Abstract
Neurotrophic receptor tyrosine kinase 2 (NTRK2) is a member of the tropomyosin receptor kinase family associated with the tumor development. However, the detailed function of NTRK2 in lung cancer, especially in lung adenocarcinoma (LUAD), is still not fully understood. Here, we investigated the effects of NTRK2 on LUAD biology. Through analyzing bioinformatics data derived from several databases, such as Oncomine, Gene Expression Profiling Interactive Analysis and UALCAN, we found that NTRK2 expression was significantly decreased in LUAD tissues. Clinical data acquired from Wanderer database, which is linked to The Cancer Genome Atlas database, demonstrated that the expression and methylation site of NTRK2 were significantly related to the clinical characteristics and prognosis of LUAD. Furthermore, NTRK2 expression was increased remarkably after treatment with the protein kinase B (AKT) inhibitor MK2206 and the anticancer agent actinomycin D. Functional enrichment analysis of NTRK2-associated coexpression genes was further conducted. Together, our results suggested that downregulated NTRK2 might be used in the diagnostic and prognostic evaluation of LUAD patients, or as a potential therapeutic target for the treatment of LUAD.
Keywords: NTRK2, Lung adenocarcinoma, Expression, Diagnosis, Prognosis
Introduction
Lung adenocarcinoma (LUAD) is the most frequent subtype of lung cancer, with incidence and mortality rates rising in both Western and Asian countries (Yan et al., 2019). Because of late diagnoses, the 5-year overall survival (OS) rate LUAD varies from 4 to 17% in line with the differences of stage and region, which is still very poor (Yan et al., 2018). At present, there is still no effective early diagnosis method for patients to receive timely treatment (Zheng et al., 2018). Therefore, it is necessary to search for novel target molecules for improving the early diagnosis and treatment of LUAD.
Previous studies have found a strong link between neurotrophic receptor tyrosine kinase 2 (NTRK2) and psychiatric disorders, such as schizophrenia (Spalek et al., 2016). Recent research advancement in the field revealed the relationship between NTRK2 and cancer biology. According to the ceRNA network, Gao et al. (2019) found that NTRK2 is related to the prognosis of invasive breast cancer. Through constructing the coexpression modules by WGCNA, NTRK2 was proposed to play a key role in the recurrence of uveal melanoma (Wan et al., 2018). Ni et al. (2017) demonstrated that activated NTRK2 alleles, especially the human tumor-associated QKI-NTRK2 fusion, could function together with Ink4a/Arf loss to promote astrocytoma formation. Furthermore, a recent study found that the interaction between differentiated glioblastoma cells and stem-like tumor cells via BDNF-NTRK2-VGF paracrine signaling accelerates tumor growth (Wang et al., 2018b). Nevertheless, there were few investigations about the relationship between NTRK2 and lung cancer, particularly LUAD, so the effects and mechanisms of NTRK2 in LUAD require further research.
The purpose of our study was to evaluate the role and mechanism of NTRK2 in human LUAD. Through bioinformatics data analysis, NTRK2 was found to be significantly downregulated in LUAD tissues. In addition, the expression level and methylation site of NTRK2 were notably correlated with clinical characteristics and prognosis. Moreover, based on the two datasets GSE6400 and GSE54293 from Gene Expression Omnibus (GEO), we observed the high levels of NTRK2 in the anticancer treatment group, indicating that NTRK2 could be used as a biomarker in evaluating clinical efficacy. In addition, Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa & Goto, 2000) analysis of NTRK2-associated coexpression genes further indicated that NTRK2 played an important part in LUAD treatment.
Materials and Methods
Data acquisition and reanalysis using different bioinformatics tools
The relevant bioinformatics data analysis of NTRK2 was obtained from several bioinformatics web resources, which were summarized in Table S1. The flow diagram of NTRK2 screen is shown in Fig. S1.
Oncomine is a cancer microarray or high-throughput sequencing data-mining platform, from which we can get gene expression signatures in human cancer tissues and cells (Rhodes et al., 2004). The data in Oncomine could be also linked into other public databases, such as GEO and The Cancer Genome Atlas (TCGA) (Hutter & Zenklusen, 2018). We conducted the comparison of NTRK2 expression across eight analyses between the LUAD and normal tissues. Additionally, Gene Expression Profiling Interactive Analysis (GEPIA) (Tang et al., 2017a), GE-mini (Tang et al., 2017b), Cancer RNA-Seq Nexus (CRN) (Li et al., 2016) and UALCAN (Chandrashekar et al., 2017), four additional cancer microarray or high-throughput sequencing data-mining databases, were employed to verify the results.
Wanderer is an interactive viewer, providing gene expression and DNA methylation data in human cancer (Diez-Villanueva, Mallona & Peinado, 2015), which enables us to screen for the possible methylation sites in the NTRK2 DNA sequence and to analyze the correlation between clinical characteristic of LUAD patients and NTRK2 expression and methylation sites. For the prognostic analysis, the Kaplan–Meier Plotter, a tool that can be used to assess the effect of genes on survival (Wang et al., 2018a), was utilized to describe the relationship between NTRK2 expression level, overall survival time (OS) and post-progression survival time (PPS). Further, the association between NTRK2 expression and disease-free survival (DFS) was completed through the GEPIA database.
Two datasets of the treatment-related transcriptome microarray, GSE6400 (Wang et al., 2007) and GSE54293 (Denisova et al., 2014), were acquired from the GEO database (Barrett & Edgar, 2008). Subsequently, the effects of NTRK2 expression on the chemotherapy for LUAD were analyzed.
The expression and methylation of NTRK2 correlation analysis were implemented by MethHC, which provided the information of DNA methylation and gene expression in human cancer (Huang et al., 2015). For the relevance between the disease prognosis and the methylation sites of NTRK2, the MethSurv tool was employed (Modhukur et al., 2018).
Using the cBioportal web tool (Gao et al., 2013), genes coexpressed with NTRK2 in LUAD were downloaded. Then, the STRING database (Szklarczyk et al., 2017) and Cytoscape software (Reimand et al., 2019) were used to complete the protein–protein interaction (PPI) network of these coexpression genes. Then, we utilized the DAVID bioinformatics resource (Huang, Sherman & Lempicki, 2009) to conduct the GO and KEGG pathway analysis of NTRK2 coexpression genes in LUAD samples. The web tools of WebGestalt (Wang et al., 2017) and PATHVIEW (Luo et al., 2017) were used for building a graphic.
Statistical analyses
The statistical tests were performed using SPSS 12.0 software (IBM Analytics). The results were expressed as the mean ± SD. Student t test, one-way ANOVA and K independent samples test were performed when appropriate. P < 0.05 was considered statistically significant.
Results
NTRK2 is downregulated in LUAD tissues
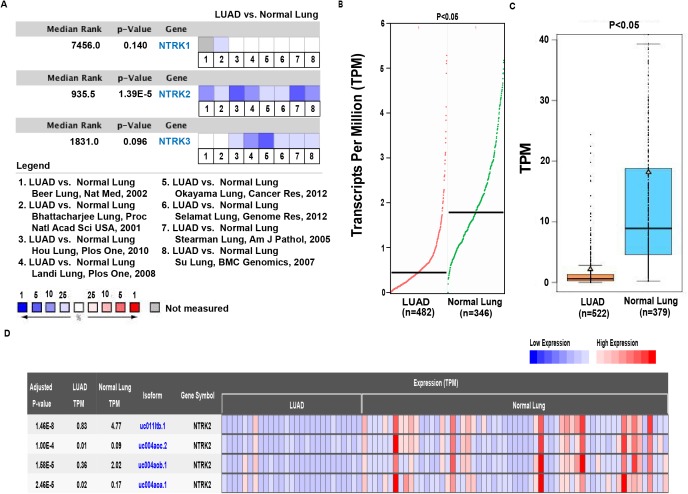
The NTRK family consists of three members, NTRK1, NTRK2 and NTRK3. Through the bioinformatics analysis of databases, we evaluated the transcriptional levels of NTRK family members in LUAD. First, we used the Oncomine database to observe the expression of NTRK1, NTRK2 and NTRK3 in eight LUAD datasets (Beer et al., 2002; Bhattacharjee et al., 2001; Hou et al., 2010; Landi et al., 2008; Okayama et al., 2012; Selamat et al., 2012; Stearman et al., 2005; Su et al., 2007). The results showed that NTRK2 had significantly lower expression in LUAD through the comparison among nine datasets, whereas NTRK1 and NTRK3 showed no statistical significance (Fig. 1A). Therefore, NTRK2 was chosen as the research target. To verify the trend, we examined the NTRK2 expression in LUAD by GEPIA and GE-mini, and we discovered the NTRK2 expression was clearly reduced in LUAD compared with the normal tissues (Figs. 1B and 1C). In addition, the heatmap from CRN database further indicated the low expression of NTRK2 in LUAD tissues (Fig. 1D). Next, given some activated oncogenes, such as Erb-B2 receptor tyrosine kinase 2 (ERBB2) and MET, have been demonstrated the driver roles in LUAD (The Cancer Genome Atlas Research Network, 2014), we want to evaluate the association between NTRK2 and these oncogenes. The data from UALCAN revealed the significantly downregulated NTRK2 (P < 0.01), upregulated ERBB2 (P < 0.01) and upregulated MET (P < 0.01) in LUAD tissues (Fig. S2A). Spearman correlation analysis showed the negative association between the expression of NTRK2 and ERBB2 or MET (Fig. S2B). Taken together, all of the above data suggested that the decreased expression of NTRK2 contributed to LUAD tumorigenesis, supporting its tumor-inhibiting function in LUAD.
Figure 1. Analysis of NTRK2 expression levels in LUAD tissues.
(A) The comparison of the messenger RNA (mRNA) expression of NTRK (NTRK1, NTRK2 and NTRK3) among eight datasets by comparing the surrounding normal lung tissues and LUAD. (B–D) The mRNA expression of NTRK2 was evaluated from the database GEPIA, GE-mini and CRN, respectively.
NTRK2 expression is associated with the clinical characteristics of LUAD patients
After determining the expression of NTRK2 in LUAD, we further analyzed the correlation between the NTRK2 expression level and the clinical characteristics of patients. Using the Wanderer database, we obtained a series of clinical data, and a summary of clinical characteristic parameters is provided in Table 1. As shown in this table, NTRK2 expression was significantly associated with gender (P = 0.007), pathologic T (P = 0.021), pathologic M (P = 0.006) and age (P = 0.036). Then, the Kaplan–Meier Plotter tool was used to evaluate the effects of NTRK2 expression on OS and PPS, confirming that the downregulated of NTRK2 expression was significantly related to shorter OS (P = 0.00029) (Fig. 2A) and PPS (P = 0.021) (Fig. 2B). Furthermore, we found that low NTRK2 expression was associated with RFS (P = 0.012) through using the GEPIA database (Fig. 2C). In conclusion, NTRK2 could be as a potential biomarker both for diagnosis and prognosis.
Table 1. The correlation between clinical characteristic parameters and the expression of NTRK2 in LUAD.
| Variables | Number | Mean ± SD | P |
|---|---|---|---|
| Gender | 0.007 | ||
| Male | 179 | 5.25 ± 1.90 | |
| Female | 212 | 5.78 ± 1.95 | |
| Radiation therapy | 0.640 | ||
| Yes | 6 | 5.12 ± 1.31 | |
| No | 89 | 5.48 ± 1.82 | |
| Kras mutation found | 0.454 | ||
| Yes | 14 | 5.48 ± 1.89 | |
| No | 34 | 5.88 ± 1.54 | |
| Pathologic T | 0.021 | ||
| T1/T1a/T1b | 122 | 6.01 ± 1.94 | |
| T2/T2a/T2b | 218 | 5.35 ± 2.01 | |
| T3 | 34 | 5.29 ± 1.43 | |
| T4 | 15 | 5.09 ± 1.52 | |
| TX | 2 | 4.36 ± 1.56 | |
| Pathologic N | 0.875 | ||
| N0 | 252 | 5.52 ± 1.86 | |
| N1 | 71 | 5.59 ± 1.89 | |
| N2 | 61 | 5.55 ± 2.32 | |
| NX | 5 | 4.85 ± 2.40 | |
| Pathologic M | 0.006 | ||
| M0 | 255 | 5.38 ± 1.85 | |
| M1/M1a/M1b | 16 | 4.86 ± 2.20 | |
| MX | 117 | 5.99 ± 2.04 | |
| Pathologic stage | 0.471 | ||
| Stage I/IA/IB | 211 | 5.63 ± 1.89 | |
| Stage IIA/IIB | 94 | 5.45 ± 1.78 | |
| Stage IIIA/IIIB | 68 | 5.52 ± 2.25 | |
| Stage IV | 17 | 4.89 ± 2.14 | |
| Race | 0.758 | ||
| White | 314 | 5.60 ± 1.92 | |
| Black or African American | 23 | 5.41 ± 2.26 | |
| Asian | 5 | 5.07 ± 1.12 | |
| Tobacco smoking history | 0.097 | ||
| Current reformed smoker for > 15 years | 94 | 5.84 ± 2.06 | |
| Current reformed smoker for < or = 15 years | 131 | 5.38 ± 1.95 | |
| Current reformed smoker, duration not specified | 2 | 5.15 ± 1.34 | |
| Lifelong non-smoker | 61 | 5.91 ± 1.75 | |
| Current smoker | 91 | 5.21 ± 1.97 | |
| Age at initial pathologic diagnosis | 0.036 | ||
| ≤60 | 125 | 5.25 ± 1.86 | |
| >60 | 248 | 5.69 ± 1.94 | |
| EGFR mutation result | 0.303 | ||
| Exon 19 deletion | 7 | 5.07 ± 1.26 | |
| L858R | 3 | 6.47 ± 1.00 | |
| Other | 9 | 5.94 ± 1.58 |
Figure 2. The effects of NTRK2 expression on prognosis in LUAD patients.
(A–B) The relationship between NTRK2 expression and OS and PPS, described by Kaplan–Meier Plotter. (C) The association between NTRK2 expression and RFS within the GEPIA database.
The roles of NTRK2 in LUAD therapies
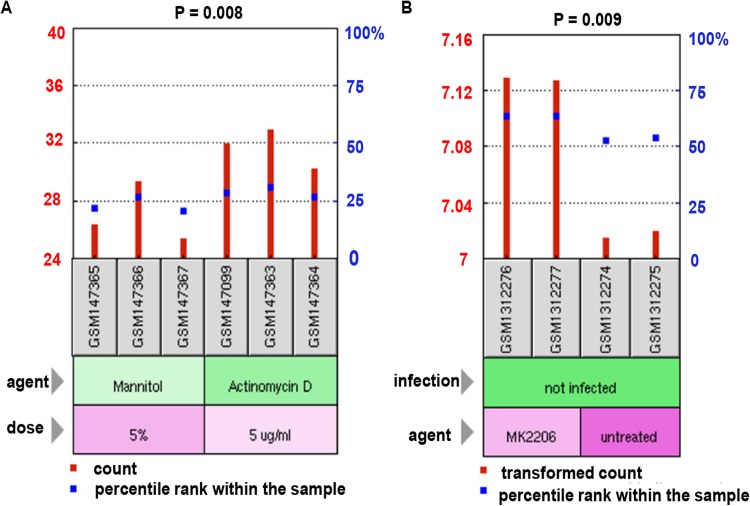
For the purpose of identifying the exact function of NTRK2 in LUAD chemotherapy, two treatment-related transcriptome microarray datasets, GSE6400 and GSE54293, were obtained from the GEO database. Previous studies have demonstrated that actinomycin D (Bai et al., 2019) and MK2206 (Dai et al., 2017) were two promising antitumor drugs. In the GSE6400 dataset, we discovered that the expression of NTRK2 was apparently higher in the actinomycin D treatment group than in the mannitol-control group (P = 0.008) (Fig. 3A). In addition, for the GSE54293 dataset, the AKT inhibitor MK2206 could enhance the NTRK2 expression levels significantly (P = 0.009) (Fig. 3B). Collectively, the findings observed above suggested that NTRK2 might enhance the response of cancer cells to the chemotherapeutics.
Figure 3. The influence of NTRK2 on the therapeutic response of LUAD patients.
(A) The GSE6400 dataset acquired from the GEO database was employed to estimate the impacts of NTRK2 expression on LUAD therapy both in the actinomycin D treatment group and the mannitol-control group. (B) In the treatment-related microarray GSE54293 dataset, the influence of NTRK2 expression on AKT inhibitor MK2206 treatment was evaluated.
The relationship between NTRK2 methylation and the clinical characteristics of LUAD patients
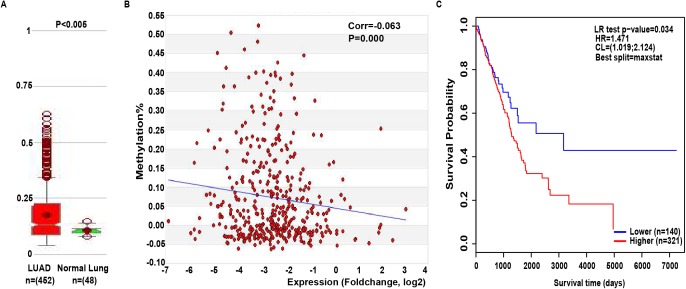
It is well-known that there is a negative correlation between DNA methylation and gene expression (Shi et al., 2017; Zhou et al., 2019). From the MethHC database, we observed that global NTRK2 methylation was significantly higher in LUAD samples compared with normal samples (P < 0.005) (Fig. 4A) and was negatively related to its expression (P = 0.000) (Fig. 4B), which gives further support for the low expression of NTRK2 in LUAD. Subsequently, the methylation site cg03628748 was screened out of the data (P = 4.35E-12) (Table S2) acquired from the Wanderer database. Then, the relationship between cg03628748 and the clinical characteristics of LUAD patients was examined, and results showed that cg03628748 was significantly related to Kras mutation (P = 0.038) and pathologic T (P = 0.000) (Table 2). Moreover, there was a significant negative correlation between higher methylation value of cg03628748 and shorter OS in LUAD patients (P = 0.034), which was analyzed by using the web tool of MethSurv (Fig. 4C).
Figure 4. The relationship between NTRK2 methylation and the clinical characteristics of LUAD patients.
(A) Global NTRK2 methylation in LUAD samples compared with the normal samples analyzed by MethHC database. (B) The association between global NTRK2 methylation and its expression in LUAD samples using the MethHC database. (C) The impact of the methylation site cg03628748 in NTRK2 on OS in LUAD patients as analyzed by the MethSurv web tool.
Table 2. The correlation between clinical characteristics of patients and the methylation site cg03628748 in NTRK2 in LUAD.
| Variables | Number | Mean ± SD | P |
|---|---|---|---|
| Gender | 0.123 | ||
| Male | 189 | 0.32 ± 0.14 | |
| Female | 219 | 0.30 ± 0.14 | |
| Radiation therapy | 0.112 | ||
| Yes | 7 | 0.22 ± 0.097 | |
| No | 96 | 0.31 ± 0.14 | |
| Kras mutation found | 0.038 | ||
| Yes | 16 | 0.38 ± 0.18 | |
| No | 34 | 0.28 ± 0.13 | |
| Pathologic T | 0.000 | ||
| T1/T1a/T1b | 127 | 0.26 ± 0.11 | |
| T2/T2a/T2b | 227 | 0.32 ± 0.14 | |
| T3 | 36 | 0.35 ± 0.16 | |
| T4 | 15 | 0.30 ± 0.15 | |
| TX | 3 | 0.23 ± 0.17 | |
| Pathologic N | 0.464 | ||
| N0 | 261 | 0.31 ± 0.14 | |
| N1 | 75 | 0.30 ± 0.13 | |
| N2 | 62 | 0.30 ± 0.14 | |
| NX | 8 | 0.24 ± 0.11 | |
| Pathologic M | 0.183 | ||
| M0 | 264 | 0.31 ± 0.14 | |
| M1/M1a/M1b | 17 | 0.27 ± 0.16 | |
| MX | 123 | 0.29 ± 0.13 | |
| Pathologic stage | 0.746 | ||
| Stage I/IA/IB | 218 | 0.31 ± 0.14 | |
| Stage IIA/IIB | 102 | 0.31 ± 0.13 | |
| Stage IIIA/IIIB | 68 | 0.31 ± 0.14 | |
| Stage IV | 19 | 0.27 ± 0.16 | |
| Race | 0.214 | ||
| White | 325 | 0.30 ± 0.13 | |
| Black or African American | 29 | 0.27 ± 0.12 | |
| Asian | 5 | 0.37 ± 0.14 | |
| Tobacco smoking history | 0.075 | ||
| Current reformed smoker for > 15 years | 101 | 0.31 ± 0.15 | |
| Current reformed smoker for < or = 15 years | 135 | 0.32 ± 0.14 | |
| Current reformed smoker, duration not specified | 2 | 0.39 ± 0.022 | |
| Lifelong non-smoker | 62 | 0.26 ± 0.12 | |
| Current smoker | 96 | 0.31 ± 0.14 | |
| Age at initial pathologic diagnosis | 0.644 | ||
| ≤60 | 131 | 0.30 ± 0.14 | |
| >60 | 259 | 0.31 ± 0.13 | |
| Residual tumor | 0.542 | ||
| RX | 16 | 0.31 ± 0.15 | |
| R0 | 271 | 0.31 ± 0.14 | |
| R1 | 10 | 0.26 ± 0.093 | |
| EGFR mutation result | 0.082 | ||
| Exon 19 deletion | 7 | 0.18 ± 0.063 | |
| L858R | 3 | 0.28 ± 0.17 | |
| Other | 9 | 0.33 ± 0.13 |
Functional enrichment analysis of NTRK2-associated coexpression genes
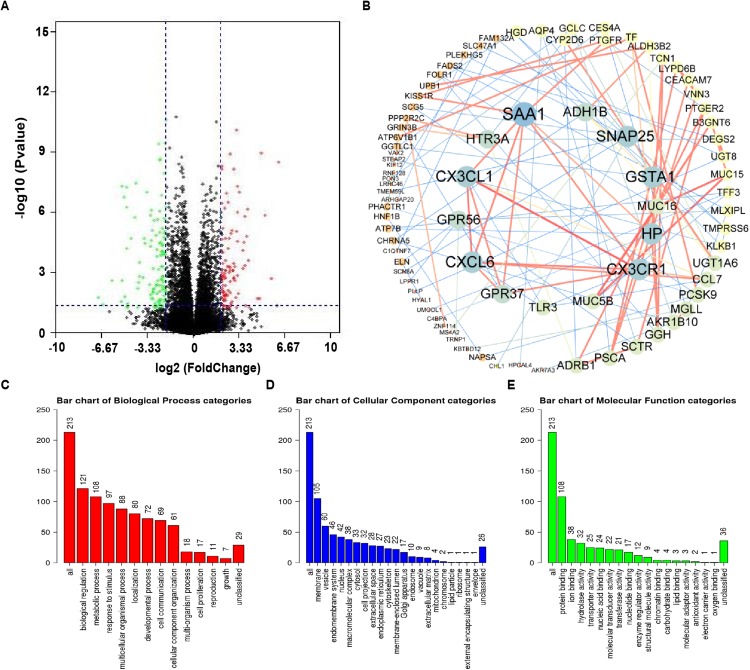
Using the cBioPortal database, 15,146 genes that were notably coexpressed with NTRK2 in the LUAD samples were acquired. The volcano plot was established for exhibiting between the altered and unaltered NTRK2 expression group (Fig. 5A). Next, we singled out 219 NTRK2-associated codifferentially expressed genes (co-DEGs) with the criteria of P value < 0.05 and |log Ratio| ≥ 2 (Table S3). Then, a PPT network of the co-DEGs was performed by using the STRING database and Cytoscape software (Fig. 5B). For the purpose of comprehending the biological function for these co-DEGs, GO and KEGG analyses were conducted by the WebGestalt and PATHVIEW web tools, respectively. The biological processes showed that these co-DEGs were mainly connected with biological regulation and metabolic processes (Fig. 5C). For the analysis of cellular components, the coexpression genes were mainly localized on cell membranes (Fig. 5D). For molecular function, protein binding was primarily enriched for these coexpression genes (Fig. 5E). Furthermore, the KEGG pathway demonstrated that these genes were involved in the process of xenobiotics and drug metabolism by cytochrome P450 (Table S4).
Figure 5. Functional enrichment analysis of NTRK2-associated co-DEGs in LUAD.
(A) The coexpression genes of NTRK2 were shown as volcano plot. (B) The PPI network of NTRK2-associated co-DEGs as completed by the STRING and Cytoscape software. (C–E) The GO analysis of NTRK2 associated co-DEGs including biological processes, cellular components and molecular function.
Discussion
This is the first study which presents comprehensive bioinformatic analysis of different public datasets that NTRK2 was identified as anti-oncogene in LUAD and could be used as a potential biomarker. Using the TCGA data from several databases, we found that NTRK2 expression was markedly decreased in LUAD tissues. The patients with downregulated NTRK2 expression and higher methylation values often had shorter OS, PPS and RFS.
Neurotrophic receptor tyrosine kinase 2 belongs to the NTRK family and has been previously shown to have an important impact on the development of the nervous system (Cocco, Scaltriti & Drilon, 2018). However, recent studies have demonstrated the possible role of NTRK2 in the development of cancer. Neurotrophic receptor tyrosine kinase 2 activation cooperates with PTEN deficiency through the activation of both the JAK–STAT3 and PI3K-AKT pathways to induce aggressiveness, resistance to current therapies and poor prognosis of T-cell acute lymphoblastic leukemia (Yuzugullu et al., 2016). Currently, NTRK fusion mutations have been reported to associate with oncogenic activation in various signaling pathways, such as AKT and MAPK, across multiple tumors (Stransky et al., 2014). Moreover, NTRK fusions were connected with poor survival in lung cancers (Rolfo & Raez, 2017). Interestingly, the reports seemed contrary to our results; this phenomenon might be explained by following reasons. First, it is known that different diseases or subtypes of tumors have diverse pathological states, which can change genes, functions. On the other hand, the structure, constitution and condition of genes may transformed, such as gene mutation, accompany with gene fusions. Furthermore, NTRK fusions are thought to occur at a low frequency across multiple tumor types (Vaishnavi, Le & Doebele, 2015). Additionally, although NTRK fusions were observed in rare cancer types, such as congenital infantile fibrosarcoma and secretory breast carcinoma, the occurrence in common cancers has been largely unexplored (Qaddoumi et al., 2016). Additionally, the difference in results might be on account of study designs or different patient populations, indicating international, multicenter randomized controlled, clinical research is needed for further study.
In the present study, GO and KEGG pathway analyses indicated that genes coexpressed with NTRK2 were mainly enriched in the processes of xenobiotics and drug metabolism. Moreover, NTRK2 expression was much higher in drug therapy groups in both the GSE6400 and GSE54293 datasets. Therefore, up-regulating NTRK2 expression to promote drug metabolism might be the mechanism that explains this phenomenon.
Nevertheless, there were several limitations to our study. First, the flow chart of analysis on the roles of NTRK2 in LUAD tumorigenesis was not strong enough, and should be further verified externally in diverse cohorts. Additionally, further validation of the roles of NTRK2 in multicenter clinical trials and prospective research is required. For the TCGA database, the included ethnicities were primarily white and black, and more studies are needed to confirm whether the findings are appropriate for other ethnic groups. Furthermore, more prognostic variables must be included to improve performance.
Conclusion
In conclusion, our study illustrated that NTRK2 was a putative cancer suppressor gene and could serve as a promising biomarker in tumorigenesis and treatment of LUAD patients. Furthermore, DNA hypermethylation has been demonstrated to be one of the mechanisms for the low-expressed NTRK2 in LADC. Understanding its detailed function and mechanisms in LUAD biological processes would provide promising insights for the prognostic and therapeutic value.
Supplemental Information
Figure S1. A Flow chart of analysis on the roles of NTRK2 in LUAD tumorigenesis.
Figure S2. The negative association between the expression of NTRK2 and ERBB2 or MET in LUAD.
Table S1: The main bioinformatics tools applied to analyze the role of NTRK2 in LUAD biological processes.
Table S2: The methylation values of CpG islands in NTRK2.
Table S3: The NTRK2-associated co-DEGs in LUAD.
Table S4: The KEGG pathway of NTRK2-associated co-DEGs.
Acknowledgments
We thank Elsevier’s English Language Editing Service for assistance with the language editing.
Funding Statement
The study was supported by the grants from National Natural Science Foundation of China (81803035, 81703036, 81572946), Natural Science Foundation of Hunan Province (No. 2019JJ50932), China Postdoctoral Science Foundation (2017M610510), Open-End Fund for the Valuable and Precision Instruments of Central South University (CSUZC201836), Youth Fund of Xiangya Hospital (2017Q17) and Postdoctoral Science Foundation of Central South University (185702). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
Xiang Wang performed the experiments, analyzed the data, prepared figures and/or tables.
Zhijie Xu conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Xi Chen contributed reagents/materials/analysis tools.
Xinxin Ren analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables.
Jie Wei contributed reagents/materials/analysis tools.
Shuyi Zhou contributed reagents/materials/analysis tools.
Xue Yang contributed reagents/materials/analysis tools.
Shuangshuang Zeng contributed reagents/materials/analysis tools.
Long Qian contributed reagents/materials/analysis tools.
Geting Wu contributed reagents/materials/analysis tools.
Zhicheng Gong conceived and designed the experiments.
Yuanliang Yan conceived and designed the experiments, performed the experiments, authored or reviewed drafts of the paper, approved the final draft.
Data Availability
The following information was supplied regarding data availability:
The raw measurements are available in the Supplemental Tables. The raw data shows the main bioinformatics tools, methylation values of CpG islands in NTRK2 and NTRK2-associated codifferentially expressed genes in lung adenocarcinoma.
References
- Bai et al. (2019).Bai L, Chen P, Xiang J, Sun J, Lei X. Enantiomeric NMR discrimination of carboxylic acids using actinomycin D as a chiral solvating agent. Organic & Biomolecular Chemistry. 2019;17(6):1466–1470. doi: 10.1039/C8OB03012J. [DOI] [PubMed] [Google Scholar]
- Barrett & Edgar (2008).Barrett T, Edgar R. Reannotation of array probes at NCBI’s GEO database. Nature Methods. 2008;5(2):117–117. doi: 10.1038/nmeth0208-117b. [DOI] [PubMed] [Google Scholar]
- Beer et al. (2002).Beer DG, Kardia SLR, Huang C-C, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, Lizyness ML, Kuick R, Hayasaka S, Taylor JMG, Iannettoni MD, Orringer MB, Hanash S. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nature Medicine. 2002;8(8):816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee et al. (2001).Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, Loda M, Weber G, Mark EJ, Lander ES, Wong W, Johnson BE, Golub TR, Sugarbaker DJ, Meyerson M. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(24):13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar et al. (2017).Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocco, Scaltriti & Drilon (2018).Cocco E, Scaltriti M, Drilon A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nature Reviews Clinical Oncology. 2018;15(12):731–747. doi: 10.1038/s41571-018-0113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai et al. (2017).Dai S, Yan Y, Xu Z, Zeng S, Qian L, Huo L, Li X, Sun L, Gong Z. SCD1 confers temozolomide resistance to human glioma cells via the Akt/GSK3β/β-catenin signaling axis. Frontiers in Pharmacology. 2017;8:960. doi: 10.3389/fphar.2017.00960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisova et al. (2014).Denisova OV, Söderholm S, Virtanen S, Von Schantz C, Bychkov D, Vashchinkina E, Desloovere J, Tynell J, Ikonen N, Theisen LL, Nyman TA, Matikainen S, Kallioniemi O, Julkunen I, Muller CP, Saelens X, Verkhusha VV, Kainov DE. Akt inhibitor MK2206 prevents influenza pH1N1 virus infection in vitro. Antimicrobial Agents and Chemotherapy. 2014;58(7):3689–3696. doi: 10.1128/AAC.02798-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Villanueva, Mallona & Peinado (2015).Díez-Villanueva A, Mallona I, Peinado MA. Wanderer, an interactive viewer to explore DNA methylation and gene expression data in human cancer. Epigenetics & Chromatin. 2015;8(1):22. doi: 10.1186/s13072-015-0014-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao et al. (2013).Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science Signaling. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao et al. (2019).Gao C, Li H, Zhuang J, Zhang HX, Wang K, Yang J, Liu C, Liu L, Zhou C, Sun C. The construction and analysis of ceRNA networks in invasive breast cancer: a study based on The Cancer Genome Atlas. Cancer Management and Research. 2019;11:1–11. doi: 10.2147/CMAR.S182521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou et al. (2010).Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens JA, Hoogsteden HC, Grosveld F, Philipsen S. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLOS ONE. 2010;5(4):e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang et al. (2015).Huang W-Y, Hsu S-D, Huang H-Y, Sun Y-M, Chou C-H, Weng S-L, Huang H-D. MethHC: a database of DNA methylation and gene expression in human cancer. Nucleic Acids Research. 2015;43(D1):D856–D861. doi: 10.1093/nar/gku1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Sherman & Lempicki (2009).Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hutter & Zenklusen (2018).Hutter C, Zenklusen JC. The cancer genome atlas: creating lasting value beyond its data. Cell. 2018;173(2):283–285. doi: 10.1016/j.cell.2018.03.042. [DOI] [PubMed] [Google Scholar]
- Kanehisa & Goto (2000).Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi et al. (2008).Landi MT, Dracheva T, Rotunno M, Figueroa JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW, Murphy SE, Yang P, Pesatori AC, Consonni D, Bertazzi PA, Wacholder S, Shih JH, Caporaso NE, Jen J. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLOS ONE. 2008;3(2):e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2016).Li J-R, Sun C-H, Li W, Chao R-F, Huang C-C, Zhou XJ, Liu C-C. Cancer RNA-Seq Nexus: a database of phenotype-specific transcriptome profiling in cancer cells. Nucleic Acids Research. 2016;44(D1):D944–D951. doi: 10.1093/nar/gkv1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo et al. (2017).Luo W, Pant G, Bhavnasi YK, Blanchard SG, Jr, Brouwer C. Pathview Web: user friendly pathway visualization and data integration. Nucleic Acids Research. 2017;45(W1):W501–W508. doi: 10.1093/nar/gkx372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modhukur et al. (2018).Modhukur V, Iljasenko T, Metsalu T, Lokk K, Laisk-Podar T, Vilo J. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics. 2018;10(3):277–288. doi: 10.2217/epi-2017-0118. [DOI] [PubMed] [Google Scholar]
- Ni et al. (2017).Ni J, Xie S, Ramkissoon SH, Luu V, Sun Y, Bandopadhayay P, Beroukhim R, Roberts TM, Stiles CD, Segal RA, Ligon KL, Hahn WC, Zhao JJ. Tyrosine receptor kinase B is a drug target in astrocytomas. Neuro-Oncology. 2017;19(1):22–30. doi: 10.1093/neuonc/now139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okayama et al. (2012).Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S, Watanabe S-I, Sakamoto H, Kumamoto K, Takenoshita S, Gotoh N, Mizuno H, Sarai A, Kawano S, Yamaguchi R, Miyano S, Yokota J. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Research. 2012;72(1):100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- Qaddoumi et al. (2016).Qaddoumi I, Orisme W, Wen J, Santiago T, Gupta K, Dalton JD, Tang B, Haupfear K, Punchihewa C, Easton J, Mulder H, Boggs K, Shao Y, Rusch M, Becksfort J, Gupta P, Wang S, Lee RP, Brat D, Peter Collins V, Dahiya S, George D, Konomos W, Kurian KM, McFadden K, Serafini LN, Nickols H, Perry A, Shurtleff S, Gajjar A, Boop FA, Klimo PD, Jr, Mardis ER, Wilson RK, Baker SJ, Zhang J, Wu G, Downing JR, Tatevossian RG, Ellison DW. Genetic alterations in uncommon low-grade neuroepithelial tumors: BRAF, FGFR1, and MYB mutations occur at high frequency and align with morphology. Acta Neuropathologica. 2016;131(6):833–845. doi: 10.1007/s00401-016-1539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand et al. (2019).Reimand J, Isserlin R, Voisin V, Kucera M, Tannus-Lopes C, Rostamianfar A, Wadi L, Meyer M, Wong J, Xu C, Merico D, Bader GD. Pathway enrichment analysis and visualization of omics data using g: Profiler, GSEA, Cytoscape and EnrichmentMap. Nature Protocols. 2019;14:482–517. doi: 10.1038/s41596-018-0103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes et al. (2004).Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfo & Raez (2017).Rolfo C, Raez L. New targets bring hope in squamous cell lung cancer: neurotrophic tyrosine kinase gene fusions. Laboratory Investigation. 2017;97(11):1268–1270. doi: 10.1038/labinvest.2017.91. [DOI] [PubMed] [Google Scholar]
- Selamat et al. (2012).Selamat SA, Chung BS, Girard L, Zhang W, Zhang Y, Campan M, Siegmund KD, Koss MN, Hagen JA, Lam WL, Lam S, Gazdar AF, Laird-Offringa IA. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Research. 2012;22(7):1197–1211. doi: 10.1101/gr.132662.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi et al. (2017).Shi Y-X, Wang Y, Li X, Zhang W, Zhou H-H, Yin J-Y, Liu Z-Q. Genome-wide DNA methylation profiling reveals novel epigenetic signatures in squamous cell lung cancer. BMC Genomics. 2017;18(1):901. doi: 10.1186/s12864-017-4223-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalek et al. (2016).Spalek K, Coynel D, Freytag V, Hartmann F, Heck A, Milnik A, de Quervain D, Papassotiropoulos A. A common NTRK2 variant is associated with emotional arousal and brain white-matter integrity in healthy young subjects. Translational Psychiatry. 2016;6(3):e758. doi: 10.1038/tp.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearman et al. (2005).Stearman RS, Dwyer-Nield L, Zerbe L, Blaine SA, Chan Z, Bunn PA, Jr, Johnson GL, Hirsch FR, Merrick DT, Franklin WA, Baron AE, Keith RL, Nemenoff RA, Malkinson AM, Geraci MW. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. American Journal of Pathology. 2005;167(6):1763–1775. doi: 10.1016/S0002-9440(10)61257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stransky et al. (2014).Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nature Communications. 2014;5(1):4846. doi: 10.1038/ncomms5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su et al. (2007).Su L-J, Chang C-W, Wu Y-C, Chen K-C, Lin C-J, Liang S-C, Lin C-H, Whang-Peng J, Hsu S-L, Chen C-H, Huang C-YF. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics. 2007;8(1):140. doi: 10.1186/1471-2164-8-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk et al. (2017).Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, von Mering C. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Research. 2017;45(D1):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang et al. (2017a).Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Research. 2017a;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang et al. (2017b).Tang Z, Li C, Zhang K, Yang M, Hu X. GE-mini: a mobile APP for large-scale gene expression visualization. Bioinformatics. 2017b;33:941–943. doi: 10.1093/bioinformatics/btw775. [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network (2014).The Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnavi, Le & Doebele (2015).Vaishnavi A, Le AT, Doebele RC. TRKing down an old oncogene in a new era of targeted therapy. Cancer Discovery. 2015;5(1):25–34. doi: 10.1158/2159-8290.CD-14-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan et al. (2018).Wan Q, Tang J, Han Y, Wang D. Co-expression modules construction by WGCNA and identify potential prognostic markers of uveal melanoma. Experimental Eye Research. 2018;166:13–20. doi: 10.1016/j.exer.2017.10.007. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2018a).Wang X, Chen D, Gao J, Long H, Zha H, Zhang A, Shu C, Zhou L, Yang F, Zhu B, Wu W. Centromere protein U expression promotes non-small-cell lung cancer cell proliferation through FOXM1 and predicts poor survival. Cancer Management and Research. 2018a;10:6971–6984. doi: 10.2147/CMAR.S182852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2007).Wang Z, Lecane PS, Thiemann P, Fan Q, Cortez C, Ma X, Tonev D, Miles D, Naumovski L, Miller RA, Magda D, Cho DG, Sessler JL, Pike BL, Yeligar SM, Karaman MW, Hacia JG. Synthesis and biologic properties of hydrophilic sapphyrins, a new class of tumor-selective inhibitors of gene expression. Molecular Cancer. 2007;6(1):9. doi: 10.1186/1476-4598-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2018b).Wang X, Prager BC, Wu Q, Kim LJY, Gimple RC, Shi Y, Yang K, Morton AR, Zhou W, Zhu Z, Obara EAA, Miller TE, Song A, Lai S, Hubert CG, Jin X, Huang Z, Fang X, Dixit D, Tao W, Zhai K, Chen C, Dong Z, Zhang G, Dombrowski SM, Hamerlik P, Mack SC, Bao S, Rich JN. Reciprocal signaling between glioblastoma stem cells and differentiated tumor cells promotes malignant progression. Cell Stem Cell. 2018b;22(4):514–528.e5. doi: 10.1016/j.stem.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2017).Wang J, Vasaikar S, Shi Z, Greer M, Zhang B. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Research. 2017;45(W1):W130–W137. doi: 10.1093/nar/gkx356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan et al. (2018).Yan Y, Xu Z, Hu X, Qian L, Li Z, Zhou Y, Dai S, Zeng S, Gong Z. SNCA is a functionally low-expressed gene in lung adenocarcinoma. Genes (Basel) 2018;9(1):16. doi: 10.3390/genes9010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan et al. (2019).Yan Y, Xu Z, Qian L, Zeng S, Zhou Y, Chen X, Wei J, Gong Z. Identification of CAV1 and DCN as potential predictive biomarkers for lung adenocarcinoma. American Journal of Physiology-Lung Cellular and Molecular Physiology. 2019;316(4):L630–L643. doi: 10.1152/ajplung.00364.2018. [DOI] [PubMed] [Google Scholar]
- Yuzugullu et al. (2016).Yuzugullu H, Von T, Thorpe LM, Walker SR, Roberts TM, Frank DA, Zhao JJ. NTRK2 activation cooperates with PTEN deficiency in T-ALL through activation of both the PI3K–AKT and JAK–STAT3 pathways. Cell Discovery. 2016;2(1):16030. doi: 10.1038/celldisc.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng et al. (2018).Zheng H, Zhan Y, Liu S, Lu J, Luo J, Feng J, Fan S. The roles of tumor-derived exosomes in non-small cell lung cancer and their clinical implications. Journal of Experimental & Clinical Cancer Research. 2018;37(1):226. doi: 10.1186/s13046-018-0901-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou et al. (2019).Zhou S, Yan Y, Chen X, Wang X, Zeng S, Qian L, Wei J, Yang X, Zhou Y, Gong Z, Xu Z. Roles of highly expressed PAICS in lung adenocarcinoma. Gene. 2019;692:1–8. doi: 10.1016/j.gene.2018.12.064. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. A Flow chart of analysis on the roles of NTRK2 in LUAD tumorigenesis.
Figure S2. The negative association between the expression of NTRK2 and ERBB2 or MET in LUAD.
Table S1: The main bioinformatics tools applied to analyze the role of NTRK2 in LUAD biological processes.
Table S2: The methylation values of CpG islands in NTRK2.
Table S3: The NTRK2-associated co-DEGs in LUAD.
Table S4: The KEGG pathway of NTRK2-associated co-DEGs.
Data Availability Statement
The following information was supplied regarding data availability:
The raw measurements are available in the Supplemental Tables. The raw data shows the main bioinformatics tools, methylation values of CpG islands in NTRK2 and NTRK2-associated codifferentially expressed genes in lung adenocarcinoma.