Abstract
The capacity for tumor cells to metastasize efficiently is directly linked to their ability to colonize secondary sites. Here we identify Six2, a developmental transcription factor, as a critical regulator of a breast cancer stem cell program that enables metastatic colonization. In several triple-negative breast cancer (TNBC) models, Six2 enhanced the expression of genes associated with embryonic stem cell programs. Six2 directly bound the sox2 srr2 enhancer, promoting sox2 expression and downstream expression of nanog, which are both key pluripotency factors. Regulation of Sox2 by Six2 enhanced cancer stem cell properties and increased metastatic colonization. Six2 and Sox2 expression correlated highly in breast cancers including TNBC, where a Six2 expression signature was predictive of metastatic burden and poor clinical outcome. Our findings demonstrate that a SIX2/SOX2 axis is required for efficient metastatic colonization, underscoring a key role for stemness factors in outgrowth at secondary sites.
Keywords: Metastasis; Metastatic colonization; Stemness; Six2, Sox2; Nanog
Introduction
The molecular mechanisms behind the metastatic progression of breast cancer remain poorly understood. Although advances in therapeutics are extending the lives of patients, metastatic spread remains the major cause of death. Understanding the mechanisms driving metastasis, particularly establishment and maintenance of metastasis, could lead to development of more effective therapeutics and increased survival.
The metastatic cascade is a multistep process involving escape from the primary tumor, dissemination, extravasation, survival/establishment and colonization. The rate-limiting step of metastasis is outgrowth at the secondary site, as newly arriving tumor cells must survive a harsh new microenvironment to establish and maintain proliferation at secondary sites[1]. Outgrowth of metastatic cells is complicated by the fact that these sites initially lack sufficient vasculature to provide nutrients needed by the cells[2]. Because the microenvironment at the secondary site presents challenges for newly arriving tumor cells, only a small percentage of these cells will go on to develop overt metastases[3]. Despite increased evidence around the importance of metastatic cell outgrowth at secondary sites, the vast majority of metastasis studies continue to focus on identifying molecular mechanisms that drive the earlier stages of metastasis, such as migration and invasion. However, recent studies have established that tumor cells exit the primary site at very early stages of the diseases[4]. Thus, targeting early stages of metastasis may be less effective, because by the time the primary tumor is diagnosed, many cancer cells would have likely already left the primary site. Since the later stages of metastasis are less efficient, and often not completed at the time of diagnosis, targeting this aspect of the metastatic cascade may be more effective[1].
Recently, advancements in imaging, mouse models and genomic and single cell genomics analyses have allowed for more accurate modeling of the later stages of metastasis. This technological progress has led to the identification of several proteins that specifically alter metastatic colonization. For example, tumor cells can indirectly activate osteoprotegerin (OPG), allowing for bone degradation and release of growth factors into the bone microenvironment, which provides signals to metastatic cells to proliferate[5]. Additionally, tumor cell-derived tenascin C (TNC) promotes the survival and outgrowth of lung metastases through regulation of stem cell signaling components musashi homolog 1 (MSI1) and leucine-rich repeat–containing G protein–coupled receptor 5 (LGR5)[6]. However, even with these promising discoveries, metastatic colonization remains the major cause of mortality in breast cancer patients. In part, this continued mortality can be attributed to a dearth of targeted therapies inhibiting metastatic colonization and/or the outgrowth of metastatic clones, which are often resistant to the chemotherapy regimens to which the primary tumor responded[1].
SIX2 is a homeodomain-containing transcription factor that plays a key developmental role in the kidney. KO of six2 in mice demonstrates a critical role for the gene in maintenance of the mesenchymal phenotype, self-renewal, and survival of progenitor cells that will eventually give rise to all epithelial cell types of the developing nephron[7]. Not surprisingly then, six2 loss in mice leads to postnatal lethality due to defects caused by premature differentiation of cells in the kidney as well as kidney hypoplasia[7]. Importantly, mutations in SIX2 have been found in humans with congenital abnormalities of the kidney, such as renal hypodysplasia[7], underscoring an important role for this gene in human kidney development.
Tumor cells often hijack developmental processes to promote metastasis[8]. Recent studies have thus begun to examine the role of SIX2 in tumor progression. Overexpression of Six2, as compared to normal tissue, has been observed in esophageal and lung carcinomas[9]. SIX2 also plays a critical role in tumor cell invasion and drug resistance in colorectal cancer[10], and regulates proliferation and epithelial-to-mesenchymal transition (EMT) in hepatocellular cancer[11]. Most recently, whole exome sequencing of Wilms tumor patient samples identified that almost a fifth (18.1%) of patients with the most aggressive blastemal subtype of Wilms tumor possess a gain-of-function mutation (Q177R) in the homeodomain of SIX1 or SIX2 that correlates with high proliferation[12]. This mutation has been shown to enhance the SIX1-mediated Warburg effect, suggesting that a similar function could be attributed to SIX2[13]. Importantly, SIX2 marks the cancer stem cell population in Wilms tumor[14], suggesting conservation of its developmental function in cancer. Collectively, these data support a critical role for SIX2 across multiple different cancer types and underscore the need to better understand the molecular mechanisms by which SIX2 promotes tumor progression and metastasis.
Previous studies from our lab have demonstrated that Six2 is critical for late-stage metastasis in triple-negative mouse mammary carcinoma models[9]. Knockdown (KD) of six2 in the triple-negative, metastatic 66cl4 mouse mammary carcinoma line results in decreased metastatic burden when injected orthotopically[9]. Interestingly, six2 KD does not affect primary tumor growth or tumor-associated lymphangiogenesis, contributors to early stage metastasis that are known to be regulated by a related Six family member, Six1[8]. Conversely, Six2 overexpression (OE) in the 4T07 mammary carcinoma cell line results in increased lung metastases after tail vein injection in BALB/c mice when compared to control cells[9]. This result is of particular interest, as the 4T07 cells, which are syngeneic to the 66cl4 cells, can reach secondary sites but are unable to efficiently colonize them[15]. These data demonstrate that Six2 promotes the ability of cancer cells to colonize secondary sites. Further analysis demonstrated that Six2 OE leads to a decrease in E-cadherin (cdh1) expression, suggesting that Six2 may mediate at least a partial epithelial-to-mesenchymal transition[9]. Importantly, restoration of E-cadherin in 4T07 cells downstream of Six2 overexpression inhibited metastasis induced by Six2[9]. However, KD of six2 inhibited metastasis of 66cl4 cells without restoration of E-cadherin expression, likely due to epigenetic silencing of cdh1[9]. Taken together, these data suggest that cdh1 repression is necessary downstream of Six2 to mediate metastasis, but that additional genes regulated by Six2 are also required.
Herein, we demonstrate that SIX2 plays a critical role in metastatic colonization by promoting stemness-associated properties in triple-negative breast cancer (TNBC) and TN mouse mammary carcinoma cells. We show that Six2 is more highly expressed in patients with TNBC as compared to other subtypes, and that it regulates metastasis not only of mouse mammary carcinoma lines, but also in a human TNBC model. Using three different models of TNBC, we demonstrate that SIX2 enhances cancer stem-cell associated phenotypes and regulates a genetic stem cell program. We identify Six2 as a direct transcriptional regulator of a master pluripotency factor, sox2, further resulting in upregulation of a second master pluripotency factor, nanog. Importantly, we show that Sox2 is the critical mediator downstream of Six2 in promoting stemness-associated phenotypes in vitro and late-stage metastasis in vivo. Finally, we demonstrate for the first time that Six2 and Sox2 expression positively correlate in human breast cancer, including TNBC, and that a Six2-mediated gene signature is associated with significantly shortened distant metastasis free survival as well as relapse and recurrence-free survival. Collectively, our data suggest that a novel SIX2/SOX2 axis may promote stem cell characteristics in newly arriving breast cancer cells at secondary sites, providing insight into the regulation of metastatic colonization and outgrowth.
Methods
Cell Lines and Culture
The 4T07 and 66cl4 mouse mammary carcinoma cells were generously provided by Dr. Fred Miller[15]. Stable Six2 overexpression (OE) and knockdown (KD) cell lines were generated and grown in media as described previously[9]. MDA-MB-231 cells have been cultured long-term in the Ford laboratory, and were grown in MEM/EBSS, FBS, HEPES, non-essential amino acids, insulin, pen/strep and L-glutamine. Authentication of the human cell line MDA-MB-231 was performed using STR analysis by the University of Colorado Cancer Center Tissue Culture Shared Resource in March of 2017. All cells were regularly monitored for mycoplasma contamination every 3 months and only mycoplasma free lines were used for studies. All cell lines were cultured at 37°C in HEPA-filtered humidified air in 5% CO2. Stable Six2 knockdown in the MDA-MB-231 cells was performed by transduction of two different lentiviral shRNAs, or with a non-targeting (NT) shRNA. After transduction, cells were selected for 1 week using 3ug/ml of puromycin to obtain stable KD of Six2. Six2 levels in KD and OE cells were examined using RT-qPCR and Western blot analysis. Transient KD of sox2 and Sox2 was performed using ON-TARGETplus SMARTpool siRNA constructs (Table S1). A non-targeting siRNA pool was used as a negative control. KD levels were assessed 72h post transfection via RT-qPCR.
RNA-sequencing
4T07 CTL and Six2 cells were plated at 3×105 cells/ml in 10cm plates and allowed to attach and grow for 48h. Cells were collected and lysed by removing media, rinsing with PBS and adding 1ml of TriReagent directly to the plate. After vortexing to ensure homogeneity, total RNA was isolated using the Zymo Research RNA Isolation kit (Light Labs, R2053). RNA samples were evaluated for purity and concentration using a spectrophotometer and submitted to the Genomics and Microarray Core Facility at the University of Colorado Denver Anschutz Medical Campus. Sequencing sample preparation was performed using standard Illumina HiSeq protocols and reagents. Directional mRNA Seq was performed using Illumina HiSeq 4000 HT Mode 1X150 cycles.
RNA-sequencing and Data Analysis
The read quality and mapping were done as described previously[16]. Significantly differentially expressed genes were determined after using DESeq2 in R with batch correction, and a significance cutoff of adjusted p value < 0.1. MA plots, PCA plots and heatmaps were generated using Matplolib from Python plotting library. To find overlap between significantly differentially expressed genes and stemness-associated gene signatures, DESeq genes were input into the StemChecker database (http://stemchecker.sysbiolab.eu). Significantly enriched gene signatures were determined using a Hypergeometric test and adjusted by Bonferroni correction. Heatmaps were generated by converting reads per kilobase per million (RPKM) values to z-scores and using ComplexHeatmap and Circularize library R packages to visualize z-score normalized gene expression. Genes were grouped by hierarchical clustering of z-score normalized gene expression. The RNA-seq data was deposited into the GEO repository with the accession number GSE123489.
GSEA
Gene-Set Enrichment Analysis (GSEA) was performed by utilizing the GSEA Preranked Module of the GenePattern Webserver[17]. All significantly expressed genes were ranked by adjusted Log2 fold change values (Six2 OE vs CTL) and tested against the C2: Curated gene sets and C5: Gene Ontology gene sets to test for enrichment.
Western Blot
Whole cell and nuclear lysates were isolated as done previously[9]. Protein concentration was determined using the Lowry Protein assay (BioRad). After gel electrophoresis of protein in 10% polyacrylamide gels, proteins were transferred to a polyvinylidene difluoride membrane at 100v for 1h 30min, blocked in 5% Milk-TBST and incubated at 4°C in primary antibodies overnight (see Table S1 for list of primary antibodies). Blots were imaged using either Film radiography or the OdysseyFc imaging system after HRP substrate chemiluminescence incubation and detection.
RT-qPCR
RNA was isolated using the Zymo Research RNA isolation kit (Light Labs, R2053). cDNA was made using the QuantaraBio cDNA synthesis kit (VWR Scientific, 95047–100). PCR was performed using the SYBR green master mix per manufacturer’s instructions and protocols (Qiagen, 330500). Primers used for this study are contained in Table S1 (IDT). Gene expression was normalized to peptidylpropyl isomerase B (cyclophilin B) (Ppib/ppib) mRNA expression and fold-change is relative to the CTL (control), NS (non-silencing), or NT (non-targeting) cells. Significance was determined by t-test or one-way anova on means ± SEM of triplicate samples for a representative experiment (n=3).
Tumorsphere Formation
Tumorsphere assays were performed as previously described[18]. Briefly, cells were trypsinized and plated at either 300, 1000 or 20,000 cells/well in 6-well, ultra-low attachment plates (Corning) in 2 mls of serum-free DMEM/F12 media (Hyclone), supplemented with 20ng/ml bFGF (BD Biosciences), 20ng/ml EGF (BD Biosciences), 4 μg/ml heparin (Sigma), penicillin-streptomycin (Hyclone), and B27 (Gibco) for all cell lines tested. 500 μl of media was added every 3–4 days. On day 7, all cells/spheres were collected, washed with PBS, digested using 0.05% trypsin in HBSS with 0.2g/L EDTA (Fisher), and single cells were plated as above to perform secondary tumorsphere assays. After an additional 7 days, tumorspheres were imaged at 4x magnification and counted by the Incucyte Zoom Live-Cell Analysis System.
Flow Cytometry
Flow cytometry analysis was done as previously described[18]. Briefly, cells were incubated for 30 min in either CD24-biotin/CD49f-APC or Epcam-FITC/CD44-APC and in the case of CD24, were additionally incubated in Strep-FITC or Strep-V450 secondary for 30 min (Table S1). Positive control ultracomp beads were also stained and used as a positive control for consistent gating. Flow cytometry was performed by submitting samples to the Flow Cytometry Core Facility at the University of Colorado Anschutz Medical campus or using a C6 Flow Cytometer. Single-stained compensation controls (UltraComp Beads, eBioscience) were stained with individual antibodies (EpCAM-FITC, CD44-APC, CD24-biotin-strep-FITC, CD24-biotin-strep-V450 or CD49f-APC) to define gating for each cell-surface marker. Gates were set at the lowest edge of fluorescence for the positive stain populations relative to the negative stain populations. Additionally, negative control (unstained) cell populations were checked to make sure they overlapped <1% with the single or double positive quadrants after gating with single-stained control beads. Flow cytometry analysis was performed using the FlowJo Analysis Software.
Animal Studies
Tail vein injections were performed in 6–8 week old female immunodeficient NOD-scid-gamma (for MDA-MB-231) and BALB/c mice (for 4T07) (Jackson Laboratories). For the MDA-MB-231 cells, 4 × 105 cells in 100ul of HBSS were injected. For the 4T07 cells, 1 × 105 cells in 100ul of DMEM medium were injected. Both the MDA-MB-231 and 4T07 cells were luciferase tagged to enable imaging of metastases in vivo. IVIS imaging was used to image the mice as described previously[9]. Quantification of luciferase signal was determined by Living Image Software. Limiting dilution assays were performed by injecting 5×105, 2.5×105 or 5×104 luciferase-tagged MDA-MB-231 cells in 100ul of a 1:1 Matrigel/PBS (Corning, 354234) underneath the nipple of 4th mammary fat pad of nude mice. Tumor formation was monitored weekly via IVIS imaging. TIC capacity was determined using the ELDA software website application (http://bioinf.wehi.edu.au/software/elda/) with a 95% confidence interval setting. All animal work was approved and performed in compliance with the Institutional Animal Care and Use Protocol at the University of Colorado Anschutz Medical Campus.
Luciferase Assays
Cells were plated in 12-well plates to achieve equal confluence (4T07: 4×104 cells/ml, 66cl4: 7.5 ×104cells/ml and MDA-MB-231: 1 ×105 cells/ml) and then co-transfected with the indicated firefly-luciferase constructs (pGF-mSox2-ONL and pGF-mSox2-T27G-ONL) and renilla-luciferase constructs (pCNL-N1_3xNLS) using lipofectamine 2000 according to manufacturer instructions (Thermo Fisher Scientific). After 72h, cells were collected and analyzed for luminescence using the Dual-luciferase Reporter Assay System (Promega). To assess relative SRR2 activity, luciferase luminescence was normalized to renilla luciferase activity.
ChIP-qPCR
Cells were plated in 15cm plates to similar confluency and then fixed with 1% formaldehyde to crosslink DNA and proteins. After terminating the crosslinking with glycine, cells were collected with ice cold PBS containing protease inhibitors and pelleted by centrifugation. Cell pellets were then processed to obtain ChIP DNA by using the Zymo Research ChIP kit (Zymo Research, D5210) according to manufacturer instructions. After obtaining ChIP DNA, qPCR was performed by diluting the samples 1:5 with nuclease-free water and using primers to amplify regions of SRR2 (Table S1). Ct (quantification) values from qPCR were normalized to input values for each condition. Results are presented as relative enrichment by subtracting the %input for IgG negative control from %Input for Six2 IP to adjust for background signal.
Datamining
Publicly available datasets were analyzed using cBioportal, Oncomine, SurvExpress or KMplotter. For survival data analyzed through KMPlotter, we used the 206510_at probe for Six2 and the 228038_at probe for Sox2 (mean expression) to analyze the relationship between Six2/Sox2 gene expression and Distant Metastasis-Free Survival and Relapse-Free Survival. After removal of biased array data (patient and clinical heterogeneity, different outcome measures and size effects) and redundant samples, results were split by the “auto select best cut off” and graphed to show the approximate low vs high expression, FDR of <20% and stated p-values.
Statistical Analysis
The experiments performed were analyzed using GraphPad Prism 6. Error bars were generated by Standard Error Mean (SEM) calculations. For experiments with two conditions, an unpaired one-tailed Student’s T-test was performed. For experiments with three or more conditions, one-way anova followed by a Bonferroni comparison was used. For the animal experiments measuring luciferase signal over time, fitting of a mixed effects model for testing the main effect of KD/OE and KD/OE-time interactions were performed. Given the statistical significance of the overall mode (P<0.05), we proceeded to carry out the multiple comparisons (with Bonferroni correction) to determine significant differences in the increase of luciferase signal over time between the KD/OE groups and their corresponding CTL groups. Asterisks represent the significance of difference from control group *, P<0.05; **, P<0.01; ***, P<0.001 and ****, P<0.0001.
Results
Six2 expression is enriched in TNBC where it promotes metastasis
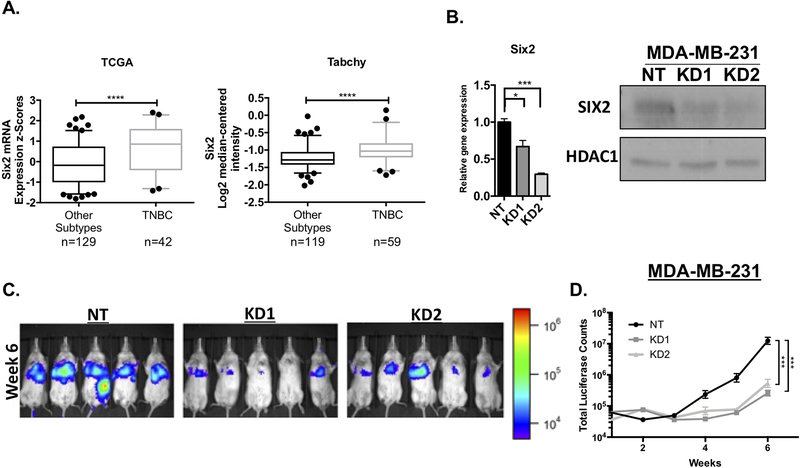
Our previous studies demonstrated that Six2 is more highly expressed in human breast carcinomas when compared to normal breast tissue, and that its expression is also increased in invasive ductal carcinomas when compared to ductal carcinoma in situ (DCIS)[9]. Building on these results, analysis of Six2 mRNA expression from multiple patient datasets, including TCGA and Tabchy datasets[19], reveals that it is more highly expressed in TNBC compared to all other subtypes of breast cancer (Fig 1A). Patients with TNBC present with higher tumor grade at diagnosis, have the highest rate of recurrence and possess the poorest prognosis overall compared to the other subtypes, making it one of the more aggressive breast cancer subtypes[20]. Given that we previously established a role for Six2 in late-stage metastasis of murine triple-negative mouse models[9] and its high expression in human TNBC, we next sought to understand the molecular mechanism through which SIX2 regulates TNBC metastasis. However, as Six2 has previously only been shown to regulate murine triple negative mammary carcinoma metastasis, we first examined whether SIX2 can also increase metastasis of human TNBC. To this end, we knocked down (KD) Six2 in luciferase tagged triple-negative MDA-MB-231 cells (Fig 1B). Tail-vein injections of non-targeting control (NT) and Six2 KD MDA-MB-231 cells into immunodeficient NOD-scid-gamma (NSG) mice demonstrate that mice injected with Six2 KD cells have significantly reduced metastatic burden when compared to mice injected with NT control cells (Fig 1C and D). These data, in conjunction with previous data in mouse mammary triple-negative models[9], demonstrate a critical role for SIX2 in the later stages of metastasis in human TNBC.
Figure 1: Six2 expression is enriched in triple-negative breast cancer (TNBC) where it promotes metastasis.
A) Six2 mRNA expression in TNBC vs all other subtypes. Expression levels of Six2 obtained from the TCGA Cell 2015 (cbioportal) (Gao et al. 2013; Cerami et al. 2012) and Tabchy breast datasets (Oncomine) (Rhodes et al. 2004) B) Levels of Six2 mRNA (left) and protein (right) in non-targeting (NT) Control MDA-MB-231 cells and in Six2 KD MDA-MB-231 cells. Gene expression was normalized to Ppib mRNA expression and fold-change is relative to the NT cells. P-values were calculated using one-way anova followed by Bonferroni multiple comparisons test of triplicate samples in a representative experiment (n=3). For western blot analysis, nuclear extracts are shown with HDAC1 used as a loading control. C) Luciferase-labeled MDA-MB-231 NT, KD1 and KD2 cells were injected into NSG mice through the tail vein. Metastatic burden was measured by IVIS imaging. Luciferase images are of mice at the week 6 timepoint (n=5 mice/condition). D) Quantification of average whole-body luciferase signal (total luciferase counts) for each condition over the duration of the animal experiment, (n=5 mice/condition). P-values to determine differences in luciferase signal over time were calculated using mixed model effects interaction analysis followed by Bonferroni correction.
Six2 regulates stem cell-associated phenotypes in TNBC and mouse mammary carcinoma cells
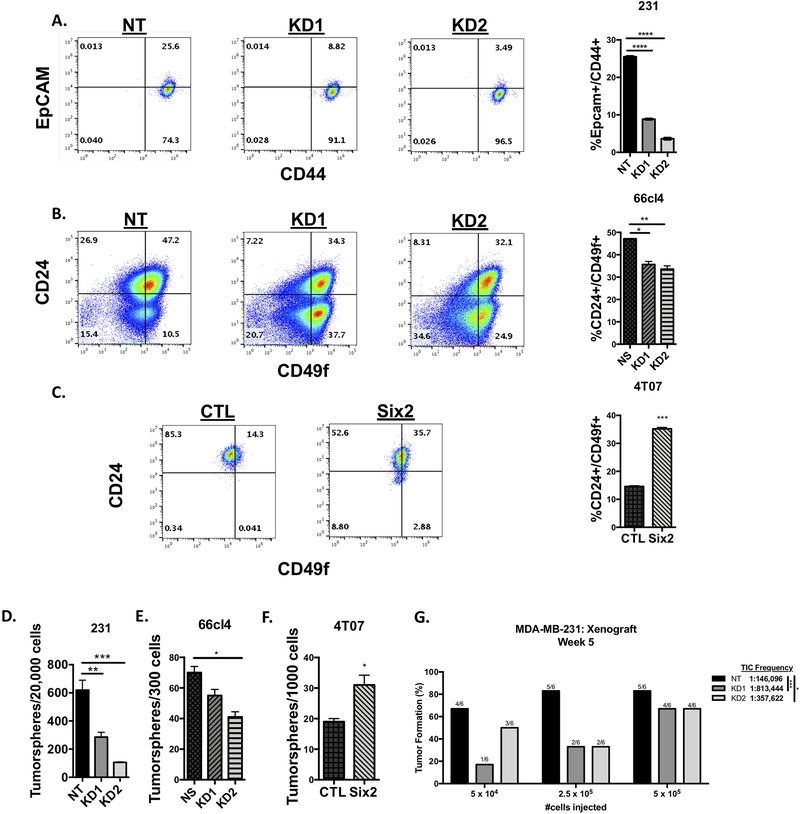
Because Six2 plays a critical role in the maintenance and self-renewal of nephron progenitor cells during kidney development, and because it promotes metastasis of a cell line (4T07) that is capable of reaching secondary sites, but cannot efficiently colonize those sites[15], we hypothesized that SIX2 mediates late-stage metastasis, particularly the survival and establishment of tumors at secondary sites, partly due to its role in stem/progenitor-like phenotypes. To test this hypothesis, we first performed flow cytometry to determine if there are differences in the stemness-associated population in response to the presence of SIX2. Because MB-MDA-231 cells express little to no CD24, we measured cell surface expression of EpCAM and CD44[21], which is known to more accurately enrich for cells with tumor-initiation and metastatic capacity[22]. Interestingly, Six2 KD results in a decrease in the EpCAM+/CD44+ population in MDA-MB-231 cells (Fig 2A). We additionally examined whether Six2 alters the cancer stem cell (CSC) population within triple-negative mouse mammary carcinoma cells where we had previously found it to mediate metastasis[9]. In this case, flow cytometry was performed to examine the CD24+/CD49f+ population, which enriches for cells with the ability to repopulate the mouse mammary gland[23], in 4T07 cells overexpressing six2 (4T07-Six2), and in 66cl4 cells with KD of six2, as compared to their control counterparts. Our data show that Six2 loss decreases, while Six2 OE increases, the percentage of CD24+/CD49f+ cells within the population (Fig 2B and C).
Figure 2: Six2 regulates stem cell-associated phenotypes in TNBC and mouse mammary carcinoma cells.
A) Six2 knockdown in MDA-MB-231 cells decreases the EpCam+/CD44+ population, as measured using flow cytometry. Quantification on the right is of triplicate samples for a representative experiment (n=3). Error bars represent the mean +/− SEM. P-values were calculated using one-way anova followed by a Bonferroni multiple comparisons test. B) six2 KD in 66cl4 cells decreases the CD24+/CD49f+ population, as measured using flow cytometry. Quantification and p-values were done as described above. C) Six2 overexpression in 4T07 cells increases the CD24+/CD49f+ population, as measured using flow cytometry. Quantification was performed as described above. P-values were calculated using an unpaired two-tailed t test. D) Six2 knockdown in MDA-MB-231 cells decreases tumorsphere formation when compared to control (NT) cells. Quantification on the right is of triplicate samples for a representative experiment (n=3). Error bars represent the mean +/− SEM. P-values were calculated using one-way anova followed by a Bonferroni multiple comparisons test. E) six2 knockdown in 66cl4 cells decreases tumorsphere formation when compared to control (NS) cells. Quantification and p-values were performed as described above. F) Six2 overexpression in 4T07 cells increases tumorsphere formation when compared to control cells. Quantification was performed as described above. P-values were calculated using an unpaired two-tailed t test. G) Six2 knockdown in MDA-MB-231 cells decreases tumor-initiating capacity compared to control (NT) cells. Cells were transplanted into the 4th mammary fat pad of nude mice at limiting dilutions. The graph displays the percentage of positive tumors at week 5 post tumor cell injection (n=6). Mean tumor-initiating cell (TIC) frequency was determined using the ELDA software program, with estimated ranges of TIC frequency being 72,956–292,560 for NT, 326,264–2,028,088 for KD1 and 175,059–730,575 for KD2.
To assess the role of SIX2 in stem cell-associated phenotypes using a more functional assay, we next performed tumorsphere formation experiments in all three models. The ability of cells to form secondary tumorspheres after dissociation and dilution to single cells is a measure of self-renewal capability, and thus serves as a surrogate CSC assay[24]. Similar to what was observed in our flow cytometry analysis, in both the MDA-MB-231 and 66cl4 cells, Six2 KD significantly decreases the ability of cells to form secondary tumorspheres (Fig 2D and E). In line with these findings, Six2 OE in the 4T07 cells significantly increases secondary tumorsphere formation (Fig 2F).
To further test whether Six2 regulates cancer stem cell frequency and functionality, we performed limiting dilution assays in vivo using the MDA-MB-231 cells with Six2 KD. We found that compared to NT control cells, Six2 KD cells have decreased tumor-initiating cell (TIC) capacity, as demonstrated by a significant decrease in TIC frequency from 1:146,096 for NT control cells to 1:813,444 and 1:357,622 for Six2 KD1 and KD2 cells, respectively (Fig 2G). Taken together, these data suggest that SIX2 may play a similar role in mediating stemness characteristics during tumor progression as it does during development[7], and that this role may allow it to mediate late-stage metastasis by enhancing the ability of cells to self-renew and survive at secondary sites.
Six2 regulates a transcriptional program associated with stemness
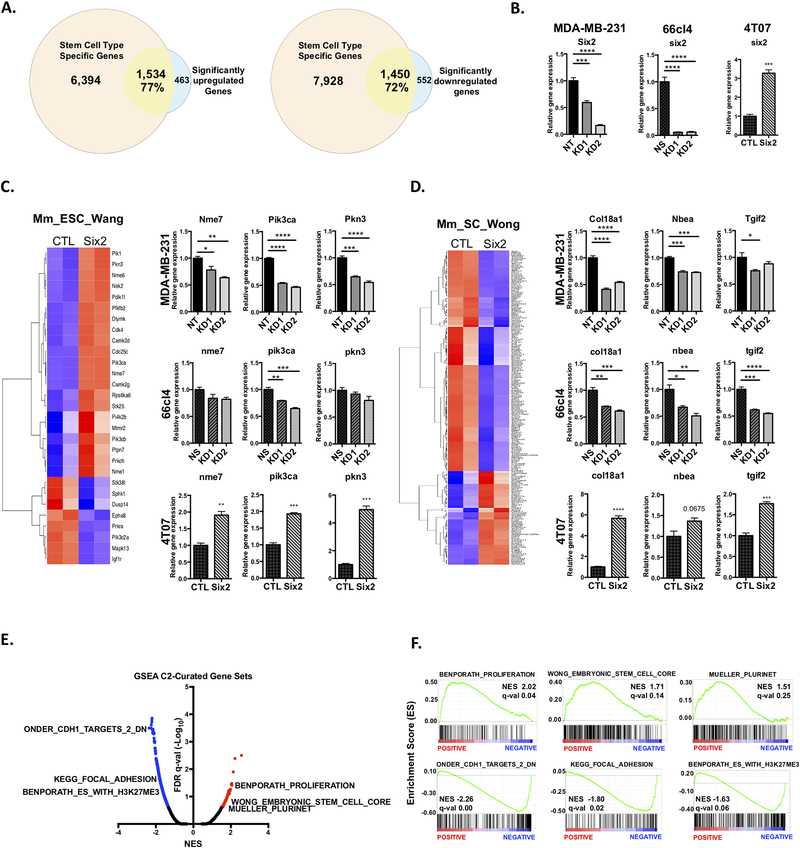
Since SIX2 is a transcription factor, we hypothesized that it directly transcriptionally regulates downstream effectors to promote CSC-like phenotypes and metastatic burden. We have previously shown that 4T07 cells express very little Six2 endogenously and that the OE levels are similar to other murine mammary triple-negative cell lines with endogenously high Six2 expression, such as the 66cl4 cells [9]. Additionally, although we observe a range of Six2 expression in human TNBC cell lines, we found that Six2 OE in 4T07 cells is comparable to, and does not artificially exceed, the levels of Six2 observed in multiple TNBC cell lines (Fig S1). Therefore, we chose this system to perform RNA-seq analysis comparing 4T07 control (4T07-CTL) and 4T07-Six2 cells. Six2 OE leads to a robust transcriptional response as evidenced by the significant increased expression of 1,997 genes and decreased expression of 2,002 genes (FDR q-value<0.1 and log2 Fold Change ± 1.0) (Fig S2A). Additionally, Principal-Component Analysis (PCA) based on the top 500 most-variable genes from each sample revealed a stark difference between 4T07-CTL and 4T07-Six2 cells, indicating that Six2 OE in 4T07 cells drives distinct changes in the transcriptional landscape (Fig S2B). Given the well-established role of Six2 as a critical transcriptional regulator of progenitor and stem cell populations[7], we used the web-server StemChecker[25] to examine whether differentially expressed genes identified from the RNA-seq analysis overlap with previously established and curated stemness-associated gene signatures. We first queried the 1,997 significantly upregulated and 2,002 significantly downregulated genes in response to Six2 OE (Adjusted p-value < 0.1 and log2 Fold Change ± 1.0) from the RNA-seq analysis to determine the overlap with gene signatures from StemChecker that represent specific normal stem cell types. We found that 77% of the Six2-upregulated genes from the RNA-seq analysis overlapped with genes from StemChecker signatures representing various stem cell types, whereas the Six2-downregulated genes contained a 72% overlap (Fig 3A). Interestingly, gene signatures representing embryonic stem cells (ESCs) were amongst the most highly enriched and represented signatures in the analysis when examining all significantly differentially expressed genes together (Fig S2C).
Figure 3: Six2 regulates a transcriptional program associated with stemness.
A) Area-proportional Venn Diagram displaying percent overlap of significantly upregulated (left) or downregulated (right) genes with normal stem cell type signatures. B) RT-qPCR analysis measuring Six2 mRNA expression after KD (MDA-MB-231 and 66cl4) and OE (4T07). Gene expression was normalized to Ppib (human) or ppib (mouse) mRNA expression and fold-change is relative to the NT cells. P-values calculated using either One-way anova followed by Bonferroni multiple comparisons test or an unpaired two-tailed t test of triplicate samples for a representative experiment are shown (n=3). Heatmap and mRNA expression of significantly expressed genes from RNA-seq analysis that overlap with the C) Mm_ESC_Wang and D) Mm_SC_Wong gene signature. Only genes that met a fold-change of > or < 1.0 (Six2 OE/CTL) and adjusted p-value of < 0.1 after batch correction (limma package) were considered for generating heatmaps. Validation of mRNA expression was performed using RT-qPCR in MDA-MB-231, 66cl4 and 4T07 cells. Gene expression was normalized to Ppib mRNA expression and fold-change is relative to the non-targeting (NT), non-silencing (NS) or control (CTL) cells respectively. P-values were calculated using either one-way anova followed by Bonferroni multiple comparisons test or unpaired T-test on means +/− SEM of triplicate samples for a representative experiment (n=3). E) Volcano plot of false discovery rate (FDR q-val) versus normalized enrichment score (NES) after GSEA from RNA-seq data (Pre-ranked GSEA module). Red dots denote significantly positively enriched pathways and blue dots denote significantly negatively enriched pathways (0.25 FDR cutoff). F) GSEA enrichment plots for EMT-, adhesion- and stemness-associated pathways selected from the C2-Curated gene set. Black bars represent genes contained within the specific pathway and are ranked from positively expressed (left) to negatively expressed (right) based on log2 fold change (Six2 OE vs CTL).
We then determined whether Six2 KD or Six2 OE (Fig 3B shows expression levels of Six2 in all three cell lines as determined in the same RNA samples used in panels C and D) could affect the expression of genes from the ESC-associated signatures. For instance, when examining a dataset that used a shRNA functional screen to identify crucial factors for embryonic stem cell proliferation and pluripotency[26], we found that the genes contained in this signature were dramatically differentially expressed between the 4T07-CTL and 4T07-Six2 cells (Fig 3C, ESC_Wang dataset[26]). The same was true for a signature containing genes that are differentially expressed between mouse embryonic stem cells and adult tissues[27] (Fig 3D, SC Wong dataset[27]), as well as signatures associated with maintaining pluripotency[28,29] (Fig S2D). To validate that SIX2 mediates gene expression alterations that are linked to stem-associated phenotypes, we performed RT-qPCR for stem cell genes identified in the different signatures on independent sets of RNA from all three cell line models. These genes were chosen based on their established roles in normal embryonic/cancer stem cell function, tumor progression, and/or metastatic colonization[30–34], and were amongst the top quartile of upregulated genes after DESeq analysis. This analysis confirmed that Six2 KD in both MDA-MB-231 and 66cl4 cells leads to a corresponding decrease in the mRNA expression of numerous genes associated with embryonic stem cells (Fig 3C and D), whereas Six2 OE leads to an increase in many of the same genes (Fig 3C and D).
To further confirm that SIX2 plays a role in a genetic stem cell program, we also performed Gene-Set Enrichment Analysis (GSEA) after converting mouse gene symbols to their human equivalents[35]. Notably, GSEA analysis using the C2-Curated gene sets revealed that Six2 OE results in negative enrichment of pathways associated with E-cadherin function (ONDER CDH1 TARGETS 2 DN) and cell focal adhesion (KEGG FOCAL ADHESION) (FDR q-value < 0.25 and Normalized p-value < 0.05) (Fig 3E and F, Table S2). This data supports previous studies from our lab demonstrating that Six2 OE in 4T07 cells results in increased metastasis in part through downregulation of E-cadherin and increased anchorage-independent growth[9]. Interestingly, we also observed enrichment of pathways associated with stemness. For instance, pathways that are upregulated to promote stemness (BENPORATH PROLIFERATION, WONG EMBRYONIC STEM CELL CORE, MUELLER PLURINET) were among the most highly positively significantly enriched pathways across all pathways analyzed in the C2-Curated gene sets (Fig 3E and F, Table S2). Additionally, we identified significantly negatively enriched pathways that contain genes that are downregulated in ESCs (BENPORATH ES WITH H3K27ME3) (Fig 3E and F, Table S2). When performing GSEA analysis using the C5-Gene Ontology gene sets, Six2 OE led to negative enrichment of pathways involved in adhesion and differentiation (Fig S2E and F, Table S2). Together, these data demonstrate that SIX2 expression broadly enhances a stem-cell gene program in both triple negative breast and mammary cancer cells.
Six2 regulates master stemness factors Sox2 and Nanog
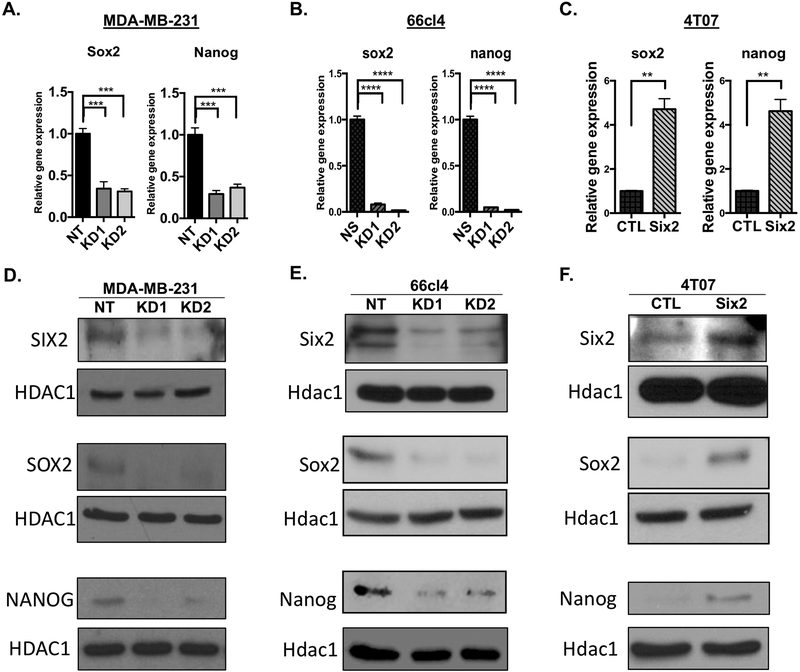
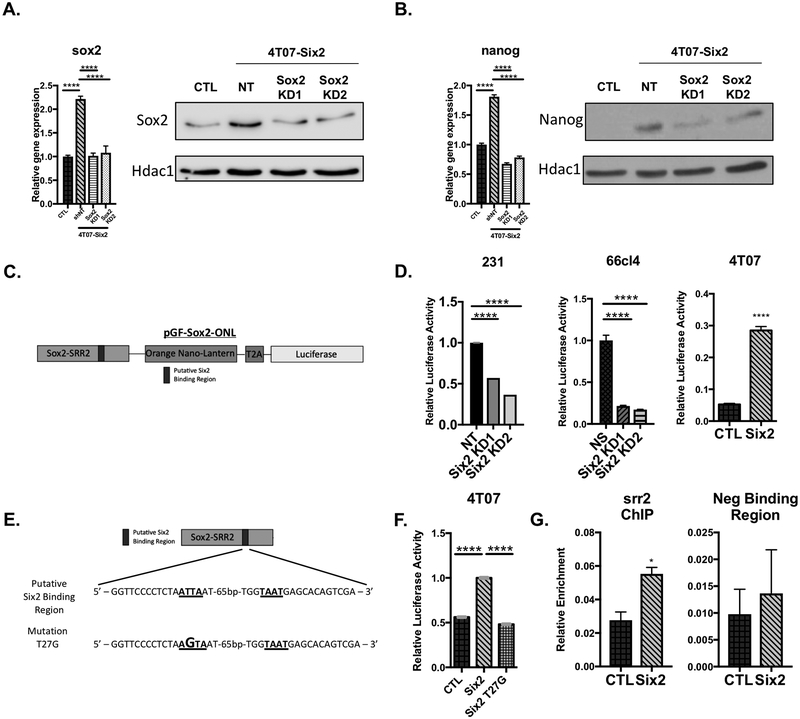
Based on the large number of stem-related genes altered by SIX2, we asked whether SIX2 promotes the expression of master regulators of stemness. To address this question, we first examined the mRNA and protein expression of well-established master regulators of stemness and pluripotency. Intriguingly, KD of Six2 in the MDA-MB-231 and 66cl4 cells resulted in corresponding decreases in both Sox2 and Nanog mRNA as measured by RT-qPCR (Fig 4A and B). In contrast, Six2 OE in the 4T07 cells resulted in upregulation of mRNA expression of both pluripotency factors (Fig 4C). Additionally, KD of Six2 leads to a decrease in both SOX2 and NANOG protein (Fig 4D and E), whereas OE of Six2 leads to corresponding increases in Sox2 and Nanog levels (Fig 4F). These data suggest that SIX2 may promote stemness-associated phenotypes through regulation of SOX2 and NANOG.
Figure 4: Six2 regulates master pluripotency factors Sox2 and Nanog.
Sox2 and Nanog mRNA expression in the A) MDA-MB-231, B) 66cl4 and C) 4T07 cells. Gene expression was determined by RT-qPCR and normalized to Ppib mRNA expression. Fold-change is relative to the NT (MDA-MB-231 cells), NS (66cl4 cells) or CTL (4T07 cells). P-values determined using an unpaired T-test or one-way anova followed by Bonferroni multiple comparisons test of means +/− SEM of triplicate samples for a representative experiment (n=3). Six2 (top), Sox2 (middle) and Nanog (bottom) protein expression in D) MDA-MB-231, E) 66cl4 and F) 4T07 cells. Protein expression after nuclear extraction was determined by Western Blot Analysis with HDAC1/Hdac1 being used as a loading control (n=3).
Previous studies have shown that in the context of breast cancer, SOX2 regulates cancer stem cell properties upstream of NANOG[36]. To determine whether Sox2 is upstream of Nanog in Six2 OE cells, we established stable KD of sox2 in the 4T07-Six2 cells (Fig 5A). Indeed, we observed that sox2 KD in 4T07-Six2 cells resulted in a corresponding decrease in Nanog mRNA and protein levels (Fig 5B), suggesting that Six2 may act more directly on sox2 to mediate alterations in Nanog levels.
Figure 5: Sox2 is upstream of Nanog and is directly regulated by Six2 via its SRR2 enhancer.
A) sox2 mRNA expression in 4T07-CTL, 4T07-Six2 and 4T07-Six2/Sox2 KD (KD1 and KD2) cells measured by RT-qPCR (left). Western blot analysis of Sox2 protein in 4T07-CTL, 4T07-Six2, and 4T07-Six2/Sox2 KD cells (right). Gene expression was determined using RT-qPCR and normalized to ppib mRNA expression. Fold-change is relative to 4T07-CTL cells. P-values were determined using a one-way anova followed by Bonferroni multiple comparisons test of means +/− SEM of triplicate samples for a representative experiment (n=3). Sox2 protein was determined using western blot analysis after whole cell extraction with Hdac1 used as a loading control. B) nanog mRNA expression in 4T07-CTL, 4T07-Six2 and 4T07-Six2/Sox2 KD (KD1 and KD2) cells measured by RT-qPCR (left). Gene expression was determined by RT-qPCR and normalized to ppib mRNA expression. Fold-change is relative to 4T07-CTL cells. P-values were determined as described above of triplicate samples for a representative experiment (n=3). Nanog protein was determined as described above (right). C) Diagram of pGF-Sox2-ONL luciferase reporter plasmid used in luciferase assays. Darker rectangle denotes putative Six2 binding region (T2A, encodes self-cleaving peptide sequence; ONL, Orange NanoLantern). D) pGF-Sox2-ONL luciferase assays performed in MDA-MB-231 +/− Six2 KD, 66cl4 +/− Six2 KD and 4T07 +/− Six2 OE cells 72h after transfection. P-values determined using an unpaired T-test or one-way anova followed by Bonferroni multiple comparisons test of means +/− SEM of triplicate samples in a representative experiment (n=3). E) Sequence of WT Six2 binding region (underlined) within SRR2 enhancer region. Bottom sequence depicts the mutant version of the SRR2 enhancer (mutation in larger font) (T27G) used in the luciferase experiment. F) Luciferase reporter assay using WT and T27G Sox2-ONL luciferase reporter plasmids in 4T07-CTL and Six2 cells. P-values were determined as described above of triplicate samples in a representative experiment (n=3). G) ChIP-qPCR analysis of 4T07-CTL and -Six2 cells. ChIP-enriched DNA was quantified using RT-qPCR with primers surrounding the predicted Six2 binding region within the SRR2-enhancer. In a separate pulldown, Six2 binding at a downstream region that does not contain known Six2 binding sites was also examined. P-values were determined using an unpaired T-test of means +/− SEM of triplicate samples in a representative experiment (n=3).
Sox2 transcriptional regulation is, in part, under the control of two enhancer regions, SRR1 and SRR2. The SRR2 domain, located ~3kb downstream of the Sox2 gene, functions as a transcriptional enhancer of Sox2 expression in ESCs[37]. Intriguingly, further analysis of the sequence of the srr2 enhancer revealed the presence of a Six2 binding region[38]. Therefore, we performed luciferase reporter assays containing the srr2 regulatory region[39] to determine whether Six2 may regulate sox2 through the SRR2 domain (Fig 5C shows the reporter construct used in assays). After transfection of a srr2 luciferase construct (pGF-Sox2-ONL, 5C)[39], along with a Renilla-luciferase plasmid (pCNL-N1_3xNLS) to normalize for transfection efficiency, we found that Six2 KD decreases, whereas Six2 OE increases, activation of the srr2 region (Fig 5D). To determine if Six2 directly regulates the srr2 enhancer via this binding region, we introduced a single point mutation (T27G) within the putative Six2 binding region of the srr2 enhancer based on a Six2 binding site that was previously identified and characterized in the gdnf gene[38] (Fig 5E). Interestingly, although introduction of the wildtype srr2 region resulted in a similar increase in srr2 enhancer activity in 4T07-Six2 cells compared to 4T07-CTL cells, introduction of the T27G mutation into the srr2 enhancer-luciferase construct inhibited the activation of luciferase activity (Fig 5F). We next performed ChIP-qPCR and found that Six2 binding at the srr2 enhancer region is enriched in 4T07-Six2 cells compared to control cells (Fig 5G). In contrast, the enrichment of Six2 observed in the sox2 regulatory regions is lost when performing a pulldown at a region approximately 6kb downstream of the srr2 region that contains no known Six2 binding sites (Fig 5G). Together, our results suggest that Six2 plays a critical role in altering CSC characteristics through direct transcriptional activation of sox2, resulting in upregulation of nanog and induction of a stem cell-like program.
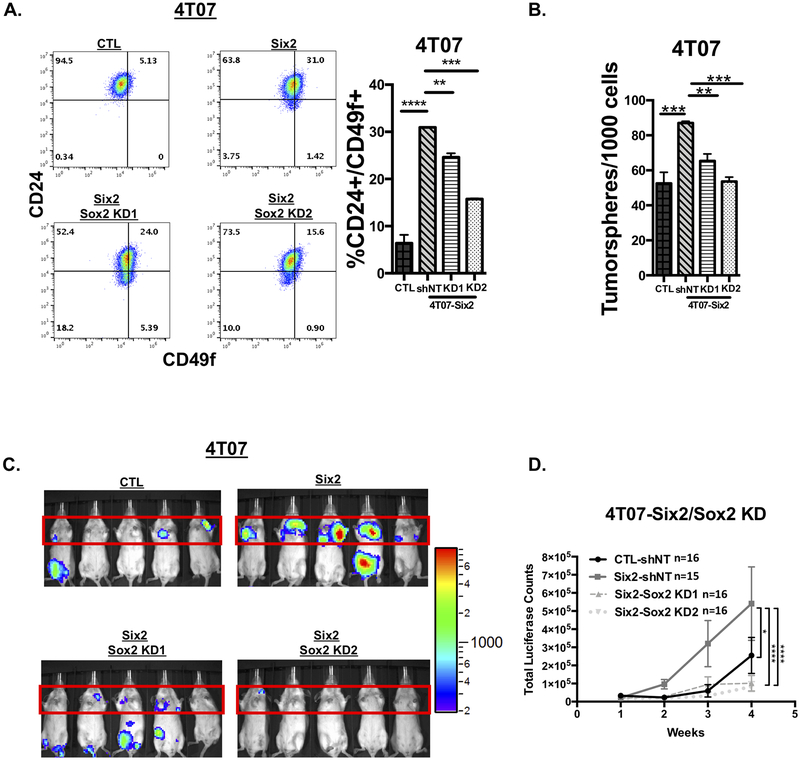
Sox2 mediates stem cell-associated phenotypes and late-stage metastasis downstream of Six2
We then asked whether Sox2 is required downstream of Six2 to mediate CSC characteristics and late-stage metastasis. To this end, we performed flow cytometry to measure the mouse mammary stem cell enriched population in response to sox2 KD downstream of Six2 in the 4T07 cells. We found that sox2 KD in 4T07-Six2 cells leads to a significant decrease in the CD24+/CD49f+ enriched CSC population when compared to control cells (4T07-Six2 shNT) (Fig 6A). Similarly, secondary tumorsphere formation was also decreased in response to sox2 KD in 4T07-Six2 cells (Fig 6B). Additionally, to better understand why we did not observe a complete reversal in the in vitro stemness-associated phenotypes after sox2 KD, we performed ChIP-qPCR in 4T07-CTL and -Six2 OE cells to examine whether Six2 was directly bound to, and therefore could transcriptionally regulate, the promoter regions of the putative targets identified in the StemChecker analysis. We found that Six2 was bound to a subset of these targets, suggesting that Six2 regulates multiple genes that a play a critical role in stemness (Fig S3). This data suggests that in order to completely reverse the in vitro stemeness-associated phenotypes regulated by Six2, multiple downstream targets, in addition to Sox2, may need to be inhibited.
Figure 6: Sox2 mediates stem cell-associated phenotypes and late-stage metastasis downstream of Six2.
A) KD of sox2 in 4T07-Six2 cells decreases the CD24+/CD49f+ mouse mammary stem cell-like population when compared to 4T07-Six2 scramble control (shNT) cells as measured by flow cytometry. Quantification on the right is of triplicate samples for a representative experiment (n=3). Error bars represent the mean +/− SEM. P-values were calculated using one-way anova followed by a Bonferroni multiple comparisons test. B) sox2 KD in 4T07-Six2 cells decreases tumorsphere formation compared to 4T07-Six2 non-targeting shRNA (4T07-Six2 shNT) cells. Quantification and calculation of p-values were done as described above. C) Luciferase-labeled 4T07-CTL shNT, -Six2 shNT, -Six2/Sox2 KD1 and -Six2/Sox2 KD2 cells were injected into Balb/c mice through the tail vein. Metastatic burden was measured using IVIS imaging. Representative pictures of animals injected (n=16 per condition at Week 2 timepoint). D) Quantification of average +/− SEM lung luciferase signal for each condition over the duration of the animal experiment (n equals at least 15 per condition, as one 4T07-Six2 shNT mouse died after week 2). P-values to determine differences in luciferase signal over time were calculated using two-way anova followed by Bonferroni correction of average luciferase signal in each condition.
To examine whether Sox2 is a critical mediator of Six2 in promoting late-stage metastasis, we performed tail-vein injections using 4T07-Six2/Sox2 KD cells (as compared to corresponding controls). As had been previously observed[9], Six2 overexpression in 4T07 cells enables metastatic outgrowth in vivo (Fig 6C, upper panels). However, KD of sox2 in 4T07 cells dramatically decreases Six2-induced metastatic outgrowth (Fig 6C, lower panels and D for quantitation), suggesting that in vivo Sox2 is the key target downstream of Six2 in promoting late-stage metastasis. Thus, our data demonstrate that Six2 regulates metastatic outgrowth in a Sox2-dependent manner.
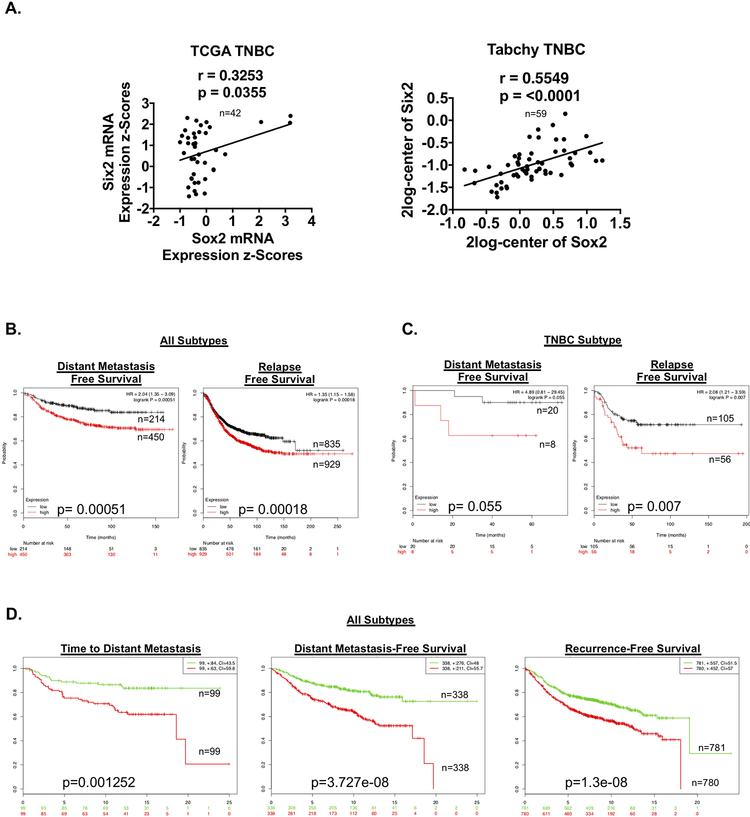
Six2 and Sox2 expression correlate in human breast cancer, and are together associated with poor prognosis
On the basis of our experimental findings that sox2 KD decreases Six2-mediated metastatic outgrowth, we next examined whether this relationship may be relevant to human breast cancer. We first performed RT-qPCR on multiple human TNBC cell lines to determine if the Six2-Sox2 axis is present in additional human TNBC cells. Indeed, we find that in both BT549 and SUM159 cells (TNBC cell lines with high endogenous Six2 expression[9]), transient KD of Six2 leads to a corresponding decrease in Sox2 (Fig S4). These data suggest that similar to what we observe in the MDA-MB-231 cell line model, Six2 also regulates Sox2 in other human TNBC models. We next performed correlation analysis of Six2 and Sox2 mRNA expression obtained from cbioportal, Oncomine and R2 Cancer Databases and demonstrate that Six2 and Sox2 positively correlate in breast cancer datasets encompassing all breast cancer subtypes (Fig S5A); a relationship that is also observed when specifically examining TNBC datasets (Fig 7A). These data suggest that the relationship between Six2 and Sox2 in our cell line models is recapitulated in human breast cancer.
Figure 7: Six2 and Sox2 expression correlate in human breast cancer, and a Six2 gene expression signature is associated with poor prognosis.
A) Linear regression analysis demonstrating that Six2 and Sox2 mRNA significantly correlate in TNBC patient samples using both the TCGA Cell 2015 dataset and the Tabchy dataset (obtained from Oncomine). B) Distant-metastasis free and relapse-free survival in patients with high or low combined expression of the Six2 gene signature in all subtypes of breast cancer (KMPlotter). C) Distant-metastasis free and relapse-free survival of TNBC patients with high or low combined expression of the Six2 gene signature (KMPlotter, auto select was used for cutoff). D) Time to distant metastasis, Distant metastasis-free survival and recurrence-free survival from the Survexpress meta-analysis dataset of the Six2 Gene Signature (median cutoff).
We then generated a Six2-gene expression signature compiled of the top 15 significantly upregulated genes in response to Six2 OE after converting mouse gene symbols to their human equivalent (Fig S5B). Of interest, the top 15 Six2-regulated genes (based on highest expression) are all associated with roles in normal and CSC function and/or tumor progression[40–51]. Importantly, high expression of the Six2-gene signature significantly correlates with a decrease in distant metastasis-free and relapse-free survival across all breast cancer patient samples as well as in TNBC-specific samples (Fig 7B and C). Additionally, the Six2-gene signature significantly correlates with a decrease in time to distant metastasis, distant metastasis-free, and recurrence free survival across all breast cancer (we were unable to query TNBC specifically in this dataset) (Fig 7D). These data demonstrate that the relationship between SIX2 and metastatic burden is not limited to our cell line models, and that the SIX2 transcriptional program is predictive of poor outcomes in human breast cancer.
Discussion
Previous data from our group, using two different triple negative mouse mammary carcinoma xenograft models, established Six2 as critical for late stage metastasis[9]. In particular, the fact that Six2 results in outgrowth of 4T07 cells, which are known to reach the lungs after orthotopic injection into the mammary gland, but to not efficiently colonize the lungs[15], demonstrates a role for Six2 in the colonization step of the metastatic cascade. Further, data within this manuscript demonstrate that SIX2 also regulates late stage metastasis in human TNBC.
Although our data suggest that a main function of SIX2 is to mediate outgrowth of cells that have newly arrived at a secondary site, given its described role in mediating mesenchymal phenotypes and anoikis resistance[9,14], SIX2 may in fact affect several additional aspects of late-stage metastasis, although this remains to be formally tested. If SIX2 regulates multiple steps of late stage metastasis (such as survival in the bloodstream and exit from the vasculature in addition to colonization at secondary sites), this finding would further emphasize the importance of developing novel means to target its function, as significant data suggests that metastases can seed additional metastatic sites[52]. In the future, it will be important to determine whether SIX2 is required for maintenance of metastatic lesions. This question is critical to address, as patients that already present with significant metastatic burden would benefit from a therapy that inhibits the continued growth of cells at the secondary site. Nonetheless, even if SIX2 is only critical for the establishment and initial stages of metastatic outgrowth, many breast cancer patients do not present with overt metastases, and thus inhibiting this step could be valuable. However, as SIX2 is a transcription factor, its direct inhibition may prove difficult. Thus, understanding the mechanism by which SIX2 mediates late stage metastasis may enable the identification of novel downstream targets or pathways that can potentially be targeted in the future.
Previous studies have shown that Six2 maintains the kidney progenitor pool during nephrogenesis, in part through impinging on stem cell-associated pathways[7]. Based on our RNA-seq analysis, we find that similarly, in the context of breast cancer, Six2 regulates a genetic stem-cell like program. We demonstrate that Six2 OE increases, whereas Six2 KD decreases, the expression of multiple genes that are enriched in normal stem cells. These data suggest that SIX2 may regulate stem cell-like properties across multiple tissue types and across normal and disease states. Interestingly, although it has mostly been studied in the context of nephrogenesis[7], previous studies from our laboratory have shown that during mammary gland development, six2 expression is increased in the mammary gland of six1 KO mice[53]. Because Six1 OE in the mammary gland results in the expansion of mammary stem/progenitor cells[8], yet six1 loss does not affect mammary gland development[53], it is possible that upregulation of six2 compensates for six1 loss to maintain mammary stem cell function. Similar to SIX1, SIX2 is rarely expressed in adult tissues, but it is re-expressed in the context of breast cancer[9]. The role of SIX2 in nephrogenesis, coupled with its re-expression in human breast cancers, suggests that SIX2 may be a key regulator of both mammary stemness and mammary tumorigenesis and/or metastasis. However, the functional role of Six2 in the normal mammary stem cell compartment and in mammary gland development has yet to functionally explored. Analysis of mRNA expression from isolated mouse fetal mammary stem and adult mouse differentiated mammary epithelial cells[23] shows that six2 mRNA is more highly expressed in the fetal mammary stem cell population compared to the adult mammary cell population, suggesting that Six2 may play a role in fetal mammary stem cells (Fig S6). Thus, investigation into whether Six2 contributes to normal mammary development may be warranted.
Previous studies using ChIP-seq analysis in the developing kidney identified gdnf, eya1 and six2 itself as direct transcriptional Six2 targets[7,38]. Interestingly, our RNA-seq analysis in 4T07 cells identify eya1 as one of the top upregulated genes in response to Six2 OE, suggesting conservation of specific downstream targets of Six2 between normal development and cancer. As outlined above, we have identified sox2 as a novel direct transcriptional target of Six2 in breast cancer cells that is critical for Six2-mediated metastasis. We further demonstrate that Six2 directly regulates sox2 transcription by binding to, and activating, the srr2 enhancer region of the sox2 gene. Thus, it will be of interest to determine whether Six2 similarly regulates Sox2 in the context of normal embryonic development. While it is important to continue to understand how SOX2, as a key stem cell factor, mediates SIX2-regulated late-stage metastasis, unfortunately, SOX2, like SIX2, is a transcription factor and thus not an ideal drug target. Moving forward, it will be important to identify more targetable downstream effectors of SIX2 that are critical in mediating its ability to promote metastatic burden.
Overall, our studies indicate that SIX2 plays an important role in promoting cancer stem cell-associated properties through a novel SIX2/SOX2 axis that is critical for metastatic colonization. Importantly, the relationship between Six2 and Sox2 is relevant in human breast cancer, and high expression of a Six2-mediated gene signature significantly correlates with poor metastatic prognosis. Although it is well established that targeting cancer stem cells at the primary tumor site can increase therapeutic efficacy[54], our findings support growing evidence that these strategies should be considered when targeting metastatic tumor cells, in which there may be different regulators of tumor outgrowth that provide an advantage in a new microenvironment[3]. Our results support this emerging concept by providing critical knowledge regarding how stem cell properties in newly arriving tumor cells promote late-stage metastasis.
Supplementary Material
Significance.
Findings provide novel mechanistic insight into stemness and the metastatic outgrowth of triple-negative breast cancer cells.
Acknowledgements
We thank Yasushi Okada for providing the pGF-mSox2-ONL and pCNL-N1_3xNLS plasmids for use in the SRR2 luciferase reporter assays. We also thank the University of Colorado Cancer Center Animal Imaging (P30CA046934), Flow Cytometry (P30CA046934), Functional Genomics (P30CA046934), and Genomics Shared Resources (P30CA046934) for aiding in experimental analysis used within this work. This work was supported by R01CA180175 (JD), R01CA095277 (HLF), The Cancer League of Colorado - 25A5976 (HLF), R01CA117907 (JME), the NCI F31 Ruth L. Kirschstein National Service Research Award 5F31CA210622–02 (MUJO), and the NCI F99/K00 Fellow Transition Award F99CA223023 (MUJO).
Footnotes
Conflict of Interest Statement: The authors have declared that no conflict of interest exists.
References
- [1].Massagué J, Obenauf AC. Metastatic colonization by circulating tumour cells. Nature 2016;529:298–306. doi: 10.1038/nature17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer 2002;2:563–72. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- [3].Chambers AF, Naumov GN, Varghese HJ, Nadkarni KV, MacDonald IC, Groom AC. Critical steps in hematogenous metastasis: an overview. Surg Oncol Clin N Am 2001;10:243–55, vii. [PubMed] [Google Scholar]
- [4].Hüsemann Y, Geigl JB, Schubert F, Musiani P, Meyer M, Burghart E, et al. Systemic Spread Is an Early Step in Breast Cancer. Cancer Cell 2008;13:58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
- [5].Fili S, Karalaki M, Schaller B. Mechanism of bone metastasis: The role of osteoprotegerin and of the host-tissue microenvironment-related survival factors. Cancer Lett 2009;283:10–9. doi: 10.1016/j.canlet.2009.01.011. [DOI] [PubMed] [Google Scholar]
- [6].Oskarsson T, Acharyya S, Zhang XH-F, Vanharanta S, Tavazoie SF, Morris PG, et al. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med 2011;17:867–74. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].McMahon AP. Development of the Mammalian Kidney. Curr. Top. Dev. Biol, vol. 117, 2016, p. 31–64. doi: 10.1016/bs.ctdb.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Christensen KL, Patrick AN, McCoy EL, Ford HL. The six family of homeobox genes in development and cancer. Adv Cancer Res 2008;101:93–126. doi: 10.1016/S0065-230X(08)00405-3. [DOI] [PubMed] [Google Scholar]
- [9].Wang C -a., Drasin D, Pham C, Jedlicka P, Zaberezhnyy V, Guney M, et al. Homeoprotein Six2 Promotes Breast Cancer Metastasis via Transcriptional and Epigenetic Control of E-Cadherin Expression. Cancer Res 2014;74:7357–70. doi: 10.1158/0008-5472.CAN-14-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wu DW, Lin PL, Wang L, Huang CC, Lee H. The YAP1/SIX2 axis is required for DDX3-mediated tumor aggressiveness and cetuximab resistance in KRAS-wild-type colorectal cancer. Theranostics 2017;7:1114–32. doi: 10.7150/thno.18175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhu SM, Chen CM, Jiang ZY, Yuan B, Ji M, Wu FH, et al. MicroRNA-185 inhibits cell proliferation and epithelial-mesenchymal transition in hepatocellular carcinoma by targeting Six2. Eur Rev Med Pharmacol Sci 2016;20:1712–9. [PubMed] [Google Scholar]
- [12].Wegert J, Ishaque N, Vardapour R, Geörg C, Gu Z, Bieg M, et al. Mutations in the SIX1/2 Pathway and the DROSHA/DGCR8 miRNA Microprocessor Complex Underlie High-Risk Blastemal Type Wilms Tumors. Cancer Cell 2015. doi: 10.1016/j.ccell.2015.01.002. [DOI] [PubMed] [Google Scholar]
- [13].Li L, Liang Y, Kang L, Liu Y, Gao S, Chen S, et al. Transcriptional Regulation of the Warburg Effect in Cancer by SIX1. Cancer Cell 2018;33:368–385.e7. doi: 10.1016/j.ccell.2018.01.010. [DOI] [PubMed] [Google Scholar]
- [14].Pierce J, Murphy AJ, Panzer A, de Caestecker C, Ayers GD, Neblett D, et al. SIX2 Effects on Wilms Tumor Biology. Transl Oncol 2014;7:800–11. doi: 10.1016/j.tranon.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].H Heppner GR Miller F, Malathy Shekhar PV. Nontransgenic models of breast cancer. Breast Cancer Res 2000;2:331–4. doi: 10.1186/bcr77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Audetat KA, Galbraith MD, Odell AT, Lee T, Pandey A, Espinosa JM, et al. A kinase-independent role for CDK19 in p53 response. Mol Cell Biol 2017:MCB.00626–16. doi: 10.1128/MCB.00626-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Reich M, Liefeld T, Gould J, Lerner J, Tamayo P. GenePattern 2.0 - Nature Genetics. Nat Genet 2006. Volume number 38: 500–1. [DOI] [PubMed] [Google Scholar]
- [18].Iwanaga R, Wang C-A, Micalizzi DS, Harrell JC, Jedlicka P, Sartorius CA, et al. Expression of Six1 in luminal breast cancers predicts poor prognosis and promotes increases in tumor initiating cells by activation of extracellular signal-regulated kinase and transforming growth factor-beta signaling pathways. Breast Cancer Res 2012;14:R100. doi: 10.1186/bcr3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Anders CK, Carey LA. Biology, metastatic patterns, and treatment of patients with triple-negative breast cancer. Clin Breast Cancer 2009;9 Suppl 2:S73–81. doi: 10.3816/CBC.2009.s.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Vazquez-Santillan K, Melendez-Zajgla J, Jimenez-Hernandez LE, Gaytan-Cervantes J, Munõz-Galindo L, Pinã-Sanchez P, et al. NF-kappa B-inducing kinase regulates stem cell phenotype in breast cancer. Sci Rep 2016;6:37340. doi: 10.1038/srep37340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hiraga T, Ito S, Nakamura H. EpCAM expression in breast cancer cells is associated with enhanced bone metastasis formation. Int J Cancer 2016;138:1698–708. doi: 10.1002/ijc.29921. [DOI] [PubMed] [Google Scholar]
- [23].Spike BT, Engle DD, Lin JC, Cheung SK, La J, Wahl GM. A Mammary Stem Cell Population Identified and Characterized in Late Embryogenesis Reveals Similarities to Human Breast Cancer. Cell Stem Cell 2012;10:183–97. doi: 10.1016/j.stem.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Weiswald LB, Bellet D, Dangles-Marie V. Spherical Cancer Models in Tumor Biology. Neoplasia (United States) 2015;17:1–15. doi: 10.1016/j.neo.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pinto JP, Kalathur RK, Oliveira DV., Barata T, Machado RSR, Machado S, et al. StemChecker: A web-based tool to discover and explore stemness signatures in gene sets. Nucleic Acids Res 2015;43:W72–7. doi: 10.1093/nar/gkv529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wang C-H, Ma N, Lin Y-T, Wu C-C, Hsiao M, Lu FL, et al. A shRNA Functional Screen Reveals Nme6 and Nme7 Are Crucial for Embryonic Stem Cell Renewal. Stem Cells 2012;30:2199–211. doi: 10.1002/stem.1203. [DOI] [PubMed] [Google Scholar]
- [27].Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell 2008;2:333–44. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yang S-H, Kalkan T, Morrisroe C, Smith A, Sharrocks AD. A genome-wide RNAi screen reveals MAP kinase phosphatases as key ERK pathway regulators during embryonic stem cell differentiation. PLoS Genet 2012;8:e1003112. doi: 10.1371/journal.pgen.1003112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Som A, Harder C, Greber B, Siatkowski M, Paudel Y, Warsow G, et al. The PluriNetworK: An electronic representation of the network underlying pluripotency in mouse, and its applications. PLoS One 2010;5:1–13. doi: 10.1371/journal.pone.0015165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wu S, He X, Li M, Shi F, Wu D, Pan M, et al. MiRNA-34a overexpression inhibits multiple myeloma cancer stem cell growth in mice by suppressing TGIF2. Am J Transl Res 2016;8:5433–43. [PMC free article] [PubMed] [Google Scholar]
- [31].Jin X, Mu P. Targeting Breast Cancer Metastasis. Breast Cancer (Auckl) 2015;9:23–34. doi: 10.4137/BCBCR.S25460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Santel A, Aleku M, Röder N, Möpert K, Durieux B, Janke O, et al. Atu027 Prevents Pulmonary Metastasis in Experimental and Spontaneous Mouse Metastasis Models. Clin Cancer Res 2010;16:5469–80. doi: 10.1158/1078-0432.CCR-10-1994. [DOI] [PubMed] [Google Scholar]
- [33].Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, et al. Identification of Selective Inhibitors of Cancer Stem Cells by High-Throughput Screening. Cell 2009;138:645–59. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Holmfeldt P, Ganuza M, Marathe H, He B, Hall T, Kang G, et al. Functional screen identifies regulators of murine hematopoietic stem cell repopulation. J Exp Med 2016;213:433–49. doi: 10.1084/jem.20150806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yates B, Braschi B, Gray KA, Seal RL, Tweedie S, Bruford EA. Genenames.org: The HGNC and VGNC resources in 2017. Nucleic Acids Res 2017;45:D619–25. doi: 10.1093/nar/gkw1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Voutsadakis I The network of pluripotency, epithelial-mesenchymal transition, and prognosis of breast cancer. Breast Cancer Targets Ther 2015;7:303. doi: 10.2147/BCTT.S71163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Miyagi S, Saito T, Mizutani K -i., Masuyama N, Gotoh Y, Iwama A, et al. The Sox-2 Regulatory Regions Display Their Activities in Two Distinct Types of Multipotent Stem Cells. Mol Cell Biol 2004;24:4207–20. doi: 10.1128/MCB.24.10.4207-4220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Brodbeck S, Besenbeck B, Englert C. The transcription factor Six2 activates expression of the Gdnf gene as well as its own promoter. Mech Dev 2004;121:1211–22. doi: 10.1016/j.mod.2004.05.019. [DOI] [PubMed] [Google Scholar]
- [39].Takai A, Nakano M, Saito K, Haruno R, Watanabe TM, Ohyanagi T, et al. Expanded palette of Nano-lanterns for real-time multicolor luminescence imaging. Proc Natl Acad Sci 2015;112:4352–6. doi: 10.1073/pnas.1418468112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wei L, Liu T-T, Wang H-H, Hong H-M, Yu AL, Feng H-P, et al. Hsp27 participates in the maintenance of breast cancer stem cells through regulation of epithelial-mesenchymal transition and nuclear factor-κB. Breast Cancer Res 2011;13:R101. doi: 10.1186/bcr3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Noman MZ, Buart S, Romero P, Ketari S, Janji B, Mari B, et al. Hypoxia-inducible miR-210 regulates the susceptibility of tumor cells to lysis by cytotoxic T cells. Cancer Res 2012;72:4629–41. doi: 10.1158/0008-5472.CAN-12-1383. [DOI] [PubMed] [Google Scholar]
- [42].Otomo R, Otsubo C, Matsushima-Hibiya Y, Miyazaki M, Tashiro F, Ichikawa H, et al. TSPAN12 is a critical factor for cancer–fibroblast cell contact-mediated cancer invasion. Proc Natl Acad Sci 2014;111:18691–6. doi: 10.1073/pnas.1412062112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Oktem G, Bilir A, Uslu R, Inan SV, Demiray SB, Atmaca H, et al. Expression profiling of stem cell signaling alters with spheroid formation in CD133(high)/CD44(high) prostate cancer stem cells. Oncol Lett 2014;7:2103–9. doi: 10.3892/ol.2014.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Tan Z, Luo X, Xiao L, Tang M, Bode AM, Dong Z, et al. The Role of PGC1 in Cancer Metabolism and its Therapeutic Implications. Mol Cancer Ther 2016;15:774–82. doi: 10.1158/1535-7163.MCT-15-0621. [DOI] [PubMed] [Google Scholar]
- [45].Xi XP, Zhuang J, Teng MJ, Xia LJ, Yang MY, Liu QG, et al. MicroRNA-17 induces epithelial-mesenchymal transition consistent with the cancer stem cell phenotype by regulating CYP7B1 expression in colon cancer. Int J Mol Med 2016;38:499–506. doi: 10.3892/ijmm.2016.2624. [DOI] [PubMed] [Google Scholar]
- [46].Inoue R, Hirohashi Y, Kitamura H, Nishida S, Murai A, Takaya A, et al. GRIK2 has a role in the maintenance of urothelial carcinoma stem-like cells, and its expression is associated with poorer prognosis. Oncotarget 2017;8:28826–39. doi: 10.18632/oncotarget.16259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wozniak AC, Anderson JE. Nitric oxide-dependence of satellite stem cell activation and quiescence on normal skeletal muscle fibers. Dev Dyn 2007;236:240–50. doi: 10.1002/dvdy.21012. [DOI] [PubMed] [Google Scholar]
- [48].Wang L, Wu J, Yuan J, Zhu X, Wu H, Li M. Midline2 is overexpressed and a prognostic indicator in human breast cancer and promotes breast cancer cell proliferation in vitro and in vivo. Front Med 2016;10:41–51. doi: 10.1007/s11684-016-0429-z. [DOI] [PubMed] [Google Scholar]
- [49].Zorniak M, Clark PA, Kuo JS. Myelin-forming cell-specific cadherin-19 is a marker for minimally infiltrative glioblastoma stem-like cells. J Neurosurg 2015;122:69–77. doi: 10.3171/2014.9.JNS132373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Takahashi M, Osumi N. Identification of a novel type II classical cadherin: Rat cadherin19 is expressed in the cranial ganglia and Schwann cell precursors during development. Dev Dyn 2005;232:200–8. doi: 10.1002/dvdy.20209. [DOI] [PubMed] [Google Scholar]
- [51].Tong Y, Zheng Y, Zhou J, Oyesiku NM, Koeffler HP, Melmed S. Genomic Characterization of Human and Rat Prolactinomas. Endocrinology 2012;153:3679–91. doi: 10.1210/en.2012-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Bross ID, Viadana E, Pickren J. Do generalized metastases occur directly from the primary? J Chronic Dis 1975;28:149–59. [DOI] [PubMed] [Google Scholar]
- [53].Coletta RD, McCoy EL, Burns V, Kawakami K, McManaman JL, Wysolmerski JJ, et al. Characterization of the Six1 homeobox gene in normal mammary gland morphogenesis. BMC Dev Biol 2010;10:4. doi: 10.1186/1471-213X-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Liu S, Wicha MS. Targeting breast cancer stem cells. J Clin Oncol 2010. doi:JCO.2009.27.5388 [pii]\r 10.1200/JCO.2009.27.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Cerami E,Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.