Abstract
A novel Tymoviridae-like virus, designated Ek Balam virus, was isolated from male Culex quinquefasciatus mosquitoes collected in Yucatan, Mexico. The genome was fully sequenced and shown to have no more than 69% nt sequence identity to its closest known relative. Mosquito cells were permissive to Ek Balam virus replication, but mammalian and avian cells were refractory, suggesting that vertebrates are not involved in the maintenance of the virus in nature.
Keywords: metagenomics, virus discovery, RNA-seq, mosquito, Tymoviridae, Mexico
All viruses currently classified into as members of the family Tymoviridae are plant viruses, and some are major agricultural pathogens (1, 2). The family includes three genera: Maculavirus, Marafivirus and Tymovirus. Viruses in the genera Marafivirus and Tymovirus are transmitted by leafhoppers and beetles, respectively, but no vectors have been identified for viruses in the genus Maculavirus. Family members contain a positive-sense, single-stranded RNA genome of 6.0 to 7.5 kb. Genomes are capped at the 5’ end, often possess a tRNA structure at the 3’ end and, except for viruses in the genus Tymovirus, are polyadenylated. The number of open reading frames (ORFs) varies according to genus and species. Viruses in the genus Maculavirus have four ORFs, and these encode a replicase, coat protein and two small proline-rich proteins, p31 and p16 (2, 3).
Mosquitoes are not recognized vectors of viruses in the family Tymoviridae, although a novel Tymoviridae-like virus, designated Culex-originated Tymoviridae-like virus (CuTLV), was recently isolated from Culex spp. mosquitoes in China (4). The genomic organization of CuTLV is similar to that of viruses in the genus Maculavirus, although notable differences were also described, including the absence of a polyadenylated tail and an ORF encoding p32. CuTLV has not yet been classified by the International Committee on Taxonomy of Viruses, and it remains to be seen whether it will be assigned to the genus Maculavirus or to a new genus or family. We report the discovery of a second novel Tymoviridae-like virus that naturally infects mosquitoes. The novel virus, designated Ek Balam virus (EkBV), was detected in male Culex quinquefasciatus collected in 2007 in the rural town of Tixkokob in Yucatan, Mexico. The virus was named after an archeological ruin in Yucatan. Descriptions of study sites and protocols for the collection, identification and homogenization of mosquitoes are provided elsewhere (5, 6). EkBV was isolated after two blind passages in Aedes albopictus (C6/36) cells, and its genome was fully sequenced by Ion Proton sequencing, reverse transcription polymerase chain reaction, Sanger sequencing, and 5’ and 3’ rapid amplification of cDNA ends as described previously (7). The genome of EkBV consists of 6516 nt (GenBank accession no. MH822863). The sequence is most closely related to the genome sequence of CuTLV (68.3% nt sequence identity with 29.6% coverage), followed by bat tymo-like virus (65.8% nt sequence identity with 41.5% coverage), an unclassified Tymoviridae-like virus detected in guano of Myotis yumanensis (GenBank accession nos. NC_030844.1 and KX580887.1; Alex Greninger, University of Washington, Seattle, WA, USA, personal communication). It is not known whether bat tymo-like virus replicates in bats or was acquired through the consumption of virus-infected material (i.e., insects), but the latter appears more likely because the virus is most closely related to EkBV and CuTLV. The next-closest relatives to EkBV are viruses in the genus Maculavirus. The genome of EkBV has a high cytosine and low guanine content (A, 23.0%; C, 31.7%, G, 18.6%; T, 26.7%), as has been reported for CuTLV and recognized members of the family Tymoviridae (1,3, 4, 8). The genome lacks a polyadenylate tail, and its 5’ and 3’ untranslated regions consist of 144 nt and 433 nt, respectively. We could not provide any convincing evidence of a tRNA-like structure in the 3’ UTR (data not shown). The genome does not possess a classical “tymobox” or “marafibox”, conserved subgenomic promoter sequences characteristic of tymoviruses and marafiviruses, respectively (1, 2).
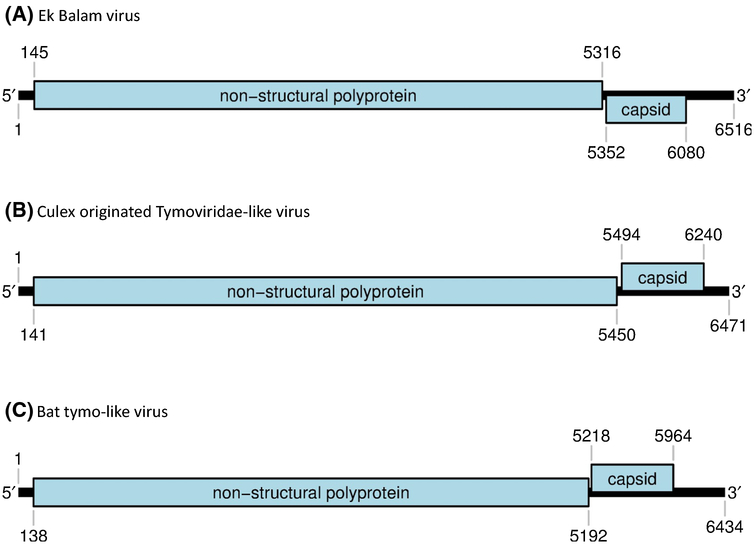
The genome of EkBV is predicted to contain at least two major ORFs (Fig. 1). The first ORF encodes a putative replicase of 1724 residues that is most closely related to the corresponding translation product of CuTLV (50.9% identity and 64.1% similarity, with 100% coverage). Predicted viral methyltransferase (pfam01660), tymovirus endopeptidase (pfam05381), helicase (pfam01443) and RdRp (pfam00978) domains are present at residues 38 to 318, 683 to 770, 870 to 1102 and 1400 to 1591, respectively. The second ORF encodes a putative coat protein of 243 residues that is most closely related to the corresponding translation product of CuTLV (61.3% identity and 75.4% similarity, with 100% coverage). A predicted tymovirus coat protein domain (pfam00983) is present at residues 113 to 196. The ORFs are separated by a 32-nt intergenic region.
Fig. 1.
Genomic organization of (A) Ek Balam virus, (B) Culex originated Tymoviridae-like virus and (C) bat tymo-like virus
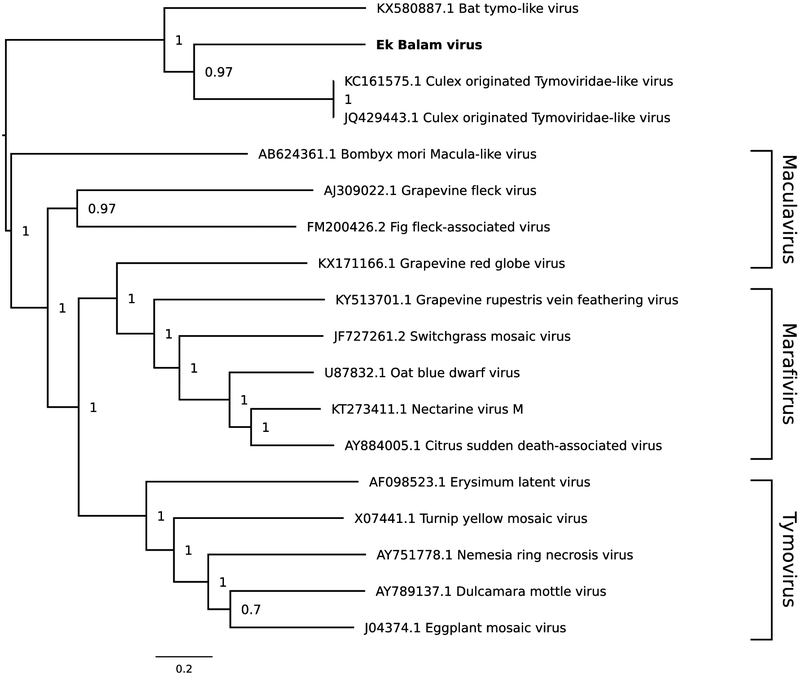
A phylogenetic tree was constructed using Bayesian methods based on the deduced amino acid sequence of the replicase of EkBV and the corresponding regions of some related sequences (Fig. 2). EkBV is most closely related phylogenetically to CuTLV, and the posterior support for this topological arrangement is 0.97. EkBV, CuTLV and bat tymo-like virus belong to a clade that is basal to recognized members of the family Tymoviridae. Viruses in this family separate into three distinct genus-specific clades.
Fig. 2.
Phylogenetic tree for EkBV and selected related reference and non-reference sequences. Amino acid sequences of the open reading frame encoding the replicase were aligned using MUSCLE (9). A maximum-likelihood phylogenetic tree was constructed using the Bayesian Markov chain Monte Carlo method implemented in MrBayes version 3.2.6 (10), sampling across the default set of fixed amino acid rate matrices with 1,000,000 generations, discarding the first 25% as burn-in. The figure was produced using FigTree (available at http://tree.bio.ed.ac.uk/software/figtree/). The tree is midpoint-rooted, and selected nodes are labelled with posterior probability values. GenBank accession numbers are shown next to virus names.
The in vitro host range of EkBV was determined by assessing its replicative ability in cell lines of avian, mammalian and mosquito origin. Six cell lines were tested: African green monkey kidney (Vero), Anopheles gambiae (Sua 4.0), baby hamster kidney (BSR-T7/5), C6/36, Culex tarsalis (CT) and duck embryonic fibroblast (DEF) cells. A cell line was considered to support EkBV replication if viral RNA was detected by RT-PCR after five passages. All mosquito cell lines were permissive to replication, indicating that the virus has a broad mosquito host range. A cytopathic effect was not observed in any mosquito cell cultures. All vertebrate cell lines were refractory to virus infection, consistent with a vertebrate replication-incompetent phenotype. CuTLV is also assumed to have a vertebrate-replication-incompetent phenotype because it cannot replicate in Vero or baby hamster kidney (BHK-21) cells (4).
The inability of EkBV to replicate in vertebrate cells and its detection in male mosquitoes suggests that the virus is not maintained in nature in a mosquito-vertebrate transmission cycle. Our findings indicate that EkBV is more likely to be maintained by vertical (i.e., transovarial) or horizontal (i.e., venereal) transmission among insect hosts. Alternatively, the virus could be maintained by mechanical (i.e., per os) transmission. Experimental infections are needed to determine whether EkBV is maintained in nature by direct mosquito-to-mosquito transmission and whether it is capable of infecting plants. The detection of EkBV in field-collected mosquitoes and C6/36 cells after five passages suggests that the virus replicates in mosquitoes and is not a surface or gut contaminant acquired after coming in contact with or feeding upon infectious material.
In conclusion, we report the isolation of a novel Tymoviridae-like virus from mosquitoes in Mexico and add to the rapidly increasing number of newly discovered viruses. Based on the genetic distance between EkBV, its closest known relative (CuTLV) and recognized members of the family Tymoviridae, we propose that EkBV and CuTLV are members of two distinct species that should be assigned to the same, yet-to-be-established genus. It remains to be determined whether these viruses will be classified as members of the family Tymoviridae or of a new family.
Acknowlegments
Mosquito collections were made possible by grant 5R21AI067281 from the National Institutes of Health. The metagenomics analysis and in vitro host range experiments were supported by an intramural grant from Iowa State University. A.E.F. and C.T. are supported by a Wellcome Trust grant [106207] and a European Research Council grant [646891] to A.E.F. The authors thank Kristina Larsen and Sean Johnston for technical assistance.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
The research reported here did not involve the use of human subjects or vertebrate animals.
Ethical statement
This study represents original work that has not been submitted to any other journal for publication. The authors have no conflict of interest to declare. No human or animal ethics approval was required for the completion of this study.
References
- 1.Martelli GP, Sabanadzovic S, Abou-Ghanem Sabanadzovic N, Edwards MC, Dreher T. The family Tymoviridae. Archives of virology. 2002. September;147(9):1837–46. [DOI] [PubMed] [Google Scholar]
- 2.King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. Virus taxonomy: classification and nomenclature of viruses: Ninth Report of the International Committee on Taxonomy of Viruses: Elsevier Academic Press; 2011. [Google Scholar]
- 3.Martelli GP, Sabanadzovic S, Abou Ghanem-Sabanadzovic N, Saldarelli P. Maculavirus, a new genus of plant viruses. Archives of virology. 2002. September;147(9):1847–53. [DOI] [PubMed] [Google Scholar]
- 4.Wang L, Lv X, Zhai Y, Fu S, Wang D, Rayner S, et al. Genomic characterization of a novel virus of the family Tymoviridae isolated from mosquitoes. PloS one. 2012;7(7):e39845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farfan-Ale JA, Lorono-Pino MA, Garcia-Rejon JE, Hovav E, Powers AM, Lin M, et al. Detection of RNA from a novel West Nile-like virus and high prevalence of an insect-specific flavivirus in mosquitoes in the Yucatan Peninsula of Mexico. The American journal of tropical medicine and hygiene. 2009. January;80(1):85–95. [PMC free article] [PubMed] [Google Scholar]
- 6.Farfan-Ale JA, Lorono-Pino MA, Garcia-Rejon JE, Soto V, Lin M, Staley M, et al. Detection of flaviviruses and orthobunyaviruses in mosquitoes in the Yucatan Peninsula of Mexico in 2008. Vector borne and zoonotic diseases. 2010. October;10(8):777–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charles J, Tangudu CS, Hurt SL, Tumescheit C, Firth AE, Garcia-Rejon JE, et al. Detection of novel and recognized RNA viruses in mosquitoes from the Yucatan Peninsula of Mexico using metagenomics and characterization of their in vitro host ranges. Journal of General Virology. accepted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morch MD, Boyer JC, Haenni AL. Overlapping open reading frames revealed by complete nucleotide sequencing of turnip yellow mosaic virus genomic RNA. Nucleic acids research. 1988. July 11;16(13):6157–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC bioinformatics. 2004. August 19;5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic biology. 2012. May;61(3):539–42. [DOI] [PMC free article] [PubMed] [Google Scholar]