Abstract
Bcl-2 family proteins control a decisive apoptotic event: mitochondrial outer membrane permeabilization (MOMP). To discover MOMP-regulating proteins, we expressed a library of intracellular single-chain variable fragments (scFvs) (“intrabodies”) and selected for those rescuing cells from apoptosis induced by BimS (the short isoform of Bim). One anti-apoptotic intrabody, intrabody 5 (IB5), recognized pyruvate kinase M2 (PKM2), which is expressed in cancer cells. PKM2 deletion ablated this clonogenic rescue; thus, IB5 activated a latent cytoprotective function of PKM2. This resulted not from pyruvate kinase activity per se but rather from the formation of an active tetrameric conformation of PKM2. A stably tetrameric PKM2 mutant, K422R, promoted cell survival even in the absence of IB5, and IB5 further increased survival. Mitochondria isolated from IB5-expressing cells were relatively resistant to MOMP in vitro. In cells, IB5 expression up-regulated Mitofusin-1 (Mfn1) and increased mitochondrial length. Importantly, Mfn1 deficiency abrogated IB5’s cytoprotective effect. PKM2’s anti-apoptotic function could help explain its preferential expression in human cancer.
Author summary
Proteins belonging to the Bcl-2 family regulate a common form of cell death known as apoptosis. Typically, these proteins function in apoptosis by controlling the formation of large pores in the mitochondrial outer membrane (MOM). While many proteins that regulate apoptosis have been identified over the years, some may still be unknown. Here, we used an unbiased approach in which we first expressed in cultured tumor cells a library of intracellular single-chain antibodies termed “intrabodies.” We then selected for intrabodies that allowed cells to evade apoptosis. We identified pyruvate kinase isoform M2 (PKM2), a major glycolytic enzyme that has been linked to cancer development, as the specific target of one such anti-apoptotic intrabody. We showed that the PKM2-specific intrabody promoted cell survival not by neutralizing its target but rather by activating an anti-apoptotic function of PKM2. While this cell survival function of PKM2 was not related to changes in the levels of Bcl-2 family proteins or to effects on the enzymatic activity of PKM2, we found that cell survival requires the increased expression of a MOM protein, Mitofusin-1 (Mfn1), known to regulate mitochondrial fusion. We conclude that this cell survival function of PKM2 could contribute to a role in cancer progression for this protein.
Introduction
Apoptosis is a cellular suicide process that is important for certain aspects of normal animal development [1] and is dysregulated in various diseases, especially cancer [e.g., 2,3]. Members of the Bcl-2 protein family act at the mitochondrial outer membrane (MOM) to regulate the central events in apoptotic cell death [4–13]. Venetoclax, a drug targeting Bcl-2, is currently approved for the treatment of a refractory form of chronic lymphocytic leukemia [14,15], and other drugs that directly target Bcl-2-family proteins are now in cancer clinical trials [16–18].
Bcl-2 family proteins function in a complex network of heterodimeric interactions that collectively decide between cell survival and death [12]. Several Bcl-2 subfamilies carry out different functions [19]. In particular, the proteins Bax and Bak comprise the effector subfamily responsible for the critical mitochondrial events in cell death. Genetic and in vitro studies [6,20–23] have shown that Bax/Bak can be activated by transient interactions with other Bcl-2 family proteins belonging to the “Bcl-2 homology domain 3 (BH3)-only” category (including Bim, Bid, Puma, and others.) Once activated, Bax/Bak undergo conformational changes to become fully integrated in the MOM. As a result, these proteins produce large lipidic membrane pores [24,25], in an event known as mitochondrial outer membrane permeabilization (MOMP) [13,19,26]. MOMP allows soluble mitochondrial proteins (e.g., cytochrome c, Smac, and Omi) to escape into the cytoplasm, where they trigger the activation of caspase proteases that carry out the cell death program. MOMP and cell death are decisively regulated by Bcl-2 family interactions [6,21,27,28], and this underlies the importance of targeting these proteins for cancer therapy. In this regard, Letai and colleagues have shown that the in vitro response of mitochondria from patient tumor samples to BH3 domain peptides can often predict the effect of therapy [29–31].
Bcl-2 family members can also be regulated by proteins outside the Bcl-2 family. For example, p53 can act at mitochondria both to activate Bax directly and to sequester Bcl-xL [32]. Similarly, the Retinoblastoma protein pRB is reported to translocate to mitochondria to promote Bax activation in a nontranscriptional manner [33], and oncogenes such as Myc and Ras also modulate the expression of key Bcl-2-family proteins [34]. The ability of proto-oncoproteins to inhibit or activate apoptosis is an important facet of their homeostatic function, inasmuch as cell death serves as a critical counterbalance to cell proliferation.
To discover molecules regulating the core mechanism of mitochondria-dependent cell death, we developed an unbiased functional selection approach that used libraries of “intrabodies”: intracellularly expressed single-chain antibodies (scFv). We found that some of the selected intrabodies specifically recognized a key metabolic regulatory protein, pyruvate kinase M2 (PKM2). This suggests that PKM2, aside from its well-documented role in glycolytic metabolism, could also have an expressly anti-apoptotic function.
PKM2 is an important regulator of tissue homeostasis as well as tumor growth and metabolism [e.g., 35] and is currently a subject of intense research [reviewed in 36,37–39]. PKM2 is a glycolytic enzyme that promotes the “Warburg effect,” also termed aerobic glycolysis, in which cells exhibit increased glucose to lactate conversion even in the presence of oxygen [40]. In cancer cells, PKM2 is typically expressed preferentially over its related isoform PKM1, even when the tissue of origin does not express PKM2. Hypothetically, cancers gain some selective advantage from the highly regulated functions of PKM2. The adaptive metabolic functions of PKM2 also come into play in some cell types that quickly transition to a proliferative state, such as lipopolysaccharide (LPS)-activated macrophages [41].
PKM1 and PKM2 are generated from transcripts of the PKM gene by alternative mRNA splicing. Both isoforms can catalyze the last step in glycolysis, in which phosphoenolpyruvate (PEP) and ADP are converted to pyruvate and ATP. Isoforms M1 and M2 are identical except for the region encoded by the one alternatively spliced exon (exon 9 for PKM1 and 10 for PKM2), yielding a difference in only 22 amino acids. PKM1 exists as a constitutively active tetramer, whereas PKM2 is subject to many forms of regulation. Various metabolites, including fructose-1,6-bisphosphate (FBP), serine, phenylalanine, and triiodo-L-thyronine (T3), can allosterically regulate PKM2’s glycolytic activity [40,42]. In vitro biochemical studies have shown that PKM2 exists in equilibrium between a glycolytically active tetramer form and less active dimer or monomer forms [43,44]. Based on crystallographic data, it has also been proposed that PKM2 tetramers can transition between inactive T-state and active R-state conformations [45].
Paradoxically, it is the ability of PKM2’s glycolytic activity to be reduced that favors rapid cell proliferation. Reduced PK activity correlates with increased biosynthesis of metabolites important for cell proliferation, potentially explaining why tumor cells prefer the M2 isoform [40,46]. Consistent with this idea, treatment of cells with small-molecule activators of PKM2 [47,48] or the replacement of PKM2 with the constitutively active isoform PKM1 [35] can reduce cell proliferation in some situations. In primary Mouse Embryonic Fibroblasts (MEFs), deletion of PKM2 results in increased PKM1 expression, and this in turn impairs nucleotide availability for DNA synthesis, thereby inhibiting cell cycle progression [49].
PKM2 is reported also to have nonglycolytic functions. Many PKM2 interaction partners have been described, including multiple transcription factors [50]. For example, PKM2 is reported to cooperate with Hif-1α to regulate the transcription of multiple glycolysis-related proteins, which contribute to metabolic remodeling and the Warburg effect [41,51–53]. These transcriptional functions require the nuclear import of PKM2 [52–55]. PKM2’s nuclear translocation can be promoted by epidermal growth factor receptor (EGFR) activation [56] and regulated by Erk1/2 and JMJD5 [57,58]. In the nucleus, PKM2 can promote β-catenin transactivation, leading to the expression of cyclin D1 and tumorigenesis [56]. A PKM2-activating compound, TEPP-46, which causes PKM2 tetramerization, inhibits Hif-1α–dependent transcriptional effects [41], supporting the idea that the dimeric form of PKM2 is responsible for transcriptional functions. Dimeric PKM2 is also reported to possess protein kinase activity, targeting multiple oncogenic factors [54,59–61]. However, PKM2 protein kinase activity is controversial, as Vander Heiden and colleagues found no evidence of protein kinase activity for PKM2 in cell lysates [62].
In some cases, PKM2 ablation can produce or enhance cell death [63–69]. Precisely how PKM2 affects apoptosis is unclear. PKM2 silencing has been reported to stabilize proapoptotic Bim [70] or down-regulate the expression of the anti-apoptotic proteins Bcl-xL or Mcl-1 [71,72]. However, PKM2 knockdown produces an artificial situation. PKM2 has multiple functions that may be regulated independently, and experiments in which this protein is ablated would involve a simultaneous loss of all of these activities, along with a compensatory up-regulation of PKM1, making interpretation difficult. In contrast to the studies just mentioned, Sabatini and colleagues showed that the inhibition of PKM2 activity under ischemic conditions had the effect of promoting cell survival rather than cell death [73]. The cells bordering necrotic foci in gliomas expressed higher levels of the enzyme SHMT2, leading to an allosteric inhibition of PKM2’s glycolytic activity. This provided a significant protection from ischemic cell death. In another ischemia model, these authors found that overexpression of PKM2 or treatment with the PKM2-activating compound TEPP-46 eliminated the increased cell viability produced by SHMT2. It is unclear whether this connection between reduced PKM2 activity and survival is a general phenomenon or only applies to certain cancer cell subsets or environments.
In contrast to studies emphasizing PKM2 loss of function, our results now show that PKM2 possesses a positive cytoprotective function that can be activated by a PKM2-specific intrabody We show that this latent function of PKM2 counteracts the central Bax/Bak-dependent mitochondrial apoptotic mechanism. Moreover, the stably tetrameric mutant PKM2 (K422R) supported intrabody 5 (IB5)’s cytoprotective effect, arguing that the anti-apoptotic function involves the cytoplasmic tetramer form rather than the nuclear dimer form of PKM2. The K422R mutant also produced BimS resistance in MEFs after expression for several passages, even in the absence of IB5. This mutant’s ability to counteract the central apoptotic pathway could provide a selective advantage for these cells, and indeed, this mutation was found to be spontaneously selected in Bloom syndrome patient tumor cells. The IB5/PKM2-induced cytoprotective function depended in part on up-regulation of the mitochondrial fusion–related protein Mitofusin-1 (Mfn1). Therefore, we propose that PKM2 can activate an Mfn1-dependent general anti-apoptotic pathway, which could help explain why human cancer cells often preferentially express the M2 isoform of PK.
Results and discussion
Molecules regulating the core apoptotic machinery could be important in a variety of pathological and physiological situations. To discover such proteins, we adapted a functional selection approach that had been developed to identify proteins with various intracellular functions [74–76]. First, we infected HEK293T (293T) cells with a lentiviral library of genes encoding “intrabodies” [76]: intracellularly expressed scFvs. These molecules consisted of the variable regions from immunoglobulin heavy (VH) and light (VL) chains, connected by a flexible peptide linker, which formed a naïve human combinatorial scFv lentiviral library (diversity 4.5 x 109). We induced apoptosis in the cells by transiently transfecting them with a cDNA encoding BimS, the most potent pro-apoptotic isoform of Bim. Bim is one of the most important BH3-only proteins and is required for cell homeostasis in numerous physiological settings. Importantly, BimS promotes cell death by acting in the central apoptotic death mechanism, both by activating Bax/Bak and by sequestering anti-apoptotic Bcl-2 family members [21,27]. Bax/Bak activation then produces MOMP, crista junction remodeling, and apoptosis [6,77].
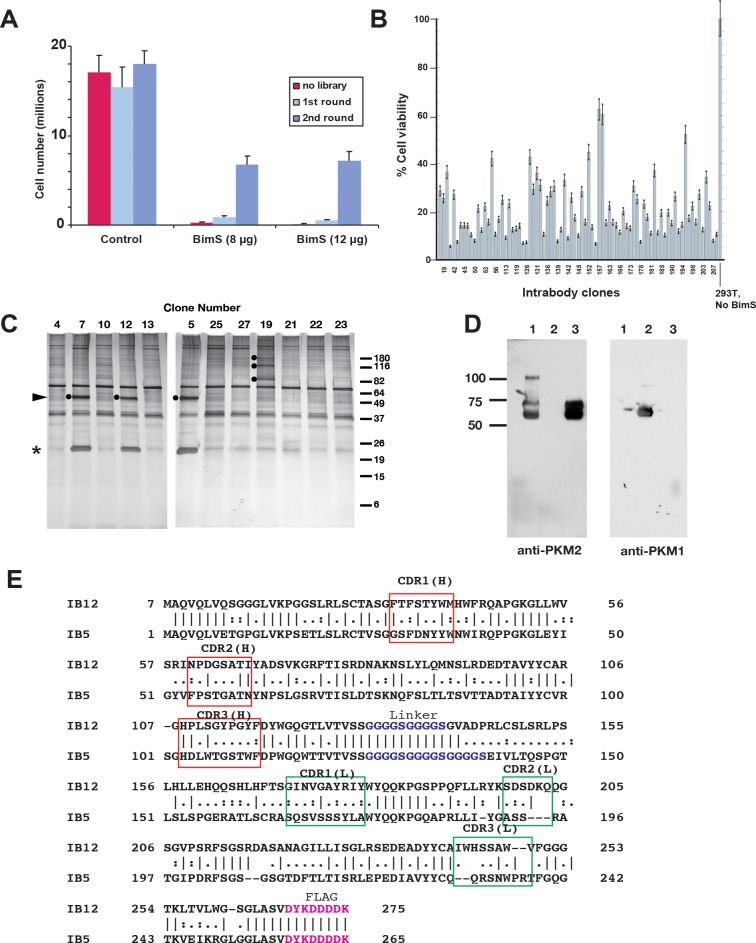
We then selected for intrabodies that rescued cells from BimS-induced death by recovering scFv-encoding DNA from the surviving cells. This selection process was efficient, as BimS transfection killed approximately 99% of the control cells, whereas expression of the lentiviral scFv library rescued a small percentage of the cells (Fig 1A). We then recovered the scFv-encoding DNA from surviving cells, with which we created a new lentiviral library for a second round of selection. In this round, intrabodies rescued approximately 40% of the cells from BimS-induced death, implying a substantial enrichment of intrabodies with prosurvival activity (Fig 1A). A third round of selection did not increase the percentage of cell survival. This was not surprising, given the likelihood that many of the recovered intrabodies would promote survival with less than 100% efficiency.
Fig 1. Selection of intrabodies that rescue cells from BimS-induced apoptosis identifies PKM2 as a target.
A. Enrichment in two rounds of selection. For the first round, 293T cells were first infected with a lentiviral human naïve scFv library. Then, 1 x 105 of the scFv-expressing cells were transiently transfected with BimS (using either 4 μg/ml or 6 μg/ml of plasmid DNA input per reaction) under the control of the EF-1α promoter, as indicated in Materials and methods. Lentiviral DNA was recovered from these rescued cells and used for a second round of selection. In the second round, many more cells (approximately 40%) were rescued from BimS-induced apoptosis. B. Individual scFv genes protected cells to various extents from BimS-induced apoptosis. After three rounds of selection, intrabody-coding sequences were amplified by PCR and subcloned into a plasmid for expression in E. coli. Individual DNA sequences were sequenced and expressed in 293T cells for testing of their ability to protect cells from apoptosis induced by transfection with BimS. The percentage of viable cells, relative to cells not transfected with BimS, was assayed. C. Some intrabodies arising from the selection procedure immunoprecipitated specific cellular proteins. Intrabodies that rescued cells from BimS-induced death were chosen for pull-down analysis, as described in Materials and methods. Left panel: Triton-X-100 (1%) cell extracts were incubated with anti-FLAG beads, then proteins were eluted with 3xFLAG peptide and separated by SDS-PAGE with silver stain. Specific bands are marked with dots; some bands (e.g., at 37 and 70 kD) are nonspecific. Clones 5, 7, and 12 (independent isolates) pulled down a 55-kD protein now identified as PKM2, while clone 19 pulled down several specific bands (not studied here). The bands near 25 kD are the scFv polypeptides, whose expression levels varied. D. IB5 recognized PKM2. Shown is an IP-western of lysates from cells expressing IB5. The lysates were incubated with anti-FLAG beads, and coprecipitating proteins were eluted with FLAG2 peptide (lane 3). Immunoblots were probed with antibody to PKM2 (left) or PKM1 (right). Purified PKM2 (lane 1) and PKM1 (lane 2) were controls for antibody specificity. E. Protein sequences of IB5 and IB12 are dissimilar, underscoring the functional importance of their common target, PKM2. Red boxes, heavy-chain CDRs; green boxes, light-chain CDRs; magenta type, FLAG tag. The selected intrabody plasmids were sequenced by Sanger sequencing. Sequences were analyzed with Vbase2. Note: underlying data are included in corresponding tabs in the accompanying supplemental Excel file, S1 Data. 293T, HEK293T; CDR, Complementarity Determining Region; EF-1α, elongation factor 1α; IB, intrabody; PKM1/M2, pyruvate kinase isoform M1/M2; scFv, single-chain variable fragment.
To identify intrabodies that inhibit apoptosis, we isolated single intrabody-encoding genes by subcloning the enriched DNA from the second round into bacteria. We introduced approximately 300 of these individual scFv genes separately into 293T cells. As expected, many of the intrabodies rescued 293T cells from apoptosis induced by BimS expression, to varying extents (Fig 1B). To identify protein targets, we performed FLAG pull-downs of some of the intrabody-target protein complexes, which we resolved on silver-stained SDS-polyacrylamide gels. We found that some intrabodies precipitated specific cellular proteins (Fig 1C). By matrix-assisted laser desorption/ionization-time-of-flight (MALDI-TOF) mass spectrometry and immunoblot analysis (Fig 1D), we identified one protein target of three different scFv-encoding DNA clones (5, 7, and 12) as PKM2. scFvs 5 and 7 had an identical DNA sequence, whereas scFv 12 was different (the light-chain Complementarity Determining Regions [CDRs] of scFv 12 were essentially distinct from those of scFv 5, and heavy-chain CDRs were only approximately 30% identical; Fig 1E). This apparent convergent selection underscores the potential importance of PKM2 as an apoptosis-regulating target protein. We chose IB5 for further study. The specific pull-down of PKM2 and not PKM1 suggests that intrabody binding might involve residues encoded in exon 10, which is unique to PKM2.
To verify that PKM2 is indeed the functional target of IB5, we first used small interfering RNA (siRNA) to silence PKM2 in 293T cells and saw a substantial reduction in the ability of IB5 to rescue cells from BimS killing (S1 Fig). Next, we expressed IB5 in MEFs genetically deficient for PKM2 [49]. Human and mouse PKM2 display approximately 98% sequence identity; thus, IB5 was expected to cross-react with the mouse protein. Indeed, IB5 rescued a percentage of the wild-type (WT) cells from BimS-induced killing, but this rescue was entirely abrogated in PKM2-deficient MEFs (Fig 2A). Moreover, reconstituting the PKM2-null MEFs with cDNA encoding WT PKM2, but not PKM1, restored the cytoprotective ability of IB5 (Fig 2A). PKM2-null MEFs are known to up-regulate expression of PKM1 [49], further confirming the specificity of the effect for PKM2. We conclude that IB5 does not neutralize PKM2 but rather acts positively to stimulate an anti-apoptotic function of PKM2. The intrabody IB5 likely mimics an unidentified physiological interaction partner of PKM2 that activates this cytoprotective function.
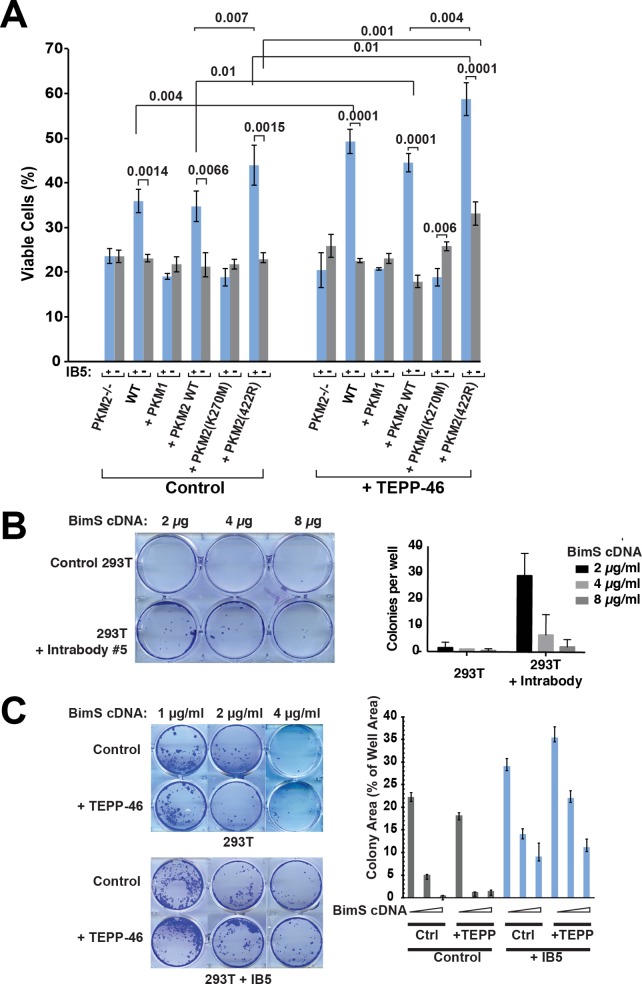
Fig 2. Genetic deletion of PKM2 abrogated the protective effect of scFv 5.
Reconstitution of PKM2-deficient MEFs with PKM2 WT or PKM2 (K422R), but not PKM1 or PKM2 (K240M), restored cell death protection. The PKM2-activating compound TEPP-46 enhanced the clonogenic survival effect of IB5. A. Viability assays with reconstituted cells. WT MEFs, PKM2-deficient MEFs, or PKM2-deficient MEFs reconstituted with WT or mutant PKM2 cDNA were infected or not with IB5, then later transfected with BimS expression plasmid. Surviving cells were counted 48 h afterwards. Note that only WT cells or PKM2-deficient MEFs reconstituted with WT PKM2 exhibited cytoprotective activity of IB5. B. IB5 produced clonogenic survival despite BimS expression. Control or IB5-expressing cells were transfected with BimS cDNA, and after 5 d, the plates were fixed with 6.0% glutaraldehyde and stained with 0.5% crystal violet. Top: example crystal violet-stained plate; bottom: average colony counts from three independent experiments, ± SEM. C. IB5-induced clonogenic rescue of 293T cells from BimS was enhanced by treatment with TEPP-46. Control 293T or IB5-expressing cells were incubated with or without 27 g/ml TEPP-46 for 3 h, then transfected with BimS cDNA in a further 24-h incubation also including TEPP-46 or vehicle. The plates were fixed and stained with crystal violet after 1 week. Left: examples of crystal violet-stained plates; right: as cells did not typically grow as discrete colonies, we quantified the total area of colonies (a measure of the total number of proliferating cells) formed in each well using ImageJ software [78]; mean ± SEM are shown from three independent experiments. Note: underlying data are included in corresponding tabs in the accompanying supplemental Excel file, S1 Data. 293T, HEK293T; IB, intrabody; MEF, Mouse Embryonic Fibroblast; PKM1/M2, pyruvate kinase isoform M1/M2; scFv, single-chain variable fragment; WT, wild-type.
IB5, like a number of our intrabody hits, rescued a moderate percentage (approximately 15%–20%) of the 293T cells from BimS-induced death. We suspect that it is a tall order for intrabodies to be very potent. These molecules are monovalent and thus do not have the enhanced avidity of IgG or IgM. Also, to have an effect revealed by selection, they would likely need to be expressed as abundantly as their target proteins and therefore might be present in limiting amounts. Consistent with this possibility, when we expressed IB5 with a weaker promoter, we found that its survival effect was reduced. In any case, this cell survival is potentially significant, given how potently and directly BimS activates the central mitochondrial pathway. Importantly, we found that the PKM2-specific intrabody produced clonogenic survival, meaning that the surviving cells were able to proliferate (Fig 2B and 2C). Thus, this latent anti-apoptotic activity of PKM2 could be consequential for physiological situations, e.g., for the progression of preneoplastic cells. If even a fraction of these survive, they could ultimately undergo further adaptations, leading to oncogenesis.
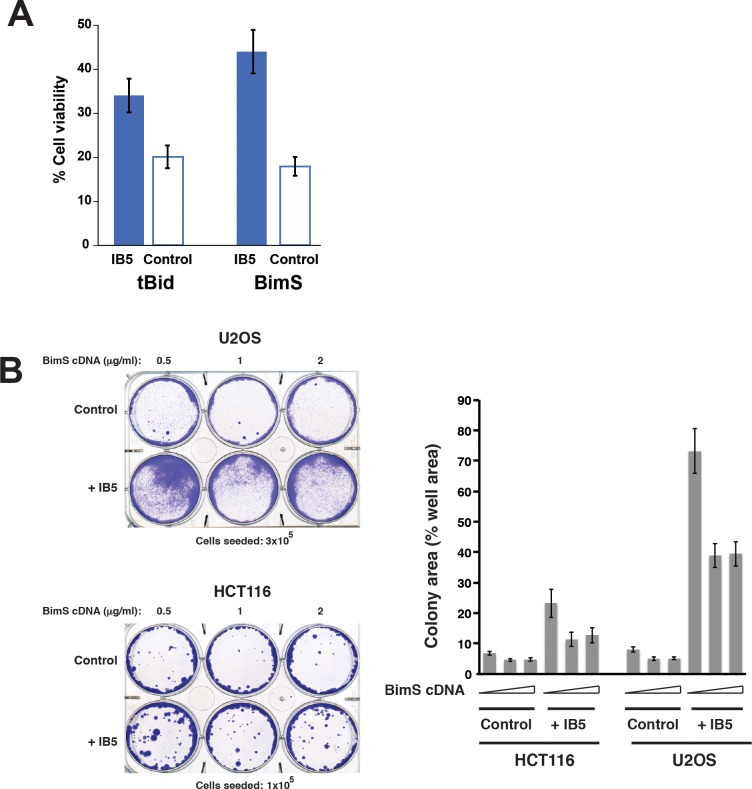
We found that the PKM2-specific intrabody protected cells from death induced by another potent BH3-only protein, truncated Bid (tBid; Fig 3A). As Bim and Bid are two major activators of the core apoptotic pathway, directly upstream of Bax/Bak activation and mitochondrial permeabilization, this suggests that PKM2 can inhibit the common apoptotic pathway at the level of Bax/Bak activation. If so, PKM2 could oppose physiological cell death triggered by multiple pro-apoptotic pathways. Indeed, we found that IB5 promoted clonogenic survival in cells treated with the DNA-damaging drug etoposide, although not with another cytotoxic agent, staurosporine (S2 Fig). The reasons for this are unclear, but we note that staurosporine is a promiscuous protein kinase inhibitor and presumably acts pleiotropically. Moreover, staurosporine was reported to activate both the intrinsic and the extrinsic (death receptor–mediated) pathways of apoptosis [79] and might therefore be able to bypass the requirement for Bax/Bak activation in these cells.
Fig 3. IB5 expression rescued 293T cells from death induced by transient expression of the potent pro-apoptotic proteins BimS and tBid.
A. IB5-expressing 293T were protected from death induced by transient expression of tBid or BimS. 293T cells were transfected treated with 1 g/ml tBid or BimS cDNA. Surviving cells were counted after 72 h. B. IB5 expression rescued U2OS and HCT116 cells clonogenically from BimS-induced death. Left: examples of crystal violet-stained plates; right: as cells did not typically grow as discrete colonies, we measured colony area as a percentage of total plate area. Note: underlying data are included in corresponding tabs in the accompanying supplemental Excel file, S1 Data. 293T, HEK293T; IB5, intrabody 5; tBid, truncated Bid.
In further experiments, we found that IB5 substantially protected other tumor cell lines, U2OS and HCT116, from BimS-induced apoptosis (Fig 3B). However, anti-apoptotic function was cell type–specific, as IB5 failed to rescue two breast cancer–derived cell lines (parental MDA-MB231 and a lung metastatic derivative MDA-MB231-LM2) from BimS-induced cell death (S3 Fig).
We ruled out the explanation that IB5 expression could alter the intracellular levels of PKM2 (S4 Fig). We could not determine whether IB5 expression altered the expression level of the exogenous BimS cDNA because the control condition, in which IB5 was absent, produced cell death in almost all of the BimS-expressing cells. However, an effect on BimS levels is unlikely, as IB5 expression did not affect levels of endogenous Bim EL and L isoforms in normal 293T cells. Moreover, Bim EL, L, and S isoforms were detectable in the BimS-resistant cell population rescued by IB5 (S4 Fig).
Enhanced PKM2 glycolytic activity is not the sole explanation for the anti-apoptotic effect of IB5
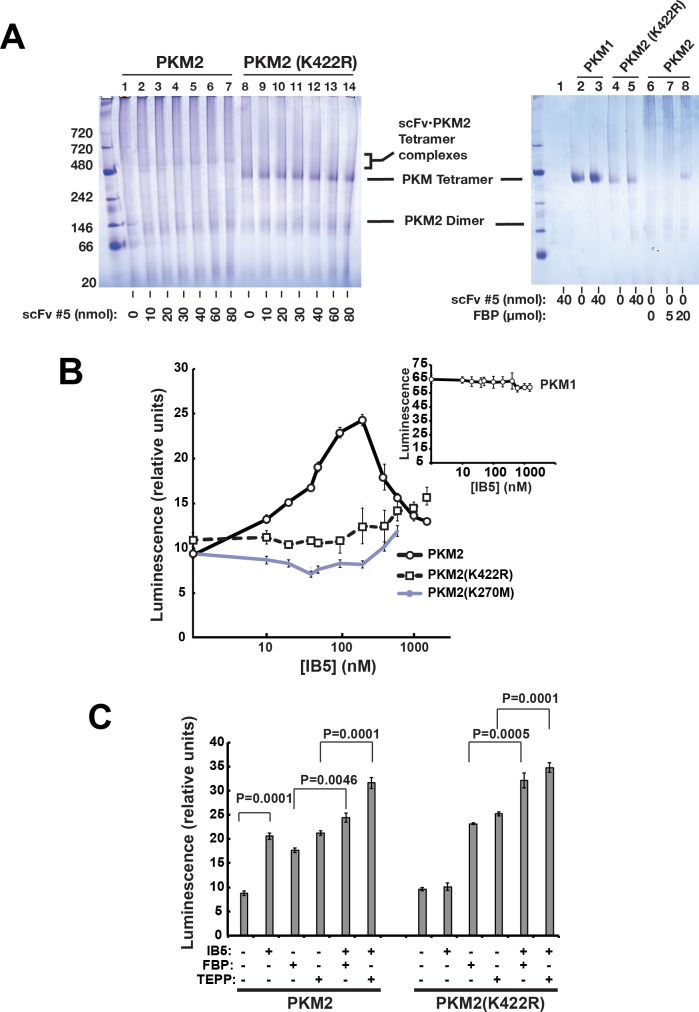
Because glucose metabolism can influence cell survival [52,80–82], we asked whether IB5 could rescue cells simply through stimulating PKM2’s glycolytic activity. First, to analyze the antibody’s interaction with PKM2 in vitro, we produced both PKM2 and a monovalent scFv corresponding to IB5 (scFv 5) as recombinant proteins in E. coli. We then mixed these proteins at various ratios and analyzed them by blue native gel electrophoresis (Fig 4A). We found that monovalent scFv 5 strongly increased the tetrameric PKM2 species and shifted up the tetramer band to a degree dependent on the molar input ratio of scFv. Thus, the antibody bound directly to PKM2, promoting its stable tetramerization. Moreover, increasing the input ratio of scFv:PKM2 gradually altered PKM2’s electrophoretic mobility, apparently reflecting the binding stoichiometry. This altered mobility in native gels might indicate either an increased mass or an altered tertiary conformation of the PKM2 tetramers.
Fig 4. Recombinant scFv 5 induced tetramerization of recombinant PKM2 and stimulated its glycolytic activity in vitro.
A. Blue native gel electrophoresis. This method resolved the dimer and tetramer forms of PKM2. Left: A monovalent form of scFv 5, produced in E. coli, induced tetramer formation in WT PKM2 along with a bandshift whose magnitude was dependent on the input amount of scFv 5. The mutant PKM2 (K422R) was constitutively tetrameric and did not exhibit a bandshift in the presence of scFv 5. Reaction volume was 20 μl. Right: scFv 5 did not produce a band shift with recombinant PKM1, which ran as a tetramer; added FBP produced the tetramer form of WT PKM2 (lanes 6–8). B. scFv 5 stimulated glycolytic activity of WT and mutant PKM2. Activity was measured by Kinase-Glo Plus Luminescent Kinase Assay kit (Promega), using ADP and PEP as substrates, with PKM2 at 50 nM. Shown are values with the basal activity of PKM2 alone subtracted out. Inset: scFv 5 did not affect PKM1’s glycolytic activity. C. IB5 stimulated the glycolytic activity of K422R mutant PKM2 to a greater extent in the presence of allosteric activators FBP or TEPP-46. Note: underlying data are included in corresponding tabs in the accompanying supplemental Excel file S1 Data. FBP, fructose-1,6-bisphosphate; IB5, intrabody 5; PEP, phosphoenolpyruvate; PKM2, pyruvate kinase isoform M2; scFv, single-chain variable fragment; WT, wild-type.
We then found that purified scFv 5 stimulated PKM2’s glycolytic activity in a concentration-dependent manner (Fig 4B), confirming indirectly that the scFv interacts with PKM2. The activity declined at higher concentrations of scFv 5. This might result from PKM2 aggregation. These data suggest that scFv 5 activates PKM2 allosterically. (Note also that scFv 5 altered neither the electrophoretic mobility nor the PK activity of PKM1, consistent with IB5’s specificity for PKM2.)
Based on these results, we considered the possibility that the anti-apoptotic effect of IB5 purely reflected an increased glycolytic activity of PKM2. However, we found that treating cells with the PKM2-activating compound TEPP-46 (Fig 2A and 2C) [47] alone did not protect cells from BimS-induced death. Similarly, reconstituting PKM2-null MEFs with the constitutively active PKM1 failed to rescue cells (Fig 2A). Thus, high PK activity by itself was insufficient to produce an anti-apoptotic effect. On the other hand, culturing 293T cells in the presence of TEPP-46 enhanced the prosurvival effect of IB5 expression to a modest but statistically significant extent (Fig 2A and 2C). This could mean that increased glycolytic activity does contribute somewhat to cell survival. Alternatively, the effect of IB5, alone or in combination with TEPP-46, might result from stabilizing a tetrameric conformation of PKM2, whose glycolytic activity may be irrelevant to the survival function.
Studies with PKM2 mutants
To help define the aspects of PKM2 function required for the IB5-induced anti-apoptotic effect, we reconstituted PKM2-null MEFs with WT or mutant forms of PKM2. As expected, WT PKM2 increased cell viability when coexpressed with IB5 (Fig 2A). We then analyzed the K270M mutation, reported to be catalytically dead [52,83,84]. This mutant indeed lacked basal glycolytic activity in vitro, but the addition of high concentrations of scFv 5 stimulated its PK activity somewhat (Fig 4B). PKM2 (K270M) failed to support the cytoprotective effect of IB5 (Fig 3A). If the K270M mutation merely inactivated the catalytic site, this would suggest that PKM2’s glycolytic activity is required for the cell survival effect. However, using blue native gel electrophoresis, we found that the K270M mutation also prevented the protein from forming stable tetramers in vitro, when incubated with scFv 5 and/or FBP (S5A Fig). These results have at least three possible explanations (which are not mutually exclusive): 1) PKM2’s glycolytic activity is required for the anti-apoptotic function, 2) a specific tetrameric conformation of PKM2 that produces a nonglycolytic activity is required, or 3) IB5 binding is reduced by the K270M mutation. We note that IB5 seems to have some affinity for PKM2 (K270M) because higher concentrations of scFv 5 stimulated this mutant’s glycolytic activity (Fig 4B).
We analyzed another mutant, PKM2 (K367M), which also was reported to be inactive for glycolysis [56]. We confirmed that recombinant PKM2 (K367M) indeed had little PK activity, even when incubated with scFv 5 or FBP (S5B Fig). Furthermore, like K270M, K367M lacked anti-apoptotic activity when coexpressed with IB5 (S5C Fig). However, blue native gel electrophoresis showed that recombinant K367M mutant protein did not form stable tetramers when mixed with scFv 5 (S5A Fig). Furthermore, when both scFv 5 and FBP were added, this mutant primarily formed an aberrantly migrating species, possibly a malformed tetramer. (We did also observe a minor species migrating with authentic tetramers.) Thus, because the K270M and K367M mutations impaired both PKM2’s glycolytic activity and tetramerization in vitro, the data did not resolve whether PKM2’s glycolytic activity is required for the IB5-induced cytoprotective effect.
In any case, taking together our observations that treating cells with the PKM2-activating compound TEPP-46 or replacing PKM2 with the constitutively active PKM1 failed to rescue cells from BimS-induced death, we conclude that high PK activity alone is insufficient for cell rescue by IB5. In addition, we found that the addition of 2-deoxyglucose (20 mM) to the cultures did not prevent cell rescue by IB5 (S6 Fig), suggesting that glycolysis in general was unnecessary for the cytoprotective effect of IB5.
We next considered the possibility that IB5-induced PKM2 tetramerization per se is important for the cell survival activity. To test this, we reconstituted PKM2-deficient MEFs with the stably tetrameric PKM2 (K422R) mutant. As we confirmed in Fig 4C, this mutant is glycolytically inactive, unless an allosteric activator such as FBP is added, causing a quaternary conformational change from T-state to R-state [45]. Blue native gel electrophoresis confirmed the spontaneous formation of K422R tetramers in the absence of FBP (Fig 4A). It is unclear why IB5 did not shift up the PKM2 (K422R) tetramer band, whereas it did shift up the WT tetramer. We speculate that the tertiary structure of this mutant tetramer is more rigid than that of WT PKM2. Nevertheless, it does appear that IB5 interacts with PKM2 (K422R), as the recombinant scFv stimulated this mutant’s glycolytic activity (Fig 4B), especially upon the addition of FBP or TEPP-46 (Fig 4C). It should be noted that within cells, the allosteric activator FBP is likely to be present, at concentrations dependent on the metabolic state.
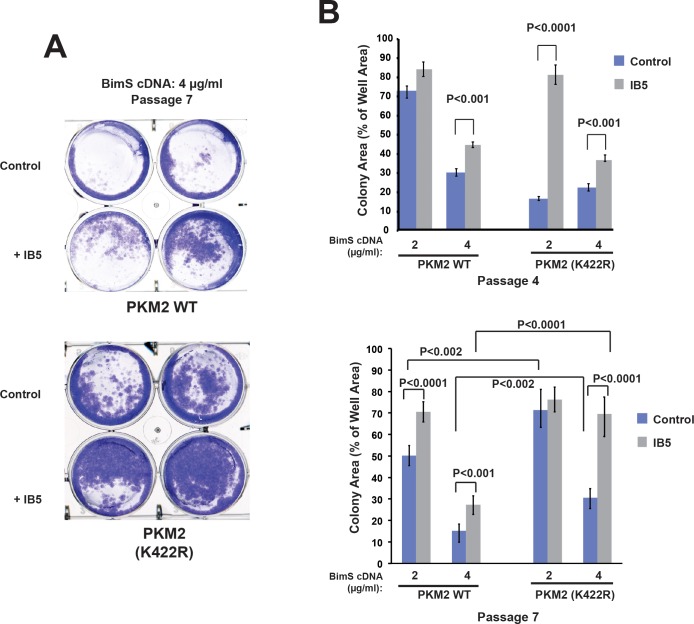
In MEFs reconstituted with PKM2 variants for a short time (3–4 passages), the K422R mutant slightly increased the numbers of viable cells compared with WT PKM2, and IB5 expression further increased viability (Fig 5). At this early time, IB5 expression enhanced clonogenic survival with PKM2 (K422R) in a manner similar to WT PKM2. (For unknown reasons, survival in the absence of IB5 was somewhat reduced with this mutant, compared with WT PKM2.) However, in MEFs that had expressed the K422R mutant for a longer time (passage 7), clonogenic survival was increased even in the absence of IB5 and was further enhanced by IB5 expression (Fig 5). This argues that stably tetrameric PKM2 promoted a cell survival function that developed over time (see below).
Fig 5. The stably tetrameric mutant PKM2 (K422R) produced cell rescue in response to expression of IB5; in long-term culture, this mutant also enhanced BimS-resistant clonogenic survival even in the absence of IB5.
PKM2-deficient MEFs reconstituted with WT or mutant PKM2 cDNA were infected or not with IB5, then 1 x 105 cells were plated and transfected with BimS expression plasmid. The plates were fixed and stained with crystal violet after 1 week, and the total area of colonies were counted as above. A. Example of stained colonies at passage 7. B. Quantification of clonogenic survival for passages 4 and 7. At both early and later passages following IB5 or control infection, the K422R mutant supported the cytoprotective effect of IB5; at later passage, this mutant protected cells to a substantial degree even in the absence of IB5 expression. SD and P values were calculated from six individual plates. Note: underlying data are included in corresponding tabs in the accompanying supplemental Excel file S1 Data. IB5, intrabody 5; MEF, Mouse Embryonic Fibroblast; PKM2, pyruvate kinase isoform M2; WT, wild-type.
The nuclear form of PKM2 is thought to be dimeric, whereas tetramers are restricted to the cytoplasm [54]. If so, our data imply that the anti-apoptotic effect of IB5 involves cytoplasmic PKM2 molecules and does not require the transcriptional activities ascribed to dimeric PKM2 in the nucleus. Consistent with this, we found that the K270M mutant, which formed dimers but not stable tetramers, as seen in blue native gels, even in the presence of FBP (S5A Fig) but is reportedly competent in nuclear transactivational activity [52], failed to support IB5-induced cell rescue (Fig 2A).
A mitochondrial role in IB5-induced apoptosis resistance
Our results showed that IB5 and PKM2 inhibited apoptosis triggered by transient expression of BimS or tBid, which directly activate the “intrinsic” pathway involving Bax/Bak-dependent permeabilization of mitochondrial outer membranes. This raised the possibility that PKM2 can directly inhibit the activity or function of Bax/Bak at mitochondria. However, we observed no effect of adding recombinant scFv 5 and PKM2 to our well validated in vitro systems based on isolated mitochondria or liposomes mixed with Bax and cleaved Bid, which recapitulate the basic aspects of Bcl-2 family protein function in membranes [6,24,25,85]. Thus, we saw no evidence that PKM2 acts directly on the process of Bax/Bak-mediated MOMP.
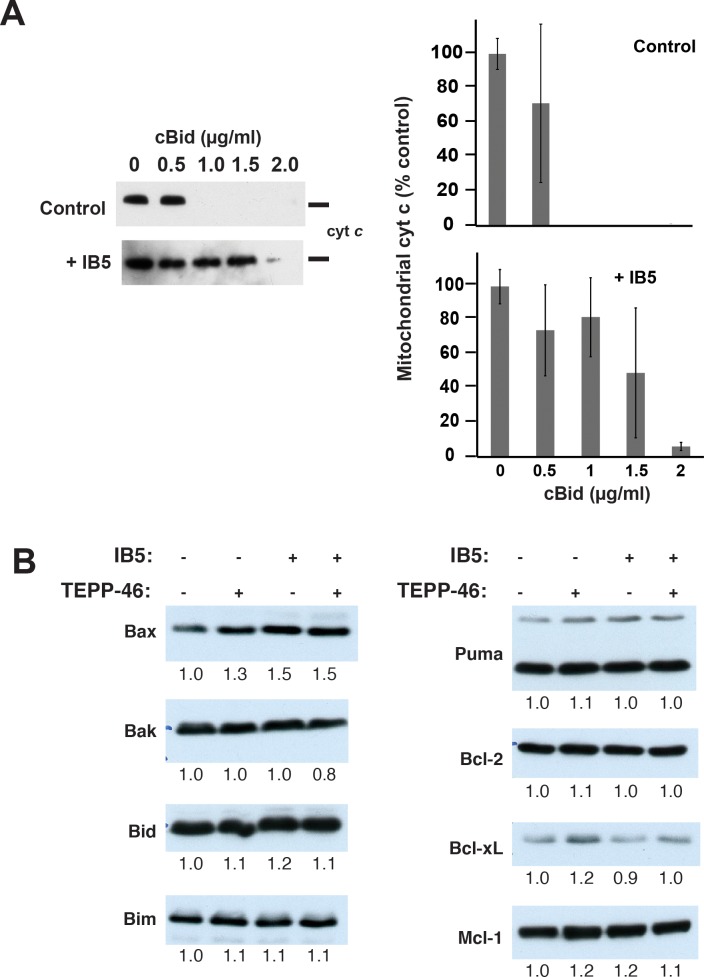
On the other hand, mitochondria isolated from 293T cells expressing IB5 were reproducibly more resistant than control mitochondria to MOMP induced by treatment with cleaved Bid protein (Fig 6A). This suggests that PKM2 could produce mitochondrial changes that could explain the cellular rescue we observed (Fig 2). As Bcl-2 family proteins are the most prominent regulators of apoptotic death at mitochondria, we first considered whether altered levels of these proteins could be responsible for MOMP resistance. In this regard, one study reported that, under conditions of oxidative stress, PKM2 can interact with and stabilize the Bcl-2 protein [86]. However, we found that IB5 expression and TEPP-46 treatment (alone or in combination) failed to change the cellular levels of Bcl-2 and other major family members Bax, Bak, Bid, Bim, Puma, Bcl-xL, and Mcl-1 (Fig 6B).
Fig 6. Mitochondria from cells expressing IB5 were relatively resistant to cBid-induced MOMP.
A. Cyt c release assay. Left panel: control (top) or intrabody-expressing (bottom) 293T cells were collected, and the mitochondrial fraction was isolated by differential centrifugation. To induce MOMP, recombinant cBid protein was added at the indicated concentrations. After incubation for 30 min at 37 ºC, samples were centrifuged, and cyt c content in mitochondrial pellet fractions was analyzed by immunoblot. A representative of three independent experiments is shown. Right panel: densitometric quantification of average cyt c content ± SEM from three independent experiments. B. Levels of several Bcl-2 family proteins were unchanged following IB5 expression or incubation with TEPP-46 or both. Cell lysates from 293T cells infected with and without IB5 and incubated with and without TEPP-46 (27 μM) were separated on SDS-12% polyacrylamide gels. Bcl-2 family proteins were detected by immunoblotting. The bands were quantified using ImageJ and normalized to the control cell lysate on the leftmost lane. Note: underlying data are included in corresponding tabs in the accompanying supplemental Excel file, S1 Data. 293T, HEK293T; cBid, cleaved Bid; cyt c, cytochrome c; IB5, intrabody 5; MOMP, mitochondrial outer membrane permeabilization.
We next used microscopy to analyze the effect of IB5 and PKM2 on mitochondrial morphology. We found that, in PKM2-null MEFs reconstituted with WT PKM2, IB5 expression increased the average mitochondrial length (in cells examined at passage 3–4 after transduction with IB5; Fig 7A and 7B). Furthermore, MEFs reconstituted with PKM2 (K422R) displayed a similar mitochondrial lengthening, even without IB5 expression. These results raised the possibility that PKM2-dependent mitochondrial lengthening and apoptosis resistance could involve alterations of proteins that regulate mitochondrial dynamics. In this regard, a recent study reported that PKM2 overexpression promoted mitochondrial fusion by binding to p53 and MDM2, promoting p53 ubiquitination and degradation, and thereby inhibiting expression of Drp1, a protein required for mitochondrial fission [87]. However, Fig 7C shows that the cytoprotective effect produced by IB5 was not accompanied by changes in the levels of Drp1 or p53, in MEFs reconstituted with PKM2 WT or PKM2 (K422R).
Fig 7. Mfn1 was required for the cytoprotective effect of IB5 and PKM2.
The effect of IB5 and PKM2 on mitochondrial morphology. PKM2-deficient MEFs reconstituted with WT PKM2 or PKM2 (K422R) cDNA were infected or not with IB5 lentivirus. A. IB5 expression with WT PKM2 increased mitochondrial length, and PKM2 (K422R) expression increased mitochondrial length even in the absence of IB5. Mitochondria were visualized by fluorescence microscopy after staining with Tom20 antibodies. Representative confocal images are shown. B. Mitochondrial length scores. Cells were analyzed 3 d after transfection with the indicated cDNA constructs (mean ± SEM of 3–5 experiments of 120–200 random selected cells). C. Immunoblots showed that IB5 expression up-regulated Mfn1 protein in MEFs reconstituted with PKM2 WT and K422R mutant. IB5 expression produced no changes in the levels of Mfn2, Drp-1, or p53. K422R expression alone produced some increase in the Mfn1 level. D. IB5 expression rescued WT and Mfn2-null MEFs from BimS-induced death but failed to rescue Mfn1-null MEFs. WT, Mfn1-, or Mfn2-deficient MEFs were infected or not with IB5 lentivirus, then 1 x 105 cells were plated and transfected with BimS expression plasmid. The plates were fixed and stained with crystal violet after 1 week and the total areas of colonies were measured as above. Mean, SD, and P values were calculated from five individual plates. Note: underlying data are included in corresponding tabs in the accompanying supplemental Excel file S1 Data. IB5, intrabody 5; MEF, Mouse Embryonic Fibroblast; Mfn, Mitofusin; PKM2, pyruvate kinase isoform M2; WT, wild-type.
We did find that IB5 expression substantially increased the levels of Mfn1, a protein involved in mitochondrial fusion (Fig 7C), but left Mfn2 levels unchanged. Importantly, reconstituting MEFs with PKM2 (K422R) alone increased Mfn1 levels, and IB5 expression elevated Mfn1 even further (Fig 7C). To determine whether Mfn1 up-regulation occurred at the transcriptional level, we used qPCR to measure MFN1 mRNA (S6B Fig). The results show that IB5 expression did not elevate MFN1 mRNA levels but rather tended to decrease them, and IB5 + PKM2 (K422R) produced the greatest decrease. We conclude that Mfn1 protein up-regulation is post-transcriptional and can even outweigh a concomitant decrease in mRNA levels. The reason for decreased MFN1 mRNA levels is unknown, but we hypothesize that it may involve either a loss of PKM2’s nuclear functions or a gain of cytoplasmic functions, as PKM2 (K422R) is mostly tetrameric and thus expected to be excluded from the nucleus.
To determine whether Mfn1 is required for the cytoprotective effect of IB5 and PKM2, we measured BimS-resistant clonogenic survival in WT and Mfn1- or Mfn2-deficient MEFs. The Mfn1 and Mfn2 deletions were confirmed by western blot (S7 Fig). Deletion of Mfn1 or Mfn2 did not affect PKM2 expression (S7 Fig). Importantly, IB5 failed to rescue Mfn1-deficient MEFs from BimS-induced clonogenic death (Fig 7D). Even when IB5 was not expressed, Mfn1-null MEFs showed greater sensitivity to BimS-induced apoptosis than WT MEFs (Fig 7D). In contrast, Mfn2-null MEFs responded similarly to WT. These results appear consistent with a previous report that Mfn1 directly inhibits mitochondria-mediated apoptosis at the step of Bax activation, downstream of Bax mitochondrial translocation [88]. This effect did not trivially arise from an unhealthiness of Mfn1-null MEFs, as they grew at approximately the same rate as WT cells. (Note: we did not show a well untreated with BimS in Fig 7D because, in the absence of BimS transfection, both the WT and mutant cells overgrew the cultures, causing many of the cells to die and detach from the substrate.)
As mentioned above, we found that MEFs expressing PKM2 (K422R) developed a significant resistance to BimS-induced death after extended culture (passage 7), even in the absence of IB5 (Fig 5B). We hypothesize that Mfn1 up-regulation gradually increases cellular resistance to apoptosis to some extent by enhancing mitochondrial fusion. Increased fusion could be expected to gradually improve the overall health of the mitochondrial network and may, for example, limit the production of reactive oxygen species. We previously observed a similarly delayed but detrimental effect on mitochondrial function in MEFs haploinsufficient for the optic atrophy 1 (Opa1) protein [89]. In that case, inefficient mitochondrial fusion caused the late-passage cells to accumulate dysfunctional mitochondria that were deficient in Complex IV subunits.
Even in cells expressing PKM2 (K422R), which had up-regulated Mfn1 to some degree, IB5 expression further elevated Mfn1 levels and enhanced cell survival (Fig 7C and Fig 5B). Thus, the extent of survival was correlated with Mfn1 levels. Moreover, our results suggest that Mfn1 promotes survival by two different mechanisms, which could be differentially engaged depending on Mfn1 levels: 1) a slowly developing process involving enhanced mitochondrial fusion, leading to healthier mitochondria and 2) an event in which Mfn1 immediately acts to inhibit Bax/Bak-mediated MOMP [88].
How IB5 cooperates with PKM2 to up-regulate Mfn1 is unknown. One possibility is that IB5, by driving PKM2 molecules into the tetramer form, could reduce the amount of dimeric nuclear PKM2 and thereby abrogate transcriptional functions of PKM2 that could down-regulate Mfn1. Alternatively, PKM2 tetramers could act in the cytoplasm to regulate the postsynthetic degradation of Mfn1. Mfn1 turnover is reported to be controlled by E3 ligases such as MARCH5, thereby regulating apoptosis [90]. However, to our knowledge, specific proteasomal degradation of Mfn1 but not Mfn2 has not been reported.
In summary, our data show that high PK activity by itself was insufficient to produce an anti-apoptotic effect, as expression of the constitutively glycolytic PKM1 or treatment of WT cells with the PKM2-stimulator TEPP-46 did not rescue cells from BimS-induced death. This argues that the anti-apoptotic effect induced by IB5 involves a nonglycolytic activity of PKM2. On the other hand, TEPP-46 significantly enhanced the cytoprotective activity of IB5, and a stably tetrameric mutant of PKM2, K422R, enhanced the effects of IB5 and TEPP-46. Taken together, these results argue that IB5’s anti-apoptotic activity involves a tetrameric conformation of PKM2.
In the absence of IB5, cells expressing PKM2 (K422R) for multiple passages displayed a degree of apoptosis resistance, and IB5 expression further enhanced this resistance. Such a cytoprotective effect of K422R may help explain why this mutation promoted oncogenesis in mice and occurred spontaneously in Bloom syndrome patient cells [91]. Bloom syndrome involves a mutation-prone mechanism and can therefore be considered an in vivo phenotypic selection process, in effect similar to our intrabody selection approach.
Finally, the anti-apoptotic activity induced by IB5 was not accompanied by changes in the levels of major Bcl-2 family proteins. In contrast, IB5 did up-regulate Mfn1, and apoptosis resistance was ablated by Mfn1 deletion. This is consistent with reports that Mfn1 protein can oppose Bax-dependent MOMP [88]. Our observation that mitochondria isolated from IB5-expressing cells were more resistant to Bax-mediated apoptosis (Fig 6) may reflect increased levels of Mfn1 in mitochondria.
Potential implications
PKM2-deficient cells can form tumors in mice. Often the rapidly proliferating subset of tumor cells remodel glucose utilization by expressing low PKM1 levels, whereas nonproliferating tumor cells are more likely to express higher levels of PKM1 [92]. These observations reinforce the idea that reduced PK activity, and not necessarily PKM2 expression per se, is important for rapid cell proliferation. However, they also pose a question: if PKM2 is not strictly required for tumor formation, why is PKM2 expression overwhelmingly favored in human cancers? Although some human cancers harbor PKM2 loss-of-function mutations, these mutations are typically heterozygous. Thus, cancer cells presumably benefit from retaining at least one WT allele of the M2 isoform, which, unlike M1, provides adaptive glycolytic regulation and nonglycolytic functions [38].
Our results suggest another potential benefit for cells expressing PKM2: resistance to apoptosis. Because PKM2 inhibits the central mechanism of apoptosis involving mitochondria, PKM2 could promote cell survival despite circumstances that would otherwise be cytotoxic. We can conjecture that particular subsets of neoplastic or preneoplastic cells could engage this mechanism to survive under adverse conditions, favoring oncogenesis. Because glycolytically active PKM2 typically corresponds with lower rates of proliferation, we could hypothesize that this cell survival function of active PKM2 tetramers might be seen primarily in slowly proliferating tumor cell subsets.
IB5 most likely promotes cell survival by altering the interaction of PKM2 with one or more protein partners. It is tempting to hypothesize that IB5 mimics a natural PKM2-interacting protein. However, the identity of such a putative ligand is still unknown, as are the circumstances under which it is potentially engaged. Perhaps this cell survival function of PKM2 occurs only under specific conditions (e.g., allosteric activation of PKM2 combined with another regulatory event), which may explain why it has not been identified through conventional approaches. An anti-apoptotic function of PKM2 could be important both in cancer cells and in normal cell populations that preferentially express PKM2, such as macrophages [41,93–96] and podocytes in the kidney [97,98].
Materials and methods
Cell culture and plasmids
Primary and immortalized WT and PKM2-deficient MEFs (PKM2Δ/Δ) were maintained in MEMα medium supplemented with 10% FBS, penicillin and streptomycin (Gibco-Invitrogen), and 0.1 mM of 2-mercaptoethanol [49]. 293T cells were maintained in DMEM containing 10% (vol/vol) FBS and antibiotics. pLHCX-Flag-mPKM2 (Plasmid #42512) was obtained from Addgene.
Intrabody library construction
The intrabody scFv library was prepared using a naïve human combinatorial scFv phage library [76]. The scFv phagemid library was digested with SfiI, and the approximately 800-bp insert scFv-coding sequence was ligated into the SfiI-digested lentiviral vector, driven by an EF1α promoter (without a secretion leader sequence), followed by a FLAG tag.
Lentiviral infection with the scFv library
Lentiviral particles were produced in 5 x 107 293T cells using pCMVD8.9 and pVSVg viral packaging vectors at a ratio of 1:1:1. For the first round of selection, culture supernatants containing lentiviral particles were collected, filtered, and used for infection of 1 × 107 293T cells per 10-mm plate. For the recloning step after rounds 2 and 3, 5 x 106 cells were used. Forty-eight h post infection, the culture medium was replaced with fresh MEMα medium supplemented with 10% FBS and penicillin/streptomycin (Gibco-Invitrogen). Immunofluorescence microscopy using anti-FLAG antibody showed that over 90% of cells expressed IB5.
Selection of intrabodies conferring BimS resistance in 293T cells
Human BimS cDNA was subcloned into pShooter mammalian expression vector (pCMV/myc/cyto; Invitrogen) to allow the expression of BimS driven by a CMV promoter. Approximately 5 x 107 293T cells were then infected with the intrabody library and then transfected with 4 μg/ml BimS plasmid using 10 μl of Lipofectamine 2000 transfection reagent (Thermo Fisher). After 24 h post infection, the culture medium was replaced with fresh MEMα medium supplemented with 10% FBS and penicillin/streptomycin (Gibco-Invitrogen).
Recovery of selected scFv from the genomic DNA by PCR and construction of intrabody libraries for the second and third rounds of selection
The integrated intrabody-coding sequences from the surviving cells were recovered after 48 h incubation and used to construct a secondary lentiviral library, as follows. Genomic DNA from the surviving 293T cells was recovered using a DNeasy Blood & Tissue kit (Qiagen). A sample of 100 ng of the genomic DNA was used as a PCR template. A pair of primers matching the regions flanking the scFv fragment was used to amplify the integrated antibody fragment from the genomic DNA. The PCR product was digested with SfiI and inserted back into the lentiviral vector for a subsequent round of BimS selection, as described above. In total, over 300 clones with distinct DNA sequences were harvested and tested individually for the ability to confer BimS resistance. Sequences were analyzed with Vbase2.
Expression of scFv in E. coli
scFv-coding sequences subcloned into pET28a plasmid were introduced into Rosetta (DE3)pLys cells (Novagen). Single colonies were picked and grown in 2 l of LB medium containing 50 μg/ml of kanamycin at 30°C for 8 h, then incubated for 12 h at 4°C with 0.2 mM IPTG under vigorous shaking. Cells were pelleted by centrifugation, frozen/thawed, resuspended in 50 ml of lysis buffer (Tris 25 mM pH 8.0, NaCl 300 mM), incubated 1 h on ice, and then lysed by sonication. The scFv was recovered from the soluble fraction by passage over a Ni++-NTA affinity column (GE Healthcare).
Target protein immunoprecipitation
FLAG-tagged intrabody was introduced along with a tandem Strep-tag by PCR into the same lentiviral vector used for selection. 293T cells infected with the intrabody lentivirus were incubated at 30°C for 72 h, as described above. After washing with cold PBS, 5 × 108 cells were lysed for 15 minutes on ice in lysis buffer (50 mM Tris HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100). Cell lysates were clarified by centrifugation for 15 min at 4°C at 16,000 x g. The total protein content of the soluble fraction was quantified using the BCA assay. For pull-down experiments, 10 mg of protein lysate was incubated with 200 μl of EZview Red anti-FLAG M2 Affinity Gel (Sigma-Aldrich) for 2 h at 4°C. Beads were washed three times in wash buffer (50 mM Tris HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% Triton X-100). Elution was performed under native conditions by competition with 3X FLAG peptide following the manufacturer’s protocol. Eluates were used for the second-step purification using Strep-Tactin Superflow Plus (Qiagen), following the manufacturer’s instructions. The final two-step purified protein was used for SDS-PAGE analysis. Bands of interest were cut out from the gel and subjected to in-gel digestion with trypsin (PR omega, Fitchburg, WI, USA), followed by MALDI TOF/TOF mass spectrometry analysis (Biomolecular and Proteomics Mass Spectrometry Facility, University of California at San Diego).
Cell viability and clonogenic survival assays
Cell viability was measured with a Countess Automated Cell Counter (Invitrogen) using trypan blue. For the clonogenic survival assay, 293T or MEFs cells were seeded in 6-well plates at 1 x 105 cells/well in a 2-ml volume, transfected with BimS cDNA, and incubated for 24 h. Afterwards, the medium was replaced and the cells cultured for 3–4 d. Thereafter, the plates were rinsed with PBS and fixed and stained with a solution containing crystal violet (0.5% w/v) and glutaraldehyde (6% v/v), as described [99]. The results were quantified using ImageJ software (either total cell area or number of colonies, as indicated in the figure legends).
PKM2 protein expression and purification
pET28a-His-hPKM2 plasmid was obtained from Addgene. The PKM2 mutants pET28a-His-hPKM2 (C358S) and pET28a-His-hPKM2 (K270M) were generated by Quick-Change mutagenesis (Stratagene). All plasmids were verified by DNA sequencing and transformed into E. coli strain BL21 (DE3). WT and mutant PKM2 proteins were overexpressed in LB medium at 30°C with 200 mM IPTG for 3 h. Cells were harvested and lysed in buffer containing 25 mM Tris (pH 8.0), 300mM NaCl. The supernatants were loaded on a Ni++-NTA affinity column (GE Healthcare) for protein purification.
PK assay
PK activity was measured by using Kinase-Glo Plus Luminescent Kinase Assay kit (Promega Corporation, Madison, WI, USA). Purified WT or mutant PKM2 (50–100 nM) was added in 100 μl assay buffer containing 50 mM Tris pH 7.5, 100 mM KCl, 10 mM MgCl2, 200 μM PEP, 200 μM ADP, and 3% DMSO. After a 15-min incubation, Kinase-Glo Plus reagent was added, according to the manufacturer’s instructions. In some cases, 0–40 μM FBP and 0–150 nM scFv 5 were added.
Mitochondria isolation and cytochrome c release assay
Mitochondria were isolated from 5 x 108 cells, as described [100]. The freshly isolated mitochondria (100 mg protein/ml) were then incubated with recombinant cleaved Bid protein [6] at the indicated concentrations in the presence or absence of purified scFv 5. After incubation for 30 min at 37 °C, mitochondria were collected by centrifugation at 10,000 × g and analyzed by immunoblotting, as described [100].
Immunoblots of Bcl-2 family proteins
293T cells were washed with PBS and lysed with PBS containing 1% NP-40. Protein concentration was determined using Pierce BCA reagent (ThermoFisher; 23221, 23224). 35 μg protein was loaded in each lane of 12% SDS-PAGE gels. Proteins were transferred to nitrocellulose membrane and immunoblotted with the following primary antibodies: anti-Bax antibody (Santa Cruz N20), anti-Bak antibody (Cell Signaling 3814), anti-Bid antibody (R&D Systems AF860), anti-Bim antibody (Sigma B7929), anti-Puma antibody (Cell Signaling 4976), anti-Bcl-2 antibody (Abcam 32124), anti-Bcl-xL antibody (Cell Signaling 2764), and anti-Mcl-1 (Santa Cruz S19) at 1:1,000 dilution. The secondary anti-rabbit and mouse antibodies, conjugated with HRP, were obtained from Santa Cruz and were used at 1:2,000 dilution. The luminescence signal was detected using ECL reagent (ThermoFisher 32106).
siRNA silencing of PKM2
Untreated or IB5-infected 293T cells (5 × 105 per well) were seeded into 6-well plates (Falcon) and transfected with 30 nM siRNA. Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was used for transient transfection, according to the manufacturer’s protocol. After a 30-h incubation, fresh medium containing 30 nM siRNA and 4 μg/ml BimS expression plasmid was added. Cell viability assay was assayed after a further 36-h incubation. The PKM2-siRNA and control siRNA were purchased from Dharmacon (SiGENOME SMART pool hPKM2, Si156, and ON-TARGET plus nontargeting siRNA 2) [101].
Quantification of Mfn1 mRNA expression by qPCR
Isolation of total RNA was performed using the RNeasy mini-kit (Qiagen, CA). After removal of contaminating DNA using DNase I (Invitrogen, CA), cDNA was synthesized using SuperScript III first-strand synthesis system for qPCR (Invitrogen). MFN1 mRNA levels were normalized against mouse GAPDH mRNA.
Supporting information
Approximately 5 x 105 cells were incubated per well for 12 h, then cells were either mock-transfected, transfected with 30 nM PKM2-specific siRNA (si M2), or as control, transfected with NF-κB p50-specific siRNA (si p50). After a further 36-h incubation, samples of the same siRNAs were added along with 4 μg of BimS cDNA in fresh medium. Viable cells were counted after another 48-h incubation. Note: underlying data are included in corresponding tabs in the accompanying supplemental Excel file S1 Data. 293T, HEK293T; IB5, intrabody 5; NF-κB, nuclear factor κB; PKM2, pyruvate kinase isoform M2; siRNA, small interfering RNA.
(TIF)
Approximately 5 x 105 HCT116 and U2OS cells were plated and transfected with BimS expression plasmid including 150 nM etoposide or 1 μM Staurosporine. The plates were fixed and stained with crystal violet after 5 d and the total areas of colonies were measured. Mean, SD, and P values were calculated from three individual plates. Note: underlying data are included in corresponding tabs in the accompanying supplemental Excel file S1 Data. IB5, intrabody 5
(TIF)
Control or IB5-expressing cells were transfected with BimS cDNA. The plates were fixed and stained with crystal violet after 12 days and the total areas of colonies were measured. Mean, SD, and P values were calculated from three individual plates. Note: underlying data are included in corresponding tabs in the accompanying supplemental Excel file S1 Data. BimS, short isoform of BimS; IB5, intrabody 5
(TIF)
293T cells were infected (lane 2, 3) or not (lane 1) with IB5 lentivirus and incubated with (lane 3) or without 2 μg of BimS cDNA (lane1, 2) in fresh medium. Cells were lysed, and total cell protein extracts were subjected to western blot analysis. BimEL (upper band), BimL (middle band) and BimS (lower band) were detected using Anti-Bim antibody (ab15184). GAPDH was used as loading control. 293T, HEK293T; GAPDH, glyceraldehyde phosphate dehydrogenase; IB5, intrabody 5; PKM2, pyruvate kinase isoform M2
(TIF)
A. PKM2-deficient MEFs reconstituted with WT or mutant PKM2 cDNA were infected or not with IB5, then 2 x 104 cells were plated and transfected with BimS expression plasmid. The plates were fixed and stained with crystal violet after 1 week and the total area of colonies were counted as above. Means, SDs, and P values were calculated from three experiments. B. Blue native gel electrophoresis of PKM2 WT and mutations. C. scFv 5 stimulated glycolytic activity of WT PKM2 and PKM2 (K367M). Activity was measured as in Fig 4. Note: underlying data are included in corresponding tabs in the accompanying supplemental Excel file S1 Data. IB5, intrabody 5; MEF, Mouse Embryonic Fibroblast; PKM2, pyruvate kinase isoform M2; scFv, single-chain variable fragment; WT, wild-type
(TIF)
A. 2-deoxy-D-glucose had no effect on 293T cell survival induced by IB5 intrabody. 293T cells were infected or not with IB5, then 2 x 104 cells were plated and transfected with BimS expression plasmid. The glycolytic inhibitor 2-deoxy-D-glucose (20 mM) was added to the MEMαmedium, and after 24 h, cells were transfected or not with 1 μg of BimS cDNA in fresh medium. The plates were fixed and stained with crystal violet after 1 week. B. IB5 reduced MFN1 mRNA levels, implying that Mfn1 protein up-regulation is post-transcriptional. PKM2-deficient MEFs reconstituted with WT or mutant PKM2 cDNA were infected or not with IB5, and MFN1 mRNA levels were quantified by qPCR. Means, SDs, and P values based on four independent experiments are indicated. Note: underlying data are included in corresponding tabs in the accompanying supplemental Excel file S1 Data. 293T, HEK293T; IB5, intrabody 5; MEMα; PKM2, pyruvate kinase isoform M2; WT, wild-type
(TIF)
Lysates from the indicated MEF strains were analyzed by immunoblotting with antibodies directed against Mfn1, Mfn2, and PKM2, as indicated. Actin and GAPDH were used as loading controls. GAPDH, glyceraldehyde phosphate dehydrogenase; MEF, Mouse Embryonic Fibroblast; Mfn, Mitofusin; PKM2, pyruvate kinase isoform M2
(TIF)
(XLSX)
Acknowledgments
We thank Yulia Kushnareva for useful discussions and Angela Gao for valuable assistance. We are grateful to David Chan (Caltech) for the gift of Mfn1- and Mfn2-null MEFs.
Abbreviations
- 293T
HEK293T; BH3, Bcl-2 homology 3 domain
- EGFR
epidermal growth factor receptor
- FBP
fructose-1,6-bisphosphate
- IB5
intrabody 5
- LPS
lipopolysaccharide
- MEF
Mouse Embryonic Fibroblast
- MALDI-TOF
matrix-assisted laser desorption/ionization-time-of-flight
- Mfn1
Mitofusin-1
- MOM
mitochondrial outer membrane
- MOMP
mitochondrial outer membrane permeabilization
- PEP
phosphoenolpyruvate
- PKM1/M2
pyruvate kinase isoform M1/M2
- scFv
single-chain variable fragment
- siRNA
small interfering RNA
- T3
triiodo-L-thyronine
- VH
immunoglobulin heavy chain
- VL
immunoglobulin light chain
- WT
wild-type
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
HHMI (grant number). MGVH is an HHMI Faculty Scholar. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. NIH (grant number R01 GM62289 and R01 CA179087). Received by DDN. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Tuzlak S, Kaufmann T, Villunger A. Interrogating the relevance of mitochondrial apoptosis for vertebrate development and postnatal tissue homeostasis. Genes & development. 2016;30(19):2133–51. 10.1101/gad.289298.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown JM, Attardi LD. The role of apoptosis in cancer development and treatment response. Nature reviews Cancer. 2005;5(3):231–7. 10.1038/nrc1560 . [DOI] [PubMed] [Google Scholar]
- 3.Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35(4):495–516. 10.1080/01926230701320337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newmeyer DD, Farschon DM, Reed JC. Cell-free apoptosis in Xenopus egg extracts: inhibition by Bcl-2 and requirement for an organelle fraction enriched in mitochondria. Cell. 1994;79:353–64. [DOI] [PubMed] [Google Scholar]
- 5.Kluck RM, Bossy-Wetzel E, Green DR, Newmeyer DD. The release of cytochrome c from mitochondria: a primary site for Bcl-2 regulation of apoptosis. Science. 1997;275(5303):1132–6. Epub 1997/02/21. . [DOI] [PubMed] [Google Scholar]
- 6.Kuwana T, Mackey MR, Perkins G, Ellisman MH, Latterich M, Schneiter R, Green DR, Newmeyer DD. Bid, bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell. 2002;111(3):331–42. . [DOI] [PubMed] [Google Scholar]
- 7.Newmeyer DD, Ferguson-Miller S. Mitochondria: releasing power for life and unleashing the machineries of death. Cell. 2003;112(4):481–90. . [DOI] [PubMed] [Google Scholar]
- 8.Lopez J, Tait SW. Mitochondrial apoptosis: killing cancer using the enemy within. British journal of cancer. 2015;112(6):957–62. 10.1038/bjc.2015.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li MX, Dewson G. Mitochondria and apoptosis: emerging concepts. F1000Prime Rep. 2015;7:42 10.12703/P7-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gillies LA, Kuwana T. Apoptosis regulation at the mitochondrial outer membrane. Journal of cellular biochemistry. 2014;115(4):632–40. 10.1002/jcb.24709 . [DOI] [PubMed] [Google Scholar]
- 11.Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol. 2014;15(1):49–63. 10.1038/nrm3722 . [DOI] [PubMed] [Google Scholar]
- 12.Volkmann N, Marassi FM, Newmeyer DD, Hanein D. The rheostat in the membrane: BCL-2 family proteins and apoptosis. Cell death and differentiation. 2014;21(2):206–15. 10.1038/cdd.2013.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bender T, Martinou JC. Where killers meet—permeabilization of the outer mitochondrial membrane during apoptosis. Cold Spring Harbor perspectives in biology. 2013;5(1):a011106 10.1101/cshperspect.a011106 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green DR. A BH3 Mimetic for Killing Cancer Cells. Cell. 2016;165(7):1560 10.1016/j.cell.2016.05.080 . [DOI] [PubMed] [Google Scholar]
- 15.Croce CM, Reed JC. Finally, An Apoptosis-Targeting Therapeutic for Cancer. Cancer research. 2016;76(20):5914–20. 10.1158/0008-5472.CAN-16-1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarosiek KA, Letai A. Directly targeting the mitochondrial pathway of apoptosis for cancer therapy using BH3 mimetics—recent successes, current challenges and future promise. The FEBS journal. 2016;283(19):3523–33. 10.1111/febs.13714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leverson JD, Zhang H, Chen J, Tahir SK, Phillips DC, Xue J, Nimmer P, Jin S, Smith M, Xiao Y, Kovar P, Tanaka A, Bruncko M, Sheppard GS, Wang L, Gierke S, Kategaya L, Anderson DJ, Wong C, Eastham-Anderson J, Ludlam MJ, Sampath D, Fairbrother WJ, Wertz I, Rosenberg SH, Tse C, Elmore SW, Souers AJ. Potent and selective small-molecule MCL-1 inhibitors demonstrate on-target cancer cell killing activity as single agents and in combination with ABT-263 (navitoclax). Cell death & disease. 2015;6:e1590 10.1038/cddis.2014.561 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swiecicki PL, Bellile E, Sacco AG, Pearson AT, Taylor JM, Jackson TL, Chepeha DB, Spector ME, Shuman A, Malloy K, Moyer J, McKean E, McLean S, Sukari A, Wolf GT, Eisbruch A, Prince M, Bradford C, Carey TE, Wang S, Nor JE, Worden FP. A phase II trial of the BCL-2 homolog domain 3 mimetic AT-101 in combination with docetaxel for recurrent, locally advanced, or metastatic head and neck cancer. Investigational new drugs. 2016;34(4):481–9. 10.1007/s10637-016-0364-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell. 2010;37(3):299–310. 10.1016/j.molcel.2010.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walensky LD, Pitter K, Morash J, Oh KJ, Barbuto S, Fisher J, Smith E, Verdine GL, Korsmeyer SJ. A stapled BID BH3 helix directly binds and activates BAX. Mol Cell. 2006;24(2):199–210. 10.1016/j.molcel.2006.08.020 . [DOI] [PubMed] [Google Scholar]
- 21.Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR, Newmeyer DD. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Molecular Cell. 2005;17(4):525–35. Epub 2005/02/22. S1097276505010774 [pii] 10.1016/j.molcel.2005.02.003 . [DOI] [PubMed] [Google Scholar]
- 22.Cheng EH, Wei MC, Weiler S, Flavell RA, Mak TW, Lindsten T, Korsmeyer SJ. BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol Cell. 2001;8(3):705–11. . [DOI] [PubMed] [Google Scholar]
- 23.Du H, Wolf J, Schafer B, Moldoveanu T, Chipuk JE, Kuwana T. BH3 domains other than Bim and Bid can directly activate Bax/Bak. The Journal of biological chemistry. 2011;286(1):491–501. 10.1074/jbc.M110.167148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gillies LA, Du H, Peters B, Knudson CM, Newmeyer DD, Kuwana T. Visual and functional demonstration of growing Bax-induced pores in mitochondrial outer membranes. Mol Biol Cell. 2015;26(2):339–49. 10.1091/mbc.E13-11-0638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schafer B, Quispe J, Choudhary V, Chipuk JE, Ajero TG, Du H, Schneiter R, Kuwana T. Mitochondrial outer membrane proteins assist Bid in Bax-mediated lipidic pore formation. Mol Biol Cell. 2009;20(8):2276–85. 10.1091/mbc.E08-10-1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9(1):47–59. Epub 2007/12/22. nrm2308 [pii] 10.1038/nrm2308 . [DOI] [PubMed] [Google Scholar]
- 27.Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, Colman PM, Day CL, Adams JM, Huang DC. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17(3):393–403. 10.1016/j.molcel.2004.12.030 . [DOI] [PubMed] [Google Scholar]
- 28.Llambi F, Moldoveanu T, Tait SW, Bouchier-Hayes L, Temirov J, McCormick LL, Dillon CP, Green DR. A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol Cell. 2011;44(4):517–31. 10.1016/j.molcel.2011.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Del Gaizo Moore V, Letai A. BH3 profiling—measuring integrated function of the mitochondrial apoptotic pathway to predict cell fate decisions. Cancer letters. 2013;332(2):202–5. 10.1016/j.canlet.2011.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montero J, Sarosiek KA, DeAngelo JD, Maertens O, Ryan J, Ercan D, Piao H, Horowitz NS, Berkowitz RS, Matulonis U, Janne PA, Amrein PC, Cichowski K, Drapkin R, Letai A. Drug-induced death signaling strategy rapidly predicts cancer response to chemotherapy. Cell. 2015;160(5):977–89. 10.1016/j.cell.2015.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suryani S, Carol H, Chonghaile TN, Frismantas V, Sarmah C, High L, Bornhauser B, Cowley MJ, Szymanska B, Evans K, Boehm I, Tonna E, Jones L, Manesh DM, Kurmasheva RT, Billups C, Kaplan W, Letai A, Bourquin JP, Houghton PJ, Smith MA, Lock RB. Cell and molecular determinants of in vivo efficacy of the BH3 mimetic ABT-263 against pediatric acute lymphoblastic leukemia xenografts. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014;20(17):4520–31. 10.1158/1078-0432.CCR-14-0259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, Green DR. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303(5660):1010–4. 10.1126/science.1092734 . [DOI] [PubMed] [Google Scholar]
- 33.Hilgendorf KI, Leshchiner ES, Nedelcu S, Maynard MA, Calo E, Ianari A, Walensky LD, Lees JA. The retinoblastoma protein induces apoptosis directly at the mitochondria. Genes & development. 2013;27(9):1003–15. 10.1101/gad.211326.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Juin P, Geneste O, Gautier F, Depil S, Campone M. Decoding and unlocking the BCL-2 dependency of cancer cells. Nature reviews Cancer. 2013;13(7):455–65. 10.1038/nrc3538 . [DOI] [PubMed] [Google Scholar]
- 35.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452(7184):181–6. 10.1038/nature06667 . [DOI] [PubMed] [Google Scholar]
- 36.Wong N, Ojo D, Yan J, Tang D. PKM2 contributes to cancer metabolism. Cancer letters. 2015;356(2 Pt A):184–91. 10.1016/j.canlet.2014.01.031 . [DOI] [PubMed] [Google Scholar]
- 37.Li Z, Yang P, Li Z. The multifaceted regulation and functions of PKM2 in tumor progression. Biochimica et biophysica acta. 2014;1846(2):285–96. 10.1016/j.bbcan.2014.07.008 . [DOI] [PubMed] [Google Scholar]
- 38.Iqbal MA, Gupta V, Gopinath P, Mazurek S, Bamezai RN. Pyruvate kinase M2 and cancer: an updated assessment. FEBS letters. 2014;588(16):2685–92. 10.1016/j.febslet.2014.04.011 . [DOI] [PubMed] [Google Scholar]
- 39.Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer discovery. 2012;2(10):881–98. 10.1158/2159-8290.CD-12-0345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K, Dong S, Lonial S, Wang X, Chen GZ, Xie J, Gu TL, Polakiewicz RD, Roesel JL, Boggon TJ, Khuri FR, Gilliland DG, Cantley LC, Kaufman J, Chen J. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Science signaling. 2009;2(97):ra73 10.1126/scisignal.2000431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palsson-McDermott EM, Curtis AM, Goel G, Lauterbach MA, Sheedy FJ, Gleeson LE, van den Bosch MW, Quinn SR, Domingo-Fernandez R, Johnston DG, Jiang JK, Israelsen WJ, Keane J, Thomas C, Clish C, Vander Heiden M, Xavier RJ, O'Neill LA. Pyruvate kinase M2 regulates Hif-1alpha activity and IL-1beta induction and is a critical determinant of the warburg effect in LPS-activated macrophages. Cell metabolism. 2015;21(1):65–80. 10.1016/j.cmet.2014.12.005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morgan HP, O'Reilly FJ, Wear MA, O'Neill JR, Fothergill-Gilmore LA, Hupp T, Walkinshaw MD. M2 pyruvate kinase provides a mechanism for nutrient sensing and regulation of cell proliferation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(15):5881–6. 10.1073/pnas.1217157110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mazurek S. Pyruvate kinase type M2: a key regulator of the metabolic budget system in tumor cells. The international journal of biochemistry & cell biology. 2011;43(7):969–80. 10.1016/j.biocel.2010.02.005 . [DOI] [PubMed] [Google Scholar]
- 44.Gui DY, Lewis CA, Vander Heiden MG. Allosteric regulation of PKM2 allows cellular adaptation to different physiological states. Science signaling. 2013;6(263):pe7. 10.1126/scisignal.2003925 . [DOI] [PubMed] [Google Scholar]
- 45.Wang P, Sun C, Zhu T, Xu Y. Structural insight into mechanisms for dynamic regulation of PKM2. Protein & cell. 2015;6(4):275–87. 10.1007/s13238-015-0132-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452(7184):230–3. 10.1038/nature06734 . [DOI] [PubMed] [Google Scholar]
- 47.Anastasiou D, Yu Y, Israelsen WJ, Jiang JK, Boxer MB, Hong BS, Tempel W, Dimov S, Shen M, Jha A, Yang H, Mattaini KR, Metallo CM, Fiske BP, Courtney KD, Malstrom S, Khan TM, Kung C, Skoumbourdis AP, Veith H, Southall N, Walsh MJ, Brimacombe KR, Leister W, Lunt SY, Johnson ZR, Yen KE, Kunii K, Davidson SM, Christofk HR, Austin CP, Inglese J, Harris MH, Asara JM, Stephanopoulos G, Salituro FG, Jin S, Dang L, Auld DS, Park HW, Cantley LC, Thomas CJ, Vander Heiden MG. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nature chemical biology. 2012;8(10):839–47. 10.1038/nchembio.1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parnell KM, Foulks JM, Nix RN, Clifford A, Bullough J, Luo B, Senina A, Vollmer D, Liu J, McCarthy V, Xu Y, Saunders M, Liu XH, Pearce S, Wright K, O'Reilly M, McCullar MV, Ho KK, Kanner SB. Pharmacologic activation of PKM2 slows lung tumor xenograft growth. Molecular cancer therapeutics. 2013;12(8):1453–60. 10.1158/1535-7163.MCT-13-0026 . [DOI] [PubMed] [Google Scholar]
- 49.Lunt SY, Muralidhar V, Hosios AM, Israelsen WJ, Gui DY, Newhouse L, Ogrodzinski M, Hecht V, Xu K, Acevedo PN, Hollern DP, Bellinger G, Dayton TL, Christen S, Elia I, Dinh AT, Stephanopoulos G, Manalis SR, Yaffe MB, Andrechek ER, Fendt SM, Vander Heiden MG. Pyruvate kinase isoform expression alters nucleotide synthesis to impact cell proliferation. Mol Cell. 2015;57(1):95–107. 10.1016/j.molcel.2014.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu S, Le H. Dual roles of PKM2 in cancer metabolism. Acta biochimica et biophysica Sinica. 2013;45(1):27–35. 10.1093/abbs/gms106 . [DOI] [PubMed] [Google Scholar]
- 51.Palsson-McDermott EM, O'Neill LA. The Warburg effect then and now: from cancer to inflammatory diseases. BioEssays: news and reviews in molecular, cellular and developmental biology. 2013;35(11):965–73. 10.1002/bies.201300084 . [DOI] [PubMed] [Google Scholar]
- 52.Luo W, Hu H, Chang R, Zhong J, Knabel M, O'Meally R, Cole RN, Pandey A, Semenza GL. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145(5):732–44. 10.1016/j.cell.2011.03.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luo W, Semenza GL. Pyruvate kinase M2 regulates glucose metabolism by functioning as a coactivator for hypoxia-inducible factor 1 in cancer cells. Oncotarget. 2011;2(7):551–6. 10.18632/oncotarget.299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao X, Wang H, Yang JJ, Liu X, Liu ZR. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell. 2012;45(5):598–609. 10.1016/j.molcel.2012.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lv L, Xu YP, Zhao D, Li FL, Wang W, Sasaki N, Jiang Y, Zhou X, Li TT, Guan KL, Lei QY, Xiong Y. Mitogenic and oncogenic stimulation of K433 acetylation promotes PKM2 protein kinase activity and nuclear localization. Mol Cell. 2013;52(3):340–52. 10.1016/j.molcel.2013.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, Gao X, Aldape K, Lu Z. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480(7375):118–22. 10.1038/nature10598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F, Lyssiotis CA, Aldape K, Cantley LC, Lu Z. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nature cell biology. 2012;14(12):1295–304. 10.1038/ncb2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang HJ, Hsieh YJ, Cheng WC, Lin CP, Lin YS, Yang SF, Chen CC, Izumiya Y, Yu JS, Kung HJ, Wang WC. JMJD5 regulates PKM2 nuclear translocation and reprograms HIF-1alpha-mediated glucose metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(1):279–84. 10.1073/pnas.1311249111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D, Aldape K, Hunter T, Alfred Yung WK, Lu Z. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012;150(4):685–96. 10.1016/j.cell.2012.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang Y, Li X, Yang W, Hawke DH, Zheng Y, Xia Y, Aldape K, Wei C, Guo F, Chen Y, Lu Z. PKM2 regulates chromosome segregation and mitosis progression of tumor cells. Mol Cell. 2014;53(1):75–87. 10.1016/j.molcel.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang Y, Wang Y, Wang T, Hawke DH, Zheng Y, Li X, Zhou Q, Majumder S, Bi E, Liu DX, Huang S, Lu Z. PKM2 phosphorylates MLC2 and regulates cytokinesis of tumour cells. Nature communications. 2014;5:5566 10.1038/ncomms6566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hosios AM, Fiske BP, Gui DY, Vander Heiden MG. Lack of Evidence for PKM2 Protein Kinase Activity. Mol Cell. 2015;59(5):850–7. 10.1016/j.molcel.2015.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chu B, Wang J, Wang Y, Yang G. Knockdown of PKM2 induces apoptosis and autophagy in human A549 alveolar adenocarcinoma cells. Molecular medicine reports. 2015;12(3):4358–63. 10.3892/mmr.2015.3943 . [DOI] [PubMed] [Google Scholar]
- 64.Li C, Zhao Z, Zhou Z, Liu R. PKM2 Promotes Cell Survival and Invasion Under Metabolic Stress by Enhancing Warburg Effect in Pancreatic Ductal Adenocarcinoma. Digestive diseases and sciences. 2016;61(3):767–73. 10.1007/s10620-015-3931-2 . [DOI] [PubMed] [Google Scholar]
- 65.Yuan S, Qiao T, Zhuang X, Chen W, Xing N, Zhang Q. Knockdown of the M2 Isoform of Pyruvate Kinase (PKM2) with shRNA Enhances the Effect of Docetaxel in Human NSCLC Cell Lines In Vitro. Yonsei Med J. 2016;57(6):1312–23. 10.3349/ymj.2016.57.6.1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang S, Ma Y, Wang P, Song Z, Liu B, Sun X, Zhang H, Yu J. Knockdown of PKM2 Enhances Radiosensitivity of Non-small cell Lung Cancer. Cell biochemistry and biophysics. 2015;73(1):21–6. 10.1007/s12013-015-0567-y . [DOI] [PubMed] [Google Scholar]
- 67.Shi HS, Li D, Zhang J, Wang YS, Yang L, Zhang HL, Wang XH, Mu B, Wang W, Ma Y, Guo FC, Wei YQ. Silencing of pkm2 increases the efficacy of docetaxel in human lung cancer xenografts in mice. Cancer Sci. 2010;101(6):1447–53. 10.1111/j.1349-7006.2010.01562.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim DJ, Park YS, Kang MG, You YM, Jung Y, Koo H, Kim JA, Kim MJ, Hong SM, Lee KB, Jang JJ, Park KC, Yeom YI. Pyruvate kinase isoenzyme M2 is a therapeutic target of gemcitabine-resistant pancreatic cancer cells. Experimental cell research. 2015;336(1):119–29. 10.1016/j.yexcr.2015.05.017 . [DOI] [PubMed] [Google Scholar]
- 69.Gines A, Bystrup S, Ruiz de Porras V, Guardia C, Musulen E, Martinez-Cardus A, Manzano JL, Layos L, Abad A, Martinez-Balibrea E. PKM2 Subcellular Localization Is Involved in Oxaliplatin Resistance Acquisition in HT29 Human Colorectal Cancer Cell Lines. PloS one. 2015;10(5):e0123830 10.1371/journal.pone.0123830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu W, Lu SX, Li M, Zhang C, Liu LL, Fu J, Jin JT, Luo RZ, Zhang CZ, Yun JP. Pyruvate kinase M2 prevents apoptosis via modulating Bim stability and associates with poor outcome in hepatocellular carcinoma. Oncotarget. 2015;6(9):6570–83. 10.18632/oncotarget.3262 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dong T, Yan Y, Chai H, Chen S, Xiong X, Sun D, Yu Y, Deng L, Cheng F. Pyruvate kinase M2 affects liver cancer cell behavior through up-regulation of HIF-1alpha and Bcl-xL in culture. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2015;69:277–84. 10.1016/j.biopha.2014.12.010 . [DOI] [PubMed] [Google Scholar]
- 72.Kwon OH, Kang TW, Kim JH, Kim M, Noh SM, Song KS, Yoo HS, Kim WH, Xie Z, Pocalyko D, Kim SY, Kim YS. Pyruvate kinase M2 promotes the growth of gastric cancer cells via regulation of Bcl-xL expression at transcriptional level. Biochemical and biophysical research communications. 2012;423(1):38–44. 10.1016/j.bbrc.2012.05.063 . [DOI] [PubMed] [Google Scholar]
- 73.Kim D, Fiske BP, Birsoy K, Freinkman E, Kami K, Possemato RL, Chudnovsky Y, Pacold ME, Chen WW, Cantor JR, Shelton LM, Gui DY, Kwon M, Ramkissoon SH, Ligon KL, Kang SW, Snuderl M, Vander Heiden MG, Sabatini DM. SHMT2 drives glioma cell survival in ischaemia but imposes a dependence on glycine clearance. Nature. 2015;520(7547):363–7. 10.1038/nature14363 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xie J, Yea K, Zhang H, Moldt B, He L, Zhu J, Lerner RA. Prevention of cell death by antibodies selected from intracellular combinatorial libraries. Chemistry & biology. 2014;21(2):274–83. 10.1016/j.chembiol.2013.12.006 . [DOI] [PubMed] [Google Scholar]
- 75.Zhang H, Torkamani A, Jones TM, Ruiz DI, Pons J, Lerner RA. Phenotype-information-phenotype cycle for deconvolution of combinatorial antibody libraries selected against complex systems. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(33):13456–61. 10.1073/pnas.1111218108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang H, Wilson IA, Lerner RA. Selection of antibodies that regulate phenotype from intracellular combinatorial antibody libraries. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(39):15728–33. 10.1073/pnas.1214275109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yamaguchi R, Lartigue L, Perkins G, Scott RT, Dixit A, Kushnareva Y, Kuwana T, Ellisman MH, Newmeyer DD. Opa1-mediated cristae opening is Bax/Bak and BH3 dependent, required for apoptosis, and independent of Bak oligomerization. Mol Cell. 2008;31(4):557–69. 10.1016/j.molcel.2008.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guzman C, Bagga M, Kaur A, Westermarck J, Abankwa D. ColonyArea: an ImageJ plugin to automatically quantify colony formation in clonogenic assays. PloS one. 2014;9(3):e92444 10.1371/journal.pone.0092444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Preta G, Fadeel B. Scythe cleavage during Fas (APO-1)-and staurosporine-mediated apoptosis. FEBS letters. 2012;586(6):747–52. Epub 2012/01/31. 10.1016/j.febslet.2012.01.034 . [DOI] [PubMed] [Google Scholar]
- 80.Fulda S, Debatin KM. HIF-1-regulated glucose metabolism: a key to apoptosis resistance? Cell cycle. 2007;6(7):790–2. 10.4161/cc.6.7.4084 . [DOI] [PubMed] [Google Scholar]
- 81.Moley KH, Mueckler MM. Glucose transport and apoptosis. Apoptosis: an international journal on programmed cell death. 2000;5(2):99–105. . [DOI] [PubMed] [Google Scholar]
- 82.Munoz-Pinedo C, El Mjiyad N, Ricci JE. Cancer metabolism: current perspectives and future directions. Cell death & disease. 2012;3:e248 10.1038/cddis.2011.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bollenbach TJ, Mesecar AD, Nowak T. Role of lysine 240 in the mechanism of yeast pyruvate kinase catalysis. Biochemistry. 1999;38(28):9137–45. 10.1021/bi990690n . [DOI] [PubMed] [Google Scholar]
- 84.Dombrauckas JD, Santarsiero BD, Mesecar AD. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry. 2005;44(27):9417–29. 10.1021/bi0474923 . [DOI] [PubMed] [Google Scholar]
- 85.Kuwana T, Olson NH, Kiosses WB, Peters B, Newmeyer DD. Pro-apoptotic Bax molecules densely populate the edges of membrane pores. Scientific reports. 2016;6:27299 10.1038/srep27299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liang J, Cao R, Wang X, Zhang Y, Wang P, Gao H, Li C, Yang F, Zeng R, Wei P, Li D, Li W, Yang W. Mitochondrial PKM2 regulates oxidative stress-induced apoptosis by stabilizing Bcl2. Cell research. 2017;27(3):329–51. Epub 2016/12/31. 10.1038/cr.2016.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu H, Yang P, Hu W, Wang Y, Lu Y, Zhang L, Fan Y, Xiao H, Li Z. Overexpression of PKM2 promotes mitochondrial fusion through attenuated p53 stability. Oncotarget. 2016. 10.18632/oncotarget.12942 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ryu SW, Choi K, Park JH, Park YM, Kim S, Choi C. Mitofusin 1 inhibits an apoptosis-associated amino-terminal conformational change in Bax, but not its mitochondrial translocation, in a GTPase-dependent manner. Cancer letters. 2012;323(1):62–8. 10.1016/j.canlet.2012.03.038 . [DOI] [PubMed] [Google Scholar]
- 89.Kushnareva Y, Seong Y, Andreyev AY, Kuwana T, Kiosses WB, Votruba M, Newmeyer DD. Mitochondrial dysfunction in an Opa1(Q285STOP) mouse model of dominant optic atrophy results from Opa1 haploinsufficiency. Cell death & disease. 2016;7:e2309 10.1038/cddis.2016.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Choudhary V, Kaddour-Djebbar I, Alaisami R, Kumar MV, Bollag WB. Mitofusin 1 degradation is induced by a disruptor of mitochondrial calcium homeostasis, CGP37157: a role in apoptosis in prostate cancer cells. International journal of oncology. 2014;44(5):1767–73. 10.3892/ijo.2014.2343 . [DOI] [PubMed] [Google Scholar]
- 91.Iqbal MA, Siddiqui FA, Chaman N, Gupta V, Kumar B, Gopinath P, Bamezai RN. Missense mutations in pyruvate kinase M2 promote cancer metabolism, oxidative endurance, anchorage independence, and tumor growth in a dominant negative manner. The Journal of biological chemistry. 2014;289(12):8098–105. 10.1074/jbc.M113.515742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Israelsen WJ, Dayton TL, Davidson SM, Fiske BP, Hosios AM, Bellinger G, Li J, Yu Y, Sasaki M, Horner JW, Burga LN, Xie J, Jurczak MJ, DePinho RA, Clish CB, Jacks T, Kibbey RG, Wulf GM, Di Vizio D, Mills GB, Cantley LC, Vander Heiden MG. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell. 2013;155(2):397–409. 10.1016/j.cell.2013.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Semba H, Takeda N, Isagawa T, Sugiura Y, Honda K, Wake M, Miyazawa H, Yamaguchi Y, Miura M, Jenkins DM, Choi H, Kim JW, Asagiri M, Cowburn AS, Abe H, Soma K, Koyama K, Katoh M, Sayama K, Goda N, Johnson RS, Manabe I, Nagai R, Komuro I. HIF-1alpha-PDK1 axis-induced active glycolysis plays an essential role in macrophage migratory capacity. Nature communications. 2016;7:11635 10.1038/ncomms11635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Barrero CA, Datta PK, Sen S, Deshmane S, Amini S, Khalili K, Merali S. HIV-1 Vpr modulates macrophage metabolic pathways: a SILAC-based quantitative analysis. PloS one. 2013;8(7):e68376 10.1371/journal.pone.0068376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shirai T, Nazarewicz RR, Wallis BB, Yanes RE, Watanabe R, Hilhorst M, Tian L, Harrison DG, Giacomini JC, Assimes TL, Goronzy JJ, Weyand CM. The glycolytic enzyme PKM2 bridges metabolic and inflammatory dysfunction in coronary artery disease. The Journal of experimental medicine. 2016;213(3):337–54. 10.1084/jem.20150900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Corcoran SE, O'Neill LA. HIF1alpha and metabolic reprogramming in inflammation. The Journal of clinical investigation. 2016;126(10):3699–707. 10.1172/JCI84431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Qi W, Keenan HA, Li Q, Ishikado A, Kannt A, Sadowski T, Yorek MA, Wu IH, Lockhart S, Coppey LJ, Pfenninger A, Liew CW, Qiang G, Burkart AM, Hastings S, Pober D, Cahill C, Niewczas MA, Israelsen WJ, Tinsley L, Stillman IE, Amenta PS, Feener EP, Vander Heiden MG, Stanton RC, King GL. Pyruvate kinase M2 activation may protect against the progression of diabetic glomerular pathology and mitochondrial dysfunction. Nature medicine. 2017. 10.1038/nm.4328 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cheon JH, Kim SY, Son JY, Kang YR, An JH, Kwon JH, Song HS, Moon A, Lee BM, Kim HS. Pyruvate Kinase M2: A Novel Biomarker for the Early Detection of Acute Kidney Injury. Toxicol Res. 2016;32(1):47–56. 10.5487/TR.2016.32.1.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nature protocols. 2006;1(5):2315–9. 10.1038/nprot.2006.339 . [DOI] [PubMed] [Google Scholar]
- 100.Waterhouse NJ, Goldstein JC, Kluck RM, Newmeyer DD, Green DR. The (Holey) study of mitochondria in apoptosis. Methods in cell biology. 2001;66:365–91. . [DOI] [PubMed] [Google Scholar]
- 101.Goldberg MS, Sharp PA. Pyruvate kinase M2-specific siRNA induces apoptosis and tumor regression. The Journal of experimental medicine. 2012;209(2):217–24. 10.1084/jem.20111487 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Approximately 5 x 105 cells were incubated per well for 12 h, then cells were either mock-transfected, transfected with 30 nM PKM2-specific siRNA (si M2), or as control, transfected with NF-κB p50-specific siRNA (si p50). After a further 36-h incubation, samples of the same siRNAs were added along with 4 μg of BimS cDNA in fresh medium. Viable cells were counted after another 48-h incubation. Note: underlying data are included in corresponding tabs in the accompanying supplemental Excel file S1 Data. 293T, HEK293T; IB5, intrabody 5; NF-κB, nuclear factor κB; PKM2, pyruvate kinase isoform M2; siRNA, small interfering RNA.
(TIF)
Approximately 5 x 105 HCT116 and U2OS cells were plated and transfected with BimS expression plasmid including 150 nM etoposide or 1 μM Staurosporine. The plates were fixed and stained with crystal violet after 5 d and the total areas of colonies were measured. Mean, SD, and P values were calculated from three individual plates. Note: underlying data are included in corresponding tabs in the accompanying supplemental Excel file S1 Data. IB5, intrabody 5
(TIF)
Control or IB5-expressing cells were transfected with BimS cDNA. The plates were fixed and stained with crystal violet after 12 days and the total areas of colonies were measured. Mean, SD, and P values were calculated from three individual plates. Note: underlying data are included in corresponding tabs in the accompanying supplemental Excel file S1 Data. BimS, short isoform of BimS; IB5, intrabody 5
(TIF)
293T cells were infected (lane 2, 3) or not (lane 1) with IB5 lentivirus and incubated with (lane 3) or without 2 μg of BimS cDNA (lane1, 2) in fresh medium. Cells were lysed, and total cell protein extracts were subjected to western blot analysis. BimEL (upper band), BimL (middle band) and BimS (lower band) were detected using Anti-Bim antibody (ab15184). GAPDH was used as loading control. 293T, HEK293T; GAPDH, glyceraldehyde phosphate dehydrogenase; IB5, intrabody 5; PKM2, pyruvate kinase isoform M2
(TIF)
A. PKM2-deficient MEFs reconstituted with WT or mutant PKM2 cDNA were infected or not with IB5, then 2 x 104 cells were plated and transfected with BimS expression plasmid. The plates were fixed and stained with crystal violet after 1 week and the total area of colonies were counted as above. Means, SDs, and P values were calculated from three experiments. B. Blue native gel electrophoresis of PKM2 WT and mutations. C. scFv 5 stimulated glycolytic activity of WT PKM2 and PKM2 (K367M). Activity was measured as in Fig 4. Note: underlying data are included in corresponding tabs in the accompanying supplemental Excel file S1 Data. IB5, intrabody 5; MEF, Mouse Embryonic Fibroblast; PKM2, pyruvate kinase isoform M2; scFv, single-chain variable fragment; WT, wild-type
(TIF)
A. 2-deoxy-D-glucose had no effect on 293T cell survival induced by IB5 intrabody. 293T cells were infected or not with IB5, then 2 x 104 cells were plated and transfected with BimS expression plasmid. The glycolytic inhibitor 2-deoxy-D-glucose (20 mM) was added to the MEMαmedium, and after 24 h, cells were transfected or not with 1 μg of BimS cDNA in fresh medium. The plates were fixed and stained with crystal violet after 1 week. B. IB5 reduced MFN1 mRNA levels, implying that Mfn1 protein up-regulation is post-transcriptional. PKM2-deficient MEFs reconstituted with WT or mutant PKM2 cDNA were infected or not with IB5, and MFN1 mRNA levels were quantified by qPCR. Means, SDs, and P values based on four independent experiments are indicated. Note: underlying data are included in corresponding tabs in the accompanying supplemental Excel file S1 Data. 293T, HEK293T; IB5, intrabody 5; MEMα; PKM2, pyruvate kinase isoform M2; WT, wild-type
(TIF)
Lysates from the indicated MEF strains were analyzed by immunoblotting with antibodies directed against Mfn1, Mfn2, and PKM2, as indicated. Actin and GAPDH were used as loading controls. GAPDH, glyceraldehyde phosphate dehydrogenase; MEF, Mouse Embryonic Fibroblast; Mfn, Mitofusin; PKM2, pyruvate kinase isoform M2
(TIF)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.