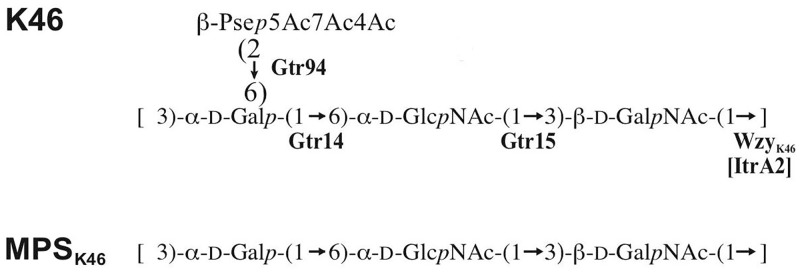Abstract
Acinetobacter baumannii isolate NIPH 329 carries a novel capsular polysaccharide (CPS) gene cluster, designated KL46, that is closely related to the KL5 locus in A. baumannii isolate SDF but includes genes for synthesis of 5,7-diacetamido-3,5,7,9-tetradeoxy-l-glycero-l-manno-non-2-ulosonic (di-N-acetylpseudaminic) acid (Pse5Ac7Ac) instead of the corresponding D-glycero-D-galacto isomer (di-N-acetyllegionaminic acid) (Leg5Ac7Ac). In agreement with the genetic content of KL46, chemical studies of the K46 CPS produced by NIPH 329 revealed a branched tetrasaccharide repeat (K unit) with an overall structure the same as K5 from SDF but with â-Pse5Ac7Ac replacing α-Leg5Ac7Ac. As for K5, the K46 unit begins with d-GalpNAc and includes α-d-GlcpNAc-(1→3)-d-GalpNAc and α-d-Galp-(1→6)-d-GlcpNAc linkages, formed by Gtr14 and Gtr15 glycosyltransferases, respectively. The Gtr94K46 glycosyltransferase, which is related to Gtr13K5, links Pse5Ac7Ac to d-Galp in the growing K unit via a â-(2→6) linkage. Nearly identical Wzy enzymes connect the K46 and K5 units via a α-D-GalpNAc-(1→3)-α-D-Galp linkage to form closely related CPSs. Both Pse5Ac7Ac in K46 and Leg5Ac7Ac in K5 are acetylated at O4 but no acetyltransferase gene is present in KL46 or KL5. Related acetyltransferases were found encoded in the NIPH 329 and SDF genomes, but not in other strains carrying an unacetylated Pse or Leg derivative in the CPS. The genes encoding the acetyltransferases were in different putative phage genomes. However, related acetyltransferases were rare among the >3000 publically available genome sequences.
Introduction
Capsular polysaccharide (CPS) is an essential virulence determinant for the globally-significant bacterial pathogen, Acinetobacter baumannii, which has been listed by the World Health Organisation as the leading critical priority pathogen for therapeutics development due to the increasing prevalence of isolates with limited treatment options [1]. A. baumannii is a nosocomial pathogen that causes respiratory tract, wound, and urinary tract infections. As the outer-most antigenic component of the cell, the CPS has been exploited in various vaccine approaches and novel phage therapies. However, these strategies have challenges due to the extreme variation of the CPS structures in different isolates; more than 125 distinct CPS biosynthesis gene clusters have been identified (J.J. Kenyon, unpublished data) at the genomic K locus (KL) that directs the synthesis of the CPS [2]. The CPS of different isolates may have different sugar compositions and include different linkages between these sugars or between oligosaccharide repeats (K units) that make up the CPS polymers extending from the cell surface (e.g. [3–8]).
Given the importance of the CPS to development of alternate therapeutics, it is critical to examine the factors that give rise to structural variation. In the majority of cases studied to date, there has been perfect correlation between the genetic content at the K locus and CPS structure produced by the same isolate. When sugars were found to be modified by the addition of acetyl, acyl or pyruvyl residues, the gene for the transferase, acetyltransferase, pyruvyltransferase, etc., generally is present in the KL gene cluster [8–13]. However, in rare cases, genes identified elsewhere in the genome contribute to CPS synthesis or modification of the final CPS structure. For example, two different genomic islands (GIs), GI-1 and GI-2, have been described in the analysis of the A. baumannii K19 and K24 CPS, respectively [14, 15]. Both GIs carry a wzy gene, encoding the polymerase that links the K units together to form the long chain CPS, and the KL19 and KL24 gene clusters both lack a wzy candidate. GI-1 was further found to include an atr gene responsible for the modification of the K19 unit with an acetyl group [14].
Recently, the K5 CPS produced by A. baumannii isolate SDF was found to be 4-O-acetylated on the non-2-ulosonic acid residue, 5,7-diacetamido-3,5,7,9-tetradeoxy-D-glycero-D-galacto-non-2-ulosonic (di-N-acetyllegionaminic) acid (Leg5Ac7Ac). However, an acetyltransferase gene was not found in the KL5 gene cluster [5], suggesting that an unidentified gene located elsewhere in the genome may be involved. In this study, we examine the CPS structure produced by A. baumannii isolate NIPH 329 [16], which carries a novel KL gene cluster related to KL5, and correlate the structure with the whole genome sequence available.
Materials and methods
Bacterial strain and cultivation
A. baumannii NIPH 329 was isolated in the Czech Republic and obtained from Prof. Alexandr Nemec [16, 17]. Bacteria were cultivated in 2TY media for 16 h; cells were harvested by centrifugation (10,000×g, 20 min), washed with phosphate-buffered saline, suspended in a 7:3 (v/v) acetone-water mixture, precipitated by centrifugation, and dried on air.
Isolation of the CPS
CPS was isolated by phenol-water extraction [18] of bacterial cells, the extract was dialyzed without layer separation and freed from insoluble contaminations by centrifugation. The resultant solution was treated with cold (4 °C) aqueous 50% CCl3CO2H; after centrifugation, the supernatant was dialyzed against distilled water and freeze-dried to give a CPS sample in a yield 9.5% of dry weight.
Sugar analysis
A CPS sample (0.5 mg) was hydrolyzed with 2 M CF3CO2H (120 °C, 2 h). Monosaccharides were identified by GLC of the alditol acetates on a Maestro (Agilent 7820) chromatograph (Interlab, Russia) equipped with an HP-5ms column (0.32 mm × 30 m) and a temperature program of 160 °C (1 min) to 290 °C at 7 °C min-1.
Mild acid hydrolysis of the CPS
A CPS sample (23 mg) was hydrolyzed with aqueous 2% HOAc (100 °C, 1 h), the products were fractionated by gel chromatography on a column (80 × 1.6 cm) of TSK HW-40 (S) in 1% HOAc using a differential refractometer (Knauer, Germany) for monitoring to give a modified polysaccharide (MPS) sample (7 mg) and Pse5Ac7Ac (2 mg).
NMR spectroscopy
Samples were deuterium-exchanged by freeze-drying from 99.9% D2O and then examined as solution in 99.95% D2O. NMR spectra were recorded on a Bruker Avance II 600 MHz spectrometer (Germany) at 50 °C for CPS and MPS or 24 °C for Pse5Ac7Ac, using sodium 3-trimethylsilylpropanoate-2,2,3,3-d4 (δH 0, δC −1.6) as the internal reference. 2D NMR spectra were obtained using standard Bruker software, and Bruker TopSpin 2.1 program was used to acquire and process the NMR data. A mixing time of 100 and 150 ms was used in TOCSY and ROESY experiments, respectively.
Bioinformatic analysis
The K locus sequence in the genome of NIPH 329 was extracted for analysis from WGS accession number APQY01000009 REGION: complement (584809–608726). Short reads for LUH5553 (KL90) and LUH5533 (KL7) were obtained from SRA accession numbers DRR006294 and DRR006297, and were assembled into contigs using the SPAdes assembly algorithm [19]. All novel gene clusters identified were annotated and characterised as described previously [2], and figures representing genetic sequences were constructed to scale using the Gene Construction Kit program. The sequence types (STs) of all isolates examined in this study were determined using the Pasteur MLST scheme (https://pubmlst.org/abaumannii/). Related KL gene clusters and phage-associated atr genes were identified using blastn (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Genome or contig sequences of interest were examined for potential prophage regions using PHASTER (http://phaster.ca/) [20].
Results
KL gene cluster in the genome of A. baumannii NIPH 329
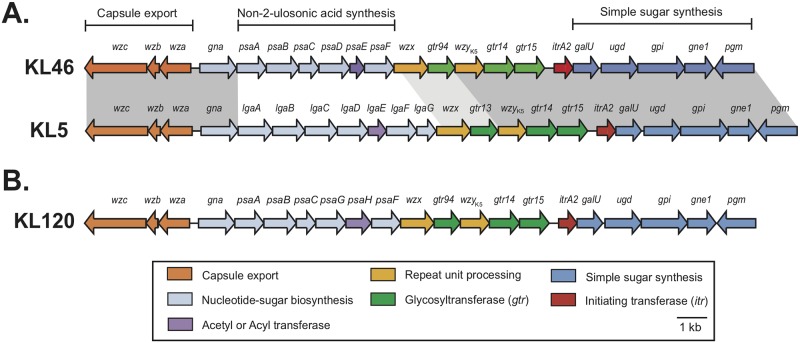
A novel CPS biosynthesis gene cluster was identified in the draft genome of A. baumannii isolate NIPH 329 (WGS accession number APQY01000009.1), annotated as described previously [2], and designated KL46 (GenBank accession number MK609549.1). The KL46 gene cluster (Fig 1A) has an arrangement that is typical for A. baumannii KL gene clusters [2] in that it includes a module of genes for CPS export (wza-wzb-wzc) and another one (galU, ugd, gpi, gne1, pgm) for synthesis of simple sugars on either side of a central region that is specific to the K46 CPS. The content of the KL46 gene cluster is similar overall to that of the KL5 gene cluster carried by A. baumannii isolate SDF (Fig 1A). However, in the central region, the KL46 gene cluster carries six genes, psaABCDEF, for synthesis of CMP-activated 5,7-di-N-acetylpseudaminic acid (Pse5Ac7Ac) [2] [21–23], whereas KL5 has lgaABCDEFG genes to make CMP-activated 5,7-di-N-acetyllegionaminic acid (Leg5Ac7Ac) [2][5].
Fig 1.
A. Comparison of the KL46 and KL5 capsular polysaccharide biosynthesis gene clusters of A. baumannii NIPH 329 and SDF, respectively. Modules of genes required for specific processes in CPS synthesis are indicated above. Dark shading between the gene clusters indicates >95% and light grey shading 80–95% nucleotide sequence identity. B. Organisation of the KL120 gene cluster that is similar to KL46. Colour scheme for the genes shown below indicates the functions of the encoded gene products. Figure is drawn to scale from GenBank accession numbers MK609549. 1 (KL46), CU468230.2 (KL5; coordinates 61721 to 89706), and LLCR01000062.1 (KL120; coordinates 12963 to 39906).
KL46 has genes for three glycosyltransferases (gtr94, gtr14 and gtr15) and the initiating transferase ItrA2 (itrA2) that transfers D-GalpNAc-P from UDP-D-GalpNAc to the UndP lipid carrier [21]. The combination of gtr14, gtr15, and itrA2 genes were found previously in the KL5 gene cluster, and the ItrA2, Gtr14 and Gtr15 proteins from KL46 are 96–99% identical to those encoded by KL5. K5 includes D-GalpNAc as the first sugar of the K unit. Gtr15 then adds D-GlcpNAc via an α-(1→3) linkage to the D-GalpNAc sugar, and Gtr14 links a D-Galp residue via an α-(1→6) linkage to the D-GlcpNAc [5]. Thus, it was expected that K46 will also include an α-D-Galp-(1→6)-α-D-GlcpNAc-(1→3)-D-GalpNAc segment. Gtr94K46 (GenPept accession number ENW42232.1) is 75% identical to Gtr13K5 (GenPept accession number CAO99488.1) that has previously been shown to link Leg5Ac7Ac to D-Galp via an α-(2→6) linkage in biosynthesis of the K5 CPS [5]. Thus, it is likely that Gtr94K46 forms a â-Pse5Ac7Ac-(2→6)-D-Galp linkage in K46.
The KL46 and KL5 gene clusters further include wzy genes that produce closely related proteins sharing 94% sequence identity. WzyK5 has been shown to form an α-D-GalpNAc-(1→3)-α-D-Galp linkage between K5 units, and WzyK46 should catalyse formation of the same linkage.
Sugar compositions of the K46 CPS
Sugar analysis of the CPSK46 by GLC of the alditol acetates revealed galactose, glucosamine, and galactosamine. The d configuration of the monosaccharides was inferred from genetic data (see below). Further studies by NMR spectroscopy indicated that the CPS also contained Pse5Ac7Ac. The CPS was hydrolyzed under mild acidic conditions to give a modified Pse5Ac7Ac-lacking polysaccharide (MPSK46) and free Pse5Ac7Ac, which were isolated by GPC on Fractogel TSK HW-40. Pse5Ac7Ac was identified using NMR spectroscopy by comparison of the 1H and 13C NMR chemical shifts and 3JH,H coupling constants with published data [24]. Formation of the MPSK46 upon mild acid hydrolysis indicates that Pse5Ac7Ac is a side-chain sugar.
Structure elucidation of the K46 CPS
The 1H NMR and 13C NMR spectra of the MPSK46 showed signals for one residue each of β-GalNAc (A), α-GlcNAc (B), and α-Gal (C), all being in the pyranose form. The 1H NMR signals were assigned by H-1/H-2,3,4,5 correlations for GlcNAc and H-1/H-2,3,4 correlations for Gal and GalNAc in the 2D 1H,1H TOCSY spectrum combined with correlations between neighbouring protons within each monosaccharide residue in the 2D 1H,1H COSY spectrum. With the 1H NMR signals assigned, the 13C NMR spectrum of the MPS was assigned using a 2D 1H,13C HSQC experiment (Table 1).
Table 1. 13C and 1H NMR chemical shifts of the capsular polysaccharide (CPS) and a modified polysaccharide (MPS) from A. baumannii NIPH 329 (δ, ppm).
| Residue | C-1 | C-2 | C-3 | C-4 | C-5 | C-6 | C-7 | C-8 | C-9 |
|---|---|---|---|---|---|---|---|---|---|
| H-1 | H-2 | H-3 (3ax,3eq) | H-4 | H-5 | H-6 (6a,6b) | H-7 | H-8 | H-9 | |
| CPS | |||||||||
| →3)-β-d-GalpNAc-(1→ | 103.9 | 52.4 | 77.0 | 65.4 | 76.1 | 62.4 | |||
| A | 4.71 | 4.07 | 3.82 | 4.07 | 3.62 | 3.74, 3.79 | |||
| →6)-α-d-GlcpNAc-(1→ | 95.6 | 54.6 | 72.5 | 70.4 | 72.6 | 66.2 | |||
| B | 5.05 | 3.96 | 3.65 | 3.74 | 3.73 | 3.64, 4.16 | |||
| →3,6)-α-d-Galp-(1→ | 99.7 | 68.7 | 80.6 | 70.5 | 70.5 | 65.2 | |||
| C | 4.97 | 3.92 | 3.95 | 4.21 | 4.05 | 3.58, 3.95 | |||
| β-Psep4Ac5Ac7Ac-(2→ | 173.3 | 101.5 | 33.8 | 70.1 | 47.0 | 74.0 | 54.8 | 69.5 | 17.8 |
| D | 1.76, 2.50 | 4.90 | 4.30 | 4.00 | 4.07 | 4.13 | 1.19 | ||
| MPS | |||||||||
| →3)-β-d-GalpNAc-(1→ | 103.8 | 52.3 | 76.9 | 65.4 | 76.1 | 62.3 | |||
| A | 4.71 | 4.07 | 3.81 | 4.07 | 3.62 | 3.73, 3.79 | |||
| →6)-α-d-GlcpNAc-(1→ | 95.5 | 54.6 | 72.5 | 70.4 | 72.6 | 66.2 | |||
| B | 5.05 | 3.96 | 3.65 | 3.72 | 3.73 | 3.68, 4.09 | |||
| →3)-α-d-Galp-(1→ | 99.7 | 68.7 | 80.6 | 70.4 | 71.9 | 62.3 | |||
| C | 4.98 | 3.91 | 3.95 | 4.21 | 3.95 | 3.73, 3.73 | |||
1H NMR chemical shifts are italicized. Chemical shifts for the N-acetyl groups are δH 1.92–2.05; δC 23.0–24.0 (Me) and 174.5–175.9 (CO); for the O-acetyl group δH 2.00; δC 21.7 (Me) and 174.1 (CO).
The signals for C-3 of units A and C and C-6 of unit B at δ 76.9, 80.6, and 66.2 were shifted significantly downfield in the MPSK46, as compared with their positions in the corresponding non-substituted monosaccharides at δ 72.4, 70.4, and 61.9, respectively [25]. These displacements are characteristic of signals for linkage carbons and showed that the MPSK46 is linear and defined the glycosylation pattern in the K unit (Fig 2).
Fig 2. Structures of the capsular polysaccharides CPSK46 and the corresponding modified polysaccharide MPSK46 from A. baumannii NIPH 329.
The order of the monosaccharide residues in the MPSK46 shown in Fig 2 was determined by the 2D 1H,1H ROESY spectrum, which showed α-Gal H-1/α-GlcNAc H-6a,6b, α-GlcNAc H-1/β-GalNAc H-3, and β-GalNAc H-1/α-Gal H-3 correlations at δ 4.98/3.68, 5.05/3.81, and 4.71/3.95, respectively. It was confirmed by α-Gal H-1/α-GlcNAc C-6, α-GlcNAc H-1/β-GalNAc H-3, and β-GalNAc H-1/α-Gal C-3 correlations at δ 4.98/66.2, 5.05/76.9, and 4.71/80.6, respectively, which were observed in the 1H,13C HMBC spectrum of the MPSK46.
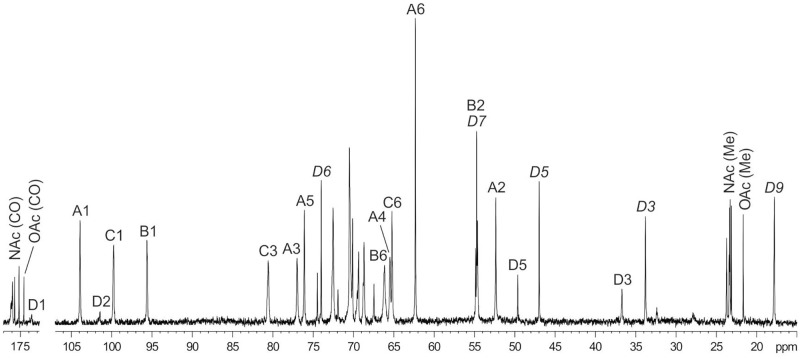
The 1H NMR and 13C NMR (Fig 3) spectra of the CPSK46 showed signals for the same three monosaccharide residues (units A-C) as present in the MPSK46 and, in addition, those for β-Psep5Ac7Ac (unit D), which was identified as described [26]. Particularly, the axial orientation of the carboxyl group, i.e. the β configuration of Pse, was inferred from a relatively large difference of 0.87 ppm between the chemical shifts of H-3ax and H-3eq in the 1H NMR spectrum [24]. The anomeric configuration of Pse was confirmed by the C-6 chemical shift of δ 74.6–74.7 (compare published data [24] δ 71.4 and 74.3 for α- and β-Psep5Ac7Ac, respectively).
Fig 3. 13C NMR spectrum of the capsular polysaccharide CPSK46 from A. baumannii NIPH 329.
Numbers refer to carbons in sugar residues denoted by letters as shown in Table 1. Signals of Pse4Ac5Ac7Ac are annotated in italics.
In the 13C NMR spectrum of the CPSK46, the signal for C-6 of α-Galp was shifted downfield to δ 65.2, as compared with its position at δ 62.3 in the spectrum of the MPSK46; hence β-Psep5Ac7Ac was attached at position 6 of α-Galp. The terminal position of pseudaminic acid in the side chain was confirmed by the results of mild acid hydrolysis of the CPSK46 giving rise to MPSK46 (see above) and a similarity of the 13C NMR chemical shifts of unit D (Table 1) with published data for free β-Psep5Ac7Ac [24].
K46 is O acetylated
The NMR spectra of the CPSK46 showed signals for an O-acetyl group (δH 2.00, δC 21.7 and 174.1). In the 13C NMR spectrum of the CPSK46, a major part of the signal for C-4 of Pse5Ac7Ac was shifted downfield to δ 70.1, and those for C-3 and C-5 were shifted upfield to δ 33.8 and 47.0, as compared with their positions in the non-O-acetylated Pse5Ac7Ac at δ 67.5, 36.8, and 49.7, respectively. These displacements were evidently due to 4-O-acetylation of Pse5Ac7Ac and thus defined the position of the O-acetyl groups in the CPSK46. As judged by the ratio of integral intensities of the signals for Pse4Ac5Ac7Ac and Pse5Ac7Ac, the degree of O-acetylation is ~75%.
Relationship of the CPSK46 structure to the KL46 gene cluster sequence
The CPS produced by A. baumannii isolate NIPH 329 (Fig 2), consists of Pse4Ac5Ac7Ac, D-Galp, D-GlcpNAc and D-GalpNAc sugars, as expected. Given the presence of itrA2 in the KL46 gene cluster, D-GalpNAc is drawn as the first sugar of the K unit in Fig 2. Like WzyK5, WzyK46 would therefore catalyse formation of a â-D-GalpNAc-(1→3)-D-Galp linkage between K units in the K46 CPS. The K46 structure further includes the expected α-D-Galp-(1→6)-â-D-GlcpNAc-(1→3)-D-GalpNAc segment, and formation of these linkages is catalysed by Gtr14K46 and Gtr15K46, respectively. Thus, Gtr94K46 would be responsible for the addition of the final Pse5Ac7Ac residue to D-Galp via a â-(2→6) linkage as predicted (Fig 2). The K46 structure is therefore largely consistent with the content of the KL46 gene cluster. However, the only atr gene in KL46 lies in the psa gene cluster (purple in Fig 1) and is responsible for the acetylation of Pse at N5 and N7. Hence, a candidate atr gene encoding an acetyltransferase (Atr) for the addition of the 4-O-acetyl group to ~75% of the Pse5Ac7Ac residues in the K46 CPS was not found at the KL46 locus.
Identification of an acetyltransferase for 4-O-acetylation of Pse5Ac7Ac in K46
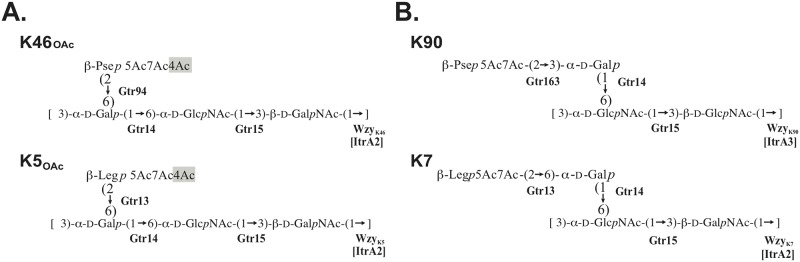
The acetylation pattern in K46 is similar to that of the K5 CPS produced by A. baumannii isolate SDF [5], which includes Leg5Ac7Ac residues that are also 4-O-acetylated (Fig 4A). Similarly, the KL5 gene cluster does not include a suitable atr gene (Fig 1). This suggests that an atr gene located elsewhere in the genome may be involved in the acetylation of the non-2-ulosonic acids in the K46 and K5 CPS units. The potential atr gene(s) should be present in the genomes of NIPH 329 and SDF but absent in strains with related but unacetylated CPS structures, such as K7 and K90 CPS produced respectively by A. baumannii isolates LUH5533 [27] and LUH5553 [28].
Fig 4.
A. CPS structures NIPH329 (this work) and SDF [5] with 4-O-acetylation of the non-2-ulosonic acid residue. B. Related CPS structures of A. baumannii LUH5553 [28] and LUH5533 [27] without 4-O-acetylation of the non-2-ulosonic acid residue. Glycosyltransferases, polymerases and acetyltrasferases are indicated in bold next the linkage they are assigned to. The 4-O-acetyl group is shaded in grey.
Assuming that the candidate Atr belongs to an established protein family, the draft genome sequence of isolate NIPH 329 was investigated for CDS features annotated as either ‘acetyltranferase’, ‘acyltranferase’, or ‘GNAT family acetytransferase’. A total of 27 gene candidates were identified (S1 Table), and each of these were further assessed to identify related genes present in the SDF genome sequence (GenBank accession number CU468230.2) but absent from the genome sequences of isolates LUH5553 and LUH5533 assembled here from available short read data (SRA numbers DRR006294 and DRR006297, respectively). Only one Atr candidate (locus tag F919_03690) encoded by the NIPH 329 genome sequence, designated Atr29 (GenPept accession number ENW41154.1), was related (80% identical) to an Atr, designated Atr30 (GenPept accession number CAP00403.1), encoded by the SDF genome, but Atr29 and Atr30 had no relative in the assembled genome sequences of LUH5533 (KL7) and LUH5553 (KL90).
To further validate the conclusion that Atr29 and Atr30 are responsible for the observed 4-O-acetylation, the genomes of all isolates producing CPS structures containing a derivative of either pseudaminic acid or legionaminic acid were examined (Table 2). None encoded a homologue of Atr29/Atr30. Furthermore, relatives of the atr gene were not widely distributed in A. baumannii, being found in only a small number of complete and draft genome sequences.
Table 2. Pseudaminic acid and legionaminic acid derivatives.
| K Type | Reference | K locus genes2 | Phage atr |
|---|---|---|---|
| Pse5Ac7Ac | |||
| K2 | [21] | psaABCDEF | - |
| K6 | [22] | psaABCDEF | - |
| K16 | [8] | psaABCDEF | - |
| K33 | [29] | psaABCDEF | - |
| K90 | [28] | psaABCDEF | - |
| Pse5Ac7Ac4OAc | |||
| K46 | This study | psaABCDEF | atr29 |
| Pse5Ac7R1 | |||
| K42 | [30] | psaABCGHF | - |
| K93 | [26] | psaABCGHF | - |
| Leg5Ac7Ac | |||
| K7 | [27] | lgaABCDEFG | - |
| K54 | Unpublished | lgaABCDEFG | - |
| Leg5Ac7Ac4OAc | |||
| K5 | [5] | lgaABCDEFG | atr30 |
| Leg5Ac7R1 | |||
| K27 | [3] | lgaABCHIFG | - |
| K44 | [3] | lgaABCHIFG | - |
| K63 | [31] | lgaABCHIFG | - |
| K8 | Unpublished | lgaABCHIFG | - |
1 R is either Ac or 3-hydroxybutanoyl (Hb)
2 Bold face genes determine the acylation/acetylation pattern of the sugar
The atr29 and atr30 genes are in phage genomes
The genetic context of the candidate atr29 gene was examined in the NIPH 329 genome sequence, and the genes located either side of atr29 were found to encode proteins related to phage proteins. The specific contig of the NIPH 329 draft genome containing the atr29 gene (WGS accession number APQY01000013.1) was therefore subjected to PHASTER analysis, which identified 44 of 59 encoded proteins of phage origin and attL and attR sites at both ends of the 42249 bp contig. As no adjacent chromosomal sequence was present, it was not possible to locate the position of the prophage in the NIPH 329 genome.
The SDF genome was also analysed using PHASTER, and the atr30 gene was found close to a potential phage region of ~23.6 kb (GenBank accession number CU468230.2; coordinates 932745–956361) at ~0.8 Mb from the K locus in the SDF chromosome (Table 3). However, this prophage sequence shared no significant identity with that in NIPH 329, suggesting that there are multiple members of the atr29/atr30 gene family, each in a different phage genome.
Table 3. A. baumannii genome sequences with phage genomes carrying an atr gene for 4-O-acetylation of the CPS.
| Strain | GenBank accession number | Phage match | Coordinates of phage genome | Coordinates of atr29/atr30 | GenPept accession number of Atr29/Atr30 | Amino acid sequence identitya |
|---|---|---|---|---|---|---|
| NIPH329 | APQY01000013.1 | PHAGE_Acinet_vB_AbaS_TRS1_NC_031098(28) [incomplete] | 1-42232 | 39593–40639 (atr29) | ENW41154.1 | 100% |
| ABBL011 | LLCR01000058.1 | PHAGE_Acinet_vB_AbaS_TRS1_NC_031098(27) [incomplete] | 97376-149013 | 141964-143010(atr29) | KRI30500.1 | 99% |
| ARLG1935 | NGIJ01000016.1 | PHAGE_Acinet_Bphi_B1251_NC_019541(11) [questionable] | 145514-178780 | 145062–146108 (atr29) | (CAT67_11335)b | 92% |
| SDF | CU468230.2 | PHAGE_Entero_mEp235 [questionable] | 932745–956361 | 959212–960231 (atr30) | CAP00403.1 | 80% |
a Amino acid sequence identity is to GenPept accession number ENW41154.1 from NIPH329.
b Translated into amino acid sequence from locus tag indicated in parentheses.
A. baumannii isolates carrying related candidate atr genes
A homologue of Atr29 with 99% amino acid (aa) sequence identity was encoded by a gene carried by A. baumannii isolate ABBL011 (WGS accession number LLCR01000058.1), and was also located in a phage-associated sequence region. However, the surrounding sequence was not closely related to the atr29-associated phage genome in NIPH 329. ABBL011 was found to carry a new KL variant, KL120 (GenBank accession number LLCR01000062.1; coordinates 12963 to 39906), a close relative of KL46 in which two genes in the psa gene cluster (psaG/H in KL120 and psaD/E in KL46) have been replaced (Fig 1B). The presence of psaG/H predicts the synthesis of Pse5Ac7Hb (where Hb is 3-hydroxybutanoyl), thus it is likely that KL120 directs the synthesis of a CPS similar to K46 but with Pse5Ac7Hb replacing Pse5Ac7Ac. ABBL011 belongs to ST6 in the Pasteur MLST scheme, whereas NIPH 329 belongs to the unrelated ST11.
A homologue of Atr29 with 92% amino acid (aa) sequence identity was also encoded by a phage-like sequence in A. baumannii isolate ARLG1935 (WGS accession numbers NGIJ01000016.1) but this phage genome was again not related to that in NIPH 329 (Table 3). This isolate also carries KL46 (WGS accession number NGIJ01000019.1; coordinates 31657 to 58229) and, like NIPH 329, belongs to ST11 indicating that more than one phage can bring in an atr gene that has the potential to modify the CPS.
4-O-Acetylation of the K46 is not universal
As close homologues of the atr gene product encoded by NIPH329 and SDF, which produce CPS with 4-O-acetylation of a non-2-ulosonic acid, are found in isolates ARLG1935 and ABBL011 that carry related KL gene clusters, it is likely that Atr29 and Atr30 also perform 4-O-acetylation of Pse and Leg residues in these CPSs. However, the KL46 gene cluster was further identified in the genome sequence of A. baumannii isolate TG22162 (WGS accession number RFCR01000020.1; coordinates 14854 to 41423). The atr29 gene was not found in the genome sequence of isolate TG22162 suggesting that 4-O-acetylation does not occur in all isolates producing a K46 CPS structure.
Discussion
The K46 CPS structure elucidated in this study includes a 4-O-acetylated variant of pseudaminic acid attached as a side chain to a á-D-Galp-(1→6)-â-D-GlcpNAc-(1→3)-D-GalpNAc backbone via a â-(2→6) linkage to D-Galp. Formation of all these linkages catalyzed by the glycosyltransferases encoded by the KL46 gene cluster could be assigned due to the close relationship to those encoded in the KL5 gene cluster. Their roles were previously assigned via comparison of the K5 [5] and K7 [27] CPS structures produced by isolates carrying the KL5 and KL7 gene clusters, a closely related pair that differ only in a segment that includes the wzy gene [2]. KL46 and KL5 are also a closely related pair of gene clusters that differ only in a central segment that includes the genes for synthesis of the non-2-ulosonic acid side-branch (Fig 1).
Though the K46 structure is largely consistent with the content of the KL46 gene cluster, the 4-O-acetylation of the Pse5Ac7Ac residue could not be explained by any gene located at the K locus, and a similar situation was observed previously for K5 [5]. In this study, we identified a potential atr candidate, atr29, in the genome of NIPH 329 in a region likely to be of phage origin, though it could not be placed in the chromosome. A homologue of the atr29 gene, atr30, was also found encoded by a potential phage genome in the SDF chromosome and the K5 CPS of SDF is also 4-O-acetylated. We also identified a further two phage-associated Atr29 homologues in the genomes of two other isolates that carry KL gene clusters with Pse5Ac7Ac biosynthesis genes. Weaker homologues of 70–80% aa sequence identity with >96% sequence coverage were also found encoded by a further 17 A. baumannii genome sequences (data not shown). However, the precise role of Atr29/Atr30 and its homologues in CPS synthesis remains to be confirmed, and in this regard determination of the structure of the CPS produced by TG22162, which also carries KL46 but does not encode any Atr29 homologue may be valuable.
The discovery of polysaccharide genes in prophage sequences, in particular acetyltransferase genes that contribute to surface polysaccharide variation in bacteria, has been reported in other Gram-negative bacteria including Salmonella [32, 33], Shigella [34, 35] and Pseudomonas [36]. Previously, it has been suggested that the incorporation of polysaccharide biosynthesis genes into phage genomes confers a potential fitness advantage for the phage, via the alteration of the CPS structure to prevent other phage that recognise the same structure from infecting the cell [35]. However, the finding of CPS genes in phage sequence demonstrates that some CPS biosynthesis genes are able to transfer between cells via mechanisms other than by homologous recombination at the K locus. Interestingly, both GI-1 and GI-2 regions with CPS genes were found to be flanked by direct repeats in A. baumannii genomes, suggesting that these islands had also inserted into the chromosome and may also be mobile. However, the exact mechanism of this transfer is still unknown. These additional evolutionary transfer mechanisms could assist with the rapid alteration of the CPS structure, and this could prevent recognition by antibodies or phage-encoded hydrolases. This would in turn potentially complicate vaccine and phage therapies used for treatment of A. baumannii infections that target specific CPS structures.
Supporting information
(DOCX)
Acknowledgments
We thank Prof. Alexandr Nemec (Laboratory of Bacterial Genetics, National Institute of Public Health, Prague, Czech Republic) for supplying A. baumannii isolate NIPH 329. Computational resources and services used in this work were provided, in part, by the HPC and Research Support Group, Queensland University of Technology, Brisbane, Australia.
Data Availability
Sequence data is available in the GenBank database under accession number MK609549.1. All other relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was financially supported by the Russian Science Foundation (project No. 19-14-00273) to YAK and an Australian Research Council (ARC) DECRA fellowship 180101563 to JJK.
References
- 1.World Health Organisation. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. 2017. https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf.
- 2.Kenyon JJ, Hall RM. Variation in the complex carbohydrate biosynthesis loci of Acinetobacter baumannii genomes. PLoS One. 2013;8(4): e62160 10.1371/journal.pone.0062160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shashkov AS, Kenyon JJ, Senchenkova SN, Shneider MM, Popova AV, Arbatsky NP, et al. Acinetobacter baumannii K27 and K44 capsular polysaccharides have the same K unit but different structures due to the presence of distinct wzy genes in otherwise closely related K gene clusters. Glycobiology. 2016;26(5): 501–8. 10.1093/glycob/cwv168 [DOI] [PubMed] [Google Scholar]
- 4.Shashkov AS, Kenyon JJ, Arbatsky NP, Shneider MM, Popova AV, Knirel YA, et al. Genetics of biosynthesis and structure of the K53 capsular polysaccharide of Acinetobacter baumannii D23 made up of a disaccharide K unit. Microbiology. 2018;164: 1289–92. 10.1099/mic.0.000710 [DOI] [PubMed] [Google Scholar]
- 5.Arbatsky NP, Kenyon JJ, Shashkov AS, Shneider MM, Popova AV, Kalinchuk N, et al. The K5 capsular polysaccharide of the bacterium Acinetobacter baumannii SDF with the same K unit containing Leg5Ac7Ac as the K7 capsular polysaccharide but a different linkage between the K units. Russ Chem Bull. Int. Ed. 2019;68(1): 163–7. [Google Scholar]
- 6.Arbatsky NP, Shneider MM, Dmitrenok A, Popova AV, Shagin D, Shelenkov A, et al. Structure and gene cluster of the K125 capsular polysaccharide from Acinetobacter baumannii MAR13-1452. Int J Biol Macromol. 2018;117: 1195–9. 10.1016/j.ijbiomac.2018.06.029 [DOI] [PubMed] [Google Scholar]
- 7.Kenyon JJ, Kasimova AA, Notaro A, Arbatsky NP, Speciale I, Shashkov AS, et al. Acinetobacter baumannii K13 and K73 capsular polysaccharides differ only in K-unit side branches of novel non-2-ulosonic acids: di-N-acetylated forms of either acinetaminic acid or 8-epiacinetaminic acid. Carbohydr Res. 2017;452: 149–55. 10.1016/j.carres.2017.10.005 [DOI] [PubMed] [Google Scholar]
- 8.Kenyon JJ, Arbatsky NP, Sweeney EL, Shashkov AS, Shneider MM, Popova AV, et al. Production of the K16 capsular polysaccharide by Acinetobacter baumannii ST25 isolate D4 involves a novel glycosyltransferase encoded in the KL16 gene cluster. Int J Biol Macromol. 2019;128: 101–6. 10.1016/j.ijbiomac.2019.01.080 [DOI] [PubMed] [Google Scholar]
- 9.Kenyon JJ, Speciale I, Hall RM, De Castro C. Structure of repeating unit of the capsular polysaccharide from Acinetobacter baumannii D78 and assignment of the K4 gene cluster. Carbohydr Res. 2016;434: 12–7. 10.1016/j.carres.2016.07.016 [DOI] [PubMed] [Google Scholar]
- 10.Kenyon JJ, Shashkov AS, Senchenkova SN, Shneider MM, Liu B, Popova AV, et al. Acinetobacter baumannii K11 and K83 capsular polysaccharides have the same 6-deoxy-L-talose-containing pentasaccharide K units but different linkages between the K units. Int J Biol Macromol. 2017;103: 648–55. 10.1016/j.ijbiomac.2017.05.082 [DOI] [PubMed] [Google Scholar]
- 11.Kasimova AA, Kenyon JJ, Arbatsky NP, Shashkov AS, Popova AV, Shneider MM, et al. Acinetobacter baumannii K20 and K21 capsular polysaccharide structures establish roles for UDP-glucose dehydrogenase Ugd2, pyruvyl transferase Ptr2 and two glycosyltransferases. Glycobiology. 2018;28(11): 876–84. 10.1093/glycob/cwy074 [DOI] [PubMed] [Google Scholar]
- 12.Kasimova AA, Kenyon JJ, Arbatsky NP, Shashkov AS, Popova AV, Knirel YA, et al. Structure of the K82 capsular polysaccharide from Acinetobacter baumannii LUH5534 containing a D-galactose 4,6-pyruvic acid acetal. Biochem (Moscow). 2018;83(7): 831–5. [DOI] [PubMed] [Google Scholar]
- 13.Shashkov AS, Liu B, Kenyon JJ, Popova AV, Shneider MM, Senchenkova SN, et al. Structures of the K35 and K15 capsular polysaccharides of Acinetobacter baumannii LUH5535 and LUH5554 containing amino and diamino uronic acids. Carbohydr Res. 2017;448: 28–34. 10.1016/j.carres.2017.05.017 [DOI] [PubMed] [Google Scholar]
- 14.Kenyon JJ, Shneider MM, Senchenkova SN, Shashkov AS, Siniagina M, Malanin S, et al. The K19 capsular polysaccharide of Acinetobacter baumannii is produced via a Wzy polymerase encoded in a small genomic island rather than the KL19 capsule gene cluster. Microbiol. 2016;162: 1479–89. [DOI] [PubMed] [Google Scholar]
- 15.Kenyon JJ, Kasimova AA, Shneider MM, Shashkov AS, Arbatsky NP, Popova AS, et al. The KL24 gene cluster and a genomic island encoding a Wzy polymerase contribute genes needed for synthesis of the K24 capsular polysaccharide by the multiply antibiotic resistant Acinetobacter baumannii isolate RCH51. Microbiol. 2017;163: 355–363. [DOI] [PubMed] [Google Scholar]
- 16.Périchon B, Goussard S, Walewski V, Krizova L, Cerqueira G, Murphy C, et al. Identification of 50 class D β-lactamases and 65 Acinetobacter-derived cephalosporinases in Acinetobacter spp. Antimicrob Agents Chemother. 2014;58(2): 936–49. 10.1128/AAC.01261-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nemec A, Janda L, Melter O, Dijkshoorn L. Genotypic and phenotypic similarity of multiresistant Acinetobacter baumannii isolates in the Czech Republic. J Med Microbiol. 1999;48(3): 287–96. 10.1099/00222615-48-3-287 [DOI] [PubMed] [Google Scholar]
- 18.Westphal O, Jann K. Bacterial lipopolysaccharides: extraction with phenol-water and further applications of the procedure In: Whistler R, editor. Methods in Carbohydrate Chemistry. New York: Academic press; 1965. p. 83–91. [Google Scholar]
- 19.Bankevich A, Nurk S, Antipov D, Gurevich A, Dvorkin M, Kulikov A, et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J Comput Biol. 2012;19(5): 455–77. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arndt D, Grant J, Marcu A, Sajed T, Pon A, Liang Y, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–21. 10.1093/nar/gkw387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kenyon JJ, Marzaioli AM, Hall RM, De Castro C. Structure of the K2 capsule associated with the KL2 gene cluster of Acinetobacter baumannii. Glycobiology. 2014;24(6): 554–63. 10.1093/glycob/cwu024 [DOI] [PubMed] [Google Scholar]
- 22.Kenyon JJ, Marzaioli AM, Hall RM, De Castro C. Structure of the K6 capsular polysaccharide from Acinetobacter baumannii isolate RBH4. Carbohydr. Res. 2015;409:30–5. 10.1016/j.carres.2015.03.016 [DOI] [PubMed] [Google Scholar]
- 23.Senchenkova SN, Shashkov AS, Shneider MM, Arbatsky NP, Popova AV, Miroshnikov KA, et al. Structure of the capsular polysaccharide of Acinetobacter baumannii ACICU containing di-N-acetylpseudaminic acid. Carbohydr. Res. 2014;391:89–92. 10.1016/j.carres.2014.04.002 [DOI] [PubMed] [Google Scholar]
- 24.Knirel YA, Shashkov AS, Tsvetkov Y, Jansson P, Zähringer U. 5,7-diamino-3,5,7,9-tetradeoxynon-2-ulosonic acids in bacterial glycopolymers: chemistry and biochemistry. Adv Carbohydr Chem Biochem. 2003;58: 371–417. [DOI] [PubMed] [Google Scholar]
- 25.Lipkind G, Shashkov AS, Mamyan S, Kochetkov N. The nuclear overhauser effect and structural factors determining the conformations of disaccharide glycosides. Carbohydr Res. 1988;181: 1–12. [Google Scholar]
- 26.Kasimova AA, Shneider MM, Arbatsky NP, Popova AV, Shashkov AS, Miroshnikov KA, et al. Structure and gene cluster of the K93 capsular polysaccharide of Acinetobacter baumannii B11911 containing 5-N-Acetyl-7-N-[(R)-3-hydroxybutanoyl]pseudaminic acid. Biochem (Moscow). 2017;82(4): 483–9. [DOI] [PubMed] [Google Scholar]
- 27.Shashkov AS, Senchenkova SN, Popova AV, Mei Z, Shneider MM, Liu B, et al. Revised structure of the capsular polysaccharide of Acinetobacter baumannii LUH5533 (serogroup O1) containing di-N-acetyllegionaminic acid. Russ Chem Bull. 2015;64(5): 1196–1199. [Google Scholar]
- 28.Senchenkova SN, Kenyon JJ, Jia T, Popova AV, Shneider MM, Kasimova AA, et al. The K90 capsular polysaccharide produced by Acinetobacter baumannii LUH5553 contains di-N-acetylpseudaminic acid and is structurally related to the K7 polysaccharide from A. baumannii LUH5533. Carbohydr. Res. 2019;479:1–5. 10.1016/j.carres.2019.04.008 [DOI] [PubMed] [Google Scholar]
- 29.Arbatsky NP, Shneider MM, Shashkov AS, Popova AV, Miroshnikov KA, Volozhantsev N, et al. Structure of the N-acetylpseudaminic acid-containing capsular polysaccharide of Acinetobacter baumannii NIPH67. Russian Chemical Bulletin. 2016;65(2):588–91. [Google Scholar]
- 30.Senchenkova SN, Popova AV, Shashkov AS, Shneider MM, Mei Z, Arbatsky NP, et al. Structure of a new pseudaminic acid-containing capsular polysaccharide of Acinetobacter baumannii LUH5550 having the KL42 capsule biosynthesis locus. Carbohydr. Res. 2015;407:154–7. 10.1016/j.carres.2015.02.006 [DOI] [PubMed] [Google Scholar]
- 31.Haseley S, Wilkinson S. Structural studies of the putative O-specific polysaccharide of Acinetobacter baumannii O24 containing 5,7-diamino-3,5,7,9-tetradeoxy-L-glycero-D-galacto-nonulosonic acid. Eur J Biochem. 1997;250:617–23. [DOI] [PubMed] [Google Scholar]
- 32.Sasaki T, Uchida T, Kurahashi K. Glucosylation of O-antigen in Salmonella carrying epsilon 15 and epsilon 34 phages. J Biol Chem. 1974;249: 761–772. [PubMed] [Google Scholar]
- 33.Wright A. Mechanism of conversion of the Salmonella O antigen by bacteriophage epsilon 34. J Bacteriol. 1971;105: 927–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verma NK, Brandt M, Verma D, Lindberg AA. Molecular characterization of the O-acetyl transferase gene of converting bacteriophage SF6 that adds group antigen 6 to Shigella flexneri. Mol Microbiol. 1991;5: 71–5. 10.1111/j.1365-2958.1991.tb01827.x [DOI] [PubMed] [Google Scholar]
- 35.Guan S, Bastin DA, Verma NK. Functional analysis of the O antigen glucosylation gene cluster of Shigella flexneri bacteriophage SfX. Microbiol. 1999;145: 1263–73. [DOI] [PubMed] [Google Scholar]
- 36.Taylor VL, Udaskin ML, Islam ST, Lam JS. The D3 bacteriophage α-Polymerase inhibitor (Iap) peptide disrupts O-antigen biosynthesis through mimicry of the chain length regulator Wzz in Pseudomonas aeruginosa. J Bacteriol. 2013;195(20): 4735–4741. 10.1128/JB.00903-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
Sequence data is available in the GenBank database under accession number MK609549.1. All other relevant data are within the manuscript and its Supporting Information files.