Abstract
Primary Sjögren's syndrome (pSS) is a complex systemic autoimmune disease with heterogeneous disease manifestations. Genetic predisposition, hormonal and environmental factors are all thought to contribute to disease etiology and pathogenesis. A better understanding of the disease pathogenesis is required in order to establish new targeted therapies. We analysed MAPK/ERK and JAK/STAT signalling networks in peripheral blood mononuclear cells (PBMCs) upon stimulation with interferon alpha 2b (IFN‐α2b) by flow cytometry to define potentially dysfunctional intracellular signalling pathways involved in disease pathogenesis. Cells derived from pSS patients displayed small but significant increases in basal phosphorylation levels of numerous signalling proteins compared to cells from healthy donors. The phosphorylation profiles following stimulation with IFNα2b differed significantly between pSS patients and healthy donors, especially regarding STAT1 Y701. PCA further grouped patients according to clinical characteristics. Type I IFN induced gene expression was found to negatively correlate with the IFN‐α2b induced phosphorylation of STAT3 S727 in T cells and positively with pSTAT1 Y701 in B cells. Increases in pSTAT1 Y701 were associated with the presence of autoantibodies. Our results indicate involvement of both STAT3 S727 and STAT1 Y701 pathways in pSS patients. Therapies targeting these pathways might therefore be beneficial for certain subgroups of patients.
Keywords: Autoantibodies, Extraglandular manifestations, Phosphoflow, Sjögren's syndrome, Type I interferon
Introduction
Sjögren's syndrome (SS) is a complex systemic autoimmune disease characterized by lymphocytic infiltrates of exocrine glands, mainly the salivary and lacrimal glands, leading to dryness of the mouth (xerostomia) and the eyes (keratoconjunctivitis sicca) 1. Hallmarks of SS include the presence of autoantibodies against anti‐Sjögren's syndrome A (SSA) and anti‐Sjögren's syndrome B (SSB) 2 which may be present in serum decades before clinical disease manifestations 3, 4. There is no cure or effective treatment for SS, with management of the disease based on the relief of symptoms. Patients suffer from a significant decrease in quality of life. A lack of effective targeted treatments is linked to the complexity of the disease, with genetic predisposition, hormonal and environmental factors all contributing to disease etiology and pathogenesis. While most SS patients display reduced tear and saliva secretion 5, there is significant heterogeneity in other disease features, pathology and clinical course. Sjögren's syndrome can present a wide range of extraglandular manifestations (EGM) including fatigue, musculoskeletal involvement (arthralgias, myalgias), skin involvement (xerosis, purpura), pulmonary involvement (bronchiectasis, obstructive airway disease), involvement of the liver and kidneys, neuropathy and lymphoma 2. This heterogeneity has been postulated to reflect distinct patient subsets, driven by unique pathophysiological mechanisms 6.
Aspects of SS pathogenesis that have gained considerable attention during the past years are features associated with type I interferon (IFN). An activated type I IFN system known as the IFN signature plays an important role in several autoimmune diseases 7, 8. In addition, polymorphisms in the genes encoding the transcription factors STAT4 and IRF5, which play a role in type I IFN signalling, have been associated with SS 9, 10. These polymorphisms have been speculated to confer a susceptibility favoring a higher IFN response which may play a role in onset or perpetuation of the disease 11. Gene expression may in addition be altered through changes in responsiveness to a given stimulus. Previous studies of peripheral blood cells from SS patients have found alterations of basal phosphorylation levels of STAT3 and STAT5 12, 13, as well as increased phosphorylation of STAT1 Y701 upon stimulation with IFN‐α, IFN‐γ and IL‐6 14.
Since approximately 50% of SS patients have an activated type I IFN system 15, we here investigated signaling networks upon stimulation with IFN‐α2b. Single cell phospho‐specific flow cytometry (phosphoflow) was used to analyse the phosphorylation status of nine different intracellular phospho‐epitopes in peripheral blood cells from primary SS patients. Moreover, basal and IFN‐α induced phosphorylation of intracellular phospho‐epitopes were correlated to expression levels of IFN responsive genes. Increased phosphorylation of STAT1 Y701 was observed in B cells following stimulation with IFN‐α, which strongly correlated with type I IFN inducible gene expression in PBMC and presence of autoantibodies. In contrast, a negative association was found in T cells for STAT3 S727 with type I IFN inducible gene expression. Signalling pathways involving these transcription factors may be involved in the aberrant induction of type I IFN inducible genes in pSS patients who might benefit from therapies targeting these processes.
Results
Patient PBMCs display altered phosphorylation levels of proteins involved in signalling pathways
In order to further stratify pSS patients and reveal new treatment targets, we here analysed MAPK/ERK and JAK/STAT signalling networks in peripheral blood cells from female pSS patients and female age‐matched healthy donors in unstimulated and IFN‐α2b stimulated PBMCs. An overview of the results are given in supplementary tables S1 (T cells), S2 (B cells) and S3 (NK cells).
We first compared basal phosphorylation levels between pSS patients and controls. Significantly increased phosphorylation of several epitopes was seen in T and NK cells of pSS patients, while B cells showed no significant differences (Fig. 1A). Principal component analysis (PCA) separated pSS patients from healthy donors (Fig. 1C). PCA was further used to concurrently relate multiple basal phosphorylation states to various clinical parameters such as presence of SSA autoantibodies (Fig. 1D), extraglandular manifestations (EGM) (Fig. 1E) and effect of prescribed medication (DMARDs and corticosteroids; Fig. 1F) within the patient cohort. Spatial groupings indicated closer similarities within the pSS and healthy donor cohorts than between the groups. Including clinical parameters in the analysis, patients without autoantibodies against SSA grouped closer to the healthy controls (Fig. 1D), while patients with EGM (Fig. 1E) and patients using prescribed medication (Fig. 1F) grouped throughout the pSS cluster.
Figure 1.
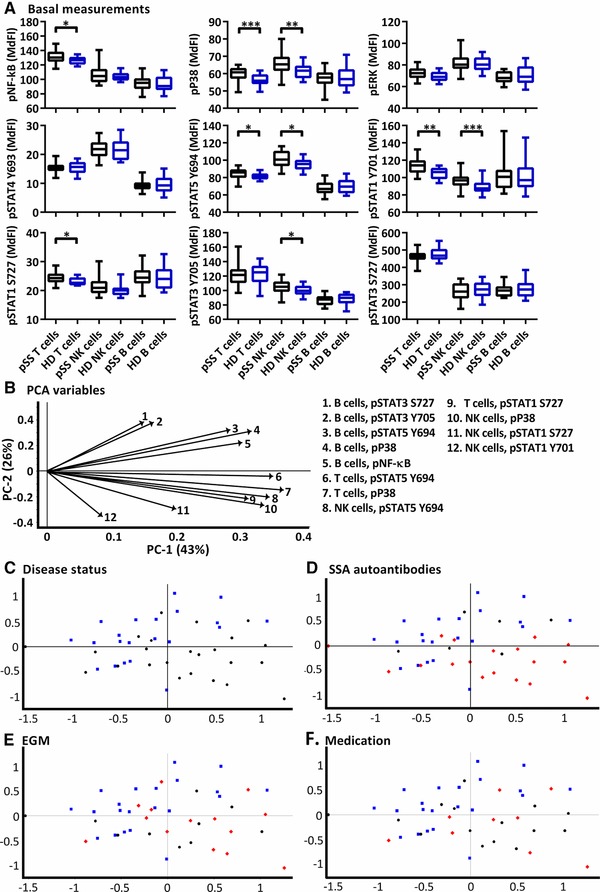
Basal phosphorylation profiles in B cells, T cells and NK cells of pSS patients compared to healthy controls. Phosphorylation levels of NF‐κB, ERK1/2, P38, STAT4 Y693, STAT5 Y694, STAT1 Y701, STAT1 S727, STAT3 Y705 and STAT3 S727 were analysed by flow cytometry. (A) Phosphorylation levels (MdFI) of various epitopes and immune cells of pSS patient (black) and healthy donors (blue). Comparisons between pairs were performed using an Unpaired Mann–Whitney test. Median, 25 to 75 percentiles and maximum and minimum are shown. Differences were considered statistically significant when p ≤ 0.05, with significance indicated as *≤0.05, **≤0.01, ***≤ = 0.001 and ****≤ = 0.0001. (B) Loading plot for the PCA containing information about the variable for the corresponding PC, with the key to the right indicating cell type and phospho protein. Variables are given by vectors, with those contributing little to the PCA plotted around the centre as denoted by the grey axis, while variables that have high contributions are plotted further from the axes. After initial calculation of principal components the model was recalculated with only variables explaining greater than 50% of the variance retained. Groupings of samples by (C) disease status, (D) presence of SSA autoantibodies, (E) presence of extraglandular manifestations (EGM), and (F) medication status. Healthy donors = blue square, pSS patients = black circles, and pSS patients positive for SSA, EGM or medication (D, E and F, respectively) = red diamonds. The data represents 21 healthy controls and 24 patients pooled from 13 independent experiments.
We next analysed phosphorylation states at different time points after stimulation with IFN‐α2b (Fig. 2). Reduced NF‐κB, P38, STAT4 Y693, STAT5 Y694 and STAT3 S727 phosphorylation was observed in cells from pSS patients. The phosphorylation profiles of IFN‐α2b stimulated T, NK and B cells were further investigated with PCA (Fig. 3). Phosphorylation levels after 15 minutes stimulation showed the strongest clustering of subgroups, while extended time course (˃15 min) gave no additional resolution (Supporting Information Fig. 1). We therefore focused all analyses on induced median fluorescence intensity (MdFI) at 15 min (MdFI15min – MdFIbasal) after stimulation with IFN‐α2b (Fig. 3A–E). Principal component analysis showed a positive shift for pSS samples along PC1 and PC2 away from healthy donor samples (Fig. 3B). PC1 explained 68% of the variation with positive movement along PC1 influenced by changes in phosphorylation of STAT1 Y701 in T cells and to a lesser degree NK and B cells (Fig. 3A). SSA negative patients and patients prescribed DMARDs or corticosteroids were distributed closer to healthy donors than SSA positive patients and patients without medication (Fig. 3C and E). Moreover, patients without EGM had little spread along PC1 (Fig. 3D). Further comparisons of variables used in the final PCA were conducted by Mann‐Whitney U tests (Fig. 3F). Comparisons were analysed with and without exclusion of patients prescribed DMARDs or corticosteroids. No statistically significant differences were observed between medicated patients and those without medication. However, although not significant, a trend of increased induction of pSTAT3 S727 in T cells was found in patients prescribed DMARDs or corticosteroids compared to patients without medication, making the phosphorylation profile more similar to that of healthy donors. Induction of pSTAT1 Y701 was found to be stronger in B cells from patients with SSA autoantibodies than SSA negative patients. Compared to healthy donors, pSS patients displayed stronger induction of pSTAT1 Y701 in T‐, B‐ and NK cells.
Figure 2.
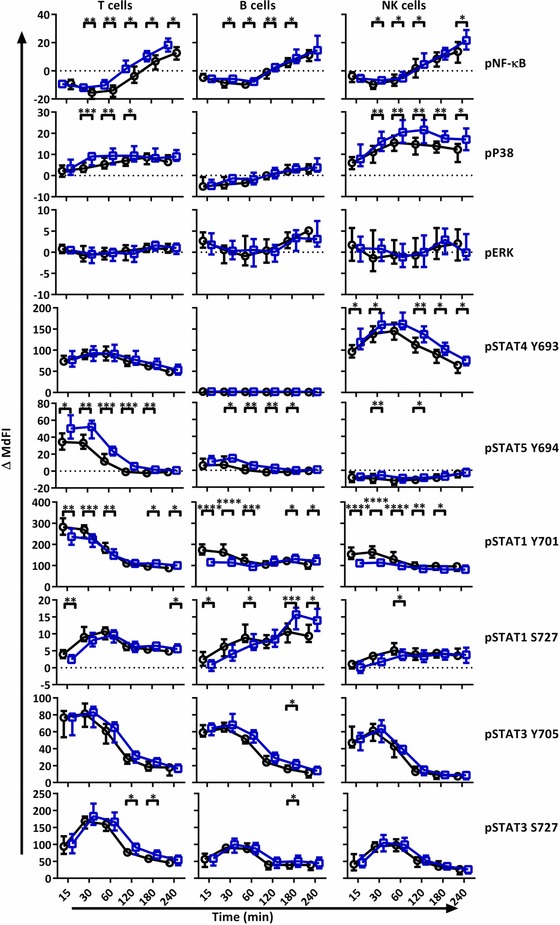
IFN‐α2b stimulation results in different phosphorylation profiles in T cells, B and NK cells of pSS patients compared to healthy controls. Phosphorylation levels of NF‐κB, p38, ERK1/2, STAT4 Y693, STAT5 Y694, STAT1 Y701, STAT1 S727, STAT3 Y705 and STAT3 S727 were analysed by flow cytometry at different time points after stimulation with IFN‐α2b. Comparisons of change of phosphorylation levels (ΔMdFI) from 0 min, between pSS patient (black) and healthy donors (blue) are given for each time point following stimulation with IFN‐α. Comparisons between pairs were done using an unpaired Mann–Whitney test. Line graphs show the median and 25 to 75 percentiles. The data represents 21 healthy controls and 24 patients pooled from 13 independent experiments. Differences were considered statistically significant when p ≤ 0.05, with significance indicated as *≤0.05, **≤0.01, ***≤0.001 and ****≤0.0001.
Figure 3.
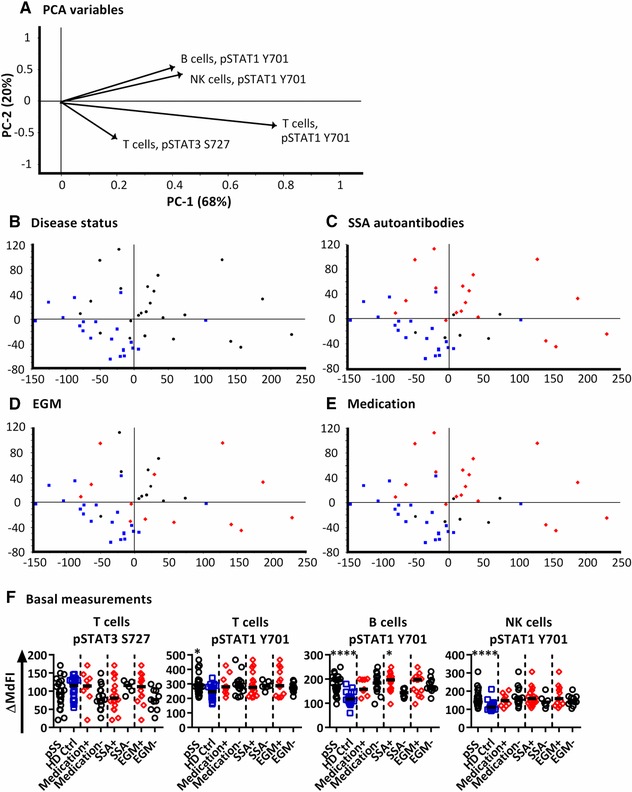
PCA analysis of induced phosphorylation in PBMCs after 15 min following stimulation with IFN‐α2b. (A) Loading plot containing information about the variables for the corresponding PCA with variables given as vectors. Variables contributing little to the PCA are plotted around the centre as denoted by the grey axis, while variables having high contributions are plotted further from the axes. After initial calculation of principal components the model was recalculated with only variable explaining greater than 50% of the variance retained. This was then followed by stepwise reduction of less significant variables with low variable leverage. Groupings of samples of by PCA are shown by (B) disease status, (C) presence of SSA autoantibodies, (D) presence of extraglandular manifestations (EGM) and (E) medication status of the pSS patients. Healthy donors = blue squares, pSS patients = black circles and pSS patients positive for SSA autoantibodies, EGM, or taking medication are shown as red diamonds (C, D, E respectively). (F) Scatterplots of variables used in the final PCA. The corresponding phospho‐protein and cell type is labelled above each plot. Groups are identified on the × axis (pSS patients (pSS) and healthy donors), and further subgrouping of the pSS patients into SSA autoantibody positive (SSA+) and negative patients (SSA−), patients with extraglandular manifestations (EGM+) and without EGM (EGM−), and patients using and not using medication (medication +/−) are indicated. Statistical comparisons were made between each of these pairs as indicated by dashed lines, with black bars representing the median. The data represents 21 healthy controls and 24 patients pooled from 13 independent experiments. Comparison between pairs was conducted using an Unpaired Mann‐Whitney test with significance indicated as *≤0.05, **≤0.01, ***≤0.001 and ****≤0.0001.
Patients with an activated IFN system have different phosphorylation profiles upon IFN‐α stimulation
To investigate whether the phosphorylation profile upon IFN‐α2b stimulation was altered in patients with an activated type I IFN system, we first determined mRNA levels of three type I IFN responsive genes (MxA, OAS1, IFI44) and 2 type II IFN responsive genes (GBP1, GBP2) in PBMCs from pSS patients (n = 19) and healthy donors (n = 14). The three type I IFN inducible genes were used to calculate IFN scores. The mean level and SD of each IFN inducible gene in the healthy control group were used to standardize expression levels of each gene for each study subject. The standardized expression levels were subsequently summed for each patient to calculate an IFN score. The threshold was set to 8.8 based on 3 × SD of healthy controls. A type I IFN signature was found in 63% of patients and 0% of controls (Fig. 4A). The IFN scores of patients prescribed DMARDs or corticosteroids did not differ significantly from those of patients without medication (Fig. 4B), but tended to be lower (median 12.10 versus 32.36).
Figure 4.
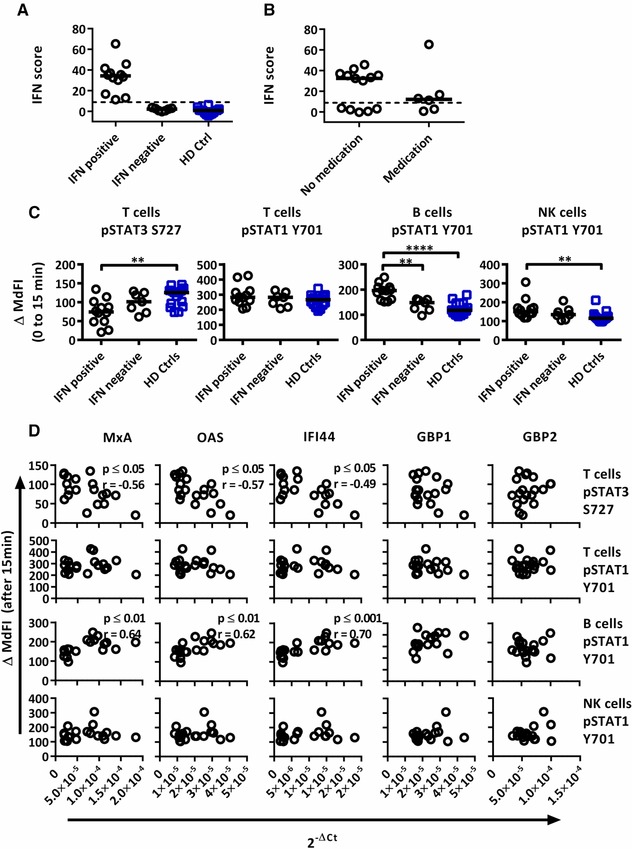
Associations of the phosphorylation profiles after IFN‐α2b stimulation with IFN inducible gene expression in pSS patients and healthy controls. Three type I IFN inducible genes (MxA, OAS1, IFI44) and two type II IFN inducible gene (GBP1, GBP2) were analysed. (A) An IFN score was calculated based on standardized expression levels of three type I IFN inducible genes (MxA, OAS1, IFI44). A threshold was set to 8.8 based on 3 × SD of healthy controls to identify patients as type I IFN signature positive or negative. (B) Comparisons between IFN score for patients prescribed DMARDs or corticosteroids and those without medication. (C) Unpaired Mann‐Whitney test comparisons of induced phosphorylation 15 min after IFN‐α stimulation for IFN signature positive patients (n = 12), IFN signature negative patients (n = 7) and healthy controls (n = 14). (D) Correlations of the phosphorylation profiles after IFN‐α2b stimulation with IFN inducible gene expression with three type I IFN inducible genes (MxA, OAS1, IFI44) and two type II IFN inducible genes (GBP1, GBP2) in pSS patients (n = 19). Outliers were removed using robust regression and outlier removal (ROUT) method and a ROUT coefficient Q of 1 was used Comparison between pairs (C) was conducted using an Unpaired Mann‐Whitney test and correlations (D) by Spearman rank test, with significance indicated as *≤0.05, **≤0.01, ***≤0.001 and ****≤0.0001.The flow cytometric data represents 15 healthy controls and 19 patients pooled from 13 independent experiments, with real time qPCR data representing a single experiment incorporating the 14 healthy controls and 19 patients.
Interestingly, the phosphorylation pattern upon in‐vitro stimulation with IFN‐α2b differed between the patient groups depending on IFN signature status (Supporting Information Table 4). Increased induction of STAT1 Y701 phosphorylation in B and NK cells was limited to type I IFN signature positive patients, while their T cells displayed reduced induction of STAT3 S727 phosphorylation (Fig. 4C). We further correlated expression levels of the individual IFN responsive genes to phosphorylation profiles of the pSS patients (Fig. 4D, Supporting Information Table 5). The observed induced phosphorylation of STAT1 Y701 in B cells correlated positively with increased expression of MxA, OAS1 and IFI44. In contrast, no correlation with GBP1 and GBP2 expression was observed.
Discussion
Little is known about the effects of an activated type I IFN system on signalling profiles in primary Sjögren's syndrome. Understanding the mechanisms that contribute to these profiles are crucial in both, understanding the pathogenesis and the development of targeted therapies. We here investigated signalling profiles in PBMCs of pSS patients under basal conditions and upon stimulation with IFN‐α2b.
An increased response to IFN‐α through STAT1 Y701 was observed in cells from pSS patients. An increased sensitivity of STAT1 Y701 activating signals in immune cells may in part drive an up‐regulation of IFN induced genes. This is further supported by the association of increased response to IFN‐α2b with upregulated mRNA levels of type I IFN regulated genes and the production of SSA autoantibodies, which have been shown to be positively associated with the upregulation of IFN regulated genes 15, 16. Type I IFN could thereby potentially induce B cell autoantibody production through a number of mechanisms including lowering of BCR signalling thresholds, upregulate surface molecules that promote antigen presentation, promoting survival and differentiation and trafficking to germinal centres 17.
Potentiated phosphorylation of STAT1 Y701 in monocytes, B cells and CD4+ T cells from pSS patients has been described previously in response to IFN‐γ and/or IL‐6 14. In addition to receptor expression, signal transduction pathways leading to phosphorylation of STAT are mediated through activation of Janus kinases and are negatively regulated by several mechanisms, including suppressor of cytokine signaling (SOCS) family members, ubiquitin carboxy‐terminal hydrolase 18 and various microRNA 18, 19. Cross regulation between STATs has also been observed, with negative regulation of STAT1 by STAT3 through competition for common docking sites 20. Previous studies have described upregulations of STAT1 and STAT3 mRNA in PBMCs from pSS patients 13, while no differences were found regarding STAT3 protein expression in CD3+ and CD19+ cells 12. Increased expression of SOCS1 and ‐3 mRNA have been found in PBMCs from pSS patients 14, and dysregulation of microRNA expression patterns have been recently observed in B and T cells from pSS patients 21.
Interestingly, we observed an NK cell specific reduction of STAT4 Y693 phosphorylation in pSS patients upon IFN‐α stimulation. NK cells have previously been shown to display high basal expression of STAT4 and reduced STAT1 compared to other cell subsets 22. This is thought to predispose the cells to STAT4 activation by type I IFN and IFN‐γ 22. Our study showed increased basal signalling in NK cells through STAT1 Y701 and STAT1 S727. Upon IFN‐α2b stimulation, pSTAT1 Y701 was greatly increased, while pSTAT4 Y693 was decreased. These results indicate that pSS patients express a NK cell phosphorylation profile represented by low relative phosphorylation ratio of STAT4 to STAT1 in response to IFN‐α, resembling those displayed by individuals with hepatitis C infections receiving IFN‐α therapy 23. Such a profile might polarize NK cells in pSS towards a low IFN‐γ producing phenotype and increased cell cytotoxicity 22, 23, 24, 25. However, no increase in NK cell killing ability has been shown on a per cell basis compared to healthy donors in pSS patients 26. Interestingly, NK cells from pSS patients have been shown to be hyporesponsive to IFN‐α induced cell cytotoxicity 27. Whether this aberrant profile is associated with polymorphisms in STAT4 commonly associated with pSS 10 is unknown.
Further IFN‐α induced induction of STAT3 S727 phosphorylation in T cells negatively correlated with expression of type I IFN inducible genes. STAT3 is known to negatively regulate type I IFN induced gene expression. Studies with STAT3 knockout or knockdown mouse embryonic fibroblasts have indicated that STAT3 can negatively regulate type I IFN induced antiviral responses and ISRE‐driven genes 28. Further, overexpression of STAT3 in THP‐1 cells downregulates IFN‐α activated STAT1 dependent genes, and knocking down STAT3 leads to elevated expression of the same genes 20. Taken together our results indicate that STAT3 S727 responses may play a role in the expression of a type I IFN signature in pSS patients. Further, patients treated with the DMARD hydroxychloroquine (HCQ) or corticosteroids displayed stronger IFN‐α induced STAT3 S727 signalling in T cells, while HCQ had little effect on induction of STAT1 Y701 signalling. In addition, patients treated with HCQ tended to have a lower interferon score. This observation is in accordance with a previous study where patients taking HCQ showed significantly reduced type I IFN scores based on expression of IFI44L, IFI44, IFIT3, LY6E and MxA in monocytes compared to patients not on HCQ 29. However, whether the effect on STAT phosphorylation and IFN signature denotes a therapeutic response is unclear.
In T and NK cells of pSS patients, the basal phosphorylation levels were increased for several of the analysed epitopes, while the phosphorylation pattern of B cells was similar to controls. Our results differ from previous studies where B cells from pSS have exhibited increased basal phosphorylation of STAT3 Y705 12 and STAT5 Y694 13, with no difference found for phosphorylation of STAT1 Y701 in T cells (CD4+ and CD4−) 13. These discrepancies are likely the result of methodological differences. For example, whole blood 13 or freshly isolated PBMCs 12 were utilized in the previous studies, while our procedure involved cryopreserved cells cultured for 6.5 h.
It should be noted that this study has a number of limitations. The number of patients included is limited, and the small sample size is further affected by the heterogeneity of the patients. Moreover, the analysis was limited to the three main subsets of lymphocytes (T, B and NK cells), and immunophenotyping studies have shown shifts in the relative frequency of cell subsets in the peripheral blood of pSS patients compared to healthy donors 30, 31, 32, 33. We therefore cannot exclude that the differences in the signalling profiles reflect these changes. The type I IFN signature was assessed in PBMCs, and assessment for each cell type would likely strengthen associations and be more informative in deriving origin of the signature. Further, associations between concentrations of proteins involved in signalling pathways and induction of phosphorylation of the protein would in addition be informative as to the mechanism behind the increased responses.
In conclusion, pSS patients show an increased response to IFN‐α through STAT1 Y701. An increased sensitivity to STAT1‐activating signals in immune cells of pSS patients may in part drive an up‐regulation of IFN induced genes and the production of SSA autoantibodies. However, the lack of an effect of HCQ on this pathway indicates a more complex relationship. Our results further suggest that the IFN signature may also in part be derived from reduced activation of STAT3 S727 which has been shown to inhibit type I IFN inducible gene expression.
Materials and methods
Blood sampling
Peripheral blood from 24 patients with pSS was collected in Lithium‐heparin tubes (BD diagnostics) at the Department of Rheumatology, Haukeland University Hospital, Bergen, Norway. Blood from 21 healthy age‐ and gender‐matched donors was collected at the blood bank at the Haukeland University Hospital in Bergen, Norway. All blood donors provided written informed consent. Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation with lymphoprep™ (Axis‐Shield, Oslo, Norway), and cryopreserved as described previously 34. The cells were stored at −150°C for approximately 12 to 16 months.
All patients fulfilled the pSS American‐European Consensus group criteria (AECC) 35 and displayed no additional autoimmune diseases or lymphoma. An overview of the cohort is shown in Table 1. The study was approved by the regional ethical committee (#2009/686).
Table 1.
Characteristics of patients and controls used in the study
| Cohort characteristics | ||
|---|---|---|
| Sjögren's syndrome | Healthy controls | |
| Females/males | 24/0 | 21/0 |
| Age, median (range) years | 57 (33–73) | 54 (42–73) |
| Clinical features | ||
| SSA antibodies (%) | 18 (75) | |
| SSB antibodies (%) | 11 (46) | |
| SSA and SSB antibodies (%) | 11 (46) | |
| ANA (%) | 18 (75) | |
| Positive Schirmer's (tear flow ˂5 mm/5 min) (%); n = 23 | 15 (65) | |
| Focus score† ≥ 1 (%); n = 14 | 11 (79) | |
| ESR, high levels‡ | 6 (25) | |
| CRP high levels (≥5 mg/L) | 3 (13) | |
| Extraglandular manifestations (%) | 14 (58) | |
| Medication | ||
| DMARDs | 6 (25) | |
| Corticosteroids | 2 (8) | |
Continuous data are expressed as median. Categorical data is expressed as frequency and percentage. †Focus score indicates the number of inflammatory foci containing more than 50 mononuclear cells per 4 mm2 biopsy tissue; ‡Age and gender dependent. DMARDs, disease‐modifying anti‐rheumatic drugs; ANA, anti‐nuclear antibodies; ESR, erythrocyte sedimentation rate; CRP, C‐reactive protein
Routine laboratory assays
Identification of anti‐Ro/SSA and anti‐La/SSB, other anti‐nuclear antibodies (ANA), erythrocyte sedimentation rate (ESR), C‐reactive protein (CRP) and extraglandular manifestations (EGM) were obtained as part of routine clinical investigation at time of blood sampling. SSA, SSB and ANA were reported as either present or absent, while other serum and blood parameters were reported as continuous values. Extraglandular manifestations were defined as disease features outside surface exocrine glands.
Real‐time quantitative PCR
Total RNA was isolated from PBMCs of 19 pSS patients and 14 healthy controls using RNeasy plus (Qiagen Nordic, Oslo, Norway). 300 ng RNA each were used in two cDNA reactions with RevertAid reverse transcriptase using Oligo(dT)18 and random nonamers, respectively. The cDNA was then pooled, diluted with 1.5 parts DNase and RNase free water (i.e., 40 μl cDNA + 60 μl water), and 5 μl were used in a real‐time PCR reaction using Taqman gene expression assays (Hs00895608_m1 (MxA); Hs00973637_m1 (OAS1); Hs00951349_m1 (IFI44); Hs00977005_m1 (GBP1); Hs00894837_m1 (GBP2); Hs03928990_g1 (18S rRNA); all Thermo Fisher Scientific, Waltham, USA). All real time PCR reactions were run in duplicates on a Light Cycler 480 (Roche Diagnostics, Oslo, Norway). 18S rRNA was used as reference gene, and relative expression levels were calculated as 2−ΔCt. The IFN score was calculated according to Feng et al. 36 by standardizing expression levels using mean and SD of the healthy controls for the respective gene and using the following formula:
where i = each of the 3 type I IFN‐inducible genes (MxA, IFI44, OAS1), gene ipSS = the gene expression level in each pSS patient, and gene iCtr = the gene expression in controls. To set a threshold, 3 × SD of healthy controls was utilized.
Antibodies used for flow cytometry
The following phospho‐specific monoclonal antibodies were used in 3 different panels during the flow cytometry protocol as described previously 34: Alexa Fluor® 647 conjugated anti‐STAT4 (pY693, clone 38/p‐STAT4, panel 1), anti‐STAT1 (pS727, clone K51‐856, panel 2) and anti‐STAT3 (pS727, clone 49/p‐STAT3, panel 3); PerCP‐Cy™5.5 conjugated anti‐ERK1/2 (pT202/pY204, clone 20A, panel 1), anti‐STAT1 (pY701, clone 4a, panel 2) and anti‐STAT3 (pY705, clone 4/P‐STAT3, panel 3); and PE‐Cy™7 conjugated anti‐NF‐κB p65 (pS529, clone K10‐895.12.50, panel 1), anti‐p38 MAPK (pT180/pY182, clone 36/p38, panel 2) and anti‐STAT5 (pY694, clone 47/ STAT5(pY694), panel 3) (all from BD Biosciences, San Jose, CA, USA). Cell surface markers incorporated in the assays were BV786 conjugated anti‐CD3 (clone SK7, BD HorizonTM), Alexa Fluor® 488 conjugated anti‐CD20 (clone H1 (FB1), BD Biosciences) and PE conjugated anti‐CD56 (clone N901, Beckmann Coulter, CA, USA).
Cell culture and stimulation
Before stimulation, cryopreserved PBMCs were rapidly thawed using a water bath set to 37°C and washed once in prewarmed (37°C) X‐vivo 20™ by centrifugation at 300 × g for 7 min. The PBMCs were then resuspended in prewarmed X‐vivo 20™ and rested at 37°C at 5% CO2 for 30 min before the cell concentration was adjusted to 3 × 106 cells/ml in X‐vivo 20™. 200 μL of PBMCs were dispensed into 7 wells of a Megablock® 96 well plate (Sarstedt, Nümbrecht, Germany), along with 2 wells of a reference cell sample that was included in every assay. The cells were rested at 37°C with 5% CO2 for 2 hours to decrease basal phosphorylation levels. Following, the cells were either left unstimulated or stimulated according to a reverse time course for 15, 30, 60, 120, 180, or 240 min with IFN‐α2b (100 ng/mL; ImmunoTools, Friesoythe, Germany).
Fluorescent cell barcoding and phospho‐epitope staining for flow cytometry
Following stimulation, PBMCs were resuspended by pipetting and immediately fixed at RT for 10 min by adding prewarmed PFA (Electron Microscopy Sciences (Hatfield, PA, USA)) at a final concentration of 1.5%. PBMCs were then centrifuged at 1000 g for 5 min 4°C and resuspended by vortexing in 50 μL PBS before drop wise addition of 1 mL ice cold methanol and incubation on ice for 30 min. The permeabilized cells were kept overnight at −80°C. After washing with PBS, the PBMCs were stained according to a 3 × 3 barcoding grid (9 stimulation conditions) using 3 levels of pacific orange (PO) and pacific blue (PB) succinimidyl ester dyes (PB 100, 25 and 6.3 ng/mL; PO 250, 70 and 0 ng/mL; Life Technologies, Grand Island, NY, USA) for 30 min in the dark at 4°C. Barcoded PBMCs were then washed once with PBS containing 1% BSA, before being combined into one sample. The sample was washed and incubated with 2 μL Fc receptor block (Miltenyi Biotec, Bergisch Gladbach, Germany) per 1 × 106 cells for 10 min on ice. Following, the sample was subdivided into three parts and incubated for 30 min at RT in the dark with the three different antibody staining panels. An aliquot of the barcoded cells was collected before addition of antibody as a barcoding only control. The samples were then washed twice and re‐suspended in PBS containing 1% BSA and 2mM EDTA (Sigma‐Aldrich) prior to analysis.
Data analysis
Samples were acquired on an LSRI Fortessa flow cytometer (BD Biosciences, San Jose, CA, USA) with BD FACSDivaTM Software (BD Biosciences) at the Bergen Flow Cytometry Core Facility, University of Bergen, Norway. The flow cytometer was equipped with 407, 488, 561 and 635 nm lasers, and emission filters for PerCP‐Cy5.5 (LP: 685, BP: 695/40), Alexa Fluor‐488 (LP: 505, BP: 530/30), PE‐Cy7 (LP: 750, BP: 780/60), PE (LP: ‐, BP: 582/15), APC (LP: ‐, BP: 670‐/‐14), Pacific blue (LP: ‐, BP:450/50), Pacific orange (LP: 570, BP: 585/42) and BV 786 (LP: 750, BP: 780/60). The cytometer was routinely calibrated with BD cytometer setup and tracking beads (BD Biosciences). A minimum of 200 000 events in the intact cell gate was collected for each sample. FACS data were analysed in FlowJo (Tree Star) and Cytobank (http://www.cytobank.org). A representative gating strategy and phosphorylation profile for a single barcoded sample is shown in Supporting Information Fig. 2. Cryopreserved PBMCs from a single donor were included in each assay as a positive control, for inter‐assay normalization and assessing assay to assay variability. MdFI of phospho‐epitopes for gated populations were exported to Microsoft excel. The raw flow cytometry data can be found at the flow data repository of the International Society for Advancement of Cytometry 37, FR‐FCM‐ZYEF. The robustness of the flow cytometry assay has been previously established 34. Relevant information for repeating the experiments is given in Supporting Information Table 6 following “The minimum information about a Flow Cytometry Experiment (MIFlowCyt)” guidelines 38.
Statistical analysis
Comparisons between categories, correlations and the production of associated graphs were done using Graphpad Prism (version 6.05). Unpaired Mann–Whitney tests were used in the comparisons between categories. Correlations were assessed by the Spearman's rank test, with outliers removed using robust regression and outlier removal (ROUT) method and a ROUT coefficient Q of 1.
Differences were considered statistically significant when p ≤ 0.05. The analysis was exploratory in nature hence no correction was made for multiple comparisons. Principle component analysis (PCA) using Unscrambler® × software (Camo software) was used to reduce dimensionality of the dataset and find clusters of patients with similar signaling profile which could be used to differentiate between disease status, presence of SSA autoantibodies, EGM and medication effect. PCA was performed using the algorithm NIPALS, the data was mean centered and run with no weighting for change of MdFI, and weighted for absolute MdFI by dividing by standard deviation. Two methods were used to remove “redundant” variables to simplify interpretation and focusing subsequent analysis. First variables that described less than 50% of the variation were removed from the initial PCA, and where appropriate stepwise reduction of less significant variables with low variable leverage was performed.
Authorship
P.V., R.J., and S.A. conceived of study and R.D., P.V., and S.A designed the study. R.D. and B.B processed PBMC samples and conducted flow cytometric analysis. S.A. conducted real‐time quantitative PCR. R.D., B.B., S.G., and S.A. analysed and processed the data. D.H. and J.G.B. selected patients and collected patient data. R.D. and S.A wrote the manuscript. All authors read and approved the manuscript.
Conflict of interest
The authors declare no commercial or financial conflict of interest.
Funding
The authors’ research is supported by the EU H2020 contract HarmonicSS (H2020‐SC1‐2016‐RTD/731944), the Broegelmann Foundation, the Western Norway Regional Health Authorities (grant nr. 912065) and the University of Bergen.
Supporting information
Peer review correspondence
Supporting Information
Acknowledgements
We thank all patients and blood donors who participated in this study. We thank Marianne Eidsheim and Kjerstin Jakobsen for excellent technical assistance, and the staff at the laboratory at the Rheumatology clinics for collection of patients’ blood samples. The flow cytometry analysis was performed at the Flow Cytometry Core Facility, Department of Clinical Science, University of Bergen. Financial support was obtained from the EU H2020 project HarmonicSS (H2020‐SC1‐2016‐RTD/731944), the Broegelmann Foundation, the Western Norway Regional Health Authorities (grant nr. 912065) and the University of Bergen.
[The copyright line of this article was changed on 18 April 2019 after original online publication.]
Contributor Information
Richard Davies, Email: richard.davies@uib.no.
Silke Appel, Email: silke.appel@uib.no.
References
- 1. Jonsson, R. , Vogelsang, P. , Volchenkov, R. , Espinosa, A. , Wahren‐Herlenius, M. and Appel, S. , The complexity of Sjogren's syndrome: novel aspects on pathogenesis. Immunol Lett. 2011. 141: 1–9. [DOI] [PubMed] [Google Scholar]
- 2. Mavragani, C. P. and Moutsopoulos, H. M. , Sjogren syndrome. Can Med Assoc J. 2014. 186: E579–E586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jonsson, R. , Theander, E. , Sjostrom, B. , Brokstad, K. and Henriksson, G. , Autoantibodies present before symptom onset in primary Sjogren syndrome. Jama‐J Am Med Assoc. 2013. 310: 1854–1855. [DOI] [PubMed] [Google Scholar]
- 4. Theander, E. , Jonsson, R. , Sjostrom, B. , Brokstad, K. , Olsson, P. and Henriksson, G. , prediction of sjogren's syndrome years before diagnosis and identification of patients with early onset and severe disease course by autoantibody profiling. Arthritis Rheumatol. 2015. 67: 2427–2436. [DOI] [PubMed] [Google Scholar]
- 5. Fox, P. C. , Autoimmune diseases and Sjogren's syndrome: an autoimmune exocrinopathy. Ann NY Acad Sci. 2007. 1098: 15–21. [DOI] [PubMed] [Google Scholar]
- 6. Hall, J. C. , Baer, A. N. , Shah, A. A. , Criswell, L. A. , Shiboski, C. H. , Rosen, A. and Casciola‐Rosen, L. , Molecular subsetting of interferon pathways in Sjogren's syndrome. Arthritis Rheumatol. 2015. 67: 2437–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yao, Y. , Liu, Z. , Jallal, B. , Shen, N. and Ronnblom, L. , Type I interferons in Sjogren's syndrome. Autoimmun Rev. 2013. 12: 558–566. [DOI] [PubMed] [Google Scholar]
- 8. Ronnblom, L. and Eloranta, M. L. , The interferon signature in autoimmune diseases. Curr Opin Rheumatol. 2013. 25: 248–253. [DOI] [PubMed] [Google Scholar]
- 9. Miceli‐Richard, C. , Comets, E. , Loiseau, P. , Puechal, X. , Hachulla, E. and Mariette, X. , Association of an IRF5 gene functional polymorphism with Sjogren's syndrome. Arthritis Rheum. 2007. 56: 3989–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Korman, B. D. , Alba, M. I. , Le, J. M. , Alevizos, I. , Smith, J. A. , Nikolov, N. P. , Kastner, D. L. et al, Variant form of STAT4 is associated with primary Sjogren's syndrome. Genes Immun. 2008. 9: 267–270. [DOI] [PubMed] [Google Scholar]
- 11. Wahren‐Herlenius, M. and Dorner, T. , Immunopathogenic mechanisms of systemic autoimmune disease. Lancet. 2013. 382: 819–831. [DOI] [PubMed] [Google Scholar]
- 12. Ramos, H. L. , Valencia‐Pacheco, G. and Alcocer‐Varela, J. , Constitutive STAT3 activation in peripheral CD3(+) cells from patients with primary Sjogren's syndrome. Scand J Rheumatol. 2008. 37: 35–39. [DOI] [PubMed] [Google Scholar]
- 13. Pertovaara, M. , Silvennoinen, O. and Isomaki, P. , STAT‐5 is activated constitutively in T cells, B cells and monocytes from patients with primary Sjogren's syndrome. Clin Exp Immunol. 2015. 181: 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pertovaara, M. , Silvennoinen, O. and Isomaki, P. , Cytokine‐induced STAT1 activation is increased in patients with primary Sjogren's syndrome. Clin Immunol. 2016. 165: 60–67. [DOI] [PubMed] [Google Scholar]
- 15. Brkic, Z. , Maria, N. I. , van Helden‐Meeuwsen, C. G. , van de Merwe, J. P. , van Daele, P. L. , Dalm, V. A. , Wildenberg, M. E. et al, Prevalence of interferon type I signature in CD14 monocytes of patients with Sjogren's syndrome and association with disease activity and BAFF gene expression. Ann Rheum Dis. 2013. 72: 728–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Emamian, E. S. , Leon, J. M. , Lessard, C. J. , Grandits, M. , Baechler, E. C. , Gaffney, P. M. , Segal, B. et al, Peripheral blood gene expression profiling in Sjogren's syndrome. Genes Immun. 2009. 10: 285–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kiefer, K. , Oropallo, M. A. , Cancro, M. P. and Marshak‐Rothstein, A. , Role of type I interferons in the activation of autoreactive B cells. Immunology and cell biology. 2012. 90: 498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ivashkiv, L. B. and Donlin, L. T. , Regulation of type I interferon responses. Nat Rev Immunol. 2014. 14: 36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gracias, D. T. , Stelekati, E. , Hope, J. L. , Boesteanu, A. C. , Doering, T. A. , Norton, J. , Mueller, Y. M. et al, The microRNA miR‐155 controls CD8(+) T cell responses by regulating interferon signaling. Nat Immunol. 2013. 14: 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ho, H. H. and Ivashkiv, L. B. , Role of STAT3 in type I interferon responses. Negative regulation of STAT1‐dependent inflammatory gene activation. J Biol Chem. 2006. 281: 14111–14118. [DOI] [PubMed] [Google Scholar]
- 21. Wang‐Renault, S. F. , Boudaoud, S. , Nocturne, G. , Roche, E. , Sigrist, N. , Daviaud, C. , Tinggaard Bugge, A. et al, Deregulation of microRNA expression in purified T and B lymphocytes from patients with primary Sjogren's syndrome. Ann Rheum Dis. 2018. 77: 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miyagi, T. , Lee, S. H. and Biron, C. A. , Intracellular staining for analysis of the expression and phosphorylation of signal transducers and activators of transcription (STATs) in NK cells. Methods Mol Biol. 2010. 612: 159–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Edlich, B. , Ahlenstiel, G. , Zabaleta Azpiroz, A. , Stoltzfus, J. , Noureddin, M. , Serti, E. , Feld, J. J. et al, Early changes in interferon signaling define natural killer cell response and refractoriness to interferon‐based therapy of hepatitis C patients. Hepatology. 2012. 55: 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee, C. K. , Rao, D. T. , Gertner, R. , Gimeno, R. , Frey, A. B. and Levy, D. E. , Distinct requirements for IFNs and STAT1 in NK cell function. J Immunol. 2000. 165: 3571–3577. [DOI] [PubMed] [Google Scholar]
- 25. Nguyen, K. B. , Salazar‐Mather, T. P. , Dalod, M. Y. , Van Deusen, J. B. , Wei, X. Q. , Liew, F. Y. , Caligiuri, M. A. et al, Coordinated and distinct roles for IFN‐alpha beta, IL‐12, and IL‐15 regulation of NK cell responses to viral infection. J Immunol. 2002. 169: 4279–4287. [DOI] [PubMed] [Google Scholar]
- 26. Izumi, Y. , Ida, H. , Huang, M. , Iwanaga, N. , Tanaka, F. , Aratake, K. , Arima, K. et al, Characterization of peripheral natural killer cells in primary Sjogren's syndrome: impaired NK cell activity and low NK cell number. J Lab Clin Med. 2006. 147: 242–249. [DOI] [PubMed] [Google Scholar]
- 27. Takeda, A. , Minato, N. and Kano, S. , Selective impairment of alpha‐interferon‐mediated natural killer augmentation in Sjogren's syndrome: differential effects of alpha‐interferon, gamma‐interferon, and interleukin 2 on cytolytic activity. Clin Exp Immunol. 1987. 70: 354–363. [PMC free article] [PubMed] [Google Scholar]
- 28. Wang, W. B. , Levy, D. E. and Lee, C. K. , STAT3 negatively regulates type I IFN‐mediated antiviral response. J Immunol. 2011. 187: 2578–2585. [DOI] [PubMed] [Google Scholar]
- 29. Maria, N. I. , Brkic, Z. , Waris, M. , van Helden‐Meeuwsen, C. G. , Heezen, K. , van de Merwe, J. P. , van Daele, P. L. et al, MxA as a clinically applicable biomarker for identifying systemic interferon type I in primary Sjogren's syndrome. Ann Rheum Dis. 2014. 73: 1052–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sudzius, G. , Mieliauskaite, D. , Siaurys, A. , Viliene, R. , Butrimiene, I. , Characiejus, D. and Dumalakiene, I. , Distribution of Peripheral Lymphocyte Populations in Primary Sjogren's Syndrome Patients. J Immunol Res. 2015. 2015: 854706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Szyszko, E. A. , Brun, J. G. , Skarstein, K. , Peck, A. B. , Jonsson, R. and Brokstad, K. A. , Phenotypic diversity of peripheral blood plasma cells in primary Sjogren's syndrome. Scand J Immunol. 2011. 73: 18–28. [DOI] [PubMed] [Google Scholar]
- 32. Bohnhorst, J. O. , Thoen, J. E. , Natvig, J. B. and Thompson, K. M. , Significantly depressed percentage of CD27+(memory) B cells among peripheral blood B cells in patients with primary Sjogren's syndrome. Scand J Immunol. 2001. 54: 421–427. [DOI] [PubMed] [Google Scholar]
- 33. Bohnhorst, J. O. , Bjorgan, M. B. , Thoen, J. E. , Natvig, J. B. and Thompson, K. M. , Bm1‐Bm5 classification of peripheral blood B cells reveals circulating germinal center founder cells in healthy individuals and disturbance in the B cell subpopulations in patients with primary Sjogren's syndrome. J Immunol. 2001. 167: 3610–3618. [DOI] [PubMed] [Google Scholar]
- 34. Davies, R. , Vogelsang, P. , Jonsson, R. and Appel, S. , An optimized multiplex flow cytometry protocol for the analysis of intracellular signaling in peripheral blood mononuclear cells. J Immunol Methods. 2016. 436: 58–63. [DOI] [PubMed] [Google Scholar]
- 35. Vitali, C. , Bombardieri, S. , Jonsson, R. , Moutsopoulos, H. M. , Alexander, E. L. , Carsons, S. E. , Daniels, T. E. et al, Classification criteria for Sjogren's syndrome: a revised version of the European criteria proposed by the American‐European Consensus Group. Ann Rheum Dis. 2002. 61: 554–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Feng, X. , Wu, H. , Grossman, J. M. , Hanvivadhanakul, P. , FitzGerald, J. D. , Park, G. S. , Dong, X. et al, Association of increased interferon‐inducible gene expression with disease activity and lupus nephritis in patients with systemic lupus erythematosus. Arthritis Rheum. 2006. 54: 2951–2962. [DOI] [PubMed] [Google Scholar]
- 37. Spidlen, J. , Breuer, K. , Rosenberg, C. , Kotecha, N. and Brinkman, R. R. , FlowRepository: a resource of annotated flow cytometry datasets associated with peer‐reviewed publications. Cytometry A. 2012. 81: 727–731. [DOI] [PubMed] [Google Scholar]
- 38. Lee, J. A. , Spidlen, J. , Boyce, K. , Cai, J. , Crosbie, N. , Dalphin, M. , Furlong, J. et al, MIFlowCyt: The Minimum Information about a Flow Cytometry Experiment. Cytometry A. 2008. 73a: 926–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Peer review correspondence
Supporting Information


