Abstract
Cancer cells from solid tumors can enter the circulatory system and survive to subsequently form distant metastases. The CellSearch® system (Menarini‐Silicon Biosystems, Huntingdon Valley, PA) was the first, FDA‐cleared system that provided a reliable tool for the investigation of circulating tumor cells (CTCs), which have been shown to be strongly associated with poor survival and therapy failure. Since that time, a number of new technologies have been introduced to improve CTC detection and/or isolation for further characterization. The continued and growing interest in the “liquid biopsy” field has spurred the development of numerous different CTC technologies. However, selecting the most appropriate CTC platform for individual applications can be challenging. No consensus has yet been reached in the community regarding which liquid biopsy technology is optimal. Here, we introduce the Parsortix™ Cell Separation System (ANGLE North America, Inc., King of Prussia, PA), a microfluidic based technology that captures rare cells based on size and deformability, offers reproducibly high capture efficiency, and produces highly enriched, viable (viability dependent on preservative used) CTCs that are amenable to a multitude of downstream analyses, including the isolation and interrogation of single cells. © 2018 The Authors. Cytometry Part A published by Wiley Periodicals, Inc. on behalf of International Society for Advancement of Cytometry.
Keywords: liquid biopsy, circulating tumor cell, microfluidics, rare cell isolation
Background and Introduction
Development of new tools and technologies is critical to advancing the application of liquid biopsy and circulating tumor cell (CTC) isolation/interrogation to precision/personalized medicine. ANGLE has developed the Parsortix™ Cell Separation System (Figure 1), a semiautomated system capable of capturing and harvesting rare cells (e.g., CTCs) from bodily fluids (e.g., blood, bone marrow, and ascites) based on cell size and deformability (or compressibility) 1, 2. The Parsortix Cell Separation System does not employ antibodies or other cell surface affinity agents to capture the target cells. Its isolation/capture principle is based on physical attributes of cells rather than chemical or biological characteristics, making it an epitope independent process and consequently agnostic to cellular genotype or immunophenotype, enabling the system to capture a variety of rare cell types, including epithelial and mesenchymal cancer cell immunophenotypes.
Figure 1.
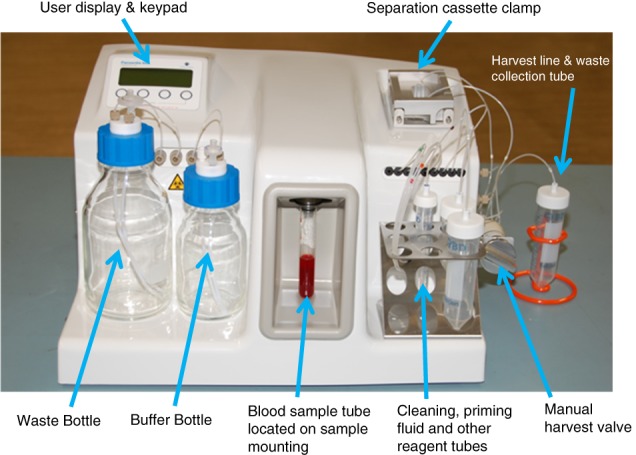
Parsortix™ PR1 Cell Separation System.
Materials and Methods
The Parsortix™ Cell Separation System
The computer controlled programmable fluidics and pneumatics of the Parsortix System enable precise control over the movement of fluids and air through a number of internal pathways, including through a single use separation cassette when mounted in the reusable cassette clamp assembly (Figure 1). The sample containing rare cells (e.g., whole blood) is routed through the separation cassette under controlled and constant pressure conditions (99 mbar) to enable separation. Using controlled pressure results in the force exerted to pass the sample remaining constant, even though the flow rate throughout the separation may vary based on the sample viscosity. Buffer, priming fluids, and cleaning fluids are drawn from external reservoirs and routed through the internal fluidic components, including, where appropriate, the separation cassette. An external manual harvest valve enables captured cells to be eluted from the cassette into an external vessel for further, user defined downstream analysis. The fluidics setup enables:
Priming of the system before use to remove air from internal components and the separation cassette;
Rinsing (with buffer) of the flow path to ensure the entire blood sample has gone through the cassette to complete a separation with minimal sample waste; and
Thorough cleaning of the system after use in preparation for the next operational cycle.
The Parsortix System is designed for use with the Parsortix GEN3 Cell Separation Cassette for the capture of rare cells. The Parsortix cassette is based on patented microfluidic particle separation technology and relies on precision plastics fabrication and inspection techniques to achieve very strict and repeatable component tolerances. The single‐use separation cassettes contain a continuous precision molded separation structure laid out in a “step” configuration yielding a final “critical gap” where the cells are captured (Figure 2A). As a result, cells of interest are preferentially captured based on their size and resistance to compression. The progressive decrease in the height of the critical gap and the use of the wider steps at the top of the structure are designed to aid in the capture of larger cells and to provide physical support above and below the captured cells to help prevent severe morphologic changes. The looped cassette design maximizes the length of the separating steps, a key factor affecting separation capability/capacity, while providing fluid paths with minimal resistance to flow. Furthermore, the stepped cassette configuration is intentionally uni‐directional, such that during a separation, the sample always flows across the step structures and through the critical gap. Parsortix cassettes are the size of a standard microscope slide and are available with different critical gap sizes, ranging from 10 μm down to 4.5 μm. The theoretical maximum capture capacity of a Parsortix cassette with a critical gap size of 6.5 μm, assuming a uniform cell size of 8 μm, would be 62,500 total cells.
Figure 2.
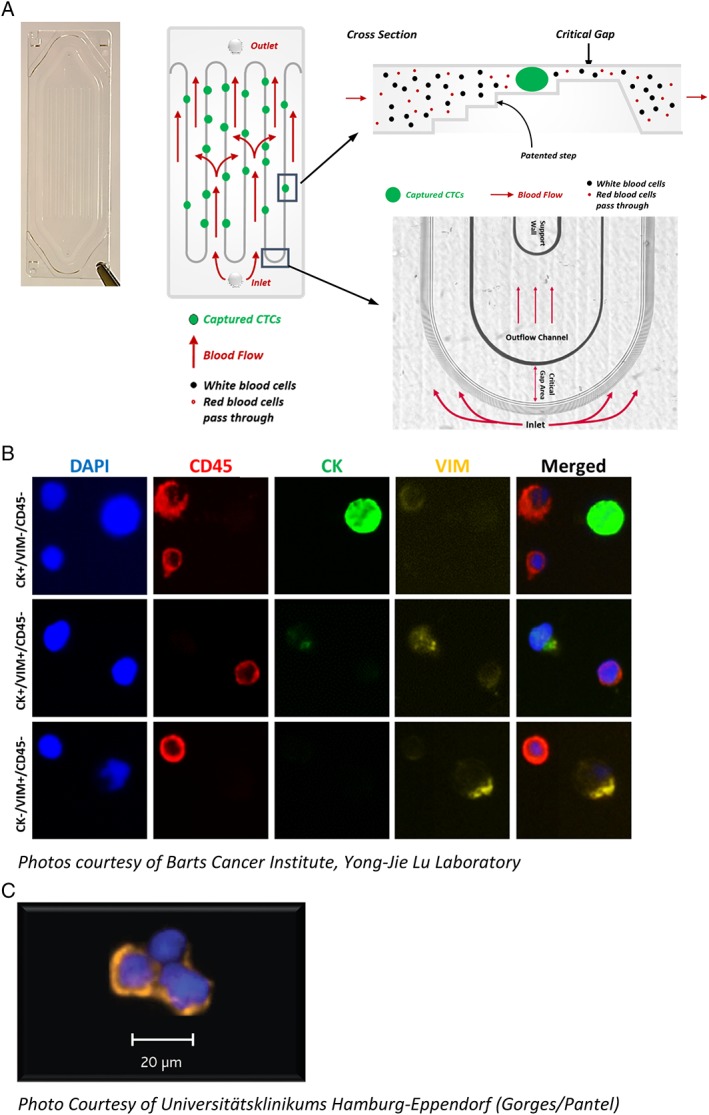
(A) Image and diagram of a Parsortix GEN3 Cell Separation Cassette showing how the blood flows into the cassette, over the step structures and through the critical gap. (B) Example images of nucleated blood cells and CTCs harvested from prostate cancer patients (7.5 ml EDTA blood samples processed within 4 h after collection) and immunofluorescently stained with DAPI (blue), CD45 (red), cytokeratin (green), and vimentin (yellow). (C) Example image of CTCs harvested from breast cancer patient (4 ml CellSave blood sample processed <24 h after collection) and immunofluorescently stained with cytokeratin (orange) and DAPI (blue).
The Parsortix System is configured to allow for the staining of cells captured inside the cassettes and/or to subsequently harvest the captured cells from the cassette into a small volume of buffer for subsequent evaluation and downstream interrogation. To harvest cells captured in the cassette, the liquid flow through the system is intentionally reversed to release the cells from the cassette's step and critical gap structures and direct them into an external collection vessel. The typical rate of separation for whole blood on the system is approximately 5 ml/h. The Parsortix System allows for the harvest of the captured cells from the separation cassette into a small volume (100–210 μl) of buffer, such as PBS, enabling a high level of versatility in how the cells could be evaluated and/or clinically interrogated 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12.
Evaluation of Parsortix System Performance in Whole Blood
Using Parsortix GEN3D6.5 Cell Separation Cassettes, which have a critical gap size of 6.5 μm, an experiment evaluating live, fluorescently labeled (CellTracker™ Green, Thermo Fisher Scientific, Waltham, MA) SKBR3 cells spiked at levels of 0, ~10, ~20, ~50, and ~100 cells into 5 ml of EDTA preserved blood drawn from healthy volunteers was performed. All healthy volunteers gave informed consent prior to the collection of the de‐identified blood samples under IRB‐approved sample collection protocols. The exact numbers of cells spiked into each blood sample was determined using limited dilution followed by direct counting of cells in the spiking volume prior to the addition of the blood. A total of five samples for each spike level were evaluated over a 5‐day period. The blood samples were spiked and processed ~18–36 h after collection, and the number of pre‐labeled cells captured inside each cassette was determined. The captured cells were then harvested from the cassette into a microtiter plate and the number of cells recovered determined. Separate experiments were performed by another laboratory evaluating live, fluorescently labeled SKBR3, Hs578T, MCF7, and BT549 cells spiked at dilution levels of approximately 1,000, 330, 100, 30, 10, and 3 cells into 5 ml of healthy volunteer blood, with a total of six samples for each spike level evaluated over a 6‐day period.
Results
Cell Line Spiking into Whole Blood
Results from the 5‐day SKBR3 spiking experiment showed high rates of capture, harvest, and recovery (Figures 3A–C) (data on file). Results from the 6‐day cell line spiking experiments showed that the Parsortix System captured ~92% (95% CI = 89–95%) of the spiked SKBR3 cells, ~87% (95% CI = 84–90%) of the spiked Hs578T cells, ~78% (95% CI = 68–87%) of the spiked MCF7 cells, and ~66% (95% CI = 64–69%) of the spiked BT549 cells (Supporting Information Figures S1A, S2A, S3A, and S4A) and harvested ~75% (95% CI = 73–77%), ~84% (95% CI = 81–86%), ~62% (95% CI = 56–67%), and ~73% (95% CI = 69–77%) of the captured cells from the cassettes, respectively (Supporting Information Figures S1B, S2B, S3B, and S4B) (data on file). Collectively, these and other data in model CTC systems (i.e., cultured cells spiked into whole blood), demonstrate reproducibly high capture and high harvest rates for viable CTCs, essential elements to the successful application of CTCs in the clinical setting. However, because actual CTCs in patient samples have been shown to be smaller than most cultured cells 13, the recovery of CTCs from actual patient blood using the Parsortix System needs to be validated on a disease by disease basis.
Figure 3.
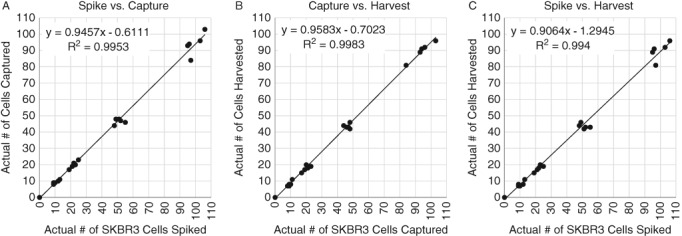
5‐Day linearity study on Parsortix System using live, fluorescently labeled SKBR3 cells spiked into 5 ml EDTA blood drawn from healthy volunteers. (A) Plot of the actual number of SKBR3 cells spiked vs. the number of SKBR3 cells captured in the separation cassette. (B) Plot of the number of SKBR3 cells captured in the separation cassette vs. the number of SKBR3 cells harvested out of the separation cassette. (C) Plot of the actual number of SKBR3 cells spiked vs. the number of SKBR3 cells harvested out of the separation cassette.
While the harvested cells from blood samples using the Parsortix System are not pure CTCs, the system affords an up to 105‐fold depletion of nucleated blood cells in a single, user‐friendly step, and thus a significant enrichment of CTCs (for EDTA blood processed within 8 h after collection using a separation cassette with a 6.5 μm critical gap size, on average, there are 200–800 residual nucleated blood cells per mL of blood which are also captured and harvested by the system). With an average capture/harvest rate of ~81 and 74%, respectively, across multiple cell lines, the system is well suited to enrich CTCs from clinical samples where the physical properties of the target rare cells (i.e., size and deformability) are different from the bulk of other cells (e.g., RBCs and WBCs) present in the samples.
Subsequent Evaluation Techniques used on Parsortix System Harvested CTCs
The isolation and harvest of live, viable CTCs (depending on the blood collection tube used) by the Parsortix System allow postharvest analyses and procedures that are not feasible with most other liquid biopsy systems. This includes such things as the ability to culture CTCs for the establishment of cell lines, the screening of therapeutic candidates to potentially tailor a therapeutic regimen to individual patients, or the screening of drug development candidates for therapeutic potential and progression through the drug development pipeline. Cell viability ensures integrity of nucleic acids, allowing molecular interrogation of the harvests with both increased sensitivity of detection (concentration effect) and decreased background (enrichment effect). The Parsortix System has been used to successfully capture and/or harvest CTCs from a number of different cancer types, with the harvested CTCs successfully evaluated by a variety of subsequent analysis techniques, including:
cytological/histological staining and/or immunostaining for identification, enumeration and detection of protein biomarkers on captured and/or harvested CTCs (Figure 2B,C) 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12;
individual gene expression (mRNA) analysis (e.g., ddPCR and qPCR) 4, 6, 7, 8, 10, 12;
multiplex nucleic acid 4, 10, 12 or protein evaluation (e.g., via Ziplex® System Flow‐Thru Chip® technology [ANGLE Biosciences, Toronto, CA]);
evaluation of DNA aberrations (e.g., aCGH and HER2 FISH) 2, 5;
whole transcriptome or genome sequencing (e.g., RNA‐seq and DNA‐seq) 6;
mouse xenograph models 11.
Additionally, the Parsortix System has a specially designed adaptor available that allows it to process samples with volumes as small as 100 μl. The Parsortix Low Volume Adaptor is ideal for blood samples from small rodent studies such as in mice, which are commonly used as oncology tumor models 11. Preliminary data have demonstrated that CTCs can be isolated from whole blood collected via tail and/or orbital vein draws from mice in xenograft models of human breast cancer (data on file).
Parsortix System Performance in the Clinical Setting
Traditionally, the isolation or analysis of CTCs has been performed/restricted to the use of blood, either whole blood or fractionated blood, as the biological fluid. However, it is well known that tumor cells, while not classically considered circulating per se, are present in other biological fluids, such as pleural effusions and ascites, as well as in “liquid tissues” such as bone marrow aspirates.
Several recent publications highlighted the ability of the Parsortix System to isolate CTCs with diverse immunophenotypes and genotypes, essential for a more comprehensive picture of disease status and etiology. Maertens et al. 7, using clear cell renal cell carcinoma (ccRCC) cell lines, compared cell capture using EpCAM‐based selection, gradient centrifugation‐based separation, and the Parsortix System. They demonstrated that not only did the Parsortix System capture more cells, it was also able to capture EpCAM negative cells, especially relevant to the known reduced EpCAM expression in ccRCC. Similarly, El‐Heliebe et al. 9 showed the ability of the Parsortix System to isolate CTCs from castration resistant prostate cancer (CRPC) patients that exhibited the spectrum of immmunophenotypes from epithelial to mesenchymal. Furthermore, the isolated CTCs were interrogatable for prognostic information through the evaluation of KRAS mutations and for therapeutic selection via assessment of ARV‐7 expression. A similar spectrum of CTC immunophenotypes was described by Xu et al. 5 in CTCs isolated from CRPC patients using the Parsortix System. These authors also identified the presence of circulating megakaryocytes among the harvested cells and showed a relationship between their presence and a positive prognosis in prostate cancer.
The Parsortix System is currently under evaluation in a multicenter clinical study (NCT 03427450) to evaluate the capture of CTCs from metastatic breast cancer patients and the use of multiple downstream analysis techniques (e.g., cytopathology, qPCR, FISH, and RNAseq) on the harvested CTCs. Formal comparisons of the Parsortix System to other existing CTC capture systems need to be conducted.
Discussion
As mentioned earlier, cells captured by the Parsortix System can be interrogated by cytological/histological staining techniques such as hematoxylin and eosin (H&E) or Wright‐Giemsa staining, as well as by immunochemical techniques, including immunofluorescence 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12. Importantly, the captured rare cells do not have to be harvested from the separation cassette in order to be evaluated by such techniques. Staining of the captured cells can be performed in the separation cassette and, given its microscope slide dimensions, visualized directly by bright‐field or fluorescence techniques. Alternatively, Parsortix System harvests can be collected directly into CytoSpin or StatSpin collectors and deposited onto slides for cytological evaluations. Direct visualization of harvested cells, particularly combined with a universal stain such as Wright‐Giemsa, allows for the identification of CTC clusters and other phenotypic features. Fluorescence in situ hybridization (FISH) techniques, such as HER‐2/Neu gene amplification 2, 5 and multiplex gene expression analyses 4, 6, 10, 12, have also been successfully applied to Parsortix System harvests. Combined, these characteristics of the Parsortix System allow the end‐user flexibility in the interrogation of captured rare cells and can help to maximize lab‐specific workflow efficiencies. For each downstream evaluation technique, the cell loss associated with each method should be carefully evaluated and characterized.
The Parsortix Cell Separation System is able to capture and harvest rare cells from biological samples of clinical relevance varying in both composition and volume obtained from subjects with numerous different cancer types. The ability to harvest live cells allows not only culturing of such cells, but ensures integrity of both nucleic acids and proteins, allowing a variety of downstream interrogation techniques to be used on the isolated rare cells, including CTCs. The published and other ongoing studies constitute a growing body of evidence in support of the clinical value in the isolation and characterization of rare cells, including CTCs, through epitope‐independent techniques such as that afforded by the Parsortix Cell Separation System.
Conflicts of Interest
M. Craig Miller, Peggy S. Robinson, Christopher Wagner, and Daniel J. O'Shannessy are paid employees of ANGLE North America, Inc. M. Craig Miller, Peggy S. Robinson, and Christopher Wagner are stock holders in ANGLE plc. Peggy S. Robinson is a corporate officer of ANGLE North America, Inc.
Supporting information
Figure S1. 6‐Day Linearity Study on Parsortix System using Live, Fluorescently Labeled SKBR3 Cells Spiked into 5mL EDTA Blood Drawn from Healthy Volunteers. A) Plot of the actual number of SKBR3 cells spiked vs. the number of SKBR3 cells captured in the separation cassette; B) Plot of the number of SKBR3 cells captured in the separation cassette vs. the number of SKBR3 cells harvested out of the separation cassette; C) Plot of the actual number of SKBR3 cells spiked vs. the number of SKBR3 cells harvested out of the separation cassette.
Figure S2. 6‐Day Linearity Study on Parsortix System using Live, Fluorescently Labeled HS578T Cells Spiked into 5mL EDTA Blood Drawn from Healthy Volunteers. A) Plot of the actual number of HS578T cells spiked vs. the number of HS578T cells captured in the separation cassette; B) Plot of the number of HS578T cells captured in the separation cassette vs. the number of HS578T cells harvested out of the separation cassette; C) Plot of the actual number of HS578T cells spiked vs. the number of HS578T cells harvested out of the separation cassette.
Figure S3. 6‐Day Linearity Study on Parsortix System using Live, Fluorescently Labeled MCF7 Cells Spiked into 5mL EDTA Blood Drawn from Healthy Volunteers. A) Plot of the actual number of MCF7 cells spiked vs. the number of MCF7 cells captured in the separation cassette; B) Plot of the number of MCF7 cells captured in the separation cassette vs. the number of MCF7 cells harvested out of the separation cassette; C) Plot of the actual number of MCF7 cells spiked vs. the number of MCF7 cells harvested out of the separation cassette.
Figure S4. 6‐Day Linearity Study on Parsortix System using Live, Fluorescently Labeled BT549 Cells Spiked into 5mL EDTA Blood Drawn from Healthy Volunteers. A) Plot of the actual number of BT549 cells spiked vs. the number of BT549 cells captured in the separation cassette; B) Plot of the number of BT549 cells captured in the separation cassette vs. the number of BT549 cells harvested out of the separation cassette; C) Plot of the actual number of BT549 cells spiked vs. the number of BT549 cells harvested out of the separation cassette.
Supplemental Figure 1. 6‐Day Linearity Study on Parsortix System using Live, Fluorescently Labeled SKBR3 Cells Spiked into 5mL EDTA Blood Drawn from Healthy Volunteers.
Supplemental Figure 2. 6‐Day Linearity Study on Parsortix System using Live, Fluorescently Labeled Hs578T Cells Spiked into 5mL EDTA Blood Drawn from Healthy Volunteers.
Supplemental Figure 3. 6‐Day Linearity Study on Parsortix System using Live, Fluorescently Labeled MCF7 Cells Spiked into 5mL EDTA Blood Drawn from Healthy Volunteers.
Supplemental Figure 4. 6‐Day Linearity Study on Parsortix System using Live, Fluorescently Labeled BT549 Cells Spiked into 5mL EDTA Blood Drawn from Healthy Volunteers.
Acknowledgment
The authors would like to thank Millie Maddocks, Lara Stevanato, Edwina Burke, and Amy Templeman from ANGLE Europe Limited for their efforts on the generation of the “Data on File” for the 6‐day cell line spiking experiments.
Literature Cited
- 1. Xu L, Mao X, Imrali A, Syed F, Mutsvangwa K, Berney D, Cathcart P, Hines J, Shamash J, Lu YJ. Optimization and evaluation of a novel size based circulating tumor cell isolation system. PLoS One 2015;10(9):e0138032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hvichia GE, Parveen Z, Wagner C, Janning M, Quidde J, Stein A, Müller V, Loges S, Neves RP, Stoecklein NH, et al. A novel microfluidic platform for size and deformability based separation and the subsequent molecular characterization of viable circulating tumor cells. Int J Cancer 2016;138(12):2894–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chudziak J, Burt DJ, Mohan S, Rothwell DG, Mesquita B, Antonello J, Dalby S, Ayub M, Priest L, Carter L, et al. Clinical evaluation of a novel microfluidic device for epitope‐independent enrichment of circulating tumour cells in patients with small cell lung cancer. Analyst 2016;141(2):669–678. [DOI] [PubMed] [Google Scholar]
- 4. Gorges TM, Kuske A, Röck K, Mauermann O, Müller V, Peine S, Verpoort K, Novosadova V, Kubista M, Riethdorf S, et al. Accession of tumor heterogeneity by multiplex transcriptome profiling of single circulating tumor cells. Clin Chem 2016;62(11):1504–1515. [DOI] [PubMed] [Google Scholar]
- 5. Xu L, Mao X, Guo T, Chan PY, Shaw G, Hines J, Stankiewicz E, Wang Y, Oliver RTD, Ahmad AS, et al. The novel association of circulating tumor cells and circulating megakaryocytes with prostate cancer prognosis. Clin Cancer Res 2017;23(17):5112–5122. [DOI] [PubMed] [Google Scholar]
- 6. Lampignano R, Yang L, Neumann MHD, Franken A, Fehm T, Niederacher D, Neubauer H. A novel workflow to enrich and isolate patient‐matched EpCAMhigh and EpCAMlow/negative CTCs enables the comparative characterization of the PIK3CA status in metastatic breast cancer. Int J Mol Sci 2017;18(1885):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maertens Y, Humberg V, Erlmeier F, Steffens S, Steinestel J, Bögemann M, Schrader AJ, Bernemann C. Comparison of isolation platforms for detection of circulating renal cell carcinoma cells. Oncotarget 2017;8(50):87710–87717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Obermayr E, Maritschnegg E, Agreiter C, Pecha N, Speiser P, Helmy‐Bader S, Danzinger S, Krainer M, Singer C, Zeillinger R. Efficient leukocyte depletion by a novel microfluidic platform enables the molecular detection and characterization of circulating tumor cells. Oncotarget 2017;9(1):812–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mayall FG, Bodger I, Pepperell J, Stevanato L, Hustler A, Mumford KM. The precious cell block. J Clin Pathol 2018;71(7):659–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. El‐Heliebi A, Hille C, Laxman N, Svedlund J, Haudum C, Ercan E, Kroneis T, Chen S, Smolle M, Rossmann C, et al. In situ detection and quantification of AR‐V7, AR‐FL, PSA, and KRAS point mutations in circulating tumor cells. Clin Chem 2018;64(3):536–546. [DOI] [PubMed] [Google Scholar]
- 11. Kitz J, Lowes LE, Goodale D, Allan AL. Circulating tumor cell analysis in preclinical mouse models of metastasis. Diagnostics (Basel) 2018;8(2); pii:E30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Porras TB, Kaur P, Ring A, Schechter N, Lang JE. Challenges in using liquid biopsies for gene expression profiling. Oncotarget 2018;9(6):7036–7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Coumans FAW, van Dalum G, Beck M, Terstappen LWMM. Filter characteristics influencing circulating tumor cell enrichment from whole blood. PLoS One 2013;8(4):e61770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. 6‐Day Linearity Study on Parsortix System using Live, Fluorescently Labeled SKBR3 Cells Spiked into 5mL EDTA Blood Drawn from Healthy Volunteers. A) Plot of the actual number of SKBR3 cells spiked vs. the number of SKBR3 cells captured in the separation cassette; B) Plot of the number of SKBR3 cells captured in the separation cassette vs. the number of SKBR3 cells harvested out of the separation cassette; C) Plot of the actual number of SKBR3 cells spiked vs. the number of SKBR3 cells harvested out of the separation cassette.
Figure S2. 6‐Day Linearity Study on Parsortix System using Live, Fluorescently Labeled HS578T Cells Spiked into 5mL EDTA Blood Drawn from Healthy Volunteers. A) Plot of the actual number of HS578T cells spiked vs. the number of HS578T cells captured in the separation cassette; B) Plot of the number of HS578T cells captured in the separation cassette vs. the number of HS578T cells harvested out of the separation cassette; C) Plot of the actual number of HS578T cells spiked vs. the number of HS578T cells harvested out of the separation cassette.
Figure S3. 6‐Day Linearity Study on Parsortix System using Live, Fluorescently Labeled MCF7 Cells Spiked into 5mL EDTA Blood Drawn from Healthy Volunteers. A) Plot of the actual number of MCF7 cells spiked vs. the number of MCF7 cells captured in the separation cassette; B) Plot of the number of MCF7 cells captured in the separation cassette vs. the number of MCF7 cells harvested out of the separation cassette; C) Plot of the actual number of MCF7 cells spiked vs. the number of MCF7 cells harvested out of the separation cassette.
Figure S4. 6‐Day Linearity Study on Parsortix System using Live, Fluorescently Labeled BT549 Cells Spiked into 5mL EDTA Blood Drawn from Healthy Volunteers. A) Plot of the actual number of BT549 cells spiked vs. the number of BT549 cells captured in the separation cassette; B) Plot of the number of BT549 cells captured in the separation cassette vs. the number of BT549 cells harvested out of the separation cassette; C) Plot of the actual number of BT549 cells spiked vs. the number of BT549 cells harvested out of the separation cassette.
Supplemental Figure 1. 6‐Day Linearity Study on Parsortix System using Live, Fluorescently Labeled SKBR3 Cells Spiked into 5mL EDTA Blood Drawn from Healthy Volunteers.
Supplemental Figure 2. 6‐Day Linearity Study on Parsortix System using Live, Fluorescently Labeled Hs578T Cells Spiked into 5mL EDTA Blood Drawn from Healthy Volunteers.
Supplemental Figure 3. 6‐Day Linearity Study on Parsortix System using Live, Fluorescently Labeled MCF7 Cells Spiked into 5mL EDTA Blood Drawn from Healthy Volunteers.
Supplemental Figure 4. 6‐Day Linearity Study on Parsortix System using Live, Fluorescently Labeled BT549 Cells Spiked into 5mL EDTA Blood Drawn from Healthy Volunteers.


