ABSTRACT
Asymptomatic carriers of Plasmodium are considered a reservoir of the parasite in humans. Therefore, in order to be effective, new malaria elimination strategies must take these targets into account. The aim of this study was to analyse genetic diversity of Plasmodium falciparum among schoolchildren in three epidemiological areas in Côte d’Ivoire. This was a cross-sectional study carried out from May 2015 to April 2016 in a primary school in rural and urban areas of San Pedro, Grand-Bassam and Abengourou, during the rainy season and the dry season. A total of 282 Plasmodium falciparum isolates were genotyped using Nested PCR of Pfmsp1 and Pfmsp2 genes. The overall frequency of K1, Mad20 and RO33 alleles was 81.6%, 53.4% and 57% for Pfmsp1 respectively. For Pfmsp2, this frequency was 84.3% and 72.2% for 3D7 and FC27. K1, Mad20 and FC27 Frequencies were significantly higher in Abengourou compared to other sites. Overall, the frequency of MIs was significantly higher in Abengourou for Pfmsp1 and Pfmsp2. However, Mad20 and RO33 alleles were significantly higher in the rainy season. No significant difference was observed between Pfmsp2 alleles in both seasons. Frequency of the 3D7 allele was significantly higher in symptomatic patients. MIs and COI increased with parasitemia for Pfmsp1and Pfmsp2. The data can be added to that available for monitoring and control of P. falciparum malaria. Further studies combining the entomological inoculation rate and the genetic diversity of P. falciparum will allow us to shed light on our understanding of the epidemiology of this parasite.
KEYWORDS: Plasmodium, asymptomatic carrier, genetic diversity, Côte d’Ivoire
Background
Despite numerous efforts to control the disease, malaria remains the most deadly parasitic disease in the world. In 2017, it was responsible for 219 million new cases, including 435,000 deaths, most of which occurred in sub-Saharan Africa [1]. In Côte d’Ivoire, malaria accounts for 43% of the reasons for consultation at health centres [2]. The transmission is perennial with some peaks during the rainy season. It is responsible for 40% of school and work absenteeism and 50% of farm income losses. People spend about 25% of their income on prevention and treatment of this disease [2].
The elimination of the malaria vector by insecticides, the use of antimalarial drugs for both curative and prophylactic treatment, the improvement of the living environment and the development of antimalarial vaccines are strategies used to control this endemic disease. To be successful, malaria elimination will require knowledge of parasite genome variation in different geographical locations and a better understanding of the factors that determine gene flow between locations. Plasmodium falciparum merozoite surface protein-1 (Pfmsp1) and Plasmodium falciparum merozoite surface protein-2 (Pfmsp2) which are asexual blood stage antigens are considered as prime candidates for the development of a malaria vaccine and are also suitable markers for the identification of genetically distinct Plasmodium falciparum parasite populations [3]. However, since the publication of the complete genome of the 3D7 reference clone of P. falciparum [4] and the sequencing of other plasmodial isolates, there have been some insights into the genetic variations responsible for phenotypes such as parasite chemo-resistance and virulence [5]. But, in these studies, parasites have been often collected from symptomatic cases. One of the difficulties of this elimination lies in the fact that most strategies concern symptomatic carriers of Plasmodium. Thus, little work on genetic polymorphism and resistance markers of P. falciparum has been done on asymptomatic infections.
We issue the following hypothesis: the populations of P. falciparum found in asymptomatic subjects present different genotypes of those found in the symptomatic.
Therefore, it would be interesting to describe the genotype of these strains compared to the profile found in symptomatic patients in different epidemiological areas. Therefore, the study of the genetic diversity of P. falciparum populations in these different epidemiological areas is an important step both towards a thorough knowledge of the strains circulating in Côte d’Ivoire and towards the development and/or evaluation of malaria vaccines. This study aimed to explore the genetic diversity of P. falciparum in asymptomatic and symptomatic malaria infections in three epidemiological areas in Côte d’Ivoire during the rainy and dry seasons.
Materials and methods
Study sites
This study was conducted in primary schools of three health districts of Côte d’Ivoire i.e Abengourou (in the east, transition forest zone), Grand-Bassam (in the south-east, coastal zone) and San Pedro (in the south-west, coast and forest zone). For each study area, sites were selected in both rural (village) and urban (city) areas. Thus, in Grand-Bassam, the urban study sites were the France, the SODECI and ‘château d’eau’ neighbourhoods while the rural study sites were Azzuretti, Mondoukou and Vitré 1A. In Abengourou, the study took place in the Agnikro, Plateau and Agnikro-extension neighbourhoods in urban areas. The rural sites were Touzoukro, CNRA and Assekro. For the San Pedro health areas, the urban sites were ‘Bardot Ouest’, Zimbabwe and SOTREF while at the rural level, Kablaké, ‘Cité Agricole’ and Baba were the selected sites (Figure 1).
Figure 1.
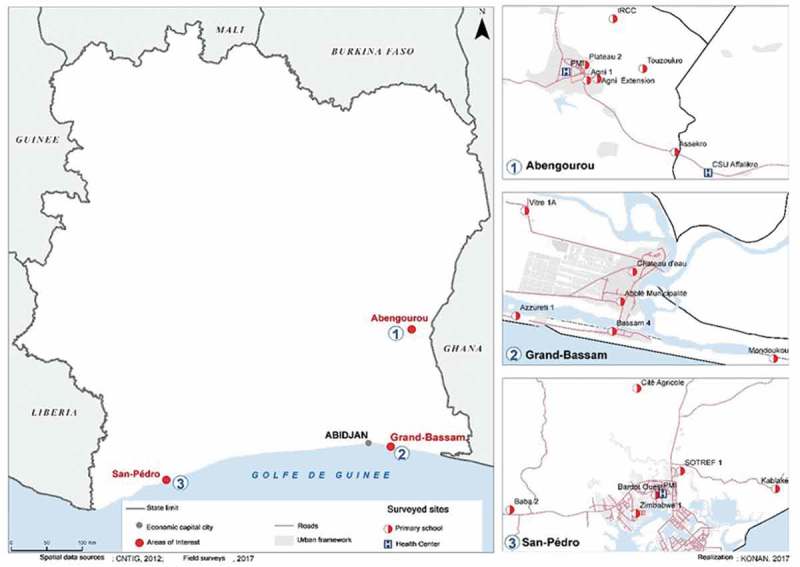
Study sites (CNTIG, 2012; Field surveys, 2017).
Type of study and samples collection
It was a cross-sectional study conducted from May 2015 to April 2016 according to the following schedule:
In Grand-Bassam: from 13 to 15 May 2015 and on 8 October 2015 during the rainy season and from 19 to 21 January 2016 during the dry season;
In Abengourou: from 17 to 19 November 2015 during the rainy season and from 1 to 3 March 2016 during the dry season;
In San Pedro: from 19 to 21 April 2016 during the rainy season and from 23 to 25 February 2016 during the dry season.
Patients were enrolled in the study in accordance with the following inclusion criteria:
primary school children aged from 4 to 16 years without gender preference,
present in the study area for at least 2 months,
single infestation by P. falciparum.
Written informed consent from the parent or legal guardian of the children was also required. The enrollment of schoolchildren was made by a two-stage cluster survey (using the no-sampling technique). Thus, in each site (rural or urban) a total of 18 classes were randomly selected. Then, in each selected class, we drew lots for 11 students, for a total of 198 students per site. Before any investigation was carried out in each locality, the health authorities, the Directorate of Primary Education Inspection, customary authorities (in rural areas), school principals and teachers were informed. After informing the parents or legal guardians of the children about the purpose, objectives and procedures of the study, their written informed consent was requested for their child’s participation the day before the survey was conducted. On the day of the survey, a questionnaire was administered to each schoolchild and school parent (with consent) to collect sociodemographic and clinical data. The clinical examination of the child by the research team doctor revealed the absence or presence of clinical signs of malaria, in case of fever, by the measurement of the temporal temperature. History of fever for 7 days prior to the survey. Malaria rapid diagnostic tests SD-Bioline Malaria-Ag-P.f/Pan™ (Lot. No.: 05ED15029) were used on site to note the detection of a malaria case. Four to five milliter (ml) of blood sample was then collected in an EDTA tube to perform the thick blood smear, thin blood smear and dried blood spots. Blood (approximately 20 μL) was spread onto filter papers (WhatmanTM), dried, and stored in individual plastic bags until DNA extraction was performed.
Sample size
The Schwartz formula was used to determine the minimum sample size in each study site:
N = Z2 *(p)*(1-p)/C2
Where:
N: sample size
Z = Z value (e.g. 1.96 for 95% confidence level)
p: Estimated proportion of the population with asexual forms of Plasmodium (p = 63%) [6]
C = confidence interval, expressed as decimal (e.g. .04 = ±4)
N = 358.19
On this basis, we obtained a sample size for each study site equal to 358 schoolchildren. To reach the sample we included 396 schoolchildren (198 in urban areas and 198 in rural areas) in each study site, taking into account a proportion of 10% of data that could not be used.
Microscopic examinations
Thick and thin blood smears were made and stained for 15 to 20 minutes with a 10% Giemsa solution and read by two independent laboratory technicians to determine the Plasmodium species and parasitemia. Plasmodial species was detected on thin blood smear. The asexual forms were counted with reference to 500 leukocytes and expressed as trophozoites/µl blood, assuming that the average leukocyte count is 8000/µl blood. The parasitemia was obtained by averaging the results of the 2 readers. In the event of a discrepancy of more than 50% in the parasitemia between the first two readers, a third reader should be involved.
Selection of samples for genotyping
In each site, 15% of the total dried blood spots collected in the site were randomized including negative and positive samples detected by microscopy.
Extraction of parasitic DNA
Plasmodial DNA was extracted from WhatmanTM N°3 paper dried blood spots using Zymo “Quick-DNA™ Universal Kit” (lot N° ZRC 186,993; the Epigenetics company; USA) according to the manufacturer’s instructions. These extracts were stored at −20 ° C until use.
Genotyping of P. falciparum
Genotyping of P falciparum consisted of a first amplification targeting a larger domain of block 2 of Pfmsp1 and that of block 3 of Pfmsp2 followed by a second more specific amplification as previously described [7]. Briefly, the first amplification used 0.25μl of the pairs of primers M1-OF/M1-OR for Pfmsp1 and M2-OF/M2-OR for Pfmsp2 in the presence of 15.875μl of water for PCR, 2.5μl of buffer 10x (standard Taq reaction buffer with MgCl2), 0.5μl dNTPs Master Mix (BioLabs® Inc.), 0.125μl of Taq DNA Polymerase (BioLabs®Inc.) and 5 μl of DNA extract as a template, i.e a final volume of 25 μl.
For the second PCR we used the amplification product of the first PCR as matrix DNA. The primer pairs specific to each Pfmsp1 and Pfmsp2 allelic family were used in the presence of the same reaction constituents as the first PCR, but with different reaction volumes. These primer pairs were respectively K1/K2 for the K1 family, MAD20-1/MAD20-2 for the MAD20 family and RO33-1/RO33-2 for the RO33 family of Pfmsp1. For the allelic families of the Pfmsp2 gene, the priming pairs FC27-1/FC27-2 and 3D7-1/3D7-2 were used, respectively for the FC27 and 3D7 family. The DNA strips were revealed by agarose gel electrophoresis prepared at 1.5% and containing Ethidium Bromide.
Ethics
The study protocol was approved by the National Research Ethics Committee (CNER) under number 020/MSLS/CNER-dkn. It was conducted in accordance with the text of the Helsinki Declaration adopted by the 18th World Medical Assembly in 1964 and its amendments, the ICH (International Conference on Harmonization) recommendations. It has been consistent with Good Clinical Practices and all applicable regulatory requirements for clinical studies as well as Côte d’Ivoire’s national laws and regulations. Free and written informed consent of patients, parents or legal guardians prior to the participant’s enrolment was obtained.
Definitions of terms
Asymptomatic subject: was considered asymptomatic any subject with asexual forms of Plasmodium, with no fever (temporal temperature below 37.8°C) and who did not show any other clinical signs of malaria during the 7 days before and after the medical examination.
Symptomatic subject: was considered symptomatic any subject with asexual forms of Plasmodium, with fever (temporal temperature greater than or equal to 37.8°C) or who showed clinical signs of malaria during the 7 days before and after the medical examination.
The allelic families of Pfmsp1 and Pfmsp2 were jointly determined during the genotyping of these markers. A sample was considered to belong to a given allelic family if at least one band appeared following amplification of the DNA of this sample with the specific primers of the family studied. If there was no band, the sample was considered not to be from this allelic family. The frequency of an allele was defined as the ratio of the number of such band to the total number of bands obtained at the locus [8]. Infections were classified as monoclonal when a single band was detected in a sample and as multiple when the isolate carried more than one band. The frequency of single infection was the number of patients with one parasite genotype over the total infected population. Frequency of multiple infections (MIs) is a percentage estimated as follows: the number of isolates with more than one parasite genotype over the total of infected individuals. The complexity of infections (COI) is defined as the mean number of different parasite genotypes per infected individual was calculated using each marker: Pfmsp1 gene, Pfmsp2 gene and by combining both markers genotyping data [9].
Statistical analysis
The data were entered using Epidata Version 3.1 and Microsoft Excel 2016 software and analyzed by SPSS (Statistical Package for Social Sciences) version 18.0 or GraphPad Prism 5 for Windows version 5.01 (August 7, 2017). Graphical representations were done using Microsoft Excel 2016.
The allelic families of Pfmsp1 and Pfmsp2 were jointly determined during the genotyping of these markers. The frequency of each allele was estimated on all detected alleles. The distribution of the allelic family and the number of genotypes detected in each infected patient were calculated by site, season, clinical status and parasitemia. The complexity of the infection was determined based on these same variables. Differences between groups were determined by t-test or variance analysis (ANOVA) for normally distributed continuous variables. The complexity of the infection and the frequency of multiple infections were calculated by combining the genotyping results of Pfmsp1 and Pfmsp2. The level of significance for statistical tests has been set at p ≤ 0.05.
Results
Characteristic of patients and malaria infection
A total of 360 blood samples (15% of 2361 samples) was obtained. The majority of the school children were under 10 years old (54.7%). The Plasmodium parasite rate was 76.1%. P. falciparum was the major species (96.4%) followed by P. ovale (0.4%) and P. malariae (0.4%). Eight mixed infections were observed, 5 with P. falciparum and P. malariae and 3 with P. falciparum and P. ovale. The parasitemia ranged between 15 and 103,040 parasites/μl of blood (SD = 8778.3) with a geometric mean of 3147.5 parasites/μl.
Pfmsp1 and Pfmsp2 genes genotyping
Allelic families frequency
The overall frequency of allelic families K1, Mad20 and RO33 was 81.6%, 53.4% and 57% respectively for the Pfmsp1 gene. Concerning the Pfmsp2 gene, the overall frequency of the 3D7 and FC27 allelic families was 84.3% and 72.2% respectively. For the Pfmsp1 gene, in general, a significant difference was observed between the frequency of the K1, Mad20 and RO33 alleles in subjects with parasite densities greater than 1000 parasites/µL of blood. However, the Mad20 and RO33 alleles were significantly higher in the rainy season than in the dry season (p-value< 0.000, p-value = 0.001) (Table 1). For Pfmsp2, the frequency of the 3D7 allele was significantly higher in the symptomatic subjects than in the asymptomatic ones. However, the frequency of the FC27 allele was significantly related to the geographical area (p-value = 0.002) (Table 2).
Table 1.
Distribution of the different allelic families of Pfmsp1 gene by site, season, clinical status and parasitemia.
| Variable | K1 n (%) | p-value | Mad20 n(%) | p-value | RO33 n (%) | p-value | |
|---|---|---|---|---|---|---|---|
| Site | Grand-Bassam | 27 (65.9) | 0.012 | 19 (46.3) | 0.009 | 21 (51.2) | 0.359 |
| Abengourou | 75 (87.2) | 57 (66.3) | 54 (62.8) | ||||
| San Pedro | 80 (83.3) | 43 (44.8) | 52 (54.2) | ||||
| Season | Rainy | 109 (84.5) | 0.193 | 82 (63.6) | < 0.000 | 86 (66.7) | 0.001 |
| Dry | 73 (77.7) | 37 (39.4) | 41 (43.6) | ||||
| Clinical status | Symptomatic | 79 (79.8) | 0.532 | 50 (50.5) | 0.445 | 63 (63.6) | 0.072 |
| Asymptomatic | 103 (83.1) | 69 (55/6) | 64 (51/6) | ||||
| Parasitemia (trophozoites/µl) | <100 | 35 (67.3) | 0.001 | 23 (44.2) | 0.005 | 26 (50) | 0.007 |
| 101–500 | 34 (72.3) | 18 (38.3) | 22 (46.8) | ||||
| 501–1000 | 19 (90.5) | 9 (42.9) | 9 (42.9) | ||||
| 1001–2000 | 28 (87.5) | 20 (62.5) | 17 (53.1) | ||||
| >2000 | 66 (93) | 49 (69) | 53 (74.6) | ||||
Table 2.
Distribution of the different allelic families of Pfmsp2 gene by site, season, clinical status and parasitemia.
| Variables | 3D7 n (%) | p-value | FC27 n (%) | p-value | |
|---|---|---|---|---|---|
| Site | Grand-Bassam | 36 (87.8) | 0.534 | 24 (58.5) | 0.002 |
| Abengourou | 74 (86) | 73 (84.9) | |||
| San Pedro | 78 (81.3) | 64 (66.7) | |||
| Season | Rainy | 107 (82.9) | 0.513 | 98 (76) | 0.141 |
| Dry | 81 (86.2) | 63 (67) | |||
| Clinical status | Symptomatic | 89 (89.9) | 0.04 | 71 (71.7) | 0.886 |
| Asymptomatic | 99 (79.8) | 90 (72.6) | |||
| Parasitemia (trophozoites/µl) | <100 | 42 (80.8) | 0.067 | 27 (51.9) | 0.000 |
| 101–500 | 35 (74.5) | 28 (59.6) | |||
| 501–1000 | 19 (90.5) | 15 (71.4) | |||
| 1001–2000 | 26 (81.3) | 31 (96.9) | |||
| >2000 | 66 (93) | 60 (84.5) | |||
Allelic families diversity
The distribution of the alleles of the Pfmsp1 and Pfmsp2 genes varied widely depending on the site. The alleles identified were classified according to their size in base pairs (bp). A total of 65 individual alleles were identified in the two genes and three sites during the two seasons in all subjects. Fourteen alleles K1 (180–900 bp), 13 Mad20 (180–800 bp) alleles and 11 alleles RO33 (160–900 bp) was found for pfmsp1 gene. Also, 15 3D7 (180–900 bp) alleles and 12 alleles FC27 (250–800 bp) was detected for pfmsp2 gene.
Overall, the allelic families K1, Mad20, RO33, 3D7 and FC27 had 1 to 4 alleles each. In symptomatic subjects, Mad20 and RO33 families had 1 to 3 alleles each while K1, 3D7 and FC27 families had 1 to 4 alleles each. However, in asymptomatic subjects each allele family had 1 to 4 alleles. During the rainy season, each allelic family had 1 to 4 alleles. In the dry season, RO33 showed only one allele, while K1 and Mad20 showed 1 to 2 alleles and the 3D7 and FC27 families had up to 4 alleles. The number of parasitic genotypes per subject varied from 1 to 6 in both asymptomatic and symptomatic patients in Grand-Bassam. In Abengourou, it was from 1 to 9 among the asymptomatic subjects and from 1 to 8 among the symptomatic ones. However, in San Pedro, it was 1to 4 among both asymptomatic and symptomatic subjects.
Prevalence of multiple infections (MI or polyclonality) and complexity of infection (COI)
The complexity of the infection showed a statistically significant difference between the 3 study sites for Pfmsp1 and Pfmsp2 (p < 0.0001). COI did not show any significant difference between seasons, but also in clinical status Pfmsp2. However, there was no significant difference between the complexity of infections obtained in asymptomatic and symptomatic patients for Pfmsp1 and Pfmsp2 (p = 0.9061). In addition, the results showed that the complexity of infection for the Pfmsp1 and Pfmsp2 allelic families increased significantly with parasitemia from 2.2 (± 0.1571) for <100 trophozoites/µl blood parasitemia to 4.2 (± 0.2272) for >2000 trophozoites/µl blood parasitemia (p < 0.0001) (Table 3).
Table 3.
COI by site, season, clinical status and parasitemia.
| Variables |
Pfmsp1 |
Pfmsp2 |
Pfmsp1 + Pfmsp2 |
||||
|---|---|---|---|---|---|---|---|
| COI (SD) | p-value | COI (SD) | p-value | COI (SD) | p-value | ||
| Site | Grand-Bassam | 2.024 (±0.2554) | < 0.0001 | 2.293 (±0.2160) | < 0.0001 | 2.902 (±0.2562) | < 0.0001 |
| Abengourou | 3.244 (±0.2508) | 3.081 (± 0.1472) | 4.047 (± 0.2181) | ||||
| San Pedro | 2.042 (± 0.09473) | 2.052 (± 0.1206) | 2.469 (± 0.1098) | ||||
| Season | Rainy | 3.023 (± 0.1827) | < 0.0001 | 2.465 (± 0.1175) | 0.7204 | 3.411 (± 0.1746) | 0.0109 |
| Dry | 1.787 (± 0.1024) | 2.532 (± 0.1472) | 2.809 (± 0.1347) | ||||
| Clinical status | Symptomatic | 2.626 (± 0.1892) | 0.3612 | 2.455 (± 0.1270) | 0.7073 | 3.141 (± 0.1833) | 0.9061 |
| Asymptomatic | 2.403 (± 0.1570) | 2.524 (± 0.1308) | 3.169 (± 0.1527) | ||||
| Parasitemia (trophozoites/µl) | <100 | 1.769 (± 0.1443) | < 0.0001 | 1.865 (± 0.1716) | < 0.0001 | 2.173 (± 0.1571) | < 0.0001 |
| 101–500 | 1.830 (± 0.1500) | 2.043 (± 0.1824) | 2.383 (± 0.1762) | ||||
| 501–1000 | 1.905 (± 0.2479) | 2.571 (± 0.2723) | 2.810 (± 0.2727) | ||||
| 1001–2000 | 2.875 (± 0.3668) | 2.813 (± 0.2352) | 3.844 (± 0.3016) | ||||
| >2000 | 3.493 (± 0.2584) | 3.085 (± 0.1576) | 4.183 (± 0.2272) | ||||
Overall, the frequency of MIs was significantly higher in Abengourou than in Grand-Bassam and San Pedro for Pfmsp1 and Pfmsp2 p = 0.018 and p = 0.031). No difference was observed between the frequency of MIs obtained in asymptomatic and symptomatic patients for either gene, while there was a significant difference during the rainy season compared to the dry season for Pfmsp1 (p = 0.017). On the other hand, a positive correlation was found between the frequency of MIs and parasitemia (p = 0.009) (Table 4).
Table 4.
Prevalence of multiple infections (MIs) Pfmsp1 and Pfmsp2 by site, season, clinical status and parasitemia.
| Variables | Pfmsp1 MIs n (%) | p-value | Pfmsp2 MIs n (%) | p-value | |
|---|---|---|---|---|---|
| Site | Grand-Bassam | 20 (48.8) | 0.018 | 29 (70.7) | 0.031 |
| Abengourou | 66 (76.7) | 72 (83.7) | |||
| SanPedro | 68 (70.8) | 60 (62.5) | |||
| Season | Rainy | 97 (75.2) | 0.017 | 92 (71.3) | 0.841 |
| Dry | 57 (60.6) | 69 (73.4) | |||
| Clinical status | Symptomatic | 72 (72.7) | 0.517 | 72 (72.7) | 0.070 |
| Asymptomatic | 154 (69.1) | 89 (71.8) | |||
| Parasitemia (trophozoites/µl) | <100 | 28 (53.8) | 0.009 | 26 (50) | 0.000 |
| 101–500 | 28 (59.6) | 30 (63.8) | |||
| 501–1000 | 13 (61.9) | 17 (81) | |||
| 1001–2000 | 23 (71.9) | 25 (78.1) | |||
| >2000 | 62 (87.3) | 63 (88.7) | |||
Discussion
P. falciparum is responsible for almost all malaria cases diagnosed in this country. This parasite has a complex genetic polymorphism that confers the ability to develop multidrug resistance or circumvent vaccines. The emergence and spread of resistant strains are hampering malaria control efforts and the situation is worsening in some regions [10,11]. The spread of drug resistance is due to gene flow and the size of the population structure of P. falciparum [12–14]. A better understanding of the population structure of P. falciparum genotypes can be an important element in adjusting control strategies for this parasites in the country [10].
In addition, genetic diversity determines the intensity of P. falciparum transmission, providing baseline data for any antimalarial drug efficacy trial and the ability to implement vaccine-based control strategies. Surface proteins 1 and 2 of merozoites (MSP1 and MSP2) are widely used to study the allelic diversity and frequency of P. falciparum, which are most often correlated with the level of transmission in the study area. The two loci were also introduced as a discriminatory tool to distinguish between new and re-emerging infections [15]. Since the parasite reservoir consists of symptomatic and asymptomatic individuals, we have evaluated the genetic diversity of P. falciparum isolates from both types of individuals. We have also estimated this diversity over time, that is, during the rainy and dry seasons, since during the latter the intensity of parasite transmission is low. This contrasts with the plasmodial prevalence of our study in the dry season. Several studies have demonstrated the transmission potential of low-density malaria infections [16–18], although their contribution to overall parasite transmission is debatable [19].
In general, the results of our study showed that the different alleles belonging to the three families K1, Mad20 and RO33 of the Pfmsp1 gene and to the two FC27 and 3D7 families of the Pfmsp2 gene were qualitatively present in the three epidemiological study sites. These alleles were found in both rural and urban areas, in the rainy season and in the dry season, as well as in symptomatic and asymptomatic individuals. Thirty-eight Pfmsp1 and 27 Pfmsp2 alleles were found. Similar results were observed by Soulama and al. in Burkina-Faso where 41 Pfmsp1 and 29 Pfmsp2 alleles were reported [20] . Moreover, it was observed in 2013 and 2014 in Côte d’Ivoire 28 Pfmsp1 and 21 Pfmsp2 alleles as well as in Gabon in Libreville and Melen in 2011 39 Pfmsp1 and 27 Pfmsp2 alleles [3]. However, in Gabon it was reported 25 Pfmsp1 and 42 Pfmsp2 alleles in Owendo (in 2008) and Libreville (in 2011) [9] while other authors noticed 33 Pfmsp1 alleles in rural and urban areas of that country including Owendo and Oyem between February 2008 and January 2009 [21]. In Benin in the coastal departments and Ouémé from May to August 2011, 25 Pfmsp1 and 28 Pfmsp2 alleles were observed [22] while in Cameroon in the localities of the eastern slope of Mount Cameroon, 34 alleles of the Pfmsp1 family were reported [23]. This variability indicates that the two genes Pfmsp1 and Pfmsp2 are highly polymorphic in P. falciparum strains, particularly in Africa with some variations linked to the malaria epidemiological areas.
Various studies have shown that the genetic diversity of P. falciparum populations varies according to the intensity of transmission in malaria-endemic areas and it is higher in hyperendemic areas than in low endemic areas [24]. Therefore, the genetic characteristics of parasites can reflect the dynamics of parasitic transmission. However, the basis of genetic diversity is the recombination that occurs during the sexual stage of the parasite’s life cycle, so one of the factors to be taken into account will be the infection rate of the vector. These high infection rates would then increase the number of possible sexual recombinations in the mosquito and, consequently, promote the circulation of a greater variety of allelic forms. In addition, the higher the transmission, the higher the recombination frequency [24–26]. Our results showed that the prevalence of Pfmsp1 allele family is significantly higher during the rainy season, particularly for the Mad20 and RO33 alleles. On the other hand, on the Island of Annobon (Equatorial Guinea) it was Pfmsp2 that showed great variability in the rainy season [25]. In contrary, the clinical status of the subject had no influence on the prevalence of Pfmsp1 block 2 alleles. Several studies have investigated the association of Pfmsp1 and Pfmsp2 with the clinical severity of the disease. For example, a study conducted to examine the relationship between the genetic diversity of P. falciparum block 2 Pfmsp1 and the clinical severity of malaria in Nigerian children showed that the presence of the K1 and MAD20 alleles was significantly associated with asymptomatic malaria and therefore would reduce the risk of developing symptomatic malaria [27]. However, it is possible that the frequency of exposure to these allelic families is also a factor associated with clinical severity. Indeed, it was reported that the association of a specific allele of Pfmsp1 (K1) with a specific var gene (var-D) was over-represented in patients with severe malaria compared to uncomplicated malaria, and this genotype combination was consistently observed [28]. In the study data series, the 3D7 allele showed a significant difference with the clinical status of the subject. However, the FC27 allele has not been associated with clinical status. Moreover, in Sudan and Uganda, authors did not find any significant difference between the frequency of 2 gene alleles detected in subjects with severe malaria and those detected in subjects with uncomplicated malaria. Therefore, no particular genotype or allelic family of Pfmsp1or Pfmsp2 was associated with severity of malaria in Uganda and sudan [29, 30].
Thus, studies on the association between genotypes and disease severity should be extended to all disease categories, including asymptomatic malaria. In addition, genetic diversity should be monitored longitudinally to ensure that such clinical associations are maintained and not biased by cross sectional selection of isolates. There are conflicting reports regarding the differential distribution of alleles in particular genes (such as Pfmsp2) according to clinical condition [31].
The FC27 Pfmsp2 genotype was twice as likely to be found in symptomatic cases as in asymptomatic cases [32], providing evidence that specific variants of Pfmsp2 can be associated with malaria morbidity. A similar association has been reported in Papua New Guinea, where the FC27 allele was linked to an increase in disease severity, but this study did not take into account asymptomatic cases [27]. Conversely, another study reported that there was no association between Pfmsp2 FC27 or 3D7 alleles and malaria symptoms [33]. However, further longitudinal studies in different geographical contexts with standardized collection and genotyping methods will be needed to clarify these results.
The different allelic families were very polymorphic in general. However, K1 was the most polymorphic allelic family for Pfmsp1 with 14 alleles in sizes ranging from 180 bp to 900 bp. On the other hand, at the level of the Pfmsp2 gene, 3D7 was the most diversified allele family with 15 alleles classified from 180 bp to 900 bp. Overall, the RO33 allele was the least polymorphic with 11 subfamilies classified from 160 to 900 bp. Similar results were found by Mwingira and al in 2011 and Soulama and al in 2009 [20,34]. However, it was previously detected in Côte d’Ivoire 13 FC27 alleles, 15 K1 alleles and 10 RO33 alleles in Gabon against 9 FC27 alleles, 12 K1 alleles and 8 RO33 alleles in Côte d’Ivoire. However, other authors have found only one allele for the RO33 family [34,35].
The alleles K1200pb, K1250pb, K1180pb, Mad20200pb, RO33160pb, 3D7300pb, 3D7400pb, 3D7250pb FC27400pb and FC27500pb were most predominant at all three sites during both seasons in asymptomatic and symptomatic patients. Among these detected genotypes, some have already been described in various malaria endemic areas, but with different frequencies. In addition, K1200pb and Mad20200pb have been prevalent in other studies in Gabon [9], in Benin [22], Cameroun [23] and in Côte d’Ivoire [3]. Indeed, genetic polymorphism is strongly affected by the level of endemicity. With high endemicity, multi-clonal infections and crosses are predominant [36]. However, the similarity of these alleles should be confirmed by sequence analysis [9]. Indeed, it has been reported that alleles of the same size and allele family are not identical [37]. Thus, one of the limitations of this study is the measurement of the genetic diversity of parasitic populations by the use of markers as a function of DNA band size. However, fragments of the same size (not identical) can be considered identical, giving a wrong impression of similarity. Within allele families, alleles of the same size may have different amino acid patterns. Nevertheless, markers such as the Pfmsp1 and Pfmsp2 genes are good markers of polymorphism and can be successfully used to characterize populations of genetic strains of P. falciparum. Allelic variations as well as the presence of common alleles should be better explored by sequencing P. falciparum isolates.
Various studies have shown that COI is used as an indicator for malaria transmission. Thus, a higher multiplicity of infections is observed in areas where transmission is particularly high [24–26,38]. From the above, it is possible to assume that that malaria transmission was much higher in Abengourou compared to San Pedro and Grand-Bassam. It has been shown in previous studies that in most hyperendemic malaria areas, COI is directly related to the intensity of transmission [39,40]. In addition, according to some authors, areas of high or intense malaria transmission are generally characterized by high parasitic diversity, and infected individuals often carry several parasite genotypes [41,42].
There was not any association between clinical status and COI. This could suggest that the genotypes found were common regardless of the severity of the malaria infestation. In contrary, the high parasitemia was linked to the heterogeneity of Plasmodium strains as previously reported [9,21,43]. However, some authors did not find any association between parasitemia and COI [15,44,45].
Overall, the frequency of MIs was significantly higher in Abengourou than in Grand-Bassam and San Pedro for Pfmsp1 and Pfmsp2. In addition, the prevalence of MIs was higher in the rainy season and in the dry season) respectively for Pfmsp1 and Pfmsp2. Indeed, the level of transmission can influence the occurrence of mixed infections; polyclonal infections being more frequent in areas of intense malaria transmission [21,46].
High level of polyclonal infections has been found in Abengourou. These data suggest an intense and persistent transmission of malaria in this locality. From 2015 to 2016, the transmission of P. falciparum malaria in the sites of our study was as follows: Grand-Bassam (Abidjan), 58 infected bites/person/year; Abengourou, urban, 84 bites/person/year and rural, 469 bites/person/year; San Pedro, urban, 09 bites/person/year and rural, bites/person/year (National Malaria Control Program, unpublished data). This could be due to the lack of impact of malaria control strategies or insufficient deployment. In addition, analysis of our results shows that the frequency of MIs increased with parasitemia for both Pfmsp1 and Pfmsp2 and Pfmsp1 + Pfmsp2 genes. These observations are consistent with those made by Mayengue et al. in Brazzaville, Democratic Republic of Congo, who found a statistically significant correlation between parasitemia and the number of genotypes [42].
Conclusion
This study provides information on the seasonal dynamics of genetic diversity of P. falciparum populations in three health districts of Côte d’Ivoire. They are interesting for the monitoring and control of P. falciparum malaria, the most deadly parasitic diseases. There was no link between the variation in P. falciparum populations, season, locality, the type of area and the clinical status. Genotypes of the populations of P. falciparum found in asymptomatic subjects were not different from those found in the symptomatic subjects. However the complexity of the infection was a function of the intensity of malaria transmission in the locality. Further studies combining the entomological inoculation rate and the genetic diversity of P. falciparum will shed light on our understanding of whether patients are infected with multiple strains contained in a single mosquito bite, or are multiply infected with single strains each harboring a different Pfmsp allele.
Acknowledgments
We would gratefully thank all the staff of the Centre de Recherche et de Lutte contre le Paludisme (CRLP) of the National Institute of Public Health of Côte d’Ivoire (INSP) in particular: DABLE Marius, TANO Konan Dominique, N’CHO Moussan, AKA Bekoin Ferdinand and COULIBALY Abdoul Karim for their technical support. We also thank the inspectors of primary education, teachers and parents and pupils of Grand-Bassam, Abengourou and San Pedro for their participation in this study. We sincerely thank Dr SOULAMA Issiaka for his technical support, AKENJI Blaise, and KOUROUMA Raissa for helping us to correct the English translation of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Financial support
This work was supported through by National Institute of Public Health of Côte d’Ivoire and the DELTAS Africa Initiative [DELGEME grant 107,740/Z/15/Z]. The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS)’s Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust [DELGEME grant 107,740/Z/15/Z] and the UK government. The views expressed in this publication are those of the author(s) and not necessarily those of AAS, NEPAD Agency, Wellcome Trust or the UK government.
References
- [1].OMS , 2018. World malaria report 2018.
- [2].Koffi D, Varela M-L, Loucoubar C, et al. Longitudinal analysis of antibody responses in symptomatic malaria cases do not mirror parasite transmission in peri-urban area of cote d’Ivoire between 2010 and 2013. PloS One. 2017;12:e0172899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Yavo W, Konaté A, Mawili-Mboumba DP, et al. Genetic Polymorphism of msp1 and msp2 in Plasmodium falciparum Isolates from Côte d’Ivoire versus Gabon. J Parasitol Res. 2016;2016:3074803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gardner MJ, Shallom SJ, Carlton JM, et al. Sequence of Plasmodium falciparum chromosomes 2, 10, 11 and 14. Nature. 2002;419:531–534. [DOI] [PubMed] [Google Scholar]
- [5].Gunawardena S, Karunaweera ND.. Advances in genetics and genomics: use and limitations in achieving malaria elimination goals. Pathog Glob Health. 2015;109:123–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yapi RB, Hürlimann E, Houngbedji CA, et al. Infection and Co-infection with helminths and plasmodium among school children in Côte d’Ivoire: results from a national cross-sectional survey. PLoS Negl Trop Dis. 2014;8:e2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Viriyakosol S, Siripoon N, Petcharapirat C, et al. Genotyping of Plasmodium falciparum isolates by the polymerase chain reaction and potential uses in epidemiological studies. Bull World Health Organ. 1995;73:11. [PMC free article] [PubMed] [Google Scholar]
- [8].Snounou G. Genotyping of Plasmodium spp. Nested PCR. Methods Mol Med. 2002;72:103–116. [DOI] [PubMed] [Google Scholar]
- [9].Ndong Ngomo JM, M’Bondoukwe NP, Yavo W, et al. Spatial and temporal distribution of Pfmsp1 and Pfmsp2 alleles and genetic profile change of Plasmodium falciparum populations in Gabon. Acta Trop. 2018;178:27–33. [DOI] [PubMed] [Google Scholar]
- [10].Bogreau H, Renaud F, Bouchiba H, et al. Genetic diversity and structure of African Plasmodium falciparum populations in urban and rural areas. Am J Trop Med Hyg. 2006;74:953–959. [PubMed] [Google Scholar]
- [11].Driss A, Hibbert JM, Wilson NO, et al. Genetic polymorphisms linked to susceptibility to malaria. Malar J. 2011;10:271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dwivedi A, Khim N, Reynes C, et al. Plasmodium falciparum parasite population structure and gene flow associated to anti-malarial drugs resistance in Cambodia. Malar J. 2016;15:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Aydemir O, Janko M, Hathaway NJ, et al. Drug-resistance and population structure of plasmodium falciparum across the democratic republic of congo using high-throughput molecular inversion probes. J Infect Dis. 2018;218:946–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mulenge FM, Hunja CW, Magiri E, et al. Genetic diversity and population structure of plasmodium falciparum in Lake Victoria islands, a region of intense transmission. Am J Trop Med Hyg. 2016;95:1077–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Atroosh WM, Al-Mekhlafi HM, Mahdy MA, et al. Genetic diversity of Plasmodium falciparum isolates from Pahang, Malaysia based on MSP-1 and MSP-2 genes. Parasit Vectors. 2011;4:233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mawili-Mboumba DP, Nikiéma R, Bouyou-Akotet MK, et al. Sub-microscopic gametocyte carriage in febrile children living in different areas of Gabon. Malar J. 2013;12:375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bousema T, Okell L, Felger I, et al. Asymptomatic malaria infections: detectability, transmissibility and public health relevance. Nat Rev Microbiol. 2014;12:833–840. [DOI] [PubMed] [Google Scholar]
- [18].Morris U, Xu W, Msellem MI, et al. Characterising temporal trends in asymptomatic Plasmodium infections and transporter polymorphisms during transition from high to low transmission in Zanzibar, 2005–2013. Infect Genet Evol. 2015;33:110–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lin JT, Saunders DL, Meshnick SR. The role of submicroscopic parasitemia in malaria transmission: what is the evidence? Trends Parasitol. 2014;30:183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Soulama I, Nébié I, Ouédraogo A, et al. Plasmodium falciparum genotypes diversity in symptomatic malaria of children living in an urban and a rural setting in Burkina Faso. Malar J. 2009;8:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mawili-Mboumba DP, Mbondoukwe N, Adande E, et al. Allelic Diversity of MSP1 Gene in Plasmodium falciparum from Rural and Urban Areas of Gabon. Korean J Parasitol. 2015;53:413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ogouyèmi-Hounto A, Gazard DK, Ndam N, et al. Genetic polymorphism of merozoite surface protein-1 and merozoite surface protein-2 in Plasmodium falciparum isolates from children in South of Benin. Parasite. 2013;20:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Apinjoh TO, Tata RB, Anchang-Kimbi JK, et al. Plasmodium falciparum merozoite surface protein 1 block 2 gene polymorphism in field isolates along the slope of mount Cameroon: a cross–sectional study. BMC Infect Dis. 2015;15 DOI: 10.1186/s12879-015-1066-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mohammed H, Kassa M, Mekete K, et al. Genetic diversity of the msp-1, msp-2, and glurp genes of Plasmodium falciparum isolates in Northwest Ethiopia. Malar J. 2018;17 DOI: 10.1186/s12936-018-2540-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cano J, Berzosa P, de Lucio A, et al. Transmission of malaria and genotypic variability of Plasmodium falciparum on the Island of Annobon (Equatorial Guinea). Malar J. 2007;6 DOI: 10.1186/1475-2875-6-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Huang B, Tuo F, Liang Y, et al. Temporal changes in genetic diversity of msp-1, msp-2, and msp-3 in Plasmodium falciparum isolates from Grande Comore Island after introduction of ACT. Malar J. 2018;17 DOI: 10.1186/s12936-018-2227-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Laishram DD, Sutton PL, Nanda N, et al. The complexities of malaria disease manifestations with a focus on asymptomatic malaria. Malar J. 2012;11:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Legrand E, Yrinesi J, Ekala M-T, et al. Discordant temporal evolution of Pfcrt and Pfmdr1 genotypes and plasmodium falciparum in vitro drug susceptibility to 4-aminoquinolines after drug policy change in French Guiana. Antimicrob Agents Chemother. 2012;56:1382–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kiwuwa MS, Ribacke U, Moll K, et al. Genetic diversity of Plasmodium falciparum infections in mild and severe malaria of children from Kampala, Uganda. Parasitol Res. 2013;112:1691–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hamid MAM, Elamin AF, Albsheer MMA, et al. Multiplicity of infection and genetic diversity of Plasmodium falciparum isolates from patients with uncomplicated and severe malaria in Gezira State, Sudan. Parasit Vectors. 2016;9 DOI: 10.1186/s13071-016-1641-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Amodu OK, Oyedeji SI, Ntoumi F, et al. Complexity of the msp2 locus and the severity of childhood malaria, in south-western Nigeria. Ann Trop Med Parasitol. 2008;102:95–102. [DOI] [PubMed] [Google Scholar]
- [32].Chaorattanakawee S, Nuchnoi P, Hananantachai H, et al. Sequence variation in Plasmodium falciparum merozoite surface protein-2 is associated with virulence causing severe and cerebral malaria. Plos One. 2018;13:e0190418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Cortés A, Mellombo M, Benet A, et al. Plasmodium falciparum: distribution of msp2 genotypes among symptomatic and asymptomatic individuals from the Wosera region of Papua New Guinea. Exp Parasitol. 2004;106:22–29. [DOI] [PubMed] [Google Scholar]
- [34].Mwingira F, Nkwengulila G, Schoepflin S, et al. Plasmodium falciparum msp1, msp2 and glurp allele frequency and diversity in sub-Saharan Africa. Malar J. 2011;10:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gosi P, Lanteri CA, Tyner SD, et al. Evaluation of parasite subpopulations and genetic diversity of the msp1, msp2 and glurp genes during and following artesunate monotherapy treatment of Plasmodium falciparum malaria in Western Cambodia. Malar J. 2013;12:403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bouyou-Akotet MK, M’Bondoukwé NP, Mawili-Mboumba DP. Genetic polymorphism of merozoite surface protein-1 in Plasmodium falciparum isolates from patients with mild to severe malaria in Libreville, Gabon. Parasite. 2015;22:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Takala SL, Escalante AA, Branch OH, et al. Genetic diversity in the Block 2 region of the merozoite surface protein 1 (MSP-1) of Plasmodium falciparum: additional complexity and selection and convergence in fragment size polymorphism. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis. 2006;6:417–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kiwanuka GN, 2009. Genetic diversity in Plasmodium falciparum merozoite surface protein 1 and 2 coding genes and its implications in malaria epidemiology: a review of published studies from 1997–2007. J. Vector Borne Dis. 46, 1–12. [PubMed] [Google Scholar]
- [39].Niang M, Thiam LG, Loucoubar C, et al. Spatio-temporal analysis of the genetic diversity and complexity of Plasmodium falciparum infections in Kedougou, southeastern Senegal. Parasit Vectors. 2017;10 DOI: 10.1186/s13071-017-1976-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Arzika I, Lamine MM, Mahamadou A, et al. Etude du polymorphisme genetique des souches de Plasmodium falciparum au Niger. Rev CAMES SANTE. 2017;5:7. [Google Scholar]
- [41].Ghanchi NK, Mårtensson A, Ursing J, et al. Genetic diversity among Plasmodium falciparum field isolates in Pakistan measured with PCR genotyping of the merozoite surface protein 1 and 2. Malar J. 2010;9:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Mayengue P, Ndounga M, Malonga F, et al. Genetic polymorphism of merozoite surface protein-1 and merozoite surface protein-2 in Plasmodium falciparum isolates from Brazzaville, Republic of Congo. Malar J. 2011;10:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kateera F, Nsobya SL, Tukwasibwe S, et al. Malaria case clinical profiles and Plasmodium falciparum parasite genetic diversity: a cross sectional survey at two sites of different malaria transmission intensities in Rwanda. Malar J. 2016;15 DOI: 10.1186/s12936-016-1287-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Mohammed H, Kassa M, Assefa A, et al. Genetic polymorphism of merozoite surface protein-2 (MSP-2) in Plasmodium falciparum isolates from Pawe District, North West Ethiopia. Plos One. 2017;12:e0177559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Mohammed H, Mindaye T, Belayneh M, et al. Genetic diversity of Plasmodium falciparum isolates based on MSP-1 and MSP-2 genes from Kolla-Shele area, Arbaminch Zuria District, southwest Ethiopia. Malar J. 2015;14:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ahmedou Salem MSO, Ndiaye M, OuldAbdallahi M, et al. Polymorphism of the merozoite surface protein-1 block 2 region in Plasmodium falciparum isolates from Mauritania. Malar J. 2014;13:26. [DOI] [PMC free article] [PubMed] [Google Scholar]


