Abstract
For using counts of circulating tumor cells (CTCs) in the clinic to aid a physician's decision, its reported values will need to be accurate and comparable between institutions. Many technologies have become available to enumerate and characterize CTCs, thereby showing a large range of reported values. Here we introduce an Open Source CTC scoring tool to enable comparison of different reviewers and facilitate the reach of a consensus on assigning objects as CTCs. One hundred images generated from two different platforms were used to assess concordance between 15 reviewers and an expert panel. Large differences were observed between reviewers in assigning objects as CTCs urging the need for computer recognition of CTCs. A demonstration of a deep learning approach on the 100 images showed the promise of this technique for future CTC enumeration. © 2018 The Authors. Cytometry Part A published by Wiley Periodicals, Inc. on behalf of International Society for Advancement of Cytometry.
Keywords: CTC, consensus, scoring, Agreement, reviewers, experts, definition, deep learning, ground truth, ACCEPT
Introduction
The peripheral blood load of circulating tumor cells (CTCs) enumerated with the CellSearch® system is directly related to the survival prospects of patients with metastatic cancer and their presence in patients with primary cancers is related to an increased risk at disease recurrence as well as survival 1, 2, 3, 4, 5, 6, 7, 8. To monitor therapy based on CTC counts and to diagnose the presence of cancer beyond the primary tumor, it is of utmost importance to accurately assign objects as CTCs. To determine whether a CTC count is below or above five CTCs the inter‐reader variability is challenging, but can be quite low 9, 10, 11. However, when a significant change in CTC number will need to be assessed, the challenge increases 12. Yet, nearly all CTC isolation techniques lack a fully automated image analysis, thereby making CTC counts subjective to the reviewer. Here we evaluate the consensus between multiple reviewers in assigning objects as CTCs by introducing an open source scoring tool that enables comparisons between reviewers and improve their ability to reach consensus (https://github.com/leoniez/ACCEPT/releases). Finally, we introduce a fully automated CTC classification system based on deep learning (DL).
Methods
Selection of Fluorescent Cell Images for CTC Scoring
A set of 100 fluorescent cell images comprising of potential CTCs and leukocytes was assembled. The fluorescent images were obtained from 7.5 mL blood samples from metastatic prostate and non‐small cell lung cancer patients 13, 14, 15. The blood was processed with the CellSearch system (Menarini Silicon Biosystems, Huntingdon Valley, PA, USA) and the EpCAM depleted blood was collected, passed through microsieves (VyCap, Deventer, The Netherlands), fluorescently labeled and images taken by fluorescent microscopy 13. The images exported from the CellSearch system as well as fluorescent images after filtration of the EpCAM depleted blood were all reanalyzed with the ACCEPT toolbox (https://github.com/LeonieZ/ACCEPT). One hundred thumbnail images were selected from which 50 were obtained from CellSearch images and 50 from the microsieves.
Development of the ACCEPT Scoring Tool
For the cell scoring we developed the ACCEPT CTC Scoring tool in Matlab 2016a (Mathworks, Natick MA). This tool will be made available as part of the general ACCEPT toolbox for CTC analysis in a future release. A screenshot of the tool is shown in Figure 1. For every object, the thumbnail images of three fluorescent channels (CD45, DAPI, PE) are shown using the full range of color intensities. The red contour surrounding the object indicates its boundary, which is automatically detected by an advanced segmentation algorithm in ACCEPT 16, 17. Next to it, an overlay image is presented, showing the CD45 signal in red, the DAPI signal in blue, and the PE signal in green. Here, the intensity is scaled from the smallest to the largest intensity value found inside the red contour. The right three scatter plots show six quantitative measurements exported from the objects to facilitate the scoring. A reviewer can choose from four presented answers: 1. Definitely not a CTC; 2. Most likely not a CTC; 3. Most likely a CTC; and 4. Definitely a CTC. Once the user decides for one answer, the tool will automatically proceed to the next object and it is not possible to change an answer afterwards. All objects are presented in a randomized order. With this tool, 15 independent reviewers from six different institutes scored the set of 100 cells. These scores formed the basis of our analysis. [Color figure can be viewed at wileyonlinelibrary.com]
Figure 1.
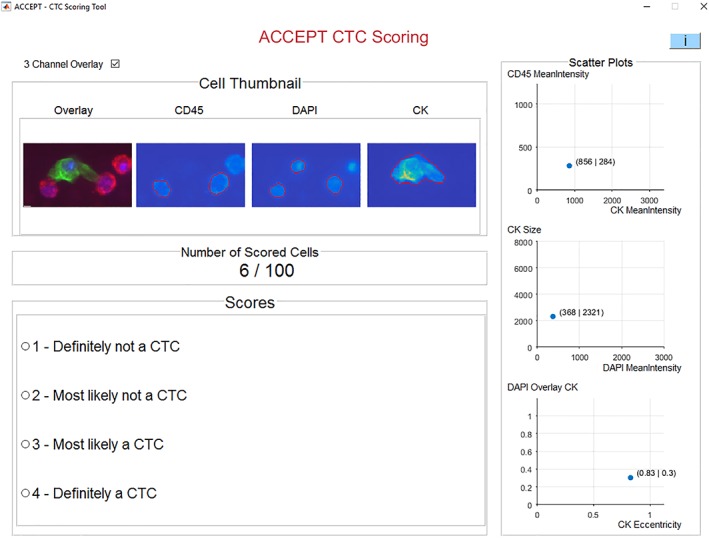
Screenshot of the ACCEPT CTC scoring tool showing a thumbnail gallery of all fluorescent channels for each presented cell, with four answers and three plots presenting measurement information of the respective cell to aid in the decision. After selecting an answer, the program automatically proceeds to the next cell. [Color figure can be viewed at wileyonlinelibrary.com]
Developing the “Ground‐Truth”
One of the major drawbacks of image‐based CTC analysis is the lack of a ground‐truth solution. While molecular information could be used to determine if a cell really is a CTC, this information is in most of the cases not available or very hard to acquire. To compensate for the lack of a real ground‐truth answer for our set, we formed an Expert Panel (EP) consisting of four experts in the field and had them score the 100 objects together in one session. Two of the experts were involved in the original definition of a CTC in the CellSearch system and the other two were trained reviewers with several years of experience in scoring CTCs. All 100 objects were discussed in an online meeting and they had to agree on one answer for each object. These answers were used as a ground‐truth for comparison with the result of the average reviewer (AR) and DL.
Automated CTC Classification by Deep Learning
A DL network approach was used to classify the 100 objects. The classifier was trained on images obtained from the CellSearch system and contained 13,123 CTC candidates, 18,820 objects with DNA staining not classified as CTC and 8,548 other objects. The details of the DL approach are described elsewhere 18.
Statistical Analysis
To rule out any agreement by chance, inter‐rater agreement between all 15 reviewers was determined with Fleiss’ kappa ĸ, whereas intra‐rater agreements between the AR, EP, and DL were determined with Cohen's kappa ĸ. The agreement was considered poor if ĸ ≤ 0.20, fair for ĸ 0.21–0.40, moderate for ĸ 0.41–0.60, substantial or good for ĸ 0.61–0.80 and almost perfect for ĸ 0.81–1.00 19, 20.
Results
Scores
The scores of all 100 objects from each reviewer, the AR score, the EP score, and the DL score are presented in Figure 2. The probability for a cell being a “CTC” returned from the DL network is scaled from 0.0 (displayed as score 1 by reviewers, visualized as dark blue) to 1.0 (displayed as score 4, visualized as yellow). Images of all 100 objects including the AR, EP, and DL score are provided in Supporting Information Figure S1.
Figure 2.
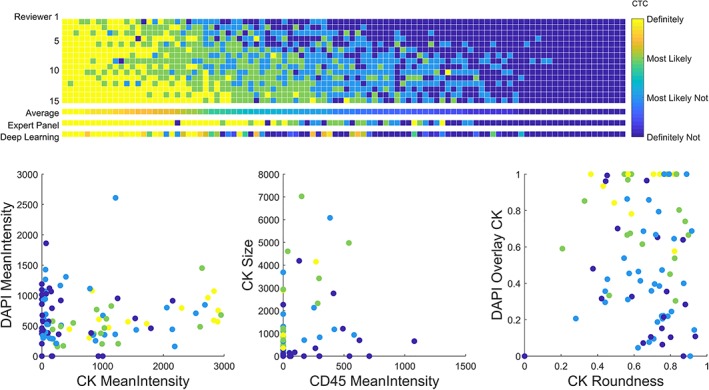
Results of the CTC scoring by 15 reviewers, summarized with the average reviewer, followed by the results of the expert panel consisting of four expert reviewers and the deep learning automated CTC scoring (upper panel). The average reviewer score of all 100 cells are presented in three scatter plots using several parameters: the mean intensity of the signal detected in all three channels (DAPI, CK, and CD45), the size and roundness of the CK signal and the overlay between the DAPI and CK signals (lower panel).
Consensus between Reviewers
Only one object out of 100 objects was scored as “Definitely a CTC” by all 15 investigators, and for only 13 more objects everyone agreed on the answer (all “Definitely not a CTC”). If we summarize answers in two classes: a “CTC” class (objects scored as 3 or 4) and “Not a CTC” class (objects scored as 1 or 2), these numbers increased to 11 objects scored as “CTC” and 30 objects scored as “Not a CTC” by all users, yet thereby 59 objects remain ambiguous. Of these 41 objects where everyone agreed on the same class, 54% cells were extracted from CellSearch and 46% cells were extracted from microsieve filtration. In total 1,500 scores were assigned; 42% for “Definitely not a CTC”, 22% for “Most likely not a CTC”, 19% for “Most likely a CTC” and 17% for “Definitely a CTC”. The inter‐rater agreement was calculated with Fleiss’ kappa and this shows a fair agreement with ĸ 0.38 for the case of four possible answers yet a moderate agreement with ĸ 0.60 if we summarize the answers in two classes.
Consensus of Expert Panel with Average Reviewer
When the results were divided into “CTC” and “Not a CTC”, the overall agreement between EP and AR score was 80% (Table 1A). The agreement between single reviewers and the EP ranged from 70% to 88%. When the EP scored “Not a CTC”, the AR agreed in 98% of the cases. When a “CTC” was scored according to the EP, the reviewers agreed only in 61%, showing that the consensus for a cell being “Not a CTC” is much higher than a positive agreement. Cohen's kappa was calculated to determine the intra‐agreement, which showed a moderate agreement of ĸ 0.60. The agreement in case of four possible answers is summarized in Supporting Information Table 1A.
Table 1.
Overview of agreement on 100 cells between (A) Average Reviewer and Expert Panel, (B) Average Reviewer and Deep Learning, and (C) Expert Panel and Deep Learning, summarizing scores as a “CTC” class and “Not a CTC” class.
| A | |||
| Agreement: 80 ĸ = 0.60 | Expert panel | ||
| Not a CTC | CTC | ||
| Average reviewers | Not a CTC | 50 | 19 |
| CTC | 1 | 30 | |
| B | |||
| Agreement: 84 ĸ = 0.64 | Deep learning | ||
| Not a CTC | CTC | ||
| Average reviewers | Not a CTC | 58 | 11 |
| CTC | 5 | 26 | |
| C | |||
| Agreement: 76 ĸ = 0.52 | Deep learning | ||
| Not a CTC | CTC | ||
| Expert panel | Not a CTC | 45 | 6 |
| CTC | 18 | 31 | |
Consensus with Deep Learning
The same set of 100 objects was reviewed by a DL network. Compared with AR, there was an overall agreement of 84%, using the two classes (see Table 1B). For objects that were scored as “Not a CTC” by the DL, the AR agreed in 92% of the cases, whereas the AR agreed on objects that were scored as “CTC” in 70%. This is in contrast to the comparison of the EP scores with DL (see Table 1C). Here, the agreement with DL is 71% of the objects scored as “Not a CTC”, but 84% on objects scored as “CTC”. The agreement between AR, EP, and DL for all four classes can be found in Supporting Information Table S1. Notably, the AR scored 13 cells as “Definitely a CTC” (score 4) and in all 13 cases both the EP and DL also scored as “Definitely a CTC”. Intra‐agreement with Cohen's kappa for AR with DL was ĸ 0.64 and for EP with DL was ĸ 0.52, which can be interpreted as a substantial (AR to DL) and moderate agreement. Calculating Fleiss’ kappa for agreement between AR, EP, and DL, showed a ĸ of 0.58 for two classes, which can be considered a moderate agreement as well.
Discussion
The demonstration that the peripheral blood tumor cell load is directly related to the clinical outcome of the patients, has led to the introduction of various platforms to enumerate and characterize CTCs (21; and all technologies presented in articles in this special issue). The morphological appearance of these CTCs is however extremely heterogeneous 22, 23, 24. This makes it difficult to arrive at a common definition of what is and what is not a “CTC”. Although genetic aberrations in the detected cells can confirm that the object indeed is a cancer cell, in practice this cannot be performed on all the CTC candidates. Moreover, the largest fraction of CTC or CTC related objects are either undergoing apoptosis or are extracellular vesicles derived from the cancer cells and might not have the complete genome represented 23, 25. However, the presence of these tumor related events is also related to poor clinical outcome 22, 26. To report a CTC count for the disease management of cancer patients, one will need to be able to rely on the truthfulness of the count. This is not as straightforward as one might think, as all the technologies presenting CTC data report different numbers of CTC and little data is available on the variability of assigning objects as CTC. Here, we introduce a tool which can help assessing the concordance between reviewers on assigning objects as CTCs. It can also be used to train the reviewers to increase concordance in reaching consensus on which objects are assigned as CTCs and which ones are not. In this study, we evaluate the consensus between 15 reviewers and an Expert Panel in assigning 100 objects as CTCs, using the ACCEPT open source scoring tool (https://github.com/leoniez/ACCEPT/releases). Agreement between reviewers on scoring all objects in four categories resulted in quite a large variation, as shown in Figure 2. No significant difference was observed between the images originating from CellSearch or from microsieve filtration. Previously, we have shown that the concordance between reviewers to score the expression of Her‐2 on CTCs can clearly be improved by the use of the ACCEPT toolbox 14. Although using a “yes” or “no” for CTC scoring improves the agreement (see Table 1 and Supporting Information Table 1), the large heterogeneity of CTC morphology makes it virtually impossible to obtain a perfect agreement between reviewers. In comparison to previous studies evaluating the consensus in scoring cells as CTC we reported lower values for Fleiss’ kappa ĸ 9, 10. The reasons for this deviation are versatile. First, the visualization used in the ACCEPT toolbox is different to the visualization in the CellSearch system and reviewers are less used to the new visualization. ACCEPT shows the true value of staining in the images, in contrast to a scaled‐up representation of the staining in CellSearch. Moreover, if we allow four possible answers the probability of agreement decreases and lowers the Fleiss’ kappa. The composition of the training set is also an important factor. We have seen that the agreement of a cell being not a CTC tends to be higher than the agreement on being a CTC. Thus, a set consisting of a lot of negative examples, is more likely to result in a high agreement 10. In our set, we tried to balance the number of positive and negative examples to account for that influence. In another comparison study cells that resulted in high disagreement in their expert panel were excluded from further analysis which most probably would have let to a lower agreement 9. Contrary to their approach, we aimed to include objects that were not directly obvious to classify as a CTC or not, since these type of cells will be most frequently present in the clinic.
Automated image analysis can eliminate any variation, since a computer will always use the same rationale to reach a conclusion. Therefore, we used a DL network that was trained on 40,491 thumbnail images obtained from CellSearch to identify CTC. For each object, the Deep Learning network provides a likelihood ratio whether the object is a CTC or not. The network performed remarkably well on the images of the 100 objects and no real difference could be observed between images obtained from CellSearch or from microsieve filtration (see Table 1 and Fig. 2). The images available from other platforms are however limited, and the future will tell whether this DL network can identify CTC independent from the platform they have been generated from or if the networks needs to be trained on images originating from various platforms.
In conclusion, we have shown that CTC agreement between reviewers can vary greatly and this presents a complication for using true CTC counts in the clinic. To reach objective agreement on CTC scoring, automated image analysis might hold the answer. We invite you to use the toolbox presented here to determine the rater variability in CTC scoring at your laboratory.
Supporting information
Supplementary Table S1 Overview of agreement on 100 cells between (A) Average Reviewer and Expert Panel; (B) Average Reviewer and Deep Learning and; (C) Expert Panel and Deep Learning scored with “1. Definitely not a CTC”; “2. Most likely not a CTC”; “3. Most likely a CTC”; and “4. Definitely a CTC”.
Supplementary Figure S1 Gallery of the 100 objects that were presented to the 15 reviewers, Expert Panel and Deep Learning network for scoring. The objects are sorted according to the AR score (high to low), followed by EP (high to low), followed by DL (high to low). The AR, EP and DL score is indicated next to the images. The scoring tool and the 100 images are available at: https://github.com/leoniez/ACCEPT/releases.
Acknowledgments
We are grateful for the expert panel consisting of Madeline Repollet, Joost Swennenhuis, Frances Tanney, Leon Terstappen, and the reviewers Marianna Alunni‐Fabbroni, Kiki Andree, Mateus Crespo, Agustin Enciso Martinez, Penny Flohr, Rita Lampignano, Leonie Majunke, Mariangela Manicone, Anouk Mentink‐Leusink, Afroditi Nanou, Marianne Oulhen, Elisabetta Rossi, Liwen Yang, and Beate Zill for scoring the images using the ACCEPT tool.
This study was supported by the EU FP7 # 305341 “CTC‐Trap” and the EU IMI # 115749‐1 “CANCER‐ID”.
Disclosure
The authors have declared no conflicts of interest.
The authors have declared no conflicts of interest.
Literature Cited
- 1. Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, Tibbe AG, Uhr JW, Terstappen LW. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res 2004;10:6897–6904. [DOI] [PubMed] [Google Scholar]
- 2. Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LWMM, Hayes DF. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. Engl J 2004;351:781–791. [DOI] [PubMed] [Google Scholar]
- 3. Cohen SJ, Punt CJA, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, et al. Relationship of circulating tumor cells to tumor response, progression‐free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:3213–3221. [DOI] [PubMed] [Google Scholar]
- 4. de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, Doyle GV, Terstappen LWWM, Pienta KJ, Raghavan D. Circulating tumor cells predict survival benefit from treatment in metastatic castration‐resistant prostate cancer. Clin Cancer Res 2008;14:6302–6309. [DOI] [PubMed] [Google Scholar]
- 5. Krebs MG, Sloane R, Priest L, Lancashire L, Hou JM, Greystoke A, Ward TH, Ferraldeschi R, Hughes A, Clack G, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non‐small‐cell lung cancer. J Clin Oncol 2011;29:1556–1563. [DOI] [PubMed] [Google Scholar]
- 6. Hiltermann TJN, Pore MM, van den Berg A, Timens W, Boezen HM, Liesker JJW, Schouwink JH, Wijnands WJA, Kerner GSMA, Kruyt FAE, et al. Circulating tumor cells in small‐cell lung cancer: A predictive and prognostic factor. Ann Oncol 2012;23:2937–2942. [DOI] [PubMed] [Google Scholar]
- 7. Janni WJ, Rack B, Terstappen LWMM, Pierga JY, Taran FA, Fehm T, Hall C, de Groot MR, Bidard FC, Friedl TWP, et al. Pooled analysis of the prognostic relevance of circulating tumor cells in primary breast cancer. Clin Cancer Res 2016;22:2583–2593. [DOI] [PubMed] [Google Scholar]
- 8. van Dalum G, Stam GJ, Scholten LF, Mastboom WJ, Vermes I, Tibbe AGJ, De Groot MR, Terstappen LWMM. Importance of circulating tumor cells in newly diagnosed colorectal cancer. Int J Oncol 2015;46:1361–1368. [DOI] [PubMed] [Google Scholar]
- 9. Ignatiadis M, Riethdorf S, Bidard FC, Vaucher I, Khazour M, Rothé F, Metallo J, Rouas G, Payne RE, Coombes RC, et al. International study on inter‐reader variability for circulating tumor cells in breast cancer. Breast Cancer Res 2014;16:R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kraan J, Sleijfer S, Strijbos MH Ignatiadis M, Peeters D, Pierga JY, Farace F, Riethdorf S, Fehm T, Zorzino L, Tibbe AGJ. External quality assurance of circulating tumor cell enumeration using the CellSearch system: A feasibility study. Cytometry Part B 2011;80(2):112–118. [DOI] [PubMed] [Google Scholar]
- 11. Tibbe AGJ, Miller MC, Terstappen LWMM. Statistical considerations for enumeration of circulating tumor cells. Cytometry A 2007;71(3):154–162. [DOI] [PubMed] [Google Scholar]
- 12. Coumans FAW, Ligthart ST, Terstappen LWMM. Interpretation of changes in circulating tumor cell counts. Transl Oncol 2012;5:486–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Wit, S , Manicone M, Rossi E, Lampignano R, Yang L, Zill B, Rengel‐Puertas A, Oulhen M, Crespo M, Berghuis AMS, Andree KC, et al EpCAMhigh and EpCAMlow circulating tumour cells in metastatic prostate and breast cancer patients. submitted (2017). [DOI] [PMC free article] [PubMed]
- 14. de Wit, S , Rossi E, Weber S, Tamminga M, Manicone M, Swennenhuis JF, Groothuis‐Oudshoorn CGM, Vidotto R, Facchinetti A, Zeune LL, et al Single tube liquid biopsy for advanced non‐small cell lung cancer. submitted (2018). [DOI] [PubMed]
- 15. de Wit S. Circulating tumor cells and beyond University of Twente, 2018. 10.3990/1.9789036545662. [DOI] [Google Scholar]
- 16. Zeune LL et al. Quantifying HER‐2 expression on circulating tumor cells by ACCEPT. PLoS One 2017;12:e0186562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zeune LL, van Dalum G, Terstappen LWMM, van Gils SA, Brune C. Multiscale segmentation via bregman distances and nonlinear spectral analysis. SIAM J Imaging Sci 2017;10:111–146. [Google Scholar]
- 18. Zeune LL, van Dalum G, Nanou A, de Wit S, Andree KC, Swennenhuis JF, Terstappen LWMM, Brune C. Deep learning to identify circulating tumor cells by ACCEPT ISMRC, 2018. [Google Scholar]
- 19. Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics 1977;33:159–174. [PubMed] [Google Scholar]
- 20. Altman, D. G. Practical statistics for medical research. (1991).
- 21. Barradas A, Terstappen L. Towards the biological understanding of ctc: capture technologies, definitions and potential to create metastasis. Cancers (Basel) 2013;5:1619–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Coumans FAW, Doggen CJM, Attard G, de Bono JS, Terstappen LWMM. All circulating EpCAM+CK+CD45‐ objects predict overall survival in castration‐resistant prostate cancer. Ann Oncol 2010;21:1851–1857. [DOI] [PubMed] [Google Scholar]
- 23. Larson CJ, Moreno JG, Pienta KJ, Gross S, Repollet M, O'Hara SM, Russell T, Terstappen LWMM. Apoptosis of circulating tumor cells in prostate cancer patients. Cytometry 2004;62A:46–53. [DOI] [PubMed] [Google Scholar]
- 24. Ligthart ST, Coumans FAW, Bidard FC, Simkens LHJ, Punt CJA, de Groot MR, Attard G, de Bono JS, Pierga JY, Terstappen LWMM. Circulating tumor cells count and morphological features in breast, colorectal and prostate cancer. PLoS One 2013;8:e67148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Swennenhuis JF, Tibbe AGJ, Levink R, Sipkema RCJ, Terstappen LWMM. Characterization of circulating tumor cells by fluorescence in situ hybridization. Cytometry A 2009;75:520–527. [DOI] [PubMed] [Google Scholar]
- 26. Nanou A, Coumans FAW, van Dalum G, Zeune LL, Dolling D, Onstenk W, Crespo M, Fontes MS, Rescigno P, Fowler G, et al. Circulating tumor cells, tumor‐derived extracellular vesicles and plasma cytokeratins in castration‐resistant prostate cancer patients. Oncotarget 2018;9:19283–19293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1 Overview of agreement on 100 cells between (A) Average Reviewer and Expert Panel; (B) Average Reviewer and Deep Learning and; (C) Expert Panel and Deep Learning scored with “1. Definitely not a CTC”; “2. Most likely not a CTC”; “3. Most likely a CTC”; and “4. Definitely a CTC”.
Supplementary Figure S1 Gallery of the 100 objects that were presented to the 15 reviewers, Expert Panel and Deep Learning network for scoring. The objects are sorted according to the AR score (high to low), followed by EP (high to low), followed by DL (high to low). The AR, EP and DL score is indicated next to the images. The scoring tool and the 100 images are available at: https://github.com/leoniez/ACCEPT/releases.


