Abstract
Malate valves act as powerful systems for balancing the ATP/NAD(P)H ratio required in various subcellular compartments in plant cells. As components of malate valves, isoforms of malate dehydrogenases (MDHs) and dicarboxylate translocators catalyse the reversible interconversion of malate and oxaloacetate and their transport. Depending on the co‐enzyme specificity of the MDH isoforms, either NADH or NADPH can be transported indirectly. Arabidopsis thaliana possesses nine genes encoding MDH isoenzymes. Activities of NAD‐dependent MDHs have been detected in mitochondria, peroxisomes, cytosol and plastids. In addition, chloroplasts possess a NADP‐dependent MDH isoform. The NADP‐MDH as part of the ‘light malate valve’ plays an important role as a poising mechanism to adjust the ATP/NADPH ratio in the stroma. Its activity is strictly regulated by post‐translational redox‐modification mediated via the ferredoxin‐thioredoxin system and fine control via the NADP +/NADP(H) ratio, thereby maintaining redox homeostasis under changing conditions. In contrast, the plastid NAD‐MDH (‘dark malate valve’) is constitutively active and its lack leads to failure in early embryo development. While redox regulation of the main cytosolic MDH isoform has been shown, knowledge about regulation of the other two cytosolic MDHs as well as NAD‐MDH isoforms from peroxisomes and mitochondria is still lacking. Knockout mutants lacking the isoforms from chloroplasts, mitochondria and peroxisomes have been characterised, but not much is known about cytosolic NAD‐MDH isoforms and their role in planta. This review updates the current knowledge on MDH isoforms and the shuttle systems for intercompartmental dicarboxylate exchange, focusing on the various metabolic functions of these valves.
Keywords: Energy supply, malate dehydrogenase, malate valve, redox balance, shuttling
Introduction
The redox state provides information on the metabolic status of a plant cell, which is needed for fast adjustment of various metabolic activities in response to continuously changing environmental conditions and developmental requirements (Dietz 2003; Foyer & Noctor 2003; Scheibe et al. 2005). Since major imbalances in the redox state can cause severe damage, plant cells possess different redox sensors such as proteins with reversibly oxidisable thiols (e.g. cytosolic glyceraldehyde‐3‐phosphate dehydrogenase, cyclophilin 20‐3) which detect deviations from redox homeostasis and activate poising mechanisms, thus avoiding the generation of excess reactive oxygen species (ROS; Scheibe & Dietz 2012; Park et al. 2013; Hildebrandt et al. 2015; Noctor & Foyer 2016).
Chloroplasts possess a light‐dependent regulatory mechanism, namely the ferredoxin‐thioredoxin system (Fd‐Trx system), controlling the activities of various key enzymes involved in assimilatory and dissimilatory pathways (Buchanan 1984; Buchanan & Balmer 2005). Reduced Trx subsequently reduces regulatory disulfide bonds of various target enzymes, thereby altering their catalytic activities. Upon illumination, various Calvin‐Benson cycle (CBC) enzymes are activated via the Fd‐Trx system, whereas oxidative pentose phosphate pathway (OPPP) enzymes, such as glucose‐6‐phosphate dehydrogenase, become inactive. The light/dark modulation of various chloroplast proteins via the Fd‐Trx system functions to control synthesis and degradation of carbohydrates to prevent futile cycling and waste of energy, by adjusting enzyme activities at the post‐translational level.
The regulation of energy metabolism is particularly complex in plants. On the one hand, ATP and NAD(P)H are generated in a number of reactions that are localised in different subcellular compartments. During illumination, photosynthesis and respiration generate the energy carrier ATP and the reducing equivalents NADPH and NADH in chloroplasts, cytosol and mitochondria, respectively, in green parts of the plant, whereas glycolysis, respiration and the OPPP produce these energy molecules in the dark and in non‐green tissues in various compartments. In turn, NAD(P)H and ATP are required for all energy‐consuming reactions, such as assimilation of C, N, S and all other cellular activities, both during photoautotrophic and heterotrophic growth. However, the delivery of NAD(P)H and ATP for various energy‐consuming reactions should occur at the required rates and in the specific compartments due to the rather limited pool sizes of these energy sources (Scheibe 2004).
Plant membranes are essentially impermeable to NAD(P)+ and NAD(P)H. Therefore, plant cells possess specific translocators enabling the direct and indirect exchange of reducing equivalents. On the one hand, NAD(P)+ can directly be shuttled between subcellular compartments via NAD+‐carrier proteins localised in mitochondria as well as in plastids (Palmieri et al. 2009). However, these transporters exhibit a low affinity for NADH and NADPH (Palmieri et al. 2009; Maurino & Engqvist 2015). On the other hand, plant cells possess specific translocators for the exchange of dicarboxylates such as malate and oxaloacetate (malate/OAA shuttles; Fig. 1). Malate is a versatile compound in plant metabolism (Lance & Rustin 1984; Maurino & Engqvist 2015) that can easily be transported across subcellular membranes and can be used as a substrate for mitochondrial ATP production or provision of NADH to the cytosol (Fig. 1). Malate/OAA shuttles acting in combination with malate dehydrogenases (MDHs) as key enzymes (also termed malate valves) connect compartments. They are powerful systems for balancing metabolic fluxes by enabling indirect transfer of reducing equivalents, and therefore, they play a key role in plant metabolism.
Figure 1.
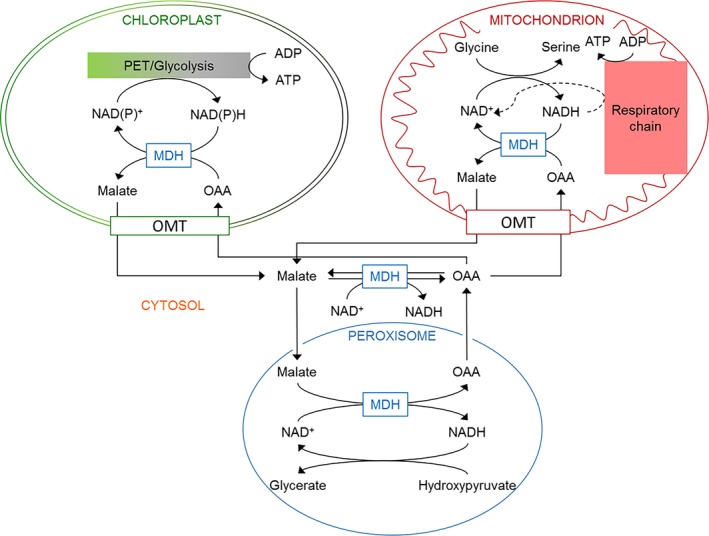
Localisation of malate dehydrogenase isoforms. Malate/OAA translocators together with malate dehydrogenases (malate valves) enable the indirect transfer of reducing equivalents between different subcellular compartments in plant cells. MDH, malate dehydrogenase; OAA, oxaloacetate; PET, photosynthetic electron transport; OMT, malate/OAA translocators.
A multigene family encodes malate dehydrogenases in Arabidopsis
The NAD‐dependent MDH activities have been detected in chloroplasts, mitochondria and peroxisomes, as well as in the cytosol (Gietl 1992; Berkemeyer et al. 1998). Besides the non‐redox regulated NAD‐MDH, chloroplasts of higher plants and green algae additionally contain a redox‐modulated NADP‐dependent MDH (Scheibe & Anderson 1981).
All eukaryotic MDH isoforms can be traced back to two ancestral genes (Ocheretina & Scheibe 1997; Ocheretina et al. 2000). One of the two ancestral genes represents the origin of cytosolic NAD‐dependent MDH isoenzymes (cyNAD‐MDH 1–3) and the NADP‐MDH in chloroplasts (Fig. 2). These isoforms arose through duplication of the ancestral gene. The second subfamily includes NAD‐MDHs of mitochondria (mtNAD‐MDH 1–2), plastids (plNAD‐MDH) and peroxisomes (pNAD‐MDH 1–2), which resulted from triplication of a pre‐existing mitochondrial MDH gene and major changes within their target sequences (Fig. 2).
Figure 2.
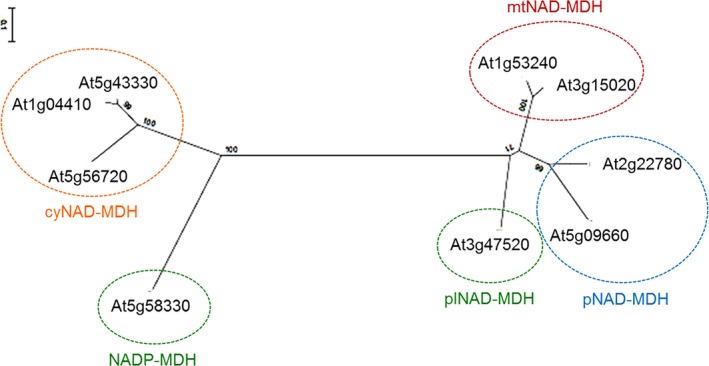
Phylogenetic tree of MDH isoforms from A. thaliana. Amino acid sequences encoding the mature MDH isoproteins were aligned via the Genedoc tool (updated version from 2007; Nicholas et al. 1997). The phylogenetic tree was calculated with the program MEGA 6 (Tamura et al. 2013) using the neighbour‐joining tree method and the statistical approach of bootstrapping. Bootstrap replications were set to 1000. cyNAD‐MDH, cytosolic malate dehydrogenase (cyNAD‐MDH 1, At1g04410; cyNAD‐MDH 2, At5g43330; cyNAD‐MDH 3, At5g56720); pNAD‐MDH, peroxisomal malate dehydrogenase (pNAD‐MDH 1, At2g22780; pNAD‐MDH 2, At5g09660); mtNAD‐MDH, mitochondrial malate dehydrogenase (mtNAD‐MDH 1, At1g53240; mtNAD‐MDH 2, At3g15020); plNAD‐MDH, plastidic malate dehydrogenase (At3g47520); NADP‐MDH, chloroplast malate dehydrogenase (At5g58330).
Within each subfamily the amino acid identity of the mature proteins is above 50%. Between the two subfamilies, amino acid identity lies at about 21–23%. However, MDHs possess similar tertiary and quaternary structures and almost identical catalytic properties (McAlister‐Henn 1988). Each precursor protein of the organellar MDH isoforms possesses a characteristic N‐terminal transit peptide (Gietl & Hock 1982). For instance, the plNAD‐MDH from Arabidopsis was originally identified by its unique transit peptide of 80 amino acids despite the high identity with coding sequences of the cytosolic isoforms (Berkemeyer et al. 1998; Imsande et al. 2001).
The light malate valve in chloroplasts is strictly regulated
The NADP‐MDH is strictly regulated by light through the Fd‐Trx system. The NADP‐MDH is present as a homodimer which possesses eight cysteine residues per subunit. Two cysteine pairs are located in the N‐ and C‐terminal extensions, whereas the other four cysteine residues are present in the core of the protein (Crétin et al. 1990). The redox‐sensitive cysteine pairs in the N‐ and C‐terminal regions are involved in the light‐dependent redox modulation of the NADP‐MDH via the Fd‐Trx system during photosynthesis; upon removal of one or the other sequence extension, the enzyme is constantly active (Fickenscher & Scheibe 1988; Ocheretina et al. 1993). This indicates that removal of the regulatory structures of the conserved globular MDH protein does not affect its activity but rather has evolved to allow for control (Scheibe 1991).
Under oxidising conditions, the N‐terminal extension of the NADP‐MDH is involved in dimerisation linking the subunits by hydrophobic interactions. In addition, the C‐terminal disulfide pulls the C‐terminal extension into the active site, blocking the place for the substrate, hence acting as an internal inhibitor of the protein. Upon reduction by the Fd‐Trx system, the C‐terminal extension of NADP‐MDH is released from the active centre while the reduction of the N‐terminal extension results in a more relaxed structure of the active site. In a two‐step reaction, finally the fully active enzyme is formed (Miginiac‐Maslow et al. 1997, 2000).
In the light, a continuous reduction by the Fd‐Trx system and the simultaneous re‐oxidation of the target enzymes by O2 allows for adjustment of the activation state of the target enzymes also in the light, since this cycling allows for flexible adjustment of the ratio between reduced to oxidised form. These redox cycles represent the basis for the fine control of enzyme activities. Activation and inactivation rates of the target enzymes are influenced by specific effectors, mainly substrates and products of the respective target enzyme, acting as a positive or negative effector (Heineke & Scheibe 2009). For instance, the activation of NADP‐MDH is only enabled when NADPH increases because it is not consumed in the CBC, e.g. due to a lack of CO2 or ATP. Under these conditions, the NADP‐MDH uses the excess NADPH generated via the photosynthetic electron transport chain to convert OAA to malate, regenerating the electron acceptor NADP+. An increased level of NADP+ in turn acts as a negative effector for reductive activation of the NADP‐MDH, in this way turning down its activity. This fine control guarantees that reducing equivalents are only exported to other subcellular compartments via the malate/OAA shuttle when they are in excess, and when they are not required in the chloroplast for assimilatory processes (Scheibe 1987, 1991; Backhausen et al. 2000). Upon a sudden increase in light intensity, NADP‐MDH activity rapidly increases within less than a minute and subsequently oscillates until the new steady‐state is reached (Scheibe & Stitt 1988). Such hysteretic response is characteristic for this type of post‐translational regulation.
Although NADP‐MDH is known to function as a poising mechanism avoiding the generation of ROS under high light, nadp‐mdh knockout mutants are indistinguishable from the wild type under various stress conditions when grown in soil. This is due to alternative systems such as the NADPH‐dependent thioredoxin reductase (NTRC)/2‐Cys‐peroxiredoxin system, as well as proline biosynthesis, photorespiration and nitrate assimilation in nadp‐mdh knock‐out mutants contributing to the maintenance of redox homeostasis when malate valve capacities are lacking (Hebbelmann et al. 2012; Selinski & Scheibe 2014).
Upon sustained high‐light intensity, an increase in the expression level of the nuclear encoded NADP‐MDH points to a signal transfer from the chloroplast to the nucleus (Becker et al. 2006). The nature of such a signal is still not yet clarified, but a transient increase in H2O2 has been put forward as a potential signal since plants lacking NADP‐MDH do not show such an increase (Heyno et al. 2014). Transfer of the signal might occur via oxidative modification of GapC (Hildebrandt et al. 2015). The glycolytic GapC, possibly in its ‘moonlighting’ function, had been found to bind to the NADP‐MDH gene (Hameister et al. 2007).
It is conceivable that in vivo kinetic effects override the thermodynamic equilibrium of the NADP‐MDH reaction. In particular, when fluctuating conditions give rise to immediate deviations from steady state. Then high pressure on the side of influx, e.g. light intensity, shifts the equilibrium towards efflux, namely the reduction of OAA. Literally, the excess electrons are then pushed through the malate valve. Malate is used for further reactions outside the chloroplasts where various options are possible, in the worst case, the dissipation of energy in the alternative oxidase pathway in the mitochondria. The flux of reductant through the valve will continue as long as the pressure from inside the chloroplast exists and keep it open. Such control is achieved by fine‐tuning of NADP‐MDH activation state by the NADP+/NADP(H) ratio in the chloroplast, with NADP+ hindering activation.
Factors that sustain a steady homeostatic flux through the CBC also include the controlled export of reducing equivalents from the chloroplast in the form of malate as required for balancing the ATP/NADPH ratio (Fridlyand et al. 1998; Fridlyand & Scheibe 1999). Due to the rapid responses of redox‐regulated enzymes, such as NADP‐GAPDH, NADP‐MDH and others, to changing metabolite levels, homeostasis is maintained in a stable non‐equilibrium state by thermodynamic buffering (Igamberdiev & Kleczkowski 2011). The system is kept in such a stable and, at the same time, flexible state as long as energy from light comes in and product is taken out. Even reactions close to the thermodynamic equilibrium are able to limit homeostatic fluxes when their total capacities are decreased in transgenic plants (Fridlyand & Scheibe 2000). Using the kinetic data available for the fine‐control of NADP‐MDH, a model has been developed that supports the function of this enzyme to control the indirect export of NADPH (Fridlyand et al. 1998).
The dark malate valve in plastids is constitutively active
In addition to the NADP‐dependent MDH, chloroplasts contain a redox‐independent activity of NAD‐MDH functioning as part of the dark malate valve and playing an important role in dark metabolism. Non‐green plastids also contain such plNAD‐MDH. In these plastids and in chloroplasts during darkness, ATP and NADPH for anabolism are generated independently via plastidial glycolysis (ATP), and plastidial OPPP (NADPH). The dark malate valve is here required to export the NADH that is formed during glycolysis at the substrate phosphorylation step catalysed by PGK and GAPDH. NADH production in this step occurs by either the bispecific CBC enzyme GapA/B in chloroplasts (Baalmann et al. 1995) or GapCp in plastids of non‐green tissues (Backhausen et al. 1998; Muñoz‐Bertomeu et al. 2009, 2010). Such oxidation of triose phosphates coupled with ATP generation requires a continuous supply of NAD+. The regeneration of NAD+ which allows for continued ATP production via glycolysis is possible due to the activity of plNAD‐MDH for indirect export of NADH in the form of malate. In addition, ATP can be imported into plastids via ATP/ADP transporters of the NTT type (Kampfenkel et al. 1995; Neuhaus et al. 1997; Möhlmann et al. 1998; Tjaden et al. 1998; Reiser et al. 2004). However, although these ATP/ADP transporters exhibit high affinities for ATP and ADP, their capacities are low (V max for ATP (NTT1) = 24 nmol·mg protein−1·h−1; V max for ATP (NTT2) = 7 nmol·mg protein−1·h−1; Möhlmann et al. 1998). Therefore, an indirect mechanism for ATP import in plastids is more favourable, e.g. via shuttling of glycolytic intermediates such as glucose 6‐phosphate (G‐6‐P) or phosphoenolpyruvate (PEP). These intermediates can be transported from the cytosol into plastids or vice versa via G‐6‐P/Pi translocators or PEP/Pi translocators (Facchinelli & Weber 2011). Homozygous plnad‐mdh knock‐out mutants do not exist. This is either due to compromised male or female gametophyte development (pollen or egg cell) in haploid stages of the plant or impaired embryo development and seed maturation in diploid stages. A total knock‐out of plNAD‐MDH that is present in 50% of the haploid pollen grains of heterozygous plnad‐mdh/plNAD‐MDH mutants causes impaired pollen tube growth in vitro, which can be overcome by adding substrates of NADH‐dependent glutamine‐2‐oxoglutarate aminotransferase (GOGAT; Selinski et al. 2014). However, pollen tube growth does not appear to be affected in vivo. This indicates that the maternal tissue in the transducing tract of the style can supply the NADH‐GOGAT substrates, in this way enabling pollen tube elongation. Although NADH‐GOGAT was shown to be an alternative pathway maintaining redox homeostasis in heterozygous plnad‐mdh/plNAD‐MDH plants and plnad‐mdh knock‐out pollen when the capacity of the dark malate valve is lacking, NADH‐GOGAT cannot compensate for the homozygous knock‐out of plnad‐mdh during embryogenesis leading to an embryo‐lethal phenotype (Beeler et al. 2014; Selinski et al. 2014). Heterozygous plnad‐mdh/plNAD‐MDH mutants are able to grow, flower and set seed, but their siliques contain green as well as white seeds during the ripening process. Microscopic analysis of green and white seeds revealed that green seeds contain developed embryos at all developmental stages whereas white seeds exclusively contain embryos in the globular stage, independent of the developmental stage of the silique. These white seeds develop into tiny, shrunken and non‐vital seeds that can easily be lost during seed cleaning procedures. Accordingly, the ratio of green to white seeds was determined as 3:1, which correlates with the theoretical ratio of 1:2:1 for wild types (green seeds) to heterozygotes (green seeds) to homozygotes (white seeds) after selfing.
The lack of chlorophyll in homozygous plnad‐mdh knock‐out seeds might be due to competition for glutamate, which is the precursor for chlorophyll biosynthesis as well as the substrate for the chloroplast 2‐oxoglutarate (2‐OG)/malate transporter AtOMT1 together with the dicarboxylate transporter DCT1. OMT1 and DCT1 play important roles in both the malate valve and the 2‐OG/malate shuttle for nitrate assimilation (Taniguchi et al. 2002; Kinoshita et al. 2011). Glutamine and 2‐OG must be provided to the plastid in exchange for glutamate to support the NADH‐GOGAT step (Fig. 4). When plNAD‐MDH is lacking, maintenance of redox homeostasis and ATP production via glycolysis then depend on the substrate glutamate. Although an increased flux through the NADH‐GOGAT pathway maintains redox homeostasis when plNAD‐MDH is lacking in homozygous progeny of the non‐lethal heterozygous mutant line, this increased flux probably leads to impaired chlorophyll biosynthesis, stopping embryo development at the globular stage (Selinski et al. 2014).
Figure 4.
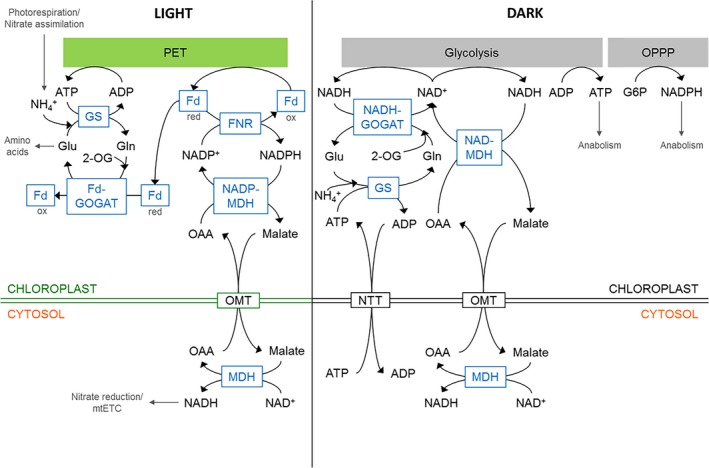
Relationship between plastidial malate valves and NH4 + assimilation. In the light, Fd‐GOGAT represents an alternative electron acceptor for Fd, and therefore, counteracts accumulation of excess NADPH generated by the Fd‐NADPH reductase (FNR). In the dark, NADH‐GOGAT represents an alternative to plNAD‐MDH, since both are able to regenerate NAD+ to maintain continuous ATP production via plastidial glycolysis. G6P, glucose 6‐phosphate; Gln, glutamine; Glu, glutamate; GS, glutamine synthetase; mtETC, mitochondrial electron transport chain; NTT, plastidial ATP/ADP transporter; OMT, malate/OAA translocators; OPPP, oxidative pentose phosphate pathway.
Cytosolic MDHs mediate intercompartmental energy exchange
As described above, excess reducing equivalents from the chloroplast are indirectly transferred as malate to the cytosol and to other subcellular compartments to avoid imbalances or lack of electron acceptors that would lead to oxidative stress (Scheibe 2004). The interconversion of malate to OAA with concomitant reduction of NAD+ to NADH can provide reducing equivalents for nitrate reduction in the cytosol or ATP from mitochondrial respiration via oxidative phosphorylation. If neither NADH nor ATP are required, energy dissipation via the alternative respiratory pathway consisting of alternative NAD(P)H dehydrogenases and the alternative oxidase (AOX) can serve to protect the cell from oxidative stress (Fig. 4). Hence, cyNAD‐MDH functions as a key player in the transfer of reducing equivalents from the chloroplast to other sinks in plant cells, and even allows for conversion of reducing equivalents into ATP or, in the worst case, energy dissipation as heat via AOX.
In Arabidopsis, three NAD‐MDH isoforms are localised in the cytosol. So far, only the main isoform (cyNAD‐MDH 1) has been well characterised and described. Using the recombinant enzyme, cyNAD‐MDH 1 is active under reducing conditions, while oxidising conditions lead to its inactivation (Hara et al. 2006). Apparently, this isoform is sensitive to H2O2 through sulphur oxidation of cysteines and methionines affecting its kinetics, secondary structure and thermodynamic stability. Furthermore, the reversible homodimerisation and reduction of cyNAD‐MDH 1 protects the protein from over‐oxidation (Huang et al. 2018).
Peroxisomal MDHs are involved in photorespiration and in fatty acid β‐oxidation
Leaf peroxisomes and glyoxysomes in lipid‐storing tissues are the two main types of microbodies in plant cells. Peroxisomes rely on reducing equivalents imported from mitochondria or chloroplasts by the malate‐OAA shuttles, since they do not possess any metabolic pathways capable of delivering NADH at the high rates required e.g. for photorespiration. Reducing equivalents are imported into peroxisomes through the uptake of malate and its conversion to OAA by pNAD‐MDH. In glyoxysomes, the pNAD‐MDH is essential for fatty acid β‐oxidation in germinating A. thaliana seeds. In leaf peroxisomes, it is needed to maintain optimal rates of photorespiration by providing NADH required for the reduction of hydroxypyruvate to glycerate by peroxisomal hydroxypyruvate reductase (HPR; Cousins et al. 2008; Pracharoenwattana et al. 2010; Timm et al. 2011). Glycerate can be transported back to the chloroplast where it is phosphorylated to yield 3‐phosphoglycerate, which can re‐enter the CBC (Ogren 1984; Leegood et al. 1995; Boldt et al. 2005; Bartsch et al. 2010; Walker et al. 2016).
Mitochondrial MDHs play an important role in photorespiration and the provision of NADH for respiration
Photorespiration results in NADH formation in plant mitochondria due to the oxidation of glycine to serine by the glycine decarboxylase complex (GDC). NADH can enter the respiratory electron transport chain, supporting ATP synthesis. Coupling of malate oxidation by mtNAD‐MDH with GDC and the pyruvate dehydrogenase complex (PDC) reaction, respectively, in mitochondria helps to maintain high fluxes, apparently by recycling NAD+ required as electron acceptors for decarboxylating glycine and pyruvate oxidation in both complexes (Lindén et al. 2016). It has been discussed that mtNAD‐MDH could play an important role in decreasing the concentration of NADH and relieving its inhibitory effect on PDC (Igamberdiev et al. 2014), or mtNAD‐MDH could recycle the NADH from glycine oxidation to keep NADH concentrations low and thus prevent substrate inhibition of GDC (Bykova et al. 2014).
Furthermore, mtNAD‐MDH was found to lower leaf respiration, probably based on cross‐talk between mtNAD‐MDH and the cytochrome bc 1 complex (Wang et al. 2010). It also appears to alter the rate of photorespiration as well as plant development, demonstrating its important role in energy metabolism and redox homeostasis (Tomaz et al. 2010; Wang et al. 2010; Lindén et al. 2016). Proteomic analyses revealed that mtNAD‐MDH 1 significantly accumulates during Arabidopsis seed germination (Fu et al. 2005), and transcript abundance of both mtNAD‐MDH isoforms significantly increases in early stages of seed germination (Weitbrecht et al. 2011). In accordance with analyses of plNAD‐MDH mutants, mtNAD‐MDH was shown to play an important role in the earliest phases of the Arabidopsis life cycle (Beeler et al. 2014; Selinski et al. 2014; Sew et al. 2016). Recently, mtNAD‐MDH 1 was shown to be insensitive to thiol‐based redox control (in vitro and in vivo) but its activity is lowered by adenine nucleotides, especially ATP (Yoshida & Hisabori 2016). Mitochondrial NAD‐MDH was demonstrated to play a significant role in buffering the redox state in the mitochondrial matrix due to its equilibrium properties (Hagedorn et al. 2004). For any calculation of in vivo concentrations of NADH, NAD+ and metabolite concentrations, it is essential to take into account the fact of ‘molecular crowding’ in the cell, resulting in binding of ‘small molecules’ to proteins and to changed concentrations of free soluble compounds (Srere 1980). In an update on the current approaches for metabolic flux analyses, factors are compiled that determine the specific properties of a metabolic network as far as its stability and capability to maintain homeostasis is concerned (Nikoloski et al. 2015). In particular, the authors point out that more data are required so that the interrelationship of metabolite levels, fluxes and enzyme properties can be integrated into mathematical models.
Plant cells possesses various dicarboxylate transporters in chloroplasts and mitochondria
The compartmentation of plant cells requires the transport of metabolites across intracellular membranes catalysed by specific membrane translocators. Especially, the inner membranes of chloroplasts and mitochondria represent the permeability barrier for metabolites and co‐enzymes and are equipped with various transporters that are specific for the various substrates. The Arabidopsis genome encodes a number of dicarboxylate transporters catalysing the transfer of dicarboxylates across the chloroplastidic or mitochondrial membranes (Heineke et al. 1991; Weber et al. 2004; Weber & Fischer 2007; Linka & Weber 2010; Facchinelli & Weber 2011; Fischer 2011; Flügge et al. 2011; Haferkamp & Schmitz‐Esser 2012; Lee & Millar 2016).
The two plastid‐localised dicarboxylate translocators (DiTs) are involved in primary ammonium assimilation and the recycling of ammonium that is lost during photorespiration. DiT1 (also termed OMT1, 2‐OG/malate‐translocator) imports the precursor of ammonium assimilation, 2‐OG, in exchange for malate. DiT2 (also termed DCT, glutamate/malate‐translocator) exports glutamate in exchange for malate (Fig. 3; Weber et al. 1995; Weber & Flügge 2002; Renne et al. 2003; Schneidereit et al. 2006; Riebeseel et al. 2010). In addition to its high affinity for 2‐OG and malate, DiT1 displays a high affinity for OAA and has been proposed as the OAA/malate valve (Taniguchi et al. 2002). The role as malate valve for DiT1 has been proven by mutant analyses, showing that cytosolic OAA is efficiently transported into chloroplasts mainly by DiT1 (Fig. 3; Kinoshita et al. 2011; Taniguchi & Miyake 2012).
Figure 3.
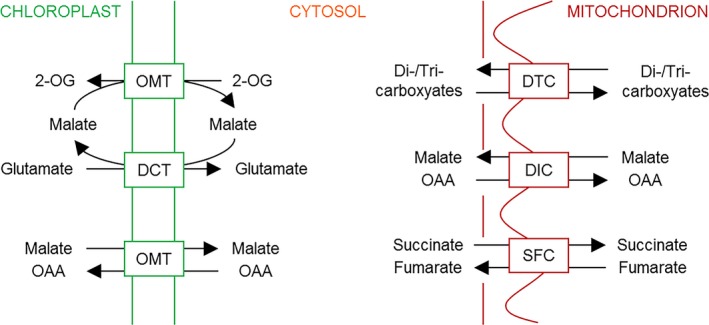
Shuttles for dicarboxylate transport. Transport processes of dicarboxylates across the inner envelope of plastids and the inner mitochondrial membrane are depicted. 2‐OG, 2‐oxoglutarate; DCT (= DiT2), glutamate/malate translocator; DIC, decarboxylate carriers; DTC, dicarboxylate/tricarboxylate carrier; OAA, oxaloacetate; OMT (= DiT1), 2‐OG/malate‐translocator, malate/OAA translocator; SFC, succinate/fumarate carrier.
In parallel to plastids, mitochondria contain various dicarboxylate translocators. The transport of specific dicarboxylates and tricarboxylates across the inner mitochondrial membrane is required for several metabolic processes, such as primary amino acid synthesis, export of reducing equivalents, fatty acid metabolism, gluconeogenesis and isoprenoid biosynthesis. The plant mitochondrial dicarboxylate/tricarboxylate carrier (DTC) mediates the transport of dicarboxylates (2‐OG, OAA, malate and succinate) and tricarboxylates (citrate, isocitrate and aconitate) by counter‐exchange (Fig. 3; Picault et al. 2002). Furthermore, plant mitochondria possess three dicarboxylate carriers (DIC1–3). These transporters facilitate the transfer of dicarboxylates, such as malate, OAA and succinate, as well as phosphate, sulphate and thiosulphate across the inner mitochondrial membrane. Accordingly, DICs may play an important role in the import of dicarboxylates into mitochondria in exchange for phosphate or sulphate to replenish the tricarboxylic acid cycle (TCAC). In addition, DICs might function as malate/OAA shuttles providing other cell compartments with reducing equivalents (Palmieri et al. 2008). Another dicarboxylate transporter, SFC, has been identified in plant mitochondria (Fig. 3). This carrier catalyses the transfer of succinate and fumarate across the inner mitochondrial membrane (Catoni et al. 2003). The main product of the glyoxylate pathway, succinate, is imported via SFC into mitochondria where it can enter the TCAC. In contrast, fumarate is exported to the cytosol in exchange for succinate entering gluconeogenesis after oxidation to malate (Catoni et al. 2003).
On both sides of the membranes carrying all of these dicarboxylate shuttle systems, the various MDH isoforms will convert OAA or malate as they arrive on the other side. Due to its equilibrium constant (K eq = 2.86 × 10−5 at pH 7.0), MDH favours the reaction towards malate formation, i.e. the reverse of what is required to regenerate OAA in the mitochondria. Therefore, only removal of the products enables the correct direction for the TCAC. On the other hand, malate is formed in chloroplasts when excess NADPH is used by NADP‐MDH for indirect export of reducing equivalents to the cytosol where it is either reconverted to OAA, when NADH is used for nitrate reduction, or transported to the mitochondria. In both examples, the direction of the enzyme reaction depends on input of substrate and removal of product in a steady‐state situation, but not its equilibrium constant. The transporters generally function as exchange carriers in both directions, driven by the concentration gradient of the respective substrates on both sides. They strictly function as shuttles, however, without any restriction as far as regulation or energy requirements are concerned, only limited by their V max and the specificity towards the transported range of substrates. They help to connect the fluxes of energy‐consuming and energy‐converting pathways between the different cellular compartments. Together with the MDH isoforms in each compartment that are operating close to equilibrium, the dicarboxylate translocators support thermodynamic buffering of the cell.
Assimilation of N is an important backup system maintaining redox homeostasis when plastidial malate valve capacities are lacking or limiting
The removal of photorespiratory ammonium (NH4 +), which is formed during the oxidation of glycine to serine, is essential for survival of a plant. The primary assimilation of NH4 + results from the collaborative activity of glutamine synthetase (GS) and GOGAT (GS/GOGAT cycle). GS catalyses the ATP‐dependent assimilation of NH4 + into glutamine, using glutamate as a substrate. Subsequently, GOGAT catalyses the reductive transfer of the amido group from glutamine to 2‐OG, resulting in the formation of two molecules of glutamate, one of them starting the assimilatory cycle again (Fig. 4). In chloroplasts of higher plants, two major classes of GOGAT enzymes are present, namely the Fd‐dependent and the NADH‐dependent GOGAT, which use either Fd or NADH as a reductant for the interconversion of glutamine and 2‐OG to glutamate (Fig. 4). While NADH‐GOGAT is crucial for NH4 + assimilation in heterotrophic tissues, Fd‐GOGAT, representing the predominant form in leaves, contributes to the assimilation of NH4 + derived from photorespiration (Lancien et al. 2002; Konishi et al. 2014).
The lack of malate valve capacities leads to improved growth when nadp‐mdh or plnad‐mdh/plNAD‐MDH Arabidopsis mutants are grown on nitrate or on ammonium, respectively, as the sole N source (Hebbelmann et al. 2012; Selinski et al. 2014). Under these conditions, Fd‐GOGAT transcripts are increased in nadp‐mdh knockout plants, and NADH‐GOGAT transcripts are more abundant in heterozygous plnad‐mdh/plNAD‐MDH mutants (Selinski & Scheibe 2014). In the case of nadp‐mdh knockout mutants, nitrite reductase as part of nitrate assimilation and Fd‐GOGAT which contributes to the assimilation of NH4 + derived from photorespiration, serve as alternative electron acceptors for reduced Fd and increase nitrate assimilation, thereby preventing, at the same time, the accumulation of excess NADPH (Hebbelmann et al. 2012; Selinski & Scheibe 2014). Correspondingly, when the dark malate valve capacity is decreased, NADH‐GOGAT, as a key enzyme of NH4 + assimilation, serves as an alternative mechanism maintaining redox homeostasis by removing NADH generated by GapCp during plastidial glycolysis (Fig. 4; Selinski et al. 2014; Selinski & Scheibe 2014).
Measurements of mitochondrial respiration in spinach leaves indicate that NADH formed inside mitochondria is oxidised by a malate/OAA shuttle to serve extra‐mitochondrial processes, e.g. reduction of nitrate in the cytosol or of hydroxypyruvate in the peroxisomes (Hanning & Heldt 1993). Furthermore, malate oxidation was shown to be one of the major sources of NADH for nitrate reduction in corn leaf blades (Neyra & Hageman 1976) that is catalysed by cytosolic NAD‐MDH isoform(s).
The malate/OAA shuttle, which itself is reversible depending on the concentrations of OAA and malate on both sides of the membrane, functions as a ‘thermodynamic buffer’ that maintains the chloroplast NADP+/NADPH ratio constant over a wide range of conditions. This corresponds to the concept put forward for the adenylate ratio for the first time by Stucki (1980). Malate valves are, therefore, important to communicate between cellular compartments and to optimise photosynthesis (Raghavendra & Padmasree 2003).
Conclusion
Malate valves, consisting of malate dehydrogenases and dicarboxylate transporters, are essential features for balancing energy metabolism and maintaining redox homeostasis in plants. In combination, they serve as thermodynamic buffers. The presence of different MDH isoforms in various subcellular compartments and their distinct roles enable plants to adapt to various stress conditions as well as to control growth and development. Although there has been much progress in unravelling roles and regulation of MDH isoforms in plants in recent years, knowledge about detailed mechanisms and controlled interplay between the systems at the various regulatory levels is still lacking in some cases.
Acknowledgements
This study was supported by a Feodor Lynen Research Fellowship (Alexander von Humboldt Foundation, Germany) to JS. Earlier work from the lab of RS has been supported by continued DFG funding (SCHE 217).
References
- Baalmann E., Backhausen J.E., Rak C., Vetter S., Scheibe R. (1995) Reductive modification and nonreductive activation of purified spinach chloroplast NADP‐dependent glyceraldehyde‐3‐phosphate dehydrogenase. Archives of Biochemistry and Biophysics, 324, 201–208. [DOI] [PubMed] [Google Scholar]
- Backhausen J.E., Vetter S., Baalmann E., Kitzmann C., Scheibe R. (1998) NAD‐dependent malate dehydrogenase and glyceraldehyde 3‐phosphate dehydrogenase isoenzymes play an important role in dark metabolism of various plastid types. Planta, 205, 359–366. [Google Scholar]
- Backhausen J.E., Kitzmann C., Horton P., Scheibe R. (2000) Electron acceptors in isolated intact spinach chloroplasts act hierarchically to prevent over‐reduction and competition for electrons. Photosynthesis Research, 64, 1–13. [DOI] [PubMed] [Google Scholar]
- Bartsch O., Mikkat S., Hagemann M., Bauwe H. (2010) An autoinhibitory domain confers redox regulation to maize glycerate kinase. Plant Physiology, 153, 832–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker B., Holtgrefe S., Jung S., Wunrau C., Kandlbinder A., Baier M., Dietz K.J., Backhausen J.E., Scheibe R. (2006) Influence of the photoperiod on redox regulation and stress responses in Arabidopsis thaliana L. (Heynh.) plants under long‐ and short‐day conditions. Planta, 224, 380–393. [DOI] [PubMed] [Google Scholar]
- Beeler S., Liu H.C., Stadler M., Schreier T., Eicke S., Lue W.L., Truernit E., Zeeman S.C., Chen J., Kötting O. (2014) Plastidial NAD‐dependent malate dehydrogenase is critical for embryo development and heterotrophic metabolism in Arabidopsis. Plant Physiology, 164, 1175–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkemeyer M., Scheibe R., Ocheretina O. (1998) A novel, non‐redox‐regulated NAD‐dependent malate dehydrogenase from chloroplasts of Arabidopsis thaliana L. Journal of Biological Chemistry, 273, 27927–27933. [DOI] [PubMed] [Google Scholar]
- Boldt R., Edner C., Kolukisaoglu U., Hagemann M., Weckwerth W., Wienkoop S., Morgenthal K., Bauwe H. (2005) D‐glycerate 3‐kinase, the last unknown enzyme in the photorespiratory cycle in Arabidopsis, belongs to a novel kinase family. The Plant Cell, 17, 2413–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan B.B. (1984) The ferredoxin/thioredoxin system: a key element in the regulatory function of light in photosynthesis. Bioscience, 34, 378–383. [PubMed] [Google Scholar]
- Buchanan B.B., Balmer Y. (2005) Redox regulation: a broadening horizon. Annual Review of Plant Biology, 56, 187–220. [DOI] [PubMed] [Google Scholar]
- Bykova N.V., Møller I.M., Gardeström P., Igamberdiev A.U. (2014) The function of glycine decarboxylase complex is optimized to maintain high photorespiratory flux via buffering of its reaction products. Mitochondrion, 19 (Pt B), 357–364. [DOI] [PubMed] [Google Scholar]
- Catoni E., Schwab R., Hilpert M., Desimone M., Schwacke R., Flügge U.I., Schumacher K., Frommer W.B. (2003) Identification of an Arabidopsis mitochondrial succinate‐fumarate translocator. FEBS Letters, 534, 87–92. [DOI] [PubMed] [Google Scholar]
- Cousins A.B., Pracharoenwattana I., Zhou W., Smith S.M., Badger M.R. (2008) Peroxisomal malate dehydrogenase is not essential for photorespiration in Arabidopsis but its absence causes an increase in the stoichiometry of photorespiratory CO2 release. Plant Physiology, 148, 786–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crétin C., Luchetta P., Joly C., Decottignies P., Lepiniec L., Gadal P., Sallantin M., Huet J.C., Pernollet J.C. (1990) Primary structure of sorghum malate dehydrogenase (NADP) deduced from cDNA sequence. Homology with malate dehydrogenase (NAD). European Journal of Biochemistry, 192, 299–303. [DOI] [PubMed] [Google Scholar]
- Dietz K.J. (2003) Redox control, redox signaling and redox homeostasis in plant cells. International Review of Cytology, 228, 141–193. [DOI] [PubMed] [Google Scholar]
- Facchinelli F., Weber A.P. (2011) The metabolite transporters of the plastid envelope: an update. Frontiers in Plant Science, 2, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fickenscher K., Scheibe R. (1988) Limited proteolysis of inactive tetrameric chloroplast NADP‐malate dehydrogenase produces active dimers. Archives of Biochemistry and Biophysics, 260, 771–779. [DOI] [PubMed] [Google Scholar]
- Fischer K. (2011) The import and export business in plastids: transport processes across the inner envelope membrane. Plant Physiology, 155, 1511–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flügge U.I., Häusler R.E., Ludewig F., Gierth M. (2011) The role of transporters in supplying energy to plant plastids. Journal of Experimental Botany, 62, 2381–2392. [DOI] [PubMed] [Google Scholar]
- Foyer C.H., Noctor G. (2003) Redox sensing and signaling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiologia Plantarum, 119, 355–364. [Google Scholar]
- Fridlyand L.E., Scheibe R. (1999) Regulation of the Calvin cycle for CO2 fixation as an example for general control mechanisms in metabolic cycles. BioSystems, 51, 79–93. [DOI] [PubMed] [Google Scholar]
- Fridlyand L.E., Scheibe R. (2000) Regulation in metabolic systems under homeostatic flux control. Archives of Biochemistry and Biophysics, 374, 198–206. [DOI] [PubMed] [Google Scholar]
- Fridlyand L.E., Backhausen J.E., Scheibe R. (1998) Flux control of the malate valve in leaf cells. Archives of Biochemistry and Biophysics, 349, 290–298. [DOI] [PubMed] [Google Scholar]
- Fu Q., Wang B.C., Jin X., Li H.B., Han P., Wei K.H., Zhang X.M., Zhu Y.X. (2005) Proteomic analysis and extensive protein identification from dry, germinating Arabidopsis seeds and young seedlings. Journal of Biochemistry and Molecular Biology, 38, 650–660. [DOI] [PubMed] [Google Scholar]
- Gietl C. (1992) Malate dehydrogenase isoenzymes: cellular locations and role in the flow of metabolites between the cytoplasm and cell organelles. Biochimica et Biophysica Acta, 1100, 217–234. [DOI] [PubMed] [Google Scholar]
- Gietl C., Hock B. (1982) Organelle‐bound malate dehydrogenase isoenzymes are synthesized as higher molecular weight precursors. Plant Physiology, 70, 483–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haferkamp I., Schmitz‐Esser S. (2012) The plant mitochondrial carrier family: functional and evolutionary aspects. Frontiers in Plant Science, 3, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagedorn P.H., Flyvbjerg H., Møller I.M. (2004) Modelling NADH turnover in plant mitochondria. Physiologia Plantarum, 120, 1–16. [DOI] [PubMed] [Google Scholar]
- Hameister S., Becker B., Holtgrefe S., Strodtkötter I., Linke V., Backhausen J.E., Scheibe R. (2007) Transcriptional regulation of NADP‐dependent malate dehydrogenase: comparative genetics and identification of DNA‐binding proteins. Journal of Molecular Evolution, 65, 437–455. [DOI] [PubMed] [Google Scholar]
- Hanning I., Heldt H.W. (1993) On the function of mitochondrial metabolism during photosynthesis in spinach (Spinacia oleracea L.) leaves (Partitioning between respiration and export of redox equivalents and precursors for nitrate assimilation products). Plant Physiology, 103, 1147–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara S., Motohashi K., Arisaka F., Romano P.G., Hosoya‐Matsuda N., Kikuchi N., Fusada N., Hisabori T. (2006) Thioredoxin‐h1 reduces and reactivates the oxidized cytosolic malate dehydrogenase dimer in higher plants. Journal of Biological Chemistry, 281, 32065–32071. [DOI] [PubMed] [Google Scholar]
- Hebbelmann I., Selinski J., Wehmeyer C., Goss T., Voss I., Mulo P., Kangasjärvi S., Aro E.M., Oelze M.L., Dietz K.J., Nunes‐Nesi A., Do P.T., Fernie A.R., Talla S.K., Raghavendra A.S., Linke V., Scheibe R. (2012) Multiple strategies to prevent oxidative stress in Arabidopsis plants lacking the malate valve enzyme NADP‐malate dehydrogenase. Journal of Experimental Botany, 63, 1445–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heineke D., Scheibe R. (2009) Photosynthesis: the Calvin cycle In: Encyclopedia of life sciences. Wiley, Chichester, UK: 10.1002/9780470015902.a0001291.pub2. [DOI] [Google Scholar]
- Heineke D., Riens B., Grosse H., Hoferichter P., Peter U., Flügge U.I., Heldt H.W. (1991) Redox transfer across the inner chloroplast envelope membrane. Plant Physiology, 95, 1131–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyno E., Innocenti G., Lemaire S.D., Issakidis‐Bourguet E., Krieger‐Liszkay A. (2014) Putative role of the malate valve enzyme NADP‐malate dehydrogenase in H2O2 signalling in Arabidopsis. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 369, 20130228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt T., Knuesting J., Berndt C., Morgan B., Scheibe R. (2015) Cytosolic thiol switches regulating basic cellular functions: GAPDH as an information hub? Biological Chemistry, 396, 523–537. [DOI] [PubMed] [Google Scholar]
- Huang J., Niazi A.K., Young D., Rosado L.A., Vertommen D., Bodra N., Abdelgawwad M.R., Vignols F., Wei B., Wahni K., Bashandy T., Bariat L., Van Breusegem F., Messens J., Reichheld J.P. (2018) Self‐protection of cytosolic malate dehydrogenase against oxidative stress in Arabidopsis. Journal of Experimental Botany, 69, 3491–3505. [DOI] [PubMed] [Google Scholar]
- Igamberdiev A.U., Kleczkowski L.A. (2011) Optimization of CO2 fixation in photosynthetic cells via thermodynamic buffering. BioSystems, 103, 224–229. [DOI] [PubMed] [Google Scholar]
- Igamberdiev A.U., Lernmark U., Gardeström P. (2014) Activity of the mitochondrial pyruvate dehydrogenase complex in plants is stimulated in the presence of malate. Mitochondrion, 19 (Pt B), 184–190. [DOI] [PubMed] [Google Scholar]
- Imsande J., Berkemeyer M., Scheibe R., Schumann U., Gietl C., Palmer R.G. (2001) A soybean plastid‐targeted NADH‐malate dehydrogenase: cloning and expression analyses. American Journal of Botany, 88, 2136–2142. [PubMed] [Google Scholar]
- Kampfenkel K., Möhlmann T., Batz O., Van Montagu M., Inzé D., Neuhaus H.E. (1995) Molecular characterization of an Arabidopsis thaliana cDNA encoding a novel putative adenylate translocator of higher plants. FEBS Letters, 374, 351–355. [DOI] [PubMed] [Google Scholar]
- Kinoshita H., Nagasaki J., Yoshikawa N., Yamamoto A., Takito S., Kawasaki M., Sugiyama T., Miyake H., Weber A.P.M., Taniguchi M. (2011) The chloroplastic 2oxoglutarate/malate transporter has dual function as the malate valve in carbon/nitrogen metabolism. The Plant Journal, 65, 15–26. [DOI] [PubMed] [Google Scholar]
- Konishi N., Ishiyama K., Matsuoka K., Maru I., Hayakawa T., Yamaya T., Kojima S. (2014) NADH‐dependent glutamate synthase plays a crucial role in assimilating ammonium in the Arabidopsis root. Physiologia Plantarum, 152, 138–151. [DOI] [PubMed] [Google Scholar]
- Lance C., Rustin P. (1984) The central role of malate in plant metabolism. Physiologie Végétale, 22, 625–641. [Google Scholar]
- Lancien M., Martin M., Hsieh M.H., Leustek T., Goodman H., Coruzzi G.M. (2002) Arabidopsis glt1‐T mutant defines a role for NADH‐GOGAT in the non‐photorespiratory ammonium assimilatory pathway. The Plant Journal, 29, 347–358. [DOI] [PubMed] [Google Scholar]
- Lee C.P., Millar A.H. (2016) The plant mitochondrial transportome: balancing metabolic demands with energetic constraints. Trends in Plant Science, 21, 662–676. [DOI] [PubMed] [Google Scholar]
- Leegood R.C., Lea P.J., Adcock M.D., Häusler R.E. (1995) The regulation and control of photorespiration. Journal of Experimental Botany, 46, 1397–1414. [Google Scholar]
- Lindén P., Keech O., Stenlund H., Gardeström P., Moritz T. (2016) Reduced mitochondrial malate dehydrogenase activity has a strong effect on photorespiratory metabolism as revealed by 13C labelling. Journal of Experimental Botany, 67, 3123–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linka N., Weber A.P. (2010) Intracellular metabolite transporters in plants. Molecular Plant, 3, 21–53. [DOI] [PubMed] [Google Scholar]
- Maurino V.G., Engqvist M.K. (2015) 2‐Hydroxy acids in plant metabolism. The Arabidopsis Book, 13, e0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlister‐Henn L. (1988) Evolutionary relationships among the malate dehydrogenases. Trends in Biochemical Sciences, 13, 178–181. [DOI] [PubMed] [Google Scholar]
- Miginiac‐Maslow M., Issakidis E., Lemaire M., Ruelland E., Jacquot J.P., Decottignies P. (1997) Light‐dependent activation of NADP‐malate dehydrogenase: a complex process. Australian Journal of Plant Physiology, 24, 529–542. [Google Scholar]
- Miginiac‐Maslow M., Johansson K., Ruelland E., Issakidis‐Bourguet E., Schepens I., Goyer A., Lemaire‐Chamley M., Jacquot J.P., Le Maréchal P., Decottignies P. (2000) Light‐activation of NADP‐malate dehydrogenase: a highly controlled process for an optimized function. Physiologia Plantarum, 110, 322–329. [Google Scholar]
- Möhlmann T., Tjaden J., Schwöppe C., Winkler H.H., Kampfenkel K., Neuhaus H.E. (1998) Occurrence of two plastidic ATP/ADP transporters in Arabidopsis thaliana L. – molecular characterisation and comparative structural analysis of similar ATP/ADP translocators from plastids and Rickettsia prowazekii . European Journal of Biochemistry, 252, 353–359. [DOI] [PubMed] [Google Scholar]
- Muñoz‐Bertomeu J., Cascales‐Miñana B., Mulet J.M., Baroja‐Fernández E., PozuetaRomero J., Kuhn J.M., Segura J., Ros R. (2009) Plastidial glyceraldehyde‐3‐phosphate dehydrogenase deficiency leads to altered root development and affects the sugar and amino acid balance in Arabidopsis. Plant Physiology, 151, 541–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz‐Bertomeu J., Cascales‐Miñana B., Irles‐Segura A., Mateu I., Nunes‐Nesi A., Fernie A.R., Segura J., Ros R. (2010) The plastidial glyceraldehyde‐3‐phosphate dehydrogenase is critical for viable pollen development in Arabidopsis. Plant Physiology, 152, 1830–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus H.E., Thom E., Möhlmann T., Steup M., Kampfenkel K. (1997) Characterization of a novel eukaryotic ATP/ADP translocator located in the plastid envelope of Arabidopsis thaliana L. The Plant Journal, 11, 73–82. [DOI] [PubMed] [Google Scholar]
- Neyra C.A., Hageman R.H. (1976) Relationships between carbon dioxide, malate, and nitrate accumulation and reduction in corn (Zea mays L.) seedlings. Plant Physiology, 58, 726–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas K.B., Nicholas H.B. Jr, Deerfield D.W. II (1997) GeneDoc: analysis and visualization of genetic variation. EMBNEW News, 4, 14. [Google Scholar]
- Nikoloski Z., Perez‐Storey R., Sweetlove L.J. (2015) Inference and prediction of metabolic network fluxes. Plant Physiology, 169, 1443–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G., Foyer C.H. (2016) Intracellular redox compartmentation and ROS‐related communication in regulation and signaling. Plant Physiology, 171, 1581–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocheretina O., Scheibe R. (1997) Cloning and sequence analysis of cDNAs encoding plant cytosolic malate dehydrogenase. Gene, 199, 145–148. [DOI] [PubMed] [Google Scholar]
- Ocheretina O., Harnecker J., Rother T., Schmid R., Scheibe R. (1993) Effects of N‐terminal truncations upon chloroplast NADP‐malate dehydrogenases from pea and spinach. Biochimica et Biophysica Acta, 1163, 10–16. [DOI] [PubMed] [Google Scholar]
- Ocheretina O., Haferkamp I., Tellioglu H., Scheibe R. (2000) Light‐modulated NADP‐malate dehydrogenases from moss, fern and green algae: insights into evolution of the enzyme's regulation. Gene, 258, 147–154. [DOI] [PubMed] [Google Scholar]
- Ogren W.L. (1984) Photorespiration: pathways, regulation, and modification. Annual Review of Plant Physiology, 35, 415–442. [Google Scholar]
- Palmieri L., Picault N., Arrigoni R., Besin E., Palmieri F., Hodges M. (2008) Molecular identification of three Arabidopsis thaliana mitochondrial dicarboxylate carrier isoforms: organ distribution, bacterial expression, reconstitution into liposomes and functional characterization. Biochemical Journal, 410, 621–629. [DOI] [PubMed] [Google Scholar]
- Palmieri F., Rieder B., Ventrella A., Blanco E., Do P.T., Nunes‐Nesi A., Trauth A.U., Fiermonte G., Tjaden J., Agrimi G., Kirchberger S., Paradies E., Fernie A.R., Neuhaus H.E. (2009) Molecular identification and functional characterization of Arabidopsis thaliana mitochondrial and chloroplastic NAD+ carrier proteins. Journal of Biological Chemistry, 284, 31249–31259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.W., Li W., Viehhauser A., He B., Kim S., Nilsson A.K., Andersson M.X., Kittle J.D., Ambavaram M.M., Luan S., Esker A.R., Tholl D., Cimini D., Ellerström M., Coaker G., Mitchell T.K., Pereira A., Dietz K.J., Lawrence C.B. (2013) Cyclophilin 20‐3 relays a 12‐oxo‐phytodienoic acid signal during stress responsive regulation of cellular redox homeostasis. Proceedings of the National Academy of Sciences of the United States of America, 110, 9559–9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picault N., Palmieri L., Pisano I., Hodges M., Palmieri F. (2002) Identification of a novel transporter for dicarboxylates and tricarboxylates in plant mitochondria. Bacterial expression, reconstitution, functional characterization, and tissue distribution. Journal of Biological Chemistry, 277, 24204–24211. [DOI] [PubMed] [Google Scholar]
- Pracharoenwattana I., Zhou W., Smith S.M. (2010) Fatty acid beta‐oxidation in germinating Arabidopsis seeds is supported by peroxisomal hydroxypyruvate reductase when malate dehydrogenase is absent. Plant Molecular Biology, 72, 101–109. [DOI] [PubMed] [Google Scholar]
- Raghavendra A.S., Padmasree K. (2003) Beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Trends in Plant Science, 8, 546–553. [DOI] [PubMed] [Google Scholar]
- Reiser J., Linka N., Lemke L., Jeblick W., Neuhaus H.E. (2004) Molecular physiological analysis of the two plastidic ATP/ADP transporters from Arabidopsis. Plant Physiology, 136, 3524–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renne P., Dressen U., Hebbeker U., Hille D., Flügge U.I., Westhoff P., Weber A.P.M. (2003) The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2. The Plant Journal, 35, 316–331. [DOI] [PubMed] [Google Scholar]
- Riebeseel E., Häusler R.E., Radchuk R., Meitzel T., Hajirezaei M.R., Emery R.J., Küster H., Nunes‐Nesi A., Fernie A.R., Weschke W., Weber H. (2010) The 2‐oxoglutarate/malate translocator mediates amino acid and storage protein biosynthesis in pea embryos. The Plant Journal, 61, 350–363. [DOI] [PubMed] [Google Scholar]
- Scheibe R. (1987) NADP+‐malate dehydrogenase in C3‐plants: regulation and role of a light‐activated enzyme. Physiologia Plantarum, 71, 393–400. [Google Scholar]
- Scheibe R. (1991) Redox‐modulation of chloroplast enzymes: a common principle for individual control. Plant Physiology, 96, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibe R. (2004) Malate valves to balance cellular energy supply. Physiologia Plantarum, 120, 21–26. [DOI] [PubMed] [Google Scholar]
- Scheibe R., Anderson L.E. (1981) Dark modulation of NADP‐dependent malate dehydrogenase and glucose‐6‐phosphate dehydrogenase in the chloroplast. Biochimica et Biophysica Acta, 636, 58–64. [DOI] [PubMed] [Google Scholar]
- Scheibe R., Dietz K.J. (2012) Reduction–oxidation network for flexible adjustment of cellular metabolism in photoautotrophic cells. Plant, Cell and Environment, 35, 202–216. [DOI] [PubMed] [Google Scholar]
- Scheibe R., Stitt M. (1988) Comparison of NADP‐malate dehydrogenase activation, QA reduction and O2 evolution in spinach leaves. Plant Physiology and Biochemistry, 26, 473–481. [Google Scholar]
- Scheibe R., Backhausen J.E., Emmerlich V., Holtgrefe S. (2005) Strategies to maintain redox homeostasis during photosynthesis under changing conditions. Journal of Experimental Botany, 56, 1481–1489. [DOI] [PubMed] [Google Scholar]
- Schneidereit J., Häusler R.E., Fiene G., Kaiser W.M., Weber A.P. (2006) Antisense repression reveals a crucial role of the plastidic 2‐oxoglutarate/malate translocator DiT1 at the interface between carbon and nitrogen metabolism. The Plant Journal, 45, 206–224. [DOI] [PubMed] [Google Scholar]
- Selinski J., Scheibe R. (2014) Lack of malate valve capacities lead to improved N‐assimilation and growth in transgenic A. thaliana plants. Plant Signaling and Behavior, 9, pii: e29057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinski J., König N., Wellmeyer B., Hanke G.T., Linke V., Neuhaus H.E., Scheibe R. (2014) The plastid‐localized NAD‐dependent malate dehydrogenase is crucial for energy homeostasis in developing Arabidopsis thaliana seeds. Molecular Plant, 7, 170–186. [DOI] [PubMed] [Google Scholar]
- Sew Y.S., Ströher E., Fenske R., Millar A.H. (2016) Loss of mitochondrial malate dehydrogenase activity alters seed metabolism impairing seed maturation and post‐germination growth in Arabidopsis. Plant Physiology, 171, 849–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srere P.A. (1980) The infrastructure of the mitochondrial matrix. Trends in Biochemical Sciences, 5, 120–121. [Google Scholar]
- Stucki J.W. (1980) The thermodynamic‐buffer enzymes. European Journal of Biochemistry, 109, 257–267. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013) MEGA 6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi M., Miyake H. (2012) Redox‐shuttling between chloroplast and cytosol: integration of intra‐chloroplast and extra‐chloroplast metabolism. Current Opinion in Plant Biology, 15, 252–260. [DOI] [PubMed] [Google Scholar]
- Taniguchi M., Taniguchi Y., Kawasaki M., Takeda S., Kato T., Sato S., Tabata S., Miyake H., Sugiyama T. (2002) Identifying and characterizing plastidic 2‐oxoglutarate/malate and dicarboxylate transporters in Arabidopsis thaliana . Plant and Cell Physiology, 43, 706–717. [DOI] [PubMed] [Google Scholar]
- Timm S., Florian A., Jahnke K., Nunes‐Nesi A., Fernie A.R., Bauwe H. (2011) The hydroxypyruvate‐reducing system in Arabidopsis: multiple enzymes for the same end. Plant Physiology, 155, 694–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjaden J., Möhlmann T., Kampfenkel K., Henrichs G., Neuhaus H.E. (1998) Altered plastidic ATP/ADP‐transporter activity influences potato (Solanum tuberosum L.) tuber morphology, yield and composition of tuber starch. The Plant Journal, 16, 531–540. [Google Scholar]
- Tomaz T., Bagard M., Pracharoenwattana I., Lindén P., Lee C.P., Carroll A.J., Ströher E., Smith S.M., Gardeström P., Millar A.H. (2010) Mitochondrial malate dehydrogenase lowers leaf respiration and alters photorespiration and plant growth in Arabidopsis. Plant Physiology, 154, 1143–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B.J., South P.F., Ort D.R. (2016) Physiological evidence for plasticity in glycolate/glycerate transport during photorespiration. Photosynthesis Research, 129, 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Yu L., Yu C.A. (2010) Cross‐talk between mitochondrial malate dehydrogenase and the cytochrome bc1 complex. Journal of Biological Chemistry, 285, 10408–10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A.P., Fischer K. (2007) Making the connections – the crucial role of metabolite transporters at the interface between chloroplast and cytosol. FEBS Letters, 581, 2215–2222. [DOI] [PubMed] [Google Scholar]
- Weber A., Flügge U.I. (2002) Interaction of cytosolic and plastidic nitrogen metabolism in plants. Journal of Experimental Botany, 53, 865–874. [DOI] [PubMed] [Google Scholar]
- Weber A., Menzlaff E., Arbinger B., Gutensohn M., Eckerskorn C., Flügge U.I. (1995) The 2‐oxoglutarate malate translocator of chloroplast envelope membranes – molecular‐cloning of a transporter containing a 12‐helix motif and expression of the functional protein in yeast‐cells. Biochemistry, 34, 2621–2627. [DOI] [PubMed] [Google Scholar]
- Weber A.P., Schneidereit J., Voll L.M. (2004) Using mutants to probe the in vivo function of plastid envelope membrane metabolite transporters. Journal of Experimental Botany, 55, 1231–1244. [DOI] [PubMed] [Google Scholar]
- Weitbrecht K., Müller K., Leubner‐Metzger G. (2011) First off the mark: early seed germination. Journal of Experimental Botany, 62, 3289–3309. [DOI] [PubMed] [Google Scholar]
- Yoshida K., Hisabori T. (2016) Adenine nucleotide‐dependent and redox‐independent control of mitochondrial malate dehydrogenase activity in Arabidopsis thaliana . Biochimica et Biophysica Acta, 1857, 810–818. [DOI] [PubMed] [Google Scholar]


