Abstract
Molecular analysis of rare single cells like circulating tumor cells (CTCs) from whole blood patient samples bears multiple challenges. One of those challenges is the efficient and ideally loss‐free isolation of CTCs over contaminating white and red blood cells. While there is a multitude of commercial and non‐commercial systems available for the enrichment of CTCs their cell output does not deliver the purity most molecular analysis methods require. Here we describe the ALS CellCelector™ which can solve this challenge allowing the retrieval of 100% pure single CTCs from blood processed by different upstream enrichment techniques. It is a multifunctional, extremely flexible system for automated screening of cell culture plates, Petri dishes, and microscope slides. Fixed or live single cells or multicellular clusters detected during screening can be picked out of those plates automatically. The complete scan and picking process is fully documented hence allowing highest standardization and reproducibility of all processes. Use of CellCelector allowed the isolation of pure single tumor cells or clusters from liquid biopsies of breast, prostate, ovarian, colorectal, lung, and brain cancers for their subsequent molecular analysis. © 2018 The Authors. Cytometry Part A published by Wiley Periodicals, Inc. on behalf of International Society for Advancement of Cytometry.
Keywords: rare single cells, liquid biopsy, single cell picking, automated image analysis, rare cell isolation, CTC isolation, purification of CTCs, circulating tumor cells, imaging cytometry, automated cell micromanipulation
Various sophisticated technologies for circulating tumor cell (CTC) enrichment from the whole blood have been developed over the last decade 1 and two dozens of industrial players are presently active in the area. The enrichment methods are usually classified as (i) antibody‐based ones (also called label‐dependent) and (ii) those based on physical properties of the cells (also called label‐independent). The latter use larger size, different mechanical deformability, and/or dielectric properties of CTCs for their separation from normal cells. CellSearch® or Isoflux™ technologies are examples of antibody‐based techniques, while Parsortix™, ScreenCell™, Vertex™, or ClearCell™ technologies can be cited as examples of label‐independent enrichment methods. Independently of the technology used the enrichment step is usually followed by a staining procedure for specific CTC detection and identification using fluorescence microscopy.
All enrichment methods provide CTC populations still contaminated by background cells at various levels (usually 1 CTC for 102–104 blood cells depending on the method) which is usually incompatible with downstream molecular analysis. Here we present the ALS CellCelector™ that enables the isolation of individual cells and clusters from CTC‐enriched cell suspensions for molecular characterization at the single cell level or re‐cultivation (e.g., in PDX models).
ALS Cell C elector™
The CellCelector™ technology (Fig. 1) represents a combination of an automated high‐content imaging cytometry platform (microscopic image detection of cells; Fig. 1A) with automated cell micromanipulation utilizing a vertical high‐precision glass micro‐capillary (Fig. 1F) fixed to a precision robotic arm (Fig. 1D) and a syringe pump (Fig. 1E) capable of handling volumes down to the nanoliter range.
Figure 1.
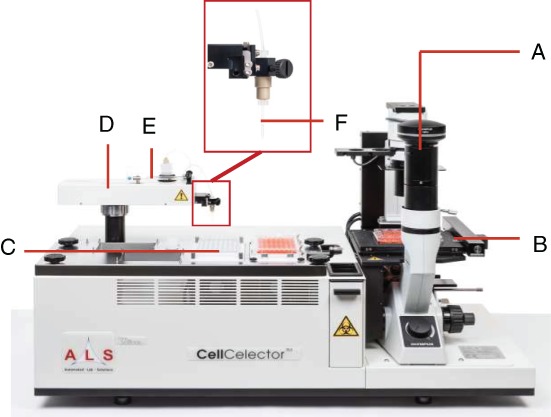
ALS CellCelector™ system with (A) inverted fluorescence microscope with up to seven fluorescence channels, 1.25×–100× objective options and scientific‐grade CCD camera; (B) high‐precision high‐speed microscope stage with autofocus capabilities for scanning of samples; (C) deck tray for placement of destination plates (can be optionally cooled to 4 °C for RNA applications); (D) exchangeable picking module (Single Cell Picking Module shown); (E) high‐precision syringe pump for aspiration/dispensing of liquids; (F) micro‐capillary for aspiration of cells (diameters 20–220 μm for single cell and cluster picking).
Scanning of the sample in bright field, phase contrast, and/or up to seven fluorescent channels is followed by an automated identification and sorting of detected cells according to positive and negative markers as wells as cell morphology. Putative CTCs are recovered into a destination vessel (PCR tubes, strips, or plates) for either downstream analysis or re‐cultivation. The cells can be picked individually or pooled together. Contrary to commercially available manual or semi‐automated cell micromanipulators which rely on user's skills and patience, the CellCelector offers the highest level of automation together with full process documentation including before and after picking images (Fig. 2A,B) and data tracing the cell from source to destination plate and well.
Figure 2.
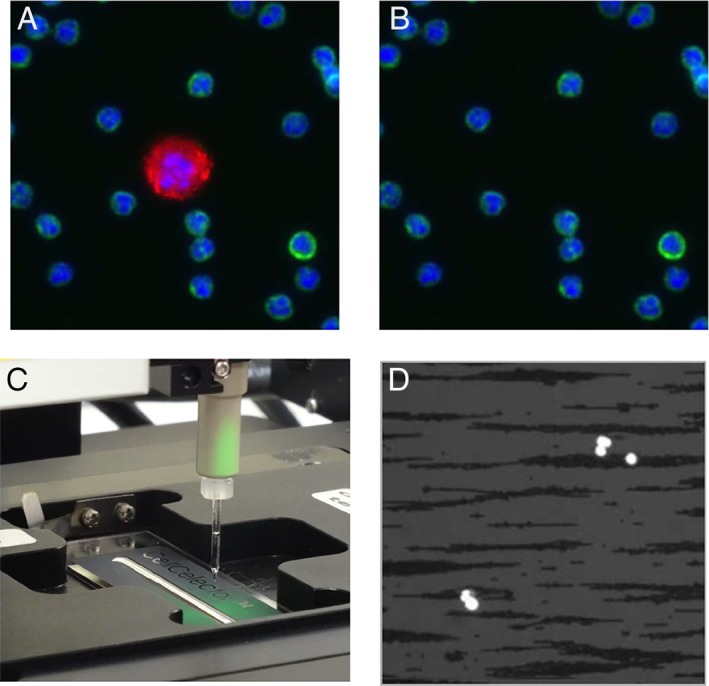
Example of (A) before and (B) after picking images acquired at 20x showing CTC recovery from enriched blood sample in the presence of contaminating WBCs. Staining: Red: CK antibody, Green: CD45; Blue: nuclei; (C) Picking of immuno‐magnetically enriched CTCs from a sample deposited onto a specifically developed ALS MagnetPick™ slide installed onto a corresponding slide adapter. The magnetic field created by the adapter prevents cells from moving during the experiment thus facilitating automatic scanning and cell picking. (D) Image of CellSearch enriched blood sample acquired at 10x with combined DAPI and BF illumination showing cell nuclei (bright) and ferromagnetic fluid particles (dark).
Thanks to its gentle stage movements and fully adjustable picking protocols and parameters (e.g., aspiration force and volume) the CellCelector can handle picking from suspension samples like typical CTC enriched cell suspensions, adherent cells, CytoSpins, tissue sections (e.g., FFPE or fresh‐frozen), and many more. It is also able to handle both fixed and viable cells. In order for the system to work fully automatically from scanning to picking the cells should be at a monolayer and stable in their position on the source vessel. This can be achieved by a variety of methods including waiting until all cells settle down on the base of the source plate, centrifugation, utilization of specific technologies (MagnetPick™ slides, Nano well plates, etc.) as described below, and various materials or coating methods to increase or decrease adhesion of the cells to the base of the source vessel.
The system is fully agnostic regarding upstream enrichment methods handling large numbers of contaminating cells (e.g., 50,000 per sample) and does not require sample volume reduction. CellCelector™ has been successfully used downstream of various CTC enrichment techniques: (i) positive immune‐magnetic: CellSearch® 2, Isoflux™ 3, 4, Illumina MagSweeper™ 5; (ii) negative immune‐magnetic: Dynal® MPC‐S 6, RosetteSep CD45 7; (iii) density gradient centrifugation 8; or (iv) size‐based separation: Parsortix™ 9, 10.
Single cell resolution can be reached using different approaches. The first one consists in the immobilization of cells using relatively sparse single‐cell distribution on a regular glass or plastic surface of standard plates or dishes. In this case cells are loosely attached to the surface which allows both automated scanning and efficient single cell picking. In the case of positive immunomagnetic enrichment (e.g., CellSearch, Isoflux, etc.) the MagnetPick™ approach developed by ALS can be used where the sample is pipetted onto a glass slide with a delimited cell‐seeding area and installed onto a special MagnetPick slide adapter (Fig. 2C,D). The latter is designed to provide a magnetic field which pulls seeded cells onto the slide effectively preventing them from moving over the glass surface. The device is compatible both with ferromagnetic nanoparticles used in CellSearch 2 or larger microscopic beads used, for example, by Isoflux technology 3.
The third method employs specific nanowell arrays which allows single cell capture and retrieval. In Lohr et al. 5 and Yao et al. 7 studies the researchers used home‐made PDMS‐based nanowell arrays with approximately 85,000 nanowells of 50 × 50 × 50 μm (Fig. 3). Presently industrially manufactured nanowell arrays are available for CellCelector users from ALS. A pre‐enriched sample loaded in such an array is scanned both in BF and fluorescence. The software automatically detects and identifies single cells, corresponding nanowell IDs and quantifies fluorescence. After pre‐defined gating and cell sorting the user can select candidate CTCs for retrieval. The cells remain unattached to the bottom of nanowells and can be recovered with 100% single cell picking efficiency. If necessary the size and the number of nanowells can be adjusted for capture of both single cells and clusters.
Figure 3.
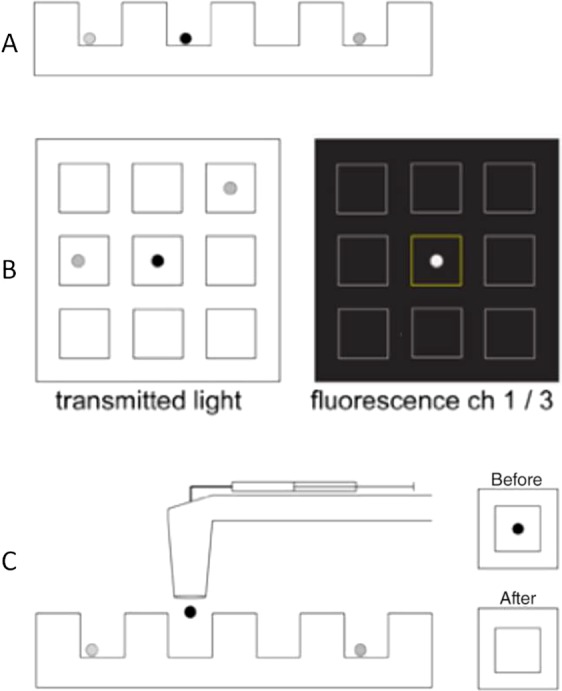
Nanowell‐based isolation of pure, single CTCs. (Workflow adapted from Lohr et al. 5, (A) Following enrichment of patient blood, the enriched sample is loaded stochastically into an array of nanowells; (B) the array is automatically scanned in BF and several fluorescence channels. Software identifies nanowells and segment cells and quantifies fluorescence levels; (C) Robotic arm retrieves single cells from individual wells and deposits into a PCR plate.
The identification and selection of multiple cell types is performed using an automated inverted microscope setup with up to seven fluorescent channels and bright field illumination (BF) as well as 10× to 40× objectives. Options for 1.25× to 100× ultra‐long‐working distance objectives are available (water or oil‐immersion objectives are not recommended). Fluorescence is created using an external, solid‐state based light source utilizing customizable filter sets based on the user's application demands. As such the CellCelector™ is compatible with most available fluorophores ranging from UV to nIR (e.g., DAPI, FITC, TRITC, Cy5, Cy7, or similar). Filters can be added at any stage should demands change. The user can review and validate automatically detected target cells one‐by‐one. A two‐step automated imaging workflow using a 10× objective for scanning and 40× for validation and documentation of detected putative CTCs provides a combination of fast scanning and high‐quality images.
The CTC recovery process is fast and very gentle for cells. The cell spends less than 10 s in the capillary and the whole single cell picking event takes around 20 s. As such a typical sample of, for example, 10 CTCs within a background of 50,000 contaminating cells can be picked in less than 5 min (utilizing single cell deposit). Pooling all 10 cells into one well can be achieved in less than 2.5 min. This includes full documentation of the process with before and after picking images as well as data tracing the cell from source to destination vessel. Aspiration without shear stress (which can be present in certain cell vacuum‐based recovery systems) allows live cell isolation. Contrary to laser microdissection systems which can damage isolated cells by excessive heat the CellCelector allows damage‐free cell recovery and thus re‐cultivation of cells, single cell RNA analysis or other demanding molecular characterization.
Variable capillary sizes allow for isolation of not only individual single cells but also multicellular CTC clusters 10, spheroids/organoids, or even working with tissue sections (FFPE/fresh frozen). Utilizing multiple picking steps (1st step picking of clusters, 2nd step picking of individual cells from the destination well of the 1st step after dissociation of the cluster) allows to study the heterogeneity of clusters on single cell level which would be otherwise impossible.
The vessels used for cell sorting and recovery by CellCelector are usually standard laboratory consumables and as such widely available, affordable and open (e.g., well plates, petri dishes, microscope slides, or CellCelector Nanowell arrays) and do not show any intrinsic cell loss like cartridge‐based dielectophoretic or flow‐sorting devices usually do because of their important dead volume. This makes the latter hardly compatible with low CTC number samples. For CellCelector practically all enriched and seeded cells can be found and retrieved. Thus Neumann et al. 2 reported that 95% of cells detected by the CellSearch imaging unit had been successfully re‐identified and isolated with the CellCelector. The authors explained the slight discrepancy by a minor cell loss during the transfer of the CellSearch cartridge content onto the ALS MagnetPick glass slide. Due to adjustable CellCelector picking parameters (e.g. pipetting volumes, pressures, waiting times etc.) the user can optimize the protocol to reach optimal picking efficiency for various cell types as shown by Choi et al. 11 reporting comparable picking rates as above. As such the CellCelector can be adjusted for samples of all kind of origin, upstream technology, and fixation.
Conclusion
The CellCelector technology represents a very open, flexible and automated research platform for scientists studying rare single cells.
As such it has been used successfully for various downstream characterization of single cell CTCs: e.g., mutation detection using targeted sequencing of cell pools and nested PCR of single cells 7, whole genome sequencing 8, whole exome sequencing and RNA sequencing of single CTCs 5, or exome sequencing of CTC clusters 10.
With the CellCelector it was possible to isolate pure single tumor cells from liquid biopsies of different cancer types including breast 2, 8, 9, 12, prostate 3, 5, ovarian 6, colorectal 13, and lung 7, 10 cancers.
Literature Cited
- 1. Joosse SA, Gorges TM, Pantel K. Biology, detection, and clinical implications of circulating tumor cells. EMBO Mol Med 2015;7:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Neumann M, Schneck H, Decker Y, Schoemer S, Franken A, Endris V, Pfarr N, Weichert W, Niederacher D, Fehm T, et al. Isolation and characterization of circulating tumor cells using a novel workflow combining CellSearch® and CellCelector™. Biotechnol Prog 2016; 33:125–132. [DOI] [PubMed] [Google Scholar]
- 3. Ma Y, Luk A, Young F, Lynch D, Chua W, Balakrishnar B, de Souza P, Becker T. Droplet digital pcr based androgen receptor variant 7 (AR‐V7) detection from prostate cancer blood biopsies. Int J Mol Sci 2016;17:E1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Agerbæk MØ, Bang‐Christensen SR, Yang MH, Clausen TM, Pereira MA, Sharma S, Ditlev SB, Nielsen MA, Choudhary S, Gustavsson T, Sorensen PH, Meyer T, Propper D, Shamash J, Theander TG, Aicher A, Daugaard M, Heeschen C, Salanti A. The VAR2CSA malaria protein efficiently retrieves circulating tumor cells in an EpCAM‐independent manner. Nat Commun 2018; 9:3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lohr J, Adalsteinsson V, Cibulskis K, Choudhury A, Rosenberg M, Cruz‐Gordillo P, Francis J, Zhang C, Shalek A, Satija R et al. Whole‐exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol 2014; 32:479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blassl C, Kuhlmann J, Weber A, Wimberger P, Fehm T, Neubauer H. Gene expression profiling of single circulating tumor cells in ovarian cancer ‐ Establishment of a multi‐marker gene panel. Mol Oncol 2016;10:1030–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yao X, Williamson C, Adalsteinsson V, D'Agostino R, Fitton T, Smaroff G, William R, Wittrup D, Love C. Tumor cells are dislodged into the pulmonary vein during lobectomy. J Thoracic Cardiovasc Surg 2014;148:3224–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Heidary M, Auer M, Ulz P, Heitzer E, Petru E, Gasch C, Riethdorf S, Mauermann O, Lafer I, Pristauz G, et al. The dynamic range of circulating tumor DNA in metastatic breast cancer. Breast Cancer Res 2014; 16:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lampignano R, Yang L, Neumann M, Franken A, Fehm T, Niederacher D, Neubauer H. A novel workflow to enrich and isolate patient‐matched EpCAMhigh and EpCAMlow/negative CTCs enables the comparative characterization of the PIK3CA status in metastatic breast cancer. Int J Mol Sci 2017;18:1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Krol I, Castro‐Giner F, Maurer M, Gkountela S, Szczerba BM, Scherrer R, Coleman N, Carreira S, Bachmann F, Anderson S, Engelhardt M, Lane H, Evans TRJ, Plummer R, Kristeleit R, Lopez J, Aceto N. Detection of circulating tumour cell clusters in human glioblastoma. Br J Cancer 2018; Aug 1 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Choi J, Ogunniyi A, Du M, Du M, Kretschmann M, Eberhardt J, Love C. Development and optimization of a process for automated recovery of single cells identified by microengraving. Biotechnol Prog 2010;26:888–895. [DOI] [PubMed] [Google Scholar]
- 12. Schneck H, Gierke B, Uppenkamp F, Behrens B, Niederacher D, Stoecklein N, Templin M, Pawlak M, Fehm T, Neubauer H. EpCAM‐independent enrichment of circulating tumor cells in metastatic breast cancer. PLoS One 2015;10:e0144535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Adalsteinsson V, Tahirova N, Tallapragada N, Yao X, Campion L, Angelini A, Douce T, Huang C, Bowman B, Williamson C, et al. Single cells from human primary colorectal tumors exhibit polyfunctional heterogeneity in secretions of ELR+ CXC chemokines. Integr Biol (Camb) 2013; 5:1272–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]


