458.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. , Zhang, Z. , Li, L. , Chen, B.‐C. , Revyakin, A. , Hajj, B. , … Liu, Z. (2014). Single‐molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell, 156(6), 1274–1285. 10.1016/j.cell.2014.01.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo, L. A. , Lin, F. R. , Cuesta, I. , Friedman, D. , Jarnik, M. , & Zaret, K. S. (2002). Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA‐4. Molecular Cell, 9(2), 279–289. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/11864602 [DOI] [PubMed] [Google Scholar]
- Cirillo, L. A. , McPherson, C. E. , Bossard, P. , Stevens, K. , Cherian, S. , Shim, E. Y. , … Zaret, K. S. (1998). Binding of the winged‐helix transcription factor HNF3 to a linker histone site on the nucleosome. The EMBO Journal, 17(1), 244–254. 10.1093/emboj/17.1.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, K. L. , Halay, E. D. , Lai, E. , & Burley, S. K. (1993). Co‐crystal structure of the HNF‐3/fork head DNA‐recognition motif resembles histone H5. Nature, 364(6436), 412–420. 10.1038/364412a0 [DOI] [PubMed] [Google Scholar]
- Cui, F. , & Zhurkin, V. B. (2014). Rotational positioning of nucleosomes facilitates selective binding of p53 to response elements associated with cell cycle arrest. Nucleic Acids Research, 42(2), 836–847. 10.1093/nar/gkt943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, J. , Xu, H. , Faiola, F. , Ma'ayan, A. , & Wang, J. (2012). Oct4 links multiple epigenetic pathways to the pluripotency network. Cell Research, 22(1), 155–167. 10.1038/cr.2011.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaghey, J. , Thakurela, S. , Charlton, J. , Chen, J. S. , Smith, Z. D. , Gu, H. , … Meissner, A. (2018). Genetic determinants and epigenetic effects of pioneer‐factor occupancy. Nature Genetics, 50(2), 250–258. 10.1038/s41588-017-0034-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelen, E. , Akinci, U. , Bryne, J. C. , Hou, J. , Gontan, C. , Moen, M. , … Poot, R. A. (2011). Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nature Genetics, 43(6), 607–611. 10.1038/ng.825 [DOI] [PubMed] [Google Scholar]
- Esch, D. , Vahokoski, J. , Groves, M. R. , Pogenberg, V. , Cojocaru, V. , Vom Bruch, H. , … Schöler, H. R. (2013). A unique Oct4 interface is crucial for reprogramming to pluripotency. Nature Cell Biology, 15(3), 295–301. 10.1038/ncb2680 [DOI] [PubMed] [Google Scholar]
- Espinosa, J. M. , & Emerson, B. M. (2001). Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site‐directed cofactor recruitment. Molecular Cell, 8(1), 57–69. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/11511360 [DOI] [PubMed] [Google Scholar]
- Goytisolo, F. A. , Gerchman, S. E. , Yu, X. , Rees, C. , Graziano, V. , Ramakrishnan, V. , & Thomas, J. O. (1996). Identification of two DNA‐binding sites on the globular domain of histone H5. The EMBO Journal, 15(13), 3421–3429. Retrieved from http://www.ncbi.nlm.nih.gov/pmc/articles/PMC451906/ [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S. , & Greally, J. M. (2016). Epigenetics, cellular memory and gene regulation. Current Biology, 26(14), R644–R648. 10.1016/j.cub.2016.06.011 [DOI] [PubMed] [Google Scholar]
- Hu, G. , Schones, D. E. , Cui, K. , Ybarra, R. , Northrup, D. , Tang, Q. , … Zhao, K. (2011). Regulation of nucleosome landscape and transcription factor targeting at tissue‐specific enhancers by BRG1. Genome Research, 21(10), 1650–1658. 10.1101/gr.121145.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado, A. , Holmes, K. A. , Ross‐Innes, C. S. , Schmidt, D. , & Carroll, J. S. (2011). FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nature Genetics, 43(1), 27–33. 10.1038/ng.730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi‐Doi, M. , Donahue, G. , Kakumanu, A. , Watts, J. A. , Mahony, S. , Pugh, B. F. , … Zaret, K. S. (2016). The pioneer transcription factor FoxA maintains an accessible nucleosome configuration at enhancers for tissue‐specific gene activation. Molecular Cell, 62(1), 79–91. 10.1016/j.molcel.2016.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi‐Doi, M. , & Zaret, K. S. (2014). Pioneer transcription factors in cell reprogramming. Genes & Development, 28(24), 2679–2692. 10.1101/gad.253443.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jozwik, K. M. , & Carroll, J. S. (2012). Pioneer factors in hormone‐dependent cancers. Nature Reviews Cancer, 12(6), 381–385. 10.1038/nrc3263 [DOI] [PubMed] [Google Scholar]
- Jozwik, K. M. , Chernukhin, I. , Serandour, A. A. , Nagarajan, S. , & Carroll, J. S. (2016). FOXA1 directs H3K4 monomethylation at enhancers via recruitment of the methyltransferase MLL3. Cell Reports, 17(10), 2715–2723. 10.1016/j.celrep.2016.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadauke, S. , Udugama, M. I. , Pawlicki, J. M. , Achtman, J. C. , Jain, D. P. , Cheng, Y. , … Blobel, G. A. (2012). Tissue‐specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell, 150(4), 725–737. 10.1016/j.cell.2012.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastenhuber, E. R. , & Lowe, S. W. (2017). Putting p53 in context. Cell, 170(6), 1062–1078. 10.1016/j.cell.2017.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. S. , Tan, Y. , Ma, W. , Merkurjev, D. , Destici, E. , Ma, Q. , … Rosenfeld, M. G. (2018). Pluripotency factors functionally premark cell‐type‐restricted enhancers in ES cells. Nature, 556(7702), 510–514. 10.1038/s41586-018-0048-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, H. W. , & Klose, R. J. (2017). The pioneer factor OCT4 requires the chromatin remodeller BRG1 to support gene regulatory element function in mouse embryonic stem cells. eLife, 6, 1–24. 10.7554/eLife.22631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laptenko, O. , Beckerman, R. , Freulich, E. , & Prives, C. (2011). P53 binding to nucleosomes within the P21 promoter in vivo leads to nucleosome loss and transcriptional activation. Proceedings of the National Academy of Sciences of the United States of America, 108(26), 10385–10390. 10.1073/pnas.1105680108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidor Nili, E. , Field, Y. , Lubling, Y. , Widom, J. , Oren, M. , & Segal, E. (2010). p53 binds preferentially to genomic regions with high DNA‐encoded nucleosome occupancy. Genome Research, 20(10), 1361–1368. 10.1101/gr.103945.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Pelham‐Webb, B. , Di Giammartino, D. C. , Li, J. , Kim, D. , Kita, K. , … Apostolou, E. (2017). Widespread mitotic bookmarking by histone marks and transcription factors in pluripotent stem cells. Cell Reports, 19(7), 1283–1293. 10.1016/j.celrep.2017.04.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z. , & Kraus, W. L. (2017). Catalytic‐independent functions of PARP‐1 determine Sox2 pioneer activity at intractable genomic loci. Molecular Cell, 65(4), 589–603.e9. 10.1016/j.molcel.2017.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien, M. , Eeckhoute, J. , Meyer, C. a. , Wang, Q. , Zhang, Y. , Li, W. , … Brown, M. (2008). FoxA1 translates epigenetic signatures into enhancer‐driven lineage‐specific transcription. Cell, 132(6), 958–970. 10.1016/j.cell.2008.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayran, A. , Khetchoumian, K. , Hariri, F. , Pastinen, T. , Gauthier, Y. , Balsalobre, A. , & Drouin, J. (2018). Pioneer factor Pax7 deploys a stable enhancer repertoire for specification of cell fate. Nature Genetics, 50(2), 259–269. 10.1038/s41588-017-0035-2 [DOI] [PubMed] [Google Scholar]
- Pardo, M. , Lang, B. , Yu, L. , Prosser, H. , Bradley, A. , Babu, M. M. , & Choudhary, J. (2010). An expanded Oct4 interaction network: Implications for stem cell biology, development, and disease. Cell Stem Cell, 6(4), 382–395. 10.1016/j.stem.2010.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne, M. (2013). Epigenetics: Core misconcept. Proceedings of the National Academy of Sciences of the United States of America, 110(18), 7101–7103. 10.1073/pnas.1305399110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan, V. , Finch, J. T. , Graziano, V. , Lee, P. L. , & Sweet, R. M. (1993). Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature, 362(6417), 219–223. 10.1038/362219a0 [DOI] [PubMed] [Google Scholar]
- Sahu, G. , Wang, D. , Chen, C. B. , Zhurkin, V. B. , Harrington, R. E. , Appella, E. , … Nagaich, A. K. (2010). p53 binding to nucleosomal DNA depends on the rotational positioning of DNA response element. Journal of Biological Chemistry, 285(2), 1321–1332. 10.1074/jbc.M109.081182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammons, M. A. , Zhu, J. , Drake, A. M. , & Berger, S. L. (2015). TP53 engagement with the genome occurs in distinct local chromatin environments via pioneer factor activity. Genome Research, 25(2), 179–188. 10.1101/gr.181883.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya, T. , Muthurajan, U. M. , Luger, K. , Tulin, A. V. , & Zaret, K. S. (2009). Nucleosome‐binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes & Development, 23(7), 804–809. 10.1101/gad.1775509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sérandour, A. A. , Avner, S. , Percevault, F. , Demay, F. , Bizot, M. , Lucchetti‐Miganeh, C. , … Eeckhoute, J. (2011). Epigenetic switch involved in activation of pioneer factor FOXA1‐dependent enhancers. Genome Research, 21(4), 555–565. 10.1101/gr.111534.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares, E. , & Zhou, H. (2018). Master regulatory role of p63 in epidermal development and disease. Cellular and Molecular Life Sciences, 75(7), 1179–1190. 10.1007/s00018-017-2701-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi, A. , Donahue, G. , & Zaret, K. S. (2012). Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell, 151(5), 994–1004. 10.1016/j.cell.2012.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi, A. , Garcia, M. F. , Jaroszewicz, A. , Osman, N. , Pellegrini, M. , & Zaret, K. S. (2015). Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell, 161(3), 555–568. 10.1016/j.cell.2015.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl, K. (1999). Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell, 98(1), 1–4. 10.1016/S0092-8674(00)80599-1 [DOI] [PubMed] [Google Scholar]
- Takahashi, K. , & Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell, 126(4), 663–676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Takaku, M. , Grimm, S. A. , Shimbo, T. , Perera, L. , Menafra, R. , Stunnenberg, H. G. , … Wade, P. A. (2016). GATA3‐dependent cellular reprogramming requires activation‐domain dependent recruitment of a chromatin remodeler. Genome Biology, 17(1), 36 10.1186/s13059-016-0897-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillo, D. , Kaplan, N. , Moore, I. K. , Fondufe‐Mittendorf, Y. , Gossett, A. J. , Field, Y. , … Hughes, T. R. (2010). High nucleosome occupancy is encoded at human regulatory sequences. PLoS One, 5(2), e9129 10.1371/journal.pone.0009129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankov, A. M. , Gu, H. , Akopian, V. , Ziller, M. J. , Donaghey, J. , Amit, I. , … Meissner, A. (2015). Transcription factor binding dynamics during human ES cell differentiation. Nature, 518(7539), 344–349. 10.1038/nature14233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg, D. L. , Snoek, T. , Mullin, N. P. , Yates, A. , Bezstarosti, K. , Demmers, J. , … Poot, R. A. (2010). An Oct4‐centered protein interaction network in embryonic stem cells. Cell Stem Cell, 6(4), 369–381. 10.1016/j.stem.2010.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen, T. , Ling, E. , Cowley, C. J. , Couch, C. H. , Wang, X. , Harmin, D. A. , … Greenberg, M. E. (2017). AP‐1 transcription factors and the BAF complex mediate signal‐dependent enhancer selection. Molecular Cell, 68(6), 1067–1082.e12. 10.1016/j.molcel.2017.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, A. , Yue, F. , Li, Y. , Xie, R. , Harper, T. , Patel, N. A. , … Sander, M. (2015). Epigenetic priming of enhancers predicts developmental competence of hESC‐derived endodermal lineage intermediates. Cell Stem Cell, 16(4), 386–399. 10.1016/j.stem.2015.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret, K. S. , Lerner, J. , & Iwafuchi‐Doi, M. (2016). Chromatin scanning by dynamic binding of pioneer factors. Molecular Cell, 62(5), 665–667. 10.1016/j.molcel.2016.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret, K. S. , & Mango, S. E. (2016). Pioneer transcription factors, chromatin dynamics, and cell fate control. Current Opinion in Genetics & Development, 37, 76–81. 10.1016/j.gde.2015.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Zhang, D. , Li, Q. , Liang, J. , Sun, L. , Yi, X. , … Shang, Y. (2016). Nucleation of DNA repair factors by FOXA1 links DNA demethylation to transcriptional pioneering. Nature Genetics, 48(9), 1003–1013. 10.1038/ng.3635 [DOI] [PubMed] [Google Scholar]


