Abstract
Failures in control of tan spot of pyrethrum, caused by Didymella tanaceti, has been associated with decreased sensitivity within the pathogen population to the succinate dehydrogenase inhibitor (SDHI) fungicide boscalid. Sequencing the SdhB, SdhC, and SdhD subunits of isolates with resistant and sensitive phenotypes identified 15 mutations, resulting in three amino acid substitutions in the SdhB (H277Y/R, I279V), six in the SdhC (S73P, G79R, H134R, H134Q, S135R and combined H134Q/S135R), and two in the SdhD (D112E, H122R). In vitro testing of their boscalid response and estimation of resistance factors (RF) identified isolates with wild-type (WT) Sdh genotypes were sensitive to boscalid. Isolates with SdhB-I279V, SdhC-H134Q and SdhD-D112E exhibited moderate resistance phenotypes (10 ≥ RF < 100) and isolates with SdhC-H134R exhibited very high resistance phenotypes (RF ≥ 1000). All other substitutions were associated with high resistance phenotypes (100 ≥ RF < 1000). High-resolution melt assays were designed and used to estimate the frequencies of substitutions in four field populations (n = 774) collected in August (pre-boscalid application) and November (post-boscalid application) 2012. The SdhB-H277Y, SdhC-H134R and SdhB-H277R genotypes were most frequently observed across populations at 56.7, 19.0, and 10.3%, respectively. In August 92.9% of D. tanaceti contained a substitution associated with decreased sensitivity. Following boscalid application, this increased to 98.9%, with no WT isolates detected in three fields. Overlaying previously obtained microsatellite and mating-type data revealed that all ten recurrent substitutions were associated with multiple genotypes. Thus, boscalid insensitivity in D. tanaceti appears widespread and not associated with clonal spread of a limited pool of individuals.
Introduction
Foliar and flower disease control in pyrethrum (Tanacetum cinerariifolium) production involves the application of a range of fungicide chemistries with timing designed to optimise the production of flowers in early austral summer. Flowers are the sole harvested product of the crop, from which pyrethrins are extracted for their insecticidal properties [1, 2]. Historically, spring fungicide applications to control ray blight, caused by Stagonosporopsis tanaceti, have been the main focus for disease management [3–5]. However, surveys conducted during 2012 and 2013 indicated that S. tanaceti had been displaced as the predominant foliar pathogen of pyrethrum in spring by Didymella tanaceti [6, 7], the causal agent of tan spot and previously regarded as of minor concern for the industry [8]. The increased prevalence of D. tanaceti coincided with failures of the spring fungicide program to effectively control foliar diseases and the detection of resistance to the succinate dehydrogenase inhibitors (SDHI) fungicide boscalid within D. tanaceti, but not S. tanaceti [7]. Boscalid has been a key component of the spring fungicide program since 2005 [5, 9]. It is theorised that fungicide resistance has provided a competitive advantage to D. tanaceti over S. tanaceti.
The development of fungicide resistance in a pathogen population is a major concern for any disease management system. Fungicides with single site modes of action, such as the SDHI, are particularly prone to the development of resistance [10, 11]. Succinate dehydrogenase is an enzyme necessary for cellular respiration [12] and is constructed from four nuclear encoded protein subunits; SdhA, SdhB, SdhC and SdhD [10, 13, 14]. The SdhB subunit contains three iron-sulphur clusters (2Fe-2S, 4Fe-4S and 3Fe-4S) which transfer electrons for the reduction of ubiquinone. The ubiquinone binding site (Q-site) is formed by the interface of the SdhB, SdhC and SdhD subunits [12, 13, 15, 16]. Boscalid inhibits ubiquinone reduction by binding to the Q-site, restricting cellular respiration and resulting in cell death. Boscalid has been used for disease control in a wide range of agricultural cropping systems, such as Alternaria alternata [17, 18], Botrytis cinerea [19, 20], and Sclerotinia sclerotiorum [21, 22] in several crops. However, use of boscalid has been associated with increasingly frequent reports of resistance in pathogen populations. Mutations within each of the subunits have been correlated with boscalid resistance within field isolates and laboratory-induced mutants of different fungal species [10, 23]. Early reports of such instances include A. alternata in pistachio [18] and Didymella bryoniae in watermelon [24]. Resistance development has also been recorded despite application strategies to minimise this risk, such as tank mixing with multi-site protectants and limiting application numbers [25].
The purpose of this study was to examine the genetic basis for boscalid resistance in D. tanaceti. It was hypothesised that boscalid resistance would be associated with mutations within the succinate dehydrogenase (Sdh) gene complex. To this end, sequencing of the SdhB, SdhC and SdhD genes was conducted for D. tanaceti individuals with known boscalid response phenotypes. A secondary objective of this study was to examine the influence of a single application of boscalid in commercial pyrethrum production on the abundance and genotype of boscalid-resistant pathogen isolates. To facilitate this, a high-resolution melt (HRM) assay was developed for the detection of known and potential unknown mutations associated with boscalid resistance.
Results
SdhB, SdhC and SdhD sequences
The predicted SdhB of D. tanaceti was 1,034-base pairs (bp) and encoded 306 amino acids (aa). The opening reading frame (ORF) was arranged into three exons, with two putative introns of 61 and 52-bp in length (Fig 1). Species sequence alignment identified that intron splice sites were identical to A. alternata, except for an intron located 6-bp upstream from the first conserved cysteine region in A. alternata, which was absent in D. tanaceti. This intron also occurred in A. solani, Zymoseptoria tritici (syn: Mycosphaerella graminicola) and Corynespora cassiicola (Fig 1). Comparison of the encoded SdhB protein sequence of D. tanaceti with other fungal species showed high amino acid conservation of the three cysteine rich clusters, associated with the iron-sulphur centres. MAFFT alignment of the SdhB protein of D. tanaceti isolates identified two polymorphic codons located within the third conserved cysteine region (Fig 2A). Histidine (CAC) to tyrosine (TAC) or arginine (CGC) substitutions occurred at codon 277 (H277Y/R) in 16 and 6 isolates, respectively. At codon 279 an isoleucine (ATT) to valine (GTT) substitution (I279V) occurred in one isolate (Fig 2A). In addition, the fourteenth nucleotide of the second intron of 15 D. tanaceti isolates exhibited an adenine (A) to guanine (G) transition.
Fig 1.
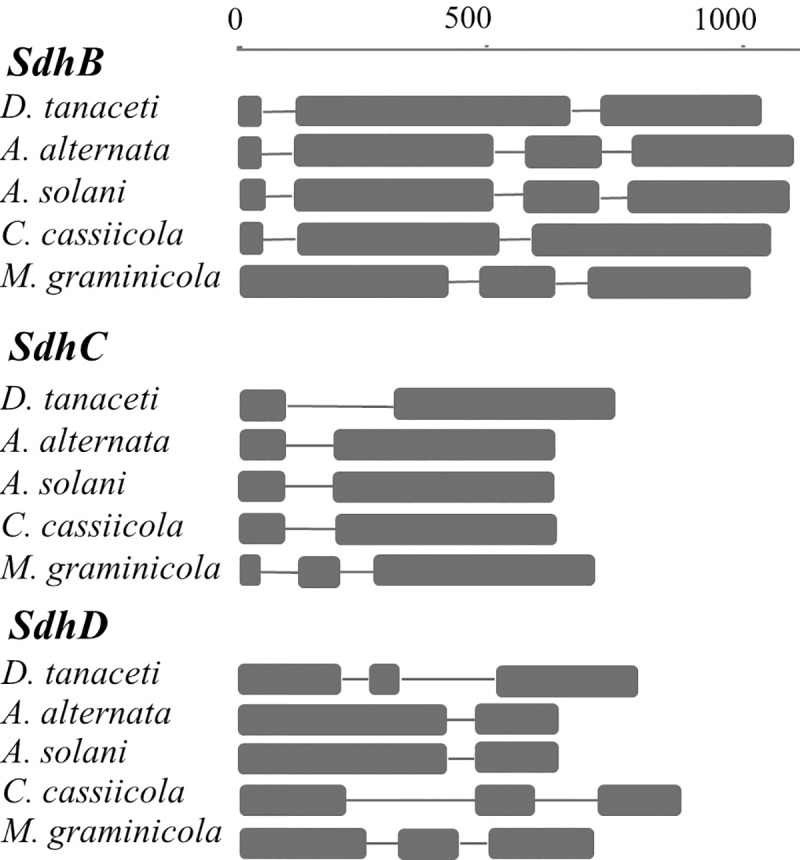
Structural arrangement of the succinate dehydrogenase subunit B, C and D genes of Didymella tanaceti, Alternaria alternata strain AR-SBL-4S [26], A. solani strain 1178-W1 [27], Corynespora cassiicola strain IbCor0008 [25], and Mycosphaerella graminicola strain R39-1 [16]. Squares indicate exons. Lines connecting the squares indicate introns. The scale bar indicates gene length (base pair).
Fig 2.
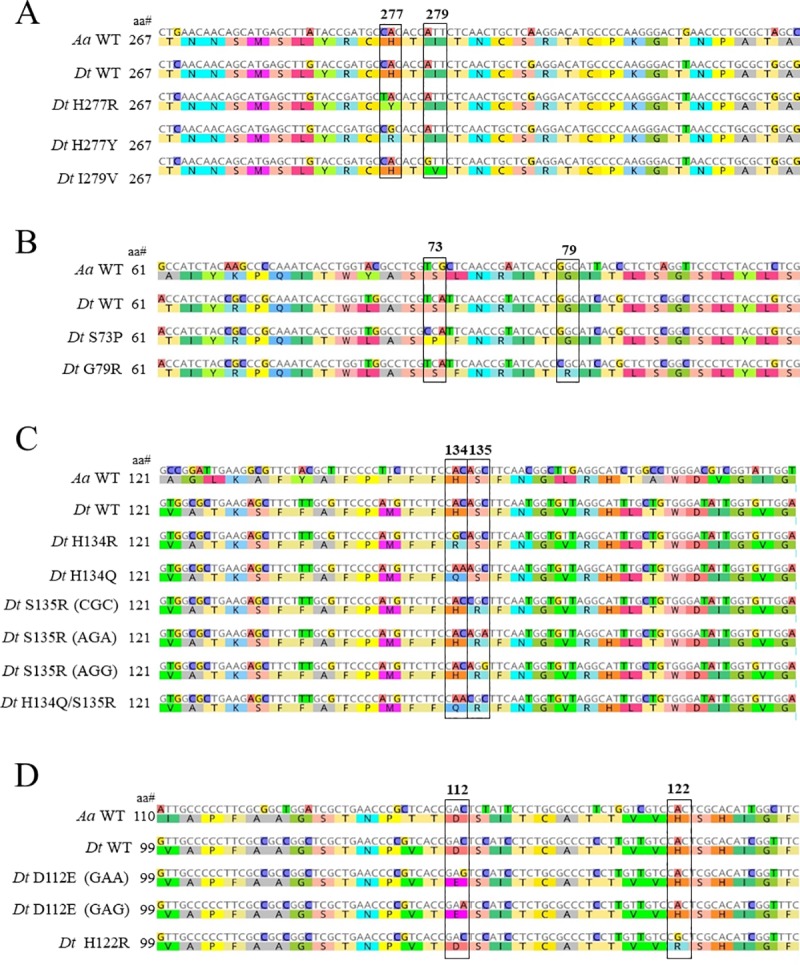
MAFFT alignment of partial succinate dehydrogenase gene sequences found in Didymella tanaceti (Dt) wild type (WT) and mutant isolates with WT sequences of Alternaria alternata (Aa) strain AR-SBL-4S [26] for (A) SdhB, (B,C) SdhC and (D) SdhD. Reference amino acid numbers relative to each species are located to the left of the alignments. Black boxes/coloured regions indicate the amino acid substitutions found in D. tanaceti.
The predicted SdhC of D. tanaceti was 746-bp and encoded 177-aa. The ORF was arranged into two exons, with a putative intron of 212-bp (Fig 1). The intron splice site was identical to A. alternata, A. solani and C. cassiicola, though the intron length was more than double in D. tanaceti (Fig 1). MAFFT alignment of the encoded SdhC protein of D. tanaceti isolates identified four polymorphic amino acid residues (Fig 2B and 2C). The most common of these was a histidine (CAC) to arginine (CGC) substitution which occurred at codon 134 (H134R) in eight isolates. At codon 88, a leucine (CTG) to methionine (ATG) substitution (L88M) occurred in four isolates. Single isolates containing either a glycine (GGC) to arginine (CGC) substitution at codon 79 (G79R) or a serine (AGC) to arginine (AGG) substitution at codon 135 (S135R) were observed (Fig 2B and 2C). A single-nucleotide polymorphism (SNP) (C/T) resulting in a synonymous substitution was also identified at codon 32 in four isolates. In addition, two G/A SNPs were identified within the putative intron. One isolate contained compound substitutions of L88M and H134R.
The predicted SdhD of D. tanaceti was 784-bp and encoded 182-aa. The ORF was arranged into three exons, with two putative introns of 50 and 185-bp (Fig 1). The first intron in D. tanaceti was not present in A. alternata, A. solani, Z. tritici and C. cassiicola, while the splice site for the second intron were identical in D. tanaceti, Z. tritici and C. cassiicola. MAFFT alignment of the SdhD protein of D. tanaceti isolates identified two polymorphic amino acid residues. An aspartate (GAC) to glutamic acid (GAG or GAA) substitution occurred at codon 112 (D112E) in three isolates and a histidine (CAC) to arginine (CGC) substitution occurred at codon 122 (H122R) in one isolate (Fig 2D). A SNP (C/T) resulting in a synonymous substitution was also identified at codon 45. In addition, three SNPs (C/T, G/A and G/A) were identified within the two putative introns. Isolates with compound substitutions at codons 112 and 122 were not found.
Three isolates with compound substitutions in two different Sdh subunits were identified. Each of these isolates contained the SdhC-L88M with either of the SdhD-D112E or SdhB-H277Y. Isolates with no substitution in any of the subunits were considered wild-type (WT). Comparison of the Sdh genotypes identified in this study and the boscalid response phenotype of isolates, previously assigned to them by Hay et al. [7], identified that all genetically WT D. tanaceti isolates and the single SdhB-I279V isolate had a sensitive phenotype[7].
HRM assay for Sdh allele detection
The developed HRM assays were able to identify the known mutations in each of the SdhB, SdhC and SdhD (Fig 3A–3D). Additional mutations were identified in the initial SdhC HRM screens by the occurrence of unique novel melt curves. The location and nature of these additional mutations was confirmed by gene sequencing. A serine (TCA) to proline (CCA) substitution occurred at codon 73 (S73P), a histidine (CAC) to glutamine (CAA) occurred at codon 134 and a combined histidine (CAC) to glutamine (CAA) and serine (AGC) to arginine (CGC) occurred at codons 134 and 135 (H134Q/S135R) (Fig 2B and 2C). Furthermore, two additional mutations (AGC to CGC and AGC to AGA) were identified which also conferred substitution of serine with arginine at codon 135 (S135R) (Fig 2B and 2C). The SdhC HRM assays were adapted, as required, to identify these additional mutations (Fig 3B and 3C). For all the HRM assays the results from isolates run in duplicate were consistent and SdhB, SdhC and SdhD sequencing results of the 92 D. tanaceti isolates selected for in vitro boscalid testing aligned with the HRM results (Table 1). Sequencing results identified the SdhC-L88M substitution in 2 isolates and in compound substitutions with SdhB-H277R (n = 1), SdhB-H277Y (n = 1), SdhC-G79R (n = 1), SdhC-H134R (n = 2), SdhC-S135R (n = 1), and SdhD-D112E (n = 3) isolates.
Fig 3. High resolution melt analysis for allele detection in the succinate dehydrogenase (Sdh) genes of Didymella tanaceti.
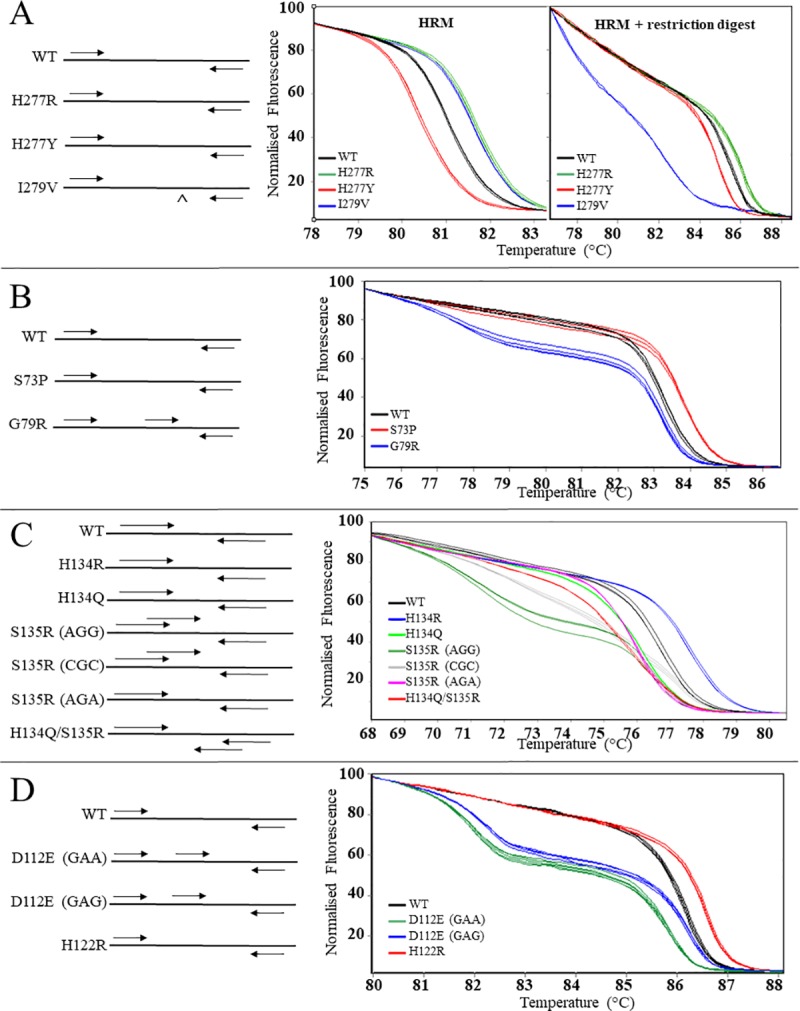
Left hand side of figure indicates primer binding regions and right-hand side shows normalised fluorescence for each of the known substitutions and wild-type in (A) SdhB codons 277 and 279; (B) SdhC codons 73 and 79; (C) SdhC codons 134 and 135 and (D) SdhD codons 112 and 122. On left hand side, black lines indicate the gene and arrows indicate the annealing position of primers (not to scale).
Table 1. EC50, resistance factor and associated phenotype of Didymella tanaceti isolates (n = 92) with known SDH substitutions.
| Isolatea | Substitutionb | Accession number | EC50c | RFd | Phenotypee | Boscalid concentrations tested (μg a.i./mL) | ||
|---|---|---|---|---|---|---|---|---|
| SdhB | SdhC | SdhD | ||||||
| 041–0002 | MK500737- MK500739 | 0.02 | 0.5 | S | 0, 0.001, 0.01, 0.05, 0.25, 0.5, 1.25 | |||
| 041–0032 | 0.04 | 0.9 | S | 0, 0.001, 0.01, 0.05, 0.25, 0.5, 1.25 | ||||
| 041–0034 | 0.05 | 1.3 | S | 0, 0.001, 0.01, 0.05, 0.25, 0.5, 1.25 | ||||
| 041–0047 | L88M | MK500740- MK500743 | 0.04 | 0.9 | S | 0, 0.001, 0.01, 0.05, 0.25, 0.5, 1.25 | ||
| 041–0049 | 0.09 | 2.2 | S | 0, 0.001, 0.01, 0.05, 0.25, 0.5, 1.25 | ||||
| 041–0076 | 0.04 | 1.0 | S | 0, 0.001, 0.01, 0.05, 0.25, 0.5, 1.25 | ||||
| 041–0077 | L88M | 0.04 | 0.9 | S | 0, 0.001, 0.01, 0.05, 0.25, 0.5, 1.25 | |||
| 041–0231 | 0.04 | 0.9 | S | 0, 0.001, 0.01, 0.05, 0.25, 0.5, 1.25 | ||||
| F50607A | 0.03 | 0.8 | S | 0, 0.001, 0.01, 0.05, 0.25, 0.5, 1.25 | ||||
| F50607B | 0.03 | 0.7 | S | 0, 0.001, 0.01, 0.05, 0.25, 0.5, 1.25 | ||||
| 041–0001 | H277Y | MK500757 | 27.60 | 673.2 | HR | 0, 2.5, 5, 10, 25, 50 | ||
| 041–0004 | H277Y | L88M | MK500758 | 15.90 | 387.8 | HR | 0, 2.5, 5, 10, 25, 50 | |
| 041–0009 | H277Y | 19.80 | 482.9 | HR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0015 | H277Y | 19.40 | 473.2 | HR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0022 | H277Y | 50.00 | 1219.5 | VHR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0041 | H277Y | 15.00 | 365.9 | HR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0088 | H277Y | MK500760 | 17.30 | 422.0 | HR | 0, 2.5, 5, 10, 25, 50 | ||
| 041–0242 | H277Y | 12.90 | 314.6 | HR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–1335 | H277Y | 2.87 | 70.0 | MR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–1338 | H277Y | 12.70 | 309.8 | HR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0018 | H277R | 9.17 | 223.7 | HR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0024 | H277R | MK500759 | 9.80 | 239.0 | HR | 0, 2.5, 5, 10, 25, 50 | ||
| 041–0031 | H277R | 5.41 | 132.0 | HR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0252 | H277R | 2.56 | 62.4 | MR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0414 | H277R | 8.28 | 202.0 | HR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0485 | H277R | 10.40 | 253.7 | HR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0586 | H277R | 8.63 | 210.5 | HR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0744 | H277R | 15.60 | 380.5 | HR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0748 | H277R | L88M | MK500762 | 11.20 | 273.2 | HR | 0, 2.5, 5, 10, 25, 50 | |
| 041–1641 | H277R | 10.20 | 248.8 | HR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0039 | I279V | 0.90 | 21.9 | MR | 0, 0.05, 0.25, 0.5, 1.25, 2.5 | |||
| 041–0227 | I279V | MK500761 | 0.38 | 9.3 | LR | 0, 0.05, 0.25, 0.5, 1.25, 2.5 | ||
| 041–0235 | I279V | 0.37 | 9.1 | LR | 0, 0.05, 0.25, 0.5, 1.25, 2.5 | |||
| 041–0253 | I279V | 0.48 | 11.8 | MR | 0, 0.05, 0.25, 0.5, 1.25, 2.5 | |||
| 041–0254 | I279V | 0.33 | 8.1 | LR | 0, 0.05, 0.25, 0.5, 1.25, 2.5 | |||
| 041–0267 | I279V | 1.87 | 45.6 | MR | 0, 0.05, 0.25, 0.5, 1.25, 2.5 | |||
| 041–0766 | I279V | 0.59 | 14.4 | MR | 0, 0.05, 0.25, 0.5, 1.25, 2.5 | |||
| 041–0902 | I279V | 0.45 | 11.0 | MR | 0, 0.05, 0.25, 0.5, 1.25, 2.5 | |||
| 041–0914 | I279V | 0.52 | 12.8 | MR | 0, 0.05, 0.25, 0.5, 1.25, 2.5 | |||
| 041–0136 | S73P | MK500751 | 19.90 | 485.4 | HR | 0, 0.5, 5, 10, 25, 50 | ||
| 041–0958 | S73P | 7.95 | 193.9 | HR | 0, 0.5, 5, 10, 25, 50 | |||
| 041–0989 | S73P | 6.22 | 151.7 | HR | 0, 0.5, 5, 10, 25, 50 | |||
| 041–0992 | S73P | 6.76 | 164.9 | HR | 0, 0.5, 5, 10, 25, 50 | |||
| 041–1029 | S73P | 20.90 | 509.8 | HR | 0, 0.5, 5, 10, 25, 50 | |||
| 041–0204 | G79R | MK500753 | 30.00 | 731.7 | HR | 0, 0.5, 5, 10, 25, 50 | ||
| 041–0245 | G79R | 31.50 | 768.3 | HR | 0, 0.5, 5, 10, 25, 50 | |||
| 041–0259 | G79R | 30.90 | 753.7 | HR | 0, 0.5, 5, 10, 25, 50 | |||
| 041–0814 | G79R/L88M | MK500756 | 46.10 | >1000 | VHR | 0, 0.5, 5, 10, 25, 50 | ||
| 041–0984 | G79R | 43.60 | >1000 | VHR | 0, 0.5, 5, 10, 25, 50 | |||
| 041–1039 | G79R | 23.90 | 582.9 | HR | 0, 0.5, 5, 10, 25, 50 | |||
| 041–1298 | G79R | 29.20 | 712.2 | HR | 0, 0.5, 5, 10, 25, 50 | |||
| CF196 | G79R | 21.30 | 519.5 | HR | 0, 0.5, 5, 10, 25, 50 | |||
| 041–0007 | H134R | MK500748 | >50 | >1000 | VHR | 0, 2.5, 5, 10, 25, 50 | ||
| 041–0016 | H134R/L88M | MK500749 | >50 | >1000 | VHR | 0, 2.5, 5, 10, 25, 50 | ||
| 041–0025 | H134R | 42.20 | 1029.3 | VHR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0027 | H134R | >50 | >1000 | VHR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0107 | H134R | >50 | >1000 | VHR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0188 | H134R/L88M | >50 | >1000 | VHR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0522 | H134R | 35.30 | 861.0 | HR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0622 | H134R | 16.60 | 404.9 | HR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–1255 | H134R | >50 | >1000 | VHR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–1345 | H134R | >50 | >1000 | VHR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0391 | H134Q | MK500754 | 3.0 | 71.9 | MR | 0, 2.5, 5, 10, 25, 50 | ||
| 041–0162 | S135R | MK500752 | 26.2 | 639.0 | HR | 0, 2.5, 5, 10, 25, 50 | ||
| 041–0328 | S135R | 16.2 | 395.1 | HR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0358 | S135R | 13.5 | 329.3 | HR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0370 | S135R | 25.6 | 624.4 | HR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0397 | S135R | 25.1 | 612.2 | HR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0455 | S135R | 13.4 | 326.8 | HR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0784 | S135R/L88M | MK500755 | >50 | >1000 | VHR | 0, 2.5, 5, 10, 25, 50 | ||
| 041–1344 | S135R | 13.5 | 329.3 | HR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–1350 | S135R | 29.4 | 717.1 | HR | 0, 2.5, 5, 10, 25, 50 | |||
| CF2 | S135R | 32.9 | 802.4 | HR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0132 | H134Q/S135R | MK500750 | 24.30 | 592.7 | HR | 0, 2.5, 5, 10, 25, 50 | ||
| 041–0163 | H134Q/S135R | 26.70 | 651.2 | HR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0193 | H134Q/S135R | 25.50 | 622.0 | HR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0201 | H134Q/S135R | 23.60 | 575.6 | HR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0875 | H134Q/S135R | 16.40 | 400.0 | HR | 0, 2.5, 5, 10, 25, 50 | |||
| 041–0035 | D112E | MK500743 | 5.52 | 134.6 | HR | 0, 0.5, 1.25, 2.5, 5, 10 | ||
| 041–0036 | L88M | D112E | MK500744 | 4.88 | 119.0 | HR | 0, 0.5, 1.25, 2.5, 5, 10 | |
| 041–0038 | L88M | D112E | MK500745 | 2.80 | 68.3 | MR | 0, 0.5, 1.25, 2.5, 5, 10 | |
| 041–0335 | D112E | 4.63 | 112.9 | HR | 0, 0.5, 1.25, 2.5, 5, 10 | |||
| 041–0340 | D112E | 9.22 | 224.9 | HR | 0, 0.5, 1.25, 2.5, 5, 10 | |||
| 041–0435 | L88M | D112E | MK500747 | 3.56 | 86.8 | MR | 0, 0.5, 1.25, 2.5, 5, 10 | |
| 041–0439 | D112E | 2.82 | 68.8 | MR | 0, 0.5, 1.25, 2.5, 5, 10 | |||
| 041–1282 | D112E | 1.88 | 45.9 | MR | 0, 0.5, 1.25, 2.5, 5, 10 | |||
| 041–1333 | D112E | 1.30 | 31.7 | MR | 0, 0.5, 1.25, 2.5, 5, 10 | |||
| F50403B | D112E | 3.02 | 73.7 | MR | 0, 0.5, 1.25, 2.5, 5, 10 | |||
| 041–0297 | H122R | MK500746 | 9.29 | 226.6 | HR | 0, 0.5, 5, 10, 25, 50 | ||
| 041–1111 | H122R | 32.20 | 785.4 | HR | 0, 0.5, 5, 10, 25, 50 | |||
| 041–1130 | H122R | 21.00 | 512.2 | HR | 0, 0.5, 5, 10, 25, 50 | |||
| CF149 | H122R | 39.10 | 953.7 | HR | 0, 0.5, 5, 10, 25, 50 | |||
a Didymella tanaceti isolates stored in the Tasmanian Institute of Agriculture fungal collection, Tasmania, Australia. All isolates originally recovered from pyrethrum leaves.
b Substitution identified via high resolution melt assay and confirmed by sequencing.
c EC50 = estimated boscalid concentration (μg/a.i./mL) to reduce radial growth by 50% of the non-amended controls. Calculated using logistic regression of the relative growth against log10 of the boscalid concentration.
d Resistance Factor (RF) = EC50X/EC50WT, where EC50X is the EC50 value of the isolate being examined, and EC50WT is the average EC50 value of the baseline isolates (WT).
e Boscalid phenotype based on the interpretation of RF values; S = sensitive (RF ≤ 3) LR = low resistance (3 > RF ≤ 10), MR = moderate resistance (10 > RF ≤ 100), HR = high resistance (100 > RF ≤ 1000) and VHR = very high resistance (RF >1000).
In vitro boscalid testing
Within the D. tanaceti isolates selected for in vitro boscalid testing (n = 92; Table 1), substitutions were associated with varying resistant phenotypes. Wild type isolates (baseline isolates; n = 10) had EC50 values ranging from 0.02 to 0.09 μg a.i/mL (average = 0.041 μg a.i/mL), resulting in a resistance factor (RF) ≤ 2.2 and a sensitive phenotype (Table 1). Of the nine isolates exhibiting a SdhB-I279V substitution, three exhibited low resistance to boscalid (EC50 = 0.33–0.38 μg a.i./mL) and six were moderately resistant (EC50 = 0.45–1.87) (Table 1). Of the ten isolates containing a SdhB-H277Y substitution, eight were highly resistant (EC50 = 12.70–27.60), one was moderately resistant (EC50 = 2.87) and one exhibited very high resistance (EC50 = 50.0; Table 1). Of the ten isolates exhibiting the other SdhB substitution at codon 277, SdhB-H277R, nine were highly resistant (EC50 = 5.40–15.60) with the other moderately resistant (EC50 = 2.56). All substitutions in the SdhC resulted in isolates with high or very high resistance. All five isolates with a SdhC-S73P substitution were highly resistant (EC50 = 6.20–20.90). Isolates with the SdhC-G79R substitution, had either high resistance (n = 6; EC50 = 21.30–31.50) or very high resistance (n = 2; EC50 = 43.60–46.10). The highest resistance to boscalid was identified in the ten isolates with the SdhC-H134R substitution with eight exhibiting very high resistance (EC50 42.20 –>50.0) and the remainder high resistance (EC50 = 16.60–35.30). The single isolate with a SdhC-H134Q substitution was moderately resistant (EC50 = 3.0). Of the ten isolates with the SdhC-S135R substitution in the adjacent codon, nine were highly resistant (EC50 13.50–32.90) while one isolate exhibited very high resistance (EC50 > 50.0). The five isolates with combined SdhC-H134Q/S135R substitutions were all highly resistant (EC50 = 16.4–26.7). The two isolates containing the SdhC-L88M substitution in solitary were as sensitive to boscalid as WT isolates. The occurrence of the SdhC-L88M substitution did not affect the fungicide response of the nine isolates containing it in combination with another substitution. Therefore, it was concluded that the SdhC-L88M substitution was not associated with any additional resistance even when in combination with other substitutions. All substitutions in the SdhD resulted in isolates with moderate or high resistance (Table 1). Of the ten isolates with a SdhD-D112E substitution, six were moderately resistant (EC50 = 1.30–3.60) and the remainder highly resistant (EC50 = 4.60–9.20). Furthermore, all four isolates with a SdhD-H122R substitution exhibited high resistance (EC50 = 9.29–39.10).
Frequency and genetic diversity of Sdh substitutions in the 2012 D. tanaceti population
In August 2012, prior to application of the spring fungicide program, 92.9% of D. tanaceti isolates from the four 2012 field populations contained a substitution associated with decreased sensitivity in the SdhB, SdhC or SdhD (Table 2). No isolates containing compound substitutions of interest were identified. Following application of the spring fungicide program the percent of isolates sampled in November 2012 containing a Sdh substitution increased to 98.9% (Table 2). This result coincided with no detection of WT isolates in three of the four fields (Table 2). The SdhB-H277Y substitution was the most common Sdh substitution in each field, irrespective of sampling period. Multinomial logistic regression of genotype frequency indicated a significant interaction between field and time of sampling (P = 0.0065). Post-hoc pairwise comparisons between sampling periods within fields and substitution found that significant reductions in WT frequency occurred from August to November in fields 12–63242 and 12–70046 (Table 2). Significant increases (P < 0.05) in Sdh substitution frequency within fields were observed for SdhB-H277R (Field 12–51907) and SdhC-S73P (Field 12–70047) from August to November (Table 2). Conversely, significant decreases (P < 0.05) over the same period within fields were observed for SdhC-H134R (Field 12–51097) and SdhD-D112E (Field 12–70046). A single isolate with the SdhC-H134Q substitution was identified across all fields and sampling periods. This substitution and three other substitution occurring at frequencies less than 1% were excluded from the analysis to prevent detrimental effects on model estimates (Table 2).
Table 2. Frequency (%) of each SDH substitution in the 2012 Didymella tanaceti population by sampling period and field.
| Subunit-substitution | 12–51907 | 12–63242 | 12–70046 | 12–70047 | All Fields | Overall | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Auga | Nova | Aug | Nov | Aug | Nov | Aug | Nov | Aug | Nov | ||
| Wildtype | 5.4 | 3.3 | 16.2b | 0.0 | 11.6 | 0.0 | 1.0 | 0.0 | 7.1 | 1.1 | 3.6 |
| SdhB-H277R | 4.5 | 16.4 | 5.9 | 17.2 | 9.3 | 11.9 | 10.8 | 7.2 | 7.4 | 12.5 | 10.3 |
| SdhB-H277Y | 58.0 | 59.9 | 38.2 | 48.4 | 46.5 | 59.7 | 57.8 | 64.5 | 52.3 | 59.9 | 56.7 |
| SdhB-I279V | 0.0 | 0.7 | 7.4 | 3.1 | 0.0 | 0.0 | 0.0 | 0.0 | 1.5 | 0.7 | 1.0 |
| SdhC-G79P | 0.0 | 0.7 | 4.4 | 0.0 | 0.0 | 1.5 | 0.0 | 0.6 | 0.9 | 0.7 | 0.8 |
| SdhC-H134Q/S135R | 2.7 | 0.7 | 1.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.2 | 0.2 | 0.6 |
| SdhC-H134R | 21.4 | 11.8 | 23.5 | 29.7 | 11.6 | 20.9 | 18.6 | 19.3 | 19.7 | 18.5 | 19.0 |
| SdhC-H134Q | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.3 | 0.0 | 0.1 |
| SdhC-S135R | 1.8 | 2.0 | 0.0 | 0.0 | 7.0 | 0.0 | 3.9 | 3.0 | 2.8 | 1.8 | 2.2 |
| SdhC-S73P | 0.9 | 0.0 | 0.0 | 0.0 | 0.0 | 4.5 | 0.0 | 2.4 | 0.3 | 1.6 | 1.0 |
| SdhD-D112E | 5.4 | 4.6 | 2.9 | 1.6 | 14.0 | 1.5 | 5.9 | 1.8 | 6.2 | 2.7 | 4.1 |
| SdhD-H122R | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.2 | 0.3 | 0.4 | 0.4 |
| Samplesc | 112 | 152 | 68 | 64 | 43 | 67 | 102 | 166 | 325 | 449 | 774 |
a Sampling period; Aug = August 2012, Nov = November 2012.
b Sampling period pairs in bold indicate significant (P < 0.05) frequency variations between periods based on multinomial logistic regression.
c Total number of individuals observed
Following combination of the microsatellite (SSR) profile and mating-type (MAT) of isolates in the 2012 D. tanaceti population, 138 genetically unique groups (Multi-Locus-Genotypes; MLG) were identified by Pearce et al. [28]. Overlaying the Sdh data with the SSR and MAT profile identified that no Sdh substitution observed in multiple individuals was associated with only a single MLG (Table 3). Furthermore, each MLG consisted of isolates containing one to eight different Sdh substitutions. Of the 70 MLGs consisting of ≥ 2 isolates, 48 MLGs were associated with > 1 Sdh substitution (average substitutions per MLG = 2.4). The most frequent substitutions, SdhB-H277Y and SdhC-H134R, were identified in 96 and 46 MLGs, respectively (Table 3). Despite occurring at a low frequency (< 1.0%), all isolates containing the SdhC-G79R and SdhD-H122R substitutions were genetically unique. When the Sdh substitutions were combined with the previously identified MLGs, 236 genetically unique groups were identified (Table 3). The 2012 population dataset is available at 10.6084/m9.figshare.7800929.
Table 3. Number of observations of each substitution (N) and associated multilocus genotypes (MLGs) broken down by sampling period, including the number shared between the two in the 2012 Didymella tanaceti population.
| Subunit-substitution | Overall | August | November | Shared MLGsb | |||
|---|---|---|---|---|---|---|---|
| N | MLGsa | N | MLGsa | N | MLGsa | ||
| Wild Type | 28 | 21 | 23 | 16 | 5 | 5 | 0 |
| SdhB-H277R | 80 | 32 | 24 | 15 | 56 | 27 | 10 |
| SdhB-H277Y | 439 | 96 | 170 | 67 | 269 | 64 | 35 |
| SdhB-I279V | 8 | 3 | 5 | 2 | 3 | 2 | 1 |
| SdhC-S73P | 8 | 3 | 1 | 1 | 7 | 2 | 0 |
| SdhC-G79R | 6 | 6 | 3 | 3 | 3 | 3 | 0 |
| SdhC-H134R | 147 | 46 | 64 | 29 | 83 | 36 | 19 |
| SdhC-H134Q | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
| SdhC-S135R | 17 | 13 | 9 | 6 | 8 | 7 | 0 |
| SdhC-H134Q/S135R | 5 | 2 | 4 | 2 | 1 | 1 | 1 |
| SdhD-D112E | 32 | 10 | 20 | 9 | 12 | 6 | 5 |
| SdhD-H122R | 3 | 3 | 1 | 1 | 2 | 2 | 0 |
| Total | 774 | 236 | 325 | 99 | 449 | 98 | |
a Multi-locus genotype derived from combining microsatellite and mating-type data [28]
b Number of multi-locus genotypes found in both sampling periods.
A breakdown of the MLGs associated with each substitution sampled in August and November identified that the WT, SdhC-S73P, SdhC-G79R, SdhC-H134Q, SdhC-S135R and SdhD-H122R isolates obtained in November were genetically different (based on SSR and MAT profile) to those occurring in August, with no MLGs shared between the two sampling periods for each substitution (Table 3). For the SdhB-H277Y and SdhC-H134R isolates only 52.2 and 65.5%, respectively, of the MLGs identified in August were present in November despite a similar number of MLGs identified in each sampling period (Table 3).
Discussion
Hay et al. [7] provided evidence that a decreased sensitivity to boscalid had developed in D. tanaceti from Tasmanian pyrethrum crops. We report that the observed resistant phenotypes are associated with mutations in the SdhB, SdhC, and SdhD subunits of the succinate dehydrogenase. Sixteen point mutations in gene sequences were detected, resulting in three substitutions in the SdhB, five substitutions in the SdhC and two substitutions in the SdhD. To our knowledge this the largest number of Sdh substitutions recovered in a solitary fungal species on one host associated with resistance. Furthermore, this study provided the first quantifiable estimates of the frequency of D. tanaceti isolates with decreased boscalid sensitivity in commercial pyrethrum fields.
Isolates with WT Sdh genotypes were sensitive to boscalid indicating this fungicide had provided sufficient control of D. tanaceti prior to the selection of resistant isolates. This also agrees with field observations following initial introduction of boscalid to the pyrethrum fungicide program. The baseline EC50 of the WT isolates established a reference point that indicates sensitivity to boscalid [29, 30]. Wild-type isolates had EC50 values less than 0.05 μg a.i./mL. Therefore, the range of 0–0.5 μg a.i./mL used by Hay et al. [7] to identify sensitive isolates may have underestimated the true shift in sensitivity of D. tanaceti. For example, based on the sensitivity range reported by Hay et al. [7] isolates with the SdhB-I279V would have been classed as sensitive to boscalid, whereas this study showed using RF, SdhB-I279V isolates were moderately resistant to boscalid. Many fungicide studies do not calculate baseline responses of WT isolates or isolates collected prior to the introduction of the fungicide, but instead infer resistance phenotypes on defined EC50 values/ranges [10, 31]. The calculation of a baseline response and subsequent RF allows greater comparison and evaluation of the response to a fungicide among different species and those collected from different hosts, and for establishment of discriminatory dose ranges for detecting major shifts in fungicide sensitivity [20, 32, 33].
Mutations in the third highly-conserved cysteine rich region of the SdhB were identified in 70.6% of the 2012 collected D. tanaceti isolates. The SdhB-H277Y was the most frequently recovered substitution and was associated with high resistance to boscalid. The recovery frequency of this substitution increased from August to November following spring fungicide applications incorporating boscalid in all fields. An additional substitution at the same codon, SdhB-H277R, also associated with high resistance, was found in a further 10% of individuals. Substitutions at homologous positions of these two substitutions in the SdhB were the first to be associated with SDHI resistance in Z. tritici [34], B. cinerea [35] and A. alternata [36]. Furthermore, genetic transformations studies have confirmed that substitutions at homologous positions of the SdhB-H277Y/R substitutions in Sclerotinia homoeocarpa and B. cinerea were determinants of decreased boscalid sensitivity [37, 38]. For many species, including A. alternata [36], B. cinerea [39–41], C. cassiicola [25], and D. bryoniae [10], the SdhB-H277Y/R are the most frequent substitutions identified from field surveys.
The high proportion of D. tanaceti isolates with SdhB-H277Y in the present study is not surprising. Lalève et al. [42] identified that homolog recombinant mutants of B. cinerea with SdhB-H272Y (corresponding to SdhB-H277Y in D. tanaceti) did not show decreased respiration or Sdh activity when compared to the WT strain. In contrast, these SdhB-H272Y transformants displayed higher initial growth than the WT strain on minimal medium at low temperature and in competition assays, in the absence of a selection pressure, the SdhB-H272Y transformants had a competitive advantage over the WT strain with increases from 40% to 70% of population after seven cycles on artificial media in vitro. No fitness penalties were found in A. alternata isolates with the SdhB-H277Y substitution [43]. If a similar lack of fitness cost is associated with SdhB-H277Y in D. tanaceti, coupled with a decreased sensitivity to boscalid, this may explain its dominance in field populations.
The only other substitution observed in the SdhB of D. tanaceti was SdhB-I279V. This substitution has also been identified in a single Stagonosporopsis citrulli isolate collected from watermelon [44]. The isolate was reported to be sensitive to boscalid and resistant to fluopyram. A substitution corresponding to SdhB-I279V has also been induced in laboratory studies in isolates of Z. tritici following UV mutagenesis and selection on media [15, 16, 45]. This substitution was associated with RF indicating low to moderate resistance to boscalid in Z. tritici (RF: 4.2–12.7) [15, 45]. Which is consistent with our findings in D. tanaceti. Additional substitutions at codon 272 in B. cinerea (SdhB-H272L and SdhB-H272V), which are associated with high resistance to boscalid [46], have not been observed in D. tanaceti to date.
Two substitutions at codon 134 of SdhC were identified in D. tanaceti with varied boscalid responses. SdhC-H134R isolates exhibited very high resistance levels, while the SdhC-H134Q exhibited moderate resistance levels. The SdhC-H134Q was identified in only one D. tanaceti isolate, though it was found in combination with SdhC-S135R in five isolates. This substitution was identified in A. solani isolates from potato in Germany in 2014 [47]. In contrast to our studies, in vitro EC50 values for the A. solani SdhC-H134Q isolates were above 100 μg boscalid/mL [47]. The SdhC-H134R was found in 19% of the 2012 D. tanaceti population and did not significantly change in frequency between the two sampling periods in three of four fields sampled. However, in the fourth field, frequency decreased nearly 50% from August to November. Fitness studies in A. alternata by Fan et al. [43] found that SdhC-H134R isolates remained competitive against sensitive isolates over the course of five successive transfers in the absence of fungicides. At the neighbouring codon, 135, three alternative point mutations encoded the SdhC-S135R substitution. Both Sierotzki et al. [48] and Lichtemberg et al. [49] have reported on the presence of SdhC-S135R in A. alternata isolate from pistachio as exhibiting resistance to carboxamides. This substitution was also reported to occur in P. teres on winter barley from Germany in 2013 [50]. As in our study, in these cases, the frequency of occurrence was low (0.6–2%). D. tanaceti isolates with the combined SdhC-H134Q/S135R had high resistance profiles, more consistent with the isolates with only the SdhC-S135R than the single tested SdhC-H134Q isolate. An association between the SdhC-L88M substitution and decreased boscalid sensitivity was not identified. These results suggest that this substitution does not alter the boscalid binding site in the succinate dehydrogenase protein.
The remaining SdhC substitutions identified in D. tanaceti were only found at a low frequency. The SdhC-S73P substitution has been identified in C. cassiicola in cucumber in Japan and was associated with moderate resistance to boscalid [25]. This substitution is homologous in position to an alanine to valine substitution at codon 84 (A84V) induced in a laboratory mutant of Z. tritici [45]. While inferring low resistance (RF = 1.7) to boscalid the Z. tritici mutant was highly resistance to other SDHI fungicides. The SdhC-G79R substitution has been identified in A. alternata isolates in peach in South Carolina [51] and in Pyrenophora teres isolates in barley in Europe [50]. In contrast to D. tanaceti, for P. teres this was the most frequent mutation identified in isolates from barley and was associated with moderate resistance to boscalid [50]. A substitution at a homologous position to G79R has been detected in laboratory strains of Z. tritici (G90R) but not in field samples [52, 53]. All the SdhC-G79R D. tanaceti isolates were genetically unique, indicating either multiple independent origins, recombination of the substitution into genetic diverse individuals, or a pathogen that has a high mutation frequency.
The SdhD-D112E substitution was encoded by two different point mutations in D. tanaceti and occurred in 4% of the 2012 population. An homologous substitution (SdhD-D133E) have also been found in A. alternata and A. solani [27, 47]. Fitness testing of D133E isolates of A. alternata from peach indicated that whilst having rapid mycelial growth, isolates were hypersensitive to oxidative stress [43], which may explain the lower prevalence of this genotype in the D. tanaceti population. A second SdhD substitution could only be found in four D. tanaceti isolates (SdhD-H122R), but homologous substitutions are also reported in S. sclerotiorum (H132R) [35, 54], B. cinerea (H132R) [39], A. solani (H133R) [27] and A. alternata (H133R) [55].
The inclusion of the SSR and mating-type data in this study provides an insight into the development of resistance within the pathogen population. The results suggested that resistance in D. tanaceti has not developed due to selection pressure favouring clonal build-up of resistant individuals. Rather, resistance is associated with a wide range of genotypes. The development of fungicide resistance in populations may have varying associations with population genetic structure. For example, Knapova and Gisi [56] found no association between SSR genotype and sensitivity to phenylamide in Phytophthora infestans, while Shrestha et al. [57] found that QoI resistant Cersospora sojina isolates clustered in a single group and were represented by a dominant clonal lineage. The development of a rapid method for allele detection provides an additional tool to analyse the impact of SDHI fungicide application on the frequency of mutations in D. tanaceti populations and for the ongoing implications and disease management options to be more greatly assessed. Previously, experiments evaluated fungicides based on infection levels pre- and post-application. However, a fungicide that appears efficacious and results in a decrease in pathogen load may be imposing a selection pressure on resistant individuals. The HRM assay will be utilised to evaluate historical changes in Sdh substitution changes in D. tanaceti populations from 2004 onwards.
The fields sampled as part of the 2012 population collection were first-harvest crops that had not received an application of boscalid, or any other SDHI fungicide, prior to the first sampling in August. Due to the high level of isolates with a mutation in the Sdh genes, associated with a decreased sensitivity to boscalid, it is proposed that these may have been introduced to, rather than produced in these fields. Didymella tanaceti can be seed-borne [58, 59]. As a perennial species seed-borne isolates may have been exposed to multiple applications of boscalid over the life of the seed crops and developed resistance prior to seed infection. However, the seed used to sow field 12–51907 was harvested in 2006/2007, and it is assumed that boscalid resistant isolates were undetectable in 2006/2007. Boscalid was first used commercially in pyrethrum crops in 2005. Thus for resistant individuals to be introduced to field 12–51907 via seed, boscalid resistant isolates would have had to be been prevalent 12 months after the introduction of boscalid use in pyrethrum and three years before fungicide control failures were identified by Hay et al. [7]. An alternative scenario, is that ascospores or other wind-blown inoculum also allows movement of resistance individuals between fields. Scott et al. [59] indicated that fields within 2 km of each other are able to contribute inoculum to each other and a sexual cycle for D. tanaceti cannot be dismissed [60]. The occurrence of the low frequency mutations in multiple fields further supported this hypothesis. Pyrethrum stubble has also been shown to be an inoculum source of D. tanaceti (T.L Pearce, Tasmanian Institute of Agriculture, Burnie, Australia, personal communication) and following termination of a pyrethrum crop it has been common practice to reincorporate the remaining plant stubble into the soil. The length of time that burned inoculum can remain viable is currently unknown, but this may also allow between season spread.
The high proportion of boscalid resistant isolates within the 2012 characterised population indicated that boscalid resistance in D. tanaceti contributed to the rapid increase in the frequency and incidence of tan spot. This resistance has occurred despite implementation of management practices by the pyrethrum industry to limit the development of fungicide resistance, including limiting single season applications and alternating mode of action fungicides. However, other pyrethrum pathogens, including S. tanaceti exhibit no evidence of a reduced sensitivity to boscalid [7], suggesting that the imposed management strategies are working for some species. Furthermore, it could be argued that S. tanaceti has greater exposure to and selection pressure imposed from boscalid than D. tanaceti, as the S. tanaceti pathogen incidence was higher relative to D. tanaceti following the period of boscalid introduction [58, 61]. Management of ray blight, caused by S. tanaceti, was the original justification for the deployment of boscalid in pyrethrum cropping [62]. This suggests that a low proportion of D. tanaceti already had resistance to SDHI fungicides or that D. tanaceti has a greater capacity to rapidly mutate to generate fungicide resistance traits. Analysis of D. tanaceti populations has identified high genetic variation among individuals, inferring a high adaptive ability [28]. Furthermore, while no evidence of a sexual cycle in S. tanaceti has been found [63], a sexual cycle in D. tanaceti cannot be dismissed [60] and would allow long range dispersal mechanisms and genetic recombination of resistant genotypes.
In 2015, boscalid was removed from the pyrethrum spring fungicide program due to concerns with control failures. However, other SDHI fungicides are commonly applied for control of tan spot and other disease in pyrethrum As cross resistance to multiple SDHI chemicals is reported for many Sdh mutations [19, 55, 64], examination of the Sdh cross resistance profile of D. tanaceti would be beneficial in identifying fungicide chemistries which will control individuals with the common SdhB-H277Y and SdhC-H134R substitutions and those that confer high resistance to boscalid. For example, B. cinerea isolates with the SdhB-H272Y/R substitutions have moderate resistance to both boscalid and penthiopyrad [19], but are sensitive or have low resistance to fluopyram [46, 65] and bixafen [65]. Similar cross-resistance patterns between SDHIs have been found in D. bryoniae [66] and A. alternata [55]. The pyrethrum industry remains reliant upon SDHI fungicides in the medium term for the control of fungal pathogens other than D. tanaceti. It is therefore recommended that alternation of SDHI chemicals across seasons, based knowledge of their cross-resistance profile, coupled with population monitoring for specific mutations be employed to mitigate the effects of fungicide resistance in D. tanaceti.
Materials and methods
SdhB, SdhC and SdhD sequences
To identify if a decreased sensitivity to boscalid was associated with mutations in the Sdh gene complex, the complete SdhB, SdhC and SdhD genes of 51 D. tanaceti isolates of known boscalid response phenotypes [7] were sequenced. This included isolates collected from 2009–2012 during which a decreased sensitivity to boscalid was first identified. The SdhB (GenBank acc: KJ426258), SdhC (GenBank acc: KJ426263) and SdhD (GenBank acc: KJ426268) gene sequences of A. alternata isolate AR-SBL-4S [26] were utilized as query sequences in a BLASTn search against a genomic database consisting of genome sequence data of D. tanaceti strain BRIP 61988 [60] in Geneious v7.1 (Biomatters Ltd., Auckland, New Zealand) to identify homologous genes in D. tanaceti.
DNA was extracted from isolates as described by Pearce et al. [28]. Entire SdhB, SdhC and SdhD gene sequences were amplified with primers Dt_SdhB_F/Dt_SdhB_R, Dt_SdhC_F/Dt_SdhC_R and Dt_SdhD_F/Dt_SdhD_R, respectively (Table 4), designed using Primer3 v2.3.4 [67]. PCR reactions were performed in a C1000 thermocycler (BioRad, Hercules, CA) in a total volume of 20 μL. Reaction mixes contained 0.05 U TopTaq polymerase (Qiagen, Hilden, Germany), 1 × PCR Buffer, 1 × CoralLoad, 0.4 μM of each primer, 200 μM of each dNTP (Bioline, Alexandria, Australia) and 3–5 ng of genomic DNA. Conditions for amplification were an initial denaturation of 5 min at 94°C, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 52°C (SdhB), 55°C (SdhC) or 56°C (SdhD) for 30 s, and extension at 72°C for 1 min, with a final elongation of 72°C for 5 min on the last cycle. Amplified products were visualised via gel electrophoresis using a 1.5% (w/v) agarose gel pre-stained with GelRed (Biotium Inc., Fremont, CA) in 1 × TAE buffer. PCR products were prepared for sequencing using the UltraClean PCR-clean up kit (Mo Bio Laboratories Inc., Carlsbad, CA), according to the manufacturer’s instructions. Sequencing in both directions was conducted at the Australian Genome Research Facility (AGRF; Melbourne, Australia) using Big Dye Terminator v3.1 chemistry and capillary separation on an AB 3730xl DNA Analyser (Applied Biosystems, Foster City, CA).
Table 4. Primers used for amplification of SdhB, SdhC and SdhD genes and High-Resolution Melt (HRM) assay.
| Primer | Sequence 5’ - 3’ | Use |
|---|---|---|
| Dt_SdhB_F | TACGTCAGGCTTTAGAAGGAAGGAG | SdhB gene |
| Dt_SdhB_R | GCCTAACTCAAAATACCAACTGC | SdhB gene |
| Dt_SdhC_F | GATACGC CCAAGACATCTACG | SdhC gene |
| Dt_SdhC_R | TACTCACTCCTCCAAATTCGTG | SdhC gene |
| Dt_SdhD_F | GCTCCACMGCTATCCTCG | SdhD gene |
| Dt_SdhD_R | TGGACACTTTAGCTTGTACTTTGCG | SdhD gene |
| SdhB _HRM_F | CTCAACAACAGCATGAGCTTG | SdhB HRM |
| SdhB _HRM_R | CAGCGCAGGGTTAAGTCC | SdhB HRM |
| SdhC_HRM_1F | CCATCTCACCATCTACCGC | SdhC HRM |
| SdhC_HRM_1R | GTAGAGGGAGCCGGAGAGC | SdhC HRM |
| SdhC_HRM_79_F | CGTCATTCAACCGTATCACCC | SdhC HRM |
| SdhC_HRM_2F | CTGAAGAGCTTCTTTGCGTTC | SdhC HRM |
| SdhC_HRM_135_F1 | CCATGTTCTTCCACAGG | SdhC HRM |
| SdhC_HRM_135_F2 | CCATGTTCTTCCACCGC | SdhC HRM |
| SdhC_HRM_2R | AGCAAATGCCTAACACCATTTGTTG | SdhC HRM |
| SdhC_HRM_QR_R | CCTAACACCATTGAAGCGTTG | SdhC HRM |
| SdhD_HRM_F | ATTCCCCTCACAGTTGCC | SdhD HRM |
| SdhD_HRM_R | TGATGCACGACTCGAAACC | SdhD HRM |
| SdhD_HRM_112_F | GCTGAACCCCGTCACCGAR | SdhD HRM |
The consensus sequences of all isolates for each gene were aligned using MAFFT v7.308 [68] to identify polymorphisms between isolates. The coding sequence of each D. tanaceti Sdh gene was inferred following alignment of the consensus sequences to A. alternata strain AR-SBL-4S, A. solani strain 1178-W1 [27], C. cassiicola strain IbCor0008 [25] and Mycosphaerella graminicola strain R39-1 [16] SdhB, SdhC and SdhD genes (GenBank acc SdhB: KC517310, AB548738, JF916687; SdhC: KC517313, AB548741, JF916699; SdhD: KC517315, AB548743, JF916693).
The Sdh genotype of each D. tanaceti isolate was compared to the boscalid EC50 range (μg a.i/mL) assigned by Hay et al. [7] from in vitro boscalid testing.
Collection of Didymella tanaceti field isolates for fungicide resistance genotyping and population responses
Additional D. tanaceti isolates were included in this study to further examine Sdh genotype variations and validate the predicted in vitro fungicide response. Firstly, a random set of D. tanaceti (n = 173) isolates collected from a range of commercial Tasmanian pyrethrum fields since 2004 was obtained for HRM assay development. Secondly, a D. tanaceti population (n = 774) collected by Pearce et al. [28] from four pyrethrum fields sown in 2011 within the pyrethrum cropping region across northern Tasmania was used to evaluate the field frequency of each Sdh substitution. Diseased leaves were sampled from each field in August and November 2012. Between the sampling periods, each field received the standard spring fungicide program, which included one application of boscalid (Filan, BASF) [28]. No other SDHI fungicides were applied in this period. Sampling in August was conducted the week prior to the application of the first fungicide in the program. Sampling of diseased material in November occurred 7 to 9 weeks after the final fungicide application of the spring program. Locations sampled within fields were recorded using GPS to allow precise re-sampling in November. Isolation, storage and DNA extraction of D. tanaceti from disease material was undertaken as outlined by Pearce et al. [28]. Mating-type idiomorph and genotype based on simple sequence repeat (SSR) markers were known for each isolate based on previously research [28], allowing for the population dynamics behind the identified Sdh substitutions to be examined.
HRM assay for Sdh allele detection
An HRM assay that detects multiple mutations within the amplified regions [69, 70] and can identify isolates at low frequency and/or novel Sdh substitutions for further in vitro boscalid testing, was designed. For assay development, the program uMELT [71] was used for each known polymorphic loci to predict the discrimination of alleles of potential PCR products of varying sizes using an in silico HRM algorithm. In instances where the melting temperature was unable to clearly differentiate the known Sdh alleles, protocols were adapted to include multiplexed HRM or HRM followed by restriction enzyme digest modifications. Prior to analysing using HRM, the differential product amplification in isolates of each allele, due to the multiplexing primers, was tested using standard PCR. The specific Sdh allele of isolates exhibiting novel melt peaks identified during HRM development were sequenced to identify new mutations and HRM assays redesigned as required.
Mutations in the SdhB were detected using a two-step process. This involved HRM analysis followed by restriction digestion and subsequent HRM (Fig 3A) due to a low melt temperature differential between two of the known alleles. The first step involved real-time PCR amplification and HRM with primers SdhB_HRM_F and SdhB_HRM_R (Table 4) that amplified an 85-bp product containing codons 277 and 279 of the SdhB. Conditions for amplification were an initial denature of 5 min at 95°C, followed by 40 cycles of denaturation at 95°C for 10 s, annealing at 50°C for 20 s and extension at 72°C for 10 s. Products were melted at 0.2°C intervals from 76 to 90°C with a 10 s hold at each temperature prior to data acquisition. Following HRM, 0.3 U of the restriction enzyme TaaI (HpyCH4III, Thermo Scientific, Waltham, MA), 1.5 uL of 10 × Buffer Tango and 3.47 uL of nuclease free water was added to each tube and tubes incubated in the Rotor-Gene for 1 hr at 65°C. TaaI recognizes CAN^GT sites. As this site was only found in individuals with a GTT (V) sequence at codon 279, the product from these individuals was digested into two fragments. To identify samples with a positive digest a re-melt protocol consisting of 95°C for 5 minutes and 40°C for 90 s was used to create heteroduplexes of all amplicons prior to melting the products over the same range as previous (Fig 3A). Isolates with a positive digest exhibited a distinct melt (Fig 3A). Analysis of the data of the two melts was undertaken and combined to give the final allele call.
Two independent multiplexed HRM reactions were used to detect concurrent substitutions within the SdhC gene (Fig 3B & 3C). For the first reaction, the primers SdhC_HRM_1F and SdhC_HRM_1R (Table 4, Fig 3B) were used to amplify a 88-bp product containing codons 73 and 79. Due to a low temperature differential between isolates with WT and mutation at codon 79, the allele-specific primer SdhC_HRM_79_F (Table 4, Fig 3B) was designed to anneal within the amplified fragment of isolates with the mutation in codon 79, resulting in two products of differing size and thus two melting events in these isolates (Fig 3B). HRM conditions included annealing at 60°C, 35 cycles and melting from 70–90°C in 0.2°C increments. For the second reaction, the primers SdhC_HRM_2F and SdhC_HRM_2R were used to amplify a 62-bp fragment containing codons 134 and 135. To discriminate all the mutations additional primers were designed to anneal within the amplified region. The allele-specific primers SdhC_HRM_135_F1 and SdhC_HRM_135_F2 were included to bind to isolates with a S135R substitution produced from an AGG and CGC mutations, respectively, resulting in two products for these isolates (Table 4, Fig 3C). Additionally, the primer SdhC_HRM_QR_R was designed to amplify a second product in isolates containing the combined H134Q/S135R substitution (Table 4, Fig 3C). HRM conditions included annealing at 55°C, 40 cycles and melting from 68 to 86°C in 0.2°C increments.
To detect mutations in the SdhD gene the primers SdhD_HRM_F and SdhD_HRM_R were designed to amplify a 112-bp product containing codons 112 and 122 (Table 4; Fig 3D). Additionally, due to a low temperature differential between alleles, the primer SdhD_HRM_112_F was included to amplify the two mutations resulting in substitution at codon 112 (Table 4. Fig 3D). HRM conditions included annealing at 58°C, 40 cycles and melting from 78 to 91°C in 0.1°C increments.
The HRM assays were conducted using the 2012 D. tanaceti population to select isolates for in vitro boscalid testing and to examine the population structure underlying the substitutions. All HRM reactions were undertaken in a Rotor-Gene Q Real-Time PCR machine (Qiagen). Reaction mixes contained 1 × Type- it HRM buffer (Qiagen), 0.35μM of each primer, 2 ng of DNA and enough nuclease free water to produce a final reaction volume of 10 μL. Within each run, three independent isolates with each known mutation were included as replicate positive controls. Data analysis of each run was undertaken in the Rotor Gene ScreenClust HRM software [72]. For analysis of each run, data was normalised within left and right boundaries of 1°C in width spanning a 10–20°C window over which melting occurred. An “unsupervised” analysis, using K-means and the known-genotype isolates (controls) employed as pseudo-unknowns, was used to identify any new putative genotypes, which separated into discrete clusters or appeared as outliers within clusters. The number of clusters and principle components (PC) were determined using the gap statistic [73]. Following the identification of all genotypes, samples were genotyped using the “supervised” mode. The known-genotype isolates were used to calculate a cluster distribution using linear discriminant analysis (LDA), with the centre of each cluster equal to the mean of the known-genotypes isolate of each identified mutation. For each isolate the probability of belonging to each genotype cluster was calculated. Isolates with low probability (<0.1) were selected for repeat HRM and analysis for confirmation. Additionally, 10% of isolates were randomly selected and screened in duplicate to evaluate reproducibility of the HRM assays.
The HRM results were used to select up to ten D. tanaceti isolates of each Sdh substitution and WT from the population for in vitro boscalid testing to confirm the phenotypic response associated with each substitution. Where more than ten isolates of each substitution were identified, SSR profile and mating-type data (based on Pearce et al. [28] or unpublished data) were used to select genetically diverse isolates for boscalid testing. Ten WT isolates were included to identify a baseline response of D. tanaceti isolates. To confirm the substitutions and further validate the HRM assay, the SdhB, SdhC and SdhD of the selected isolates (n = 92) were amplified and sequenced. Representative sequences of each mutation were deposited in GenBank (accessions: MK500737—MK500762; Table 1).
In vitro boscalid testing
For in vitro testing commercial grade boscalid (Filan; 50% a.i, BASF, Ludwigshafen, Germany) was dissolved in distilled autoclaved water and made up to 75% (v/v) in methanol to sterilise and provide an initial stock solution. Dilutions of the stock solution were amended to potato dextrose agar (PDA; Amyl Media, Dandenong, Australia) to obtain final boscalid concentrations of: 0, 0.001, 0.01, 0.05, 0.25, 0.5, 1.25, 2.5, 5, 10, 25 and 50 μg a.i/mL. The final concentration of methanol within each plate did not exceed 0.08% (v/v). Based on previously obtained EC50 range data [7], isolates of each substitution were tested on 6 or 7 boscalid concentrations in the ranges of 0 to 1.25 μg a.i/mL (WT isolates), 0 to 2.5 μg a.i/mL (SdhB-I279V isolates), 0 to 10.0 μg a.i/mL (SdhD-D112E isolates) or 0.0 to 50.0 μg a.i/mL (all remaining isolates with SdhB, SdhC and SdhD substitutions) (Table 1). For testing, a 4 mm diameter plug was excised from the margin of an actively growing 12-day old culture of each D. tanaceti isolate growing on PDA and placed onto plates of each fungicide concentration in triplicate and incubated at 21°C in the dark. Mycelial growth was measured across two perpendicular lines after 7 d incubation for each replicate plate of each fungicide concentration. Colony diameters were corrected by subtracting the diameter of the agar plug. For each isolate the average colony radius was calculated for each tested concentration. Relative growth at each concentration was calculated as the corrected colony radius divided by the averaged corrected radius of the non-amended controls (0 μg ai/ml). EC50 values were calculated using logistic regression of the relative growth against log10 of the boscalid concentration in the package NPLR [74] and calculation of the concentration that resulted in reduction of growth by 50% relative to non-amended controls. To allow inclusion of the data from the 0 μg ai/mL, the concentration was transformed, converting it to concentrations 2 log units lower than the lowest tested concentration. Data was analysed in R v3.1.3 [75]. For each isolate Resistance Factors (RFs) were also calculated using the following formula: RF = EC50X/EC50WT, where EC50X is the EC50 value of the isolate being examined, and EC50WT is the average EC50 value of the baseline isolates (WT). A phenotype was inferred based on the interpretation of RF values whereby isolates were grouped as sensitive (RF ≤ 3) or low (3 > RF ≤ 10), moderate (10 > RF ≤ 100), high (100 > RF ≤ 1000) or very high (> 1000) in resistance.
Frequency and genetic diversity of Sdh substitutions in 2012 D. tanaceti population
For the D. tanaceti population sampled in 2012, the frequency (%) of each substitution in each field was calculated for the two sampling periods and overall. Changes in the occurrence of Sdh substitutions between the two sampling periods was assessed by multinomial logistic regression where month of sampling, field of origin and the interaction between these factors were used as fixed effects. To avoid detrimental effects on model predictions, Sdh substitutions that occurred at frequencies less than 1% across the population were excluded from the data set prior to analysis. Analysis was conducted using the nnet package [76] within the R statistical language framework. Post hoc treatment comparisons were conducted with model least-squares means estimates and Tukey’s adjustment for multiple pairwise comparisons, using the package lsmeans [77]. Additionally, the Sdh substitutions were used to define sub-groups within the population. To examine the amount of genetic diversity within each of the sub-groups, the Sdh substitution data was combined with the SSR profile and mating-type data of each isolate identified by Pearce et al. [28]. For each sub-group the number of associated multilocus genotypes (MLGs), derived by combining the SSR profile and mating-type data, was calculated for the two sampling periods and overall. The number of genetically unique individuals in the overall population was also calculated by combining the Sdh, SSR and mating-type data.
Acknowledgments
The authors wish to thank Stacey Pilkington, Pattie Weichelt, Craig Palmer and Phil Gardam (Tasmanian Institute of Agriculture; University of Tasmania, Australia) for assisting with isolate collection. In addition, Drs. Sarah Pethybridge and Frank Hay are thanked for assistance in securing research funds and project development.
Data Availability
All population files are available from the Figshare database (accession: 10.6084/m9.figshare.7800929).
Funding Statement
Funding for this research was provided by the Australian Government through Australian Research Council with contribution from Botanical Resources Australia Pty. Ltd (ARC Linkage Project LP130100739), https://www.arc.gov.au/, to F. Hay, C. Wilson, S. Pethybridge and D.Gent. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Khater HF. Prospects of botanical biopesticides in insect pest management. Pharmacologia. 2012;3(12):641–56. [Google Scholar]
- 2.Casida JE, Quistad GB. Pyrethrum flower: production, chemistry, toxicology, and uses. New York: Oxford University Press; 1995. 350 p. [Google Scholar]
- 3.Pethybridge S, Wilson C. Confirmation of ray blight disease of pyrethrum in Australia. Australas Plant Pathol. 1998;27(1):45–8. [Google Scholar]
- 4.Vaghefi N, Pethybridge S, Ford R, Nicolas M, Crous P, Taylor P. Stagonosporopsis spp. associated with ray blight disease of Asteraceae. Australas Plant Pathol. 2012;41(6):675–86. [Google Scholar]
- 5.Pethybridge SJ, Hay FS, Esker PD, Gent DH, Wilson CR, Groom T, et al. Diseases of pyrethrum in Tasmania: challenges and prospects for management. Plant Dis. 2008;92(9):1260–72. 10.1094/PDIS-92-9-1260 [DOI] [PubMed] [Google Scholar]
- 6.Pearce T, Scott J, Crous P, Pethybridge S, Hay F. Tan spot of pyrethrum is caused by a Didymella species complex. Plant Pathol. 2016;65(7):1170–84. [Google Scholar]
- 7.Hay FS, Gent DH, Pilkington SJ, Pearce TL, Scott JB, Pethybridge SJ. Changes in distribution and frequency of fungi associated with a foliar disease complex of pyrethrum in Australia. Plant Dis. 2015;99(9):1227–35. 10.1094/PDIS-12-14-1357-RE [DOI] [PubMed] [Google Scholar]
- 8.Pethybridge SJ, Jones SJ, Shivas RG, Hay FS, Wilson CR, Groom T. Tan spot: A new disease of pyrethrum caused by Microsphaeropsis tanaceti sp. nov. Plant Pathol. 2008;57(6):1058–65. [Google Scholar]
- 9.Pethybridge SJ, Gent DH, Groom T, Hay FS. Minimizing crop damage through understanding relationships between pyrethrum phenology and ray blight disease severity. Plant Dis. 2013;97(11):1431–7. 10.1094/PDIS-11-12-1102-RE [DOI] [PubMed] [Google Scholar]
- 10.Avenot HF, Michailides TJ. Progress in understanding molecular mechanisms and evolution of resistance to succinate dehydrogenase inhibiting (SDHI) fungicides in phytopathogenic fungi. Crop Prot. 2010;29(7):643–51. [Google Scholar]
- 11.Deising HB, Reimann S, Pascholati SF. Mechanisms and significance of fungicide resistance. Braz J Microbiol. 2008;39(2):286–95. 10.1590/S1517-838220080002000017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hägerhäll C. Succinate: quinone oxidoreductases: variations on a conserved theme. Biochimica et Biophysica Acta 1997;1320(2):107–41. 10.1016/s0005-2728(97)00019-4 [DOI] [PubMed] [Google Scholar]
- 13.Horsefield R, Yankovskaya V, Sexton G, Whittingham W, Shiomi K, Ōmura S, et al. Structural and computational analysis of the quinone-binding site of complex II (succinate-ubiquinone oxidoreductase)—A mechanism of electron transfer and proton conduction during ubiquinone reduction. Journal of Biological Chemistry. 2006;281(11):7309–16. 10.1074/jbc.M508173200 [DOI] [PubMed] [Google Scholar]
- 14.Huang S, Millar AH. Succinate dehydrogenase: the complex roles of a simple enzyme. Current Opinion in Plant Biology. 2013;16(3):344–9. 10.1016/j.pbi.2013.02.007 [DOI] [PubMed] [Google Scholar]
- 15.Scalliet G, Bowler J, Luksch T, Kirchhofer-Allan L, Steinhauer D, Ward K, et al. Mutagenesis and functional studies with succinate dehydrogenase inhibitors in the wheat pathogen Mycosphaerella graminicola. PLoS One. 2012;7(4):e35429 10.1371/journal.pone.0035429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraaije BA, Bayon C, Atkins S, Cools HJ, Lucas JA, Fraaije MW. Risk assessment studies on succinate dehydrogenase inhibitors, the new weapons in the battle to control Septoria leaf blotch in wheat. Mol Plant Pathol. 2012;13(3):263–75. 10.1111/j.1364-3703.2011.00746.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vega B, Dewdney MM. Sensitivity of Alternaria alternata from citrus to boscalid and polymorphism in iron-sulfur and in anchored membrane subunits of succinate dehydrogenase. Plant Dis. 2015;99(2):231–9. 10.1094/PDIS-04-14-0374-RE [DOI] [PubMed] [Google Scholar]
- 18.Avenot HF, Michailides TJ. Resistance to boscalid fungicide in Alternaria alternata isolates from pistachio in California. Plant Dis. 2007;91(10):1345–50. 10.1094/PDIS-91-10-1345 [DOI] [PubMed] [Google Scholar]
- 19.Fernández-Ortuño D, Pérez-García A, Chamorro M, de la Peña E, de Vicente A, Torés JA. Resistance to the SDHI Fungicides Boscalid, Fluopyram, Fluxapyroxad, and Penthiopyrad in Botrytis cinerea from Commercial Strawberry Fields in Spain. Plant Dis. 2017:PDIS-01-17-0067-RE. [DOI] [PubMed] [Google Scholar]
- 20.Weber RW. Resistance of Botrytis cinerea to multiple fungicides in Northern German small-fruit production. Plant Dis. 2011;95(10):1263–9. 10.1094/PDIS-03-11-0209 [DOI] [PubMed] [Google Scholar]
- 21.Jones S, Pethybridge S, Gent D, Hay F. Sensitivity of Australian Sclerotinia sclerotiorum isolates from bean fields to boscalid. New Zeal J Crop Hort. 2011;39(3):203–7. [Google Scholar]
- 22.Matheron M, Porchas M. Activity of boscalid, fenhexamid, fluazinam, fludioxonil, and vinclozolin on growth of Sclerotinia minor and S. sclerotiorum and development of lettuce drop. Plant Dis. 2004;88(6):665–8. 10.1094/PDIS.2004.88.6.665 [DOI] [PubMed] [Google Scholar]
- 23.Sierotzki H, Scalliet G. A review of current knowledge of resistance aspects for the next-generation succinate dehydrogenase inhibitor fungicides. Phytopathology. 2013;103(9):880–7. 10.1094/PHYTO-01-13-0009-RVW [DOI] [PubMed] [Google Scholar]
- 24.Stevenson K, Langston D, Sanders F, editors. Baseline sensitivity and evidence of resistance to boscalid in Didymella bryoniae. APS Centennial Meeting July 26–30; 2008; Minneapolis, Minnesota.
- 25.Miyamoto T, Ishii H, Stammler G, Koch A, Ogawara T, Tomita Y, et al. Distribution and molecular characterization of Corynespora cassiicola isolates resistant to boscalid. Plant Pathol. 2010;59(5):873–81. [Google Scholar]
- 26.Vega B, Liberti D, Harmon PF, Dewdney MM. A rapid resazurin-based microtiter assay to evaluate Qol sensitivity for Alternaria alternata isolates and their molecular characterization. Plant Dis. 2012;96(9):1262–70. 10.1094/PDIS-12-11-1037-RE [DOI] [PubMed] [Google Scholar]
- 27.Mallik I, Arabiat S, Pasche J, Bolton M, Patel J, Gudmestad N. Molecular characterization and detection of mutations associated with resistance to succinate dehydrogenase-inhibiting fungicides in Alternaria solani. Phytopathology. 2014;104(1):40–9. 10.1094/PHYTO-02-13-0041-R [DOI] [PubMed] [Google Scholar]
- 28.Pearce TL, Scott JB, Pilkington SJ, Pethybridge SJ, Hay FS. Evidence for Sexual Recombination in Didymella tanaceti Populations, and Their Evolution Over Spring Production in Australian Pyrethrum Fields. Phytopathology. 2019;109(1):155–68. 10.1094/PHYTO-08-17-0280-R [DOI] [PubMed] [Google Scholar]
- 29.Brent KJ, Hollomon DW. Fungicide resistance: the assessment of risk: Global Crop Protection Federation Brussels, Belgium; 1998. [Google Scholar]
- 30.Jutsum AR, Heaney SP, Perrin BM, Wege PJ. Pesticide resistance: assessment of risk and the development and implementation of effective management strategies. Pest Manag Sci. 1998;54(4):435–46. [Google Scholar]
- 31.Gudmestad N, Arabiat S, Miller J, Pasche J. Prevalence and impact of SDHI fungicide resistance in Alternaria solani. Plant Dis. 2013;97(7):952–60. 10.1094/PDIS-12-12-1176-RE [DOI] [PubMed] [Google Scholar]
- 32.Mondal S, Bhatia A, Shilts T, Timmer L. Baseline sensitivities of fungal pathogens of fruit and foliage of citrus to azoxystrobin, pyraclostrobin, and fenbuconazole. Plant Dis. 2005;89(11):1186–94. 10.1094/PD-89-1186 [DOI] [PubMed] [Google Scholar]
- 33.Leroux P, Gredt M, Remuson F, Micoud A, Walker AS. Fungicide resistance status in French populations of the wheat eyespot fungi Oculimacula acuformis and Oculimacula yallundae. Pest Manag Sci. 2013;69(1):15–26. 10.1002/ps.3408 [DOI] [PubMed] [Google Scholar]
- 34.Skinner W, Bailey A, Renwick A, Keon J, Gurr S, Hargreaves J. A single amino-acid substitution in the iron-sulphur protein subunit of succinate dehydrogenase determines resistance to carboxin in Mycosphaerella graminicola. Curr Genet. 1998;34(5):393–8. [DOI] [PubMed] [Google Scholar]
- 35.Stammler G, Brix H, Glättli A, Semar M, Schoefl U, editors. Biological properties of the carboxamide boscalid including recent studies on its mode of action. Proc XVI Intional Plant Protection Congress, Glascow; 2007.
- 36.Avenot H, Sellam A, Karaoglanidis G, Michailides T. Characterization of mutations in the iron-sulphur subunit of succinate dehydrogenase correlating with boscalid resistance in Alternaria alternata from California pistachio. Phytopathology. 2008;98(6):736–42. 10.1094/PHYTO-98-6-0736 [DOI] [PubMed] [Google Scholar]
- 37.Popko JT Jr, Sang H, Lee J, Yamada T, Hoshino Y, Jung G. Resistance of Sclerotinia homoeocarpa field isolates to succinate dehydrogenase inhibitor fungicides. Plant Dis. 2018;102(12):2625–31. 10.1094/PDIS-12-17-2025-RE [DOI] [PubMed] [Google Scholar]
- 38.Laleve A, Gamet S, Walker A-S, Debieu D, Toquin V, Fillinger S. Site-directed mutagenesis of the P225, N230 and H272 residues of succinate dehydrogenase subunit B from Botrytis cinerea highlights different roles in enzyme activity and inhibitor binding. Environ Microbiol. 2014;16(7):2253–66. 10.1111/1462-2920.12282 [DOI] [PubMed] [Google Scholar]
- 39.Leroux P, Gredt M, Leroch M, Walker A-S. Exploring mechanisms of resistance to respiratory inhibitors in field strains of Botrytis cinerea, the causal agent of gray mold. Appl Environ Microbiol. 2010;76(19):6615–30. 10.1128/AEM.00931-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Veloukas T, Leroch M, Hahn M, Karaoglanidis GS. Detection and molecular characterization of boscalid-resistant Botrytis cinerea isolates from strawberry. Plant Dis. 2011;95(10):1302–7. 10.1094/PDIS-04-11-0317 [DOI] [PubMed] [Google Scholar]
- 41.Yin YN, Kim YK, Xiao CL. Molecular characterization of boscalid resistance in field isolates of Botrytis cinerea from apple. Phytopathology. 2011;101(8):986–95. 10.1094/PHYTO-01-11-0016 [DOI] [PubMed] [Google Scholar]
- 42.Lalève A, Fillinger S, Walker A-S. Fitness measurement reveals contrasting costs in homologous recombinant mutants of Botrytis cinerea resistant to succinate dehydrogenase inhibitors. Fungal Genet Biol. 2014;67:24–36. 10.1016/j.fgb.2014.03.006 [DOI] [PubMed] [Google Scholar]
- 43.Fan Z, Yang J-H, Fan F, Luo C-X, Schnabel G. Fitness and competitive ability of Alternaria alternata field isolates with resistance to SDHI, QoI, and MBC fungicides. Plant Dis. 2015;99(12):1744–50. 10.1094/PDIS-03-15-0354-RE [DOI] [PubMed] [Google Scholar]
- 44.Li HX, Nuckols TA, Harris D, Stevenson KL, Brewer MT. Differences in fungicide resistance profiles and multiple resistance to a quinone‐outside inhibitor (QoI), two succinate dehydrogenase inhibitors (SDHI), and a demethylation inhibitor (DMI) for two Stagonosporopsis species causing gummy stem blight of cucurbits. Pest Manag Sci. 2019. [DOI] [PubMed] [Google Scholar]
- 45.Yamashita M, Fraaije B. Non‐target site SDHI resistance is present as standing genetic variation in field populations of Zymoseptoria tritici. Pest Manag Sci. 2018;74(3):672–81. 10.1002/ps.4761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Miccolis Angelini RM, Masiello M, Rotolo C, Pollastro S, Faretra F. Molecular characterisation and detection of resistance to succinate dehydrogenase inhibitor fungicides in Botryotinia fuckeliana (Botrytis cinerea). Pest Manag Sci. 2014;70(12):1884–93. 10.1002/ps.3748 [DOI] [PubMed] [Google Scholar]
- 47.Adolf B, Volz A, Klaus A, Leiminger J, Metz N, Chaluppa N, et al., editors. Fungicide sensitivity of Alternaria solani populations in Germany. 60th German Crop Protection Conference "Crop Protection: Efficiency and Diversity"; 2016 20–23 September 2016; Halle, Germany.
- 48.Sierotzki H, Frey R, Morchoisne M, Olaya G, Mosch M, Scalliet G. Sensitivity of fungal pathogens to SDHI fungicides. In: Dehne HW, Deising HB, Gisi U, Kuck KH, Russell PE, Lyr H, editors. Modern fungicides and antifungal compounds VI 16th International Reinhardsbrunn Symposium, Friedrichroda, Germany, April 25–29, 20102011. p. 179–86.
- 49.Lichtemberg P, Puckett RD, Felts D, Luo Y, Doster L, Rodriguez D, et al. Survey of the pathogen of Alternaria late blight reveals different levels of carboxamide fungicide resistance in the main pistachio producing regions of California. Calif Agr. 2018;72(3):170–8. [Google Scholar]
- 50.Rehfus A, Miessner S, Achenbach J, Strobel D, Bryson R, Stammler G. Emergence of succinate dehydrogenase inhibitor resistance of Pyrenophora teres in Europe. Pest Manag Sci. 2016;72(10):1977–88. 10.1002/ps.4244 [DOI] [PubMed] [Google Scholar]
- 51.Yang J, Brannen P, Schnabel G. Resistance in Alternaria alternata to SDHI fungicides causes rare disease outbreak in peach orchards. Plant Dis. 2015;99(1):65–70. 10.1094/PDIS-04-14-0387-RE [DOI] [PubMed] [Google Scholar]
- 52.Heick TM, Justesen AF, Jørgensen LN. Anti-resistance strategies for fungicides against wheat pathogen Zymoseptoria tritici with focus on DMI fungicides. Crop Prot. 2017;99:108–17. [Google Scholar]
- 53.Rehfus A, Strobel D, Bryson R, Stammler G. Mutations in sdh genes in field isolates of Zymoseptoria tritici and impact on the sensitivity to various succinate dehydrogenase inhibitors. Plant Pathol. 2018;67(1):175–80. [Google Scholar]
- 54.Glattli A, Stammler G, Schlehuber S. Mutations in the target proteins of succinate-dehydrogenase inhibitors (SDHI) and 14alpha-demethylase inhibitors (DMI) conferring changes in the sensitivity—structural insights from molecular modelling 2009. 670–81 p. [Google Scholar]
- 55.Avenot HF, Van Den Biggelaar H, Morgan DP, Moral J, Joosten M, Michailides T. Sensitivities of baseline isolates and boscalid-resistant mutants of Alternaria alternata from pistachio to fluopyram, penthiopyrad, and fluxapyroxad. Plant Dis. 2014;98(2):197–205. 10.1094/PDIS-04-13-0459-RE [DOI] [PubMed] [Google Scholar]
- 56.Knapova G, Gisi U. Phenotypic and genotypic structure of Phytophthora infestans populations on potato and tomato in France and Switzerland. Plant Pathol. 2002;51(5):641–53. [Google Scholar]
- 57.Shrestha SK, Cochran A, Mengistu A, Lamour K, Castro-Rocha A, Young-Kelly H. Genetic diversity, QoI fungicide resistance, and mating type distribution of Cercospora sojina—Implications for the disease dynamics of frogeye leaf spot on soybean. PloS one. 2017;12(5):e0177220 10.1371/journal.pone.0177220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pethybridge SJ, Hay F, Jones S, Wilson C, Groom T. Seedborne infection of pyrethrum by Phoma ligulicola. Plant Dis. 2006;90(7):891–7. 10.1094/PD-90-0891 [DOI] [PubMed] [Google Scholar]
- 59.Scott J, Gent DH, Pearce T, Pethybridge SJ, Pilkington S, Hay F. Mycoflora associated with pyrethrum seed and the integration of seed steam treatment into foliar disease management strategies. Plant Dis. 2017. [DOI] [PubMed] [Google Scholar]
- 60.Pearce TL, Scott JB, Hay FS, Pethybridge SJ. Mating-type gene structure and spatial distribution of Didymella tanaceti in pyrethrum fields. Phytopathology. 2016;106(12):1521–9. 10.1094/PHYTO-01-16-0038-R [DOI] [PubMed] [Google Scholar]
- 61.Pethybridge SJ, Esker P, Dixon P, Hay F, Groom T, Wilson C, et al. Quantifying loss caused by ray blight disease in Tasmanian pyrethrum fields. Plant Dis. 2007;91(9):1116–21. 10.1094/PDIS-91-9-1116 [DOI] [PubMed] [Google Scholar]
- 62.Pethybridge SJ, Hay FS, Groom T, Wilson CR. Improving fungicide-based management of ray blight disease in Tasmanian pyrethrum fields. Plant Dis. 2008;92(6):887–95. 10.1094/PDIS-92-6-0887 [DOI] [PubMed] [Google Scholar]
- 63.Vaghefi N, Hay FS, Ades PK, Pethybridge SJ, Ford R, Taylor PWJ. Rapid changes in the genetic composition of Stagonosporopsis tanaceti population in Australian pyrethrum fields. Phytopathology. 2015;105(3):358–69. 10.1094/PHYTO-08-14-0212-R [DOI] [PubMed] [Google Scholar]
- 64.Thomas A, Langston D Jr, Stevenson K. Baseline sensitivity and cross-resistance to succinate-dehydrogenase-inhibiting and demethylation-inhibiting fungicides in Didymella bryoniae. Plant Dis. 2012;96(7):979–84. 10.1094/PDIS-09-11-0744-RE [DOI] [PubMed] [Google Scholar]
- 65.Veloukas T, Markoglou AN, Karaoglanidis GS. Differential effect of Sdh B gene mutations on the sensitivity to SDHI fungicides in Botrytis cinerea. Plant Dis. 2013;97(1):118–22. 10.1094/PDIS-03-12-0322-RE [DOI] [PubMed] [Google Scholar]
- 66.Avenot HF, Thomas A, Gitaitis RD, Langston DB Jr, Stevenson KL. Molecular characterization of boscalid and penthiopyrad resistant isolates of Didymella bryoniae and assessment of their sensitivity to fluopyram. Pest Manag Sci. 2012;68(4):645–51. 10.1002/ps.2311 [DOI] [PubMed] [Google Scholar]
- 67.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40(15):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–80. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chatzidimopoulos M, Ganopoulos I, Madesis P, Vellios E, Tsaftaris A, Pappas A. High-resolution melting analysis for rapid detection and characterization of Botrytis cinerea phenotypes resistant to fenhexamid and boscalid. Plant Pathol. 2014;63(6):1336–43. [Google Scholar]
- 70.Samaras A, Madesis P, Karaoglanidis GS. Detection of sdhB gene mutations in SDHI-resistant isolates of Botrytis cinerea using high resolution melting (HRM) analysis. Front Microbiol. 2016;7 10.3389/fmicb.2016.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dwight Z, Palais R, Wittwer CT. uMELT: prediction of high-resolution melting curves and dynamic melting profiles of PCR products in a rich web application. Bioinformatics. 2011;27(7):1019–20. 10.1093/bioinformatics/btr065 [DOI] [PubMed] [Google Scholar]
- 72.Reja V, Kwok A, Stone G, Yang L, Missel A, Menzel C, et al. ScreenClust: Advanced statistical software for supervised and unsupervised high resolution melting (HRM) analysis. Methods. 2010;50(4):S10–S4. 10.1016/j.ymeth.2010.02.006 [DOI] [PubMed] [Google Scholar]
- 73.Tibshirani R, Walther G, Hastie T. Estimating the number of clusters in a data set via the gap statistic. Journal of the Royal Statistical Society: Series B (Statistical Methodology). 2001;63(2):411–23. [Google Scholar]
- 74.Commo F, Bot BM. R package nplr n-parameter logistic regressions. 2016. [Google Scholar]
- 75.R Core Team. R: A language and environment for statistical computing. 3.2.4 ed. Vienna, Austria: R Foundation for Statistical Computing; 2016. [Google Scholar]
- 76.Venables W, Ripley B. Statistics Complements to Modern Applied Statistics with S. Fourth Edition New York, USA: Springer; 2002. [Google Scholar]
- 77.Lenth RV. Least-squares means: the R package lsmeans. J Stat Softw. 2016;69(1):1–33. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All population files are available from the Figshare database (accession: 10.6084/m9.figshare.7800929).


