Abstract
Aberrant methylation of DNA has been identified as an epigenetic biomarker for numerous cancer types. The vast majority of techniques aimed at detecting methylation require bisulfite conversion of the DNA sample prior to analysis, which until now has been a benchtop process. Although microfluidics has potential benefits of simplified operation, sample and reagent economy, and scalability, bisulfite conversion has yet to be implemented in this format. Here, we present a novel droplet microfluidic design that facilitates rapid bisulfite conversion by reducing the necessary processing steps while retaining comparable performance to existing methods. This new format has a reduced overall processing time and is readily scalable for use in high throughput DNA methylation analysis.
Keywords: Epigenetics, Bisulfite conversion, MSP, Methylation, Lab-on-chip, Magnetofluidics
1. Introduction
DNA methylation is a fundamental mechanism in the epigenetic control of gene expression (Bird 1985). Dysregulation of DNA methylation has been shown to be implicated in a number of human diseases, and is particularly prevalent in many forms of cancer (Ehrlich 2002). Hypermethylation occurring in CpG dinucleotides in the promoter region of particular genes has been shown to occur early during carcinogenesis, preventing expression and ultimately contributing to genomic instability and cancer progression (Das and Singal 2004; Ehrlich 2002). The close association between hypermethylation and cancer development has led to considerable interest in its use as a promising biomarker for cancer diagnostics, prognostics and prediction to treatment (Herman et al. 1994; Herman and Baylin 2003; Merlo et al. 1995).
A wide range of analysis techniques has been developed for the study of DNA methylation. Among these include genome-scale methylation analysis methods such as bisulfite sequencing (Dupont et al. 2004; Frommer et al. 1992) and bead probe microarrays (Dedeurwaerder et al. 2011), as well as locus-specific techniques including methylation specific PCR (MSP) (Herman et al. 1996), COBRA (Xiong and Laird 1997), and methylation-sensitive high resolution melt (MS-HRM) (Vossen et al. 2009; Wojdacz and Dobrovic 2007). A ubiquitous and essential step in these techniques is the bisulfite conversion (BSC) process, which is a biochemical technique used to differentiate between methylated and unmethylated DNA (Frommer et al. 1992). Through a series of discrete chemical processing steps, the BSC process results in the selective conversion of unmethylated cytosine residues into uracil, while leaving methylated cytosine residues unchanged. Bisulfite conversion ultimately results in the translation of DNA methylation into changes in the primary genetic sequence. The methylation-specific contrast provided by this process can be subsequently assessed through the use of standard DNA analysis techniques in order to determine the methylation status of the original templates.
Bisulfite conversion is currently performed in a benchtop environment using commercially - available BSC kits. Despite their widespread use, benchtop processes necessitate an extensive set of manual fluidic handling and centrifugation steps. As a result, these processes are generally labor-intensive and time consuming. Furthermore, the microcentrifuge tube-based protocols require relatively large sample volumes for optimal conversion, presenting a substantial constraint when processing small sample volumes. Dilution of sample volume has substantial drawbacks, as the large surface areas of microcentrifuge tube-based kits have been found to significantly reduce yields for DNA samples smaller than 1 ng, (Keeley et al. 2013). Furthermore, manual fluid transfer between reaction vessels is a highly operator-dependent process that may result in additional sample loss and contamination. Lastly, there is an inherent mismatch in scale that challenges the seamless integration of tube-based sample processing with microfluidic high-throughput analysis platforms. While some of the aforementioned analysis techniques such as sequencing utilize microfluidics to great effect in throughput enhancement, the process for bisulfite conversion has not been incorporated into the microfluidic workflow.
Historically, microfluidic devices have been employed to address numerous fundamental sample-handling issues such as these. Some key advantages attributed to microfluidic approaches include streamlined fluidic processing, reduced sample and reagent use, throughput enhancement and improved accuracy and precision (Mitchell 2001; Schulte et al. 2002). Additionally, these platforms are generally designed for automation, which addresses problems that may arise from the operator dependence of benchtop processes. Microfluidic adaptations of benchtop processes for nucleic acid analysis have been demonstrated on multiple occasions in literature. These include a streamlined chip for DNA for DNA extraction and quantitative PCR (qPCR) on the same device (Lee et al. 2006), microfluidic platform for DNA library preparation for next generation sequencing (Kim et al. 2013), and a microfluidic platform for chromatin immunoprecipitation (ChIP) that benefits from simultaneous processing of thousands of cells with reduced contamination and increased accuracy (Wu et al. 2012).
Despite its advantages, microfluidic approaches have yet to be widely adopted due to the complexities of fluidic handling which requires intricate microfluidic architecture designs as well as external fluidic interface (Mariella 2008; Park et al. 2011). To address these challenges, platforms utilizing discrete droplets for liquid control have emerged, enabling pumpless and valveless handling of reagents (Gijs 2004; Verpoorte 2003). These novel platforms utilize a number of alternative means of droplet manipulation such as magnetic bead actuation (Chiou et al. 2013; Shin et al. 2014; Zhang et al. 2011; Zhang and Wang 2013), dielectrophoresis (Velev et al. 2003) and electrowetting-on-dielectric (EWOD) (Miller and Wheeler 2008; Srinivasan et al. 2004) to perform all essential fluidic manipulations for bioanalysis, including transport, mixing, splitting, and merging of reagents. Such platforms enable efficient handling of samples and reagents with much simplified fluidic management.
Here we demonstrate a droplet microfluidic approach to the bisulfite conversion of DNA (Fig. 1). By leveraging the benefits of microfluidics, we demonstrate a parallelized and streamlined DNA processing module that is readily amenable to scaling for significantly improved sample throughput. By utilizing droplet microfluidic design, reagent handling is simplified to a series of particle translocations without requiring complex fluidic control. We validate our platform using a standard MethyLight assay (Eads et al. 2000) and demonstrate its performance and advantages over existing benchtop tube processes.
Fig. 1.
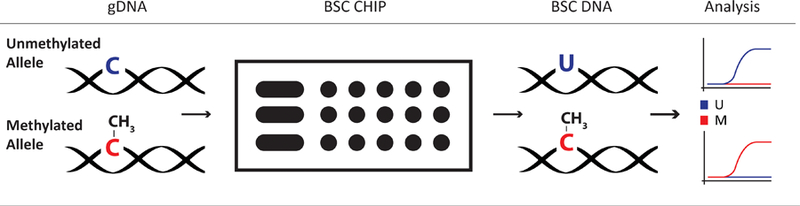
Overview of the bisulfite conversion process using the BST chip. The unmethylated cytosines (blue C) are converted by the BST process on chip into a uracil (blue U). The methylated cytosine (red C) however, is protected from the reaction and remains unchanged. The resulting unique sequence can be analyzed using qPCR or other genomic tools to identify the original methylation
2. Materials and methods
2.1. Droplet-based bisulfite conversion
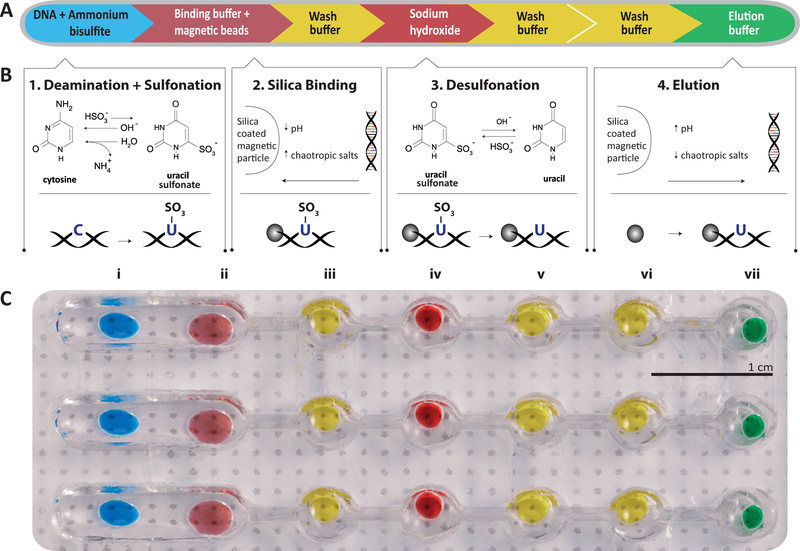
In this work, we employed our previously-developed single-tube bisulfite conversion method, methylation on beads (MOB) (Bailey et al. 2010; Keeley et al. 2013) and adapted it into a microfluidic droplet format. The bisulfite conversion process in MOB comprises four key sequential steps with intermediate washing steps (Fig. 2a). First, the DNA sample is introduced to ammonium bisulfite, resulting in sulfonation and hydrolytic deamination of unmethylated cytosine residues. By contrast, methylation protects the cytosine residue from sulfonation and prevents further chemical modification in subsequent steps. Second, as the unique feature in MOB, silica-coated superparamagnetic beads (SSBs) are employed as a solid phase substrate for tight DNA adsorption, thus allowing DNA capture and transport and facile exchange of buffers across downstream reaction steps. Third, following a brief wash, sulfonated DNA is exposed to sodium hydroxide, during which the uracil sulfonate residues undergo alkali desulfonation to yield fully converted uracil residues. Lastly, the processed DNA is purified via successive washing steps and separated from the SSBs, readying recovered DNA for downstream methylation assessment techniques, such as methylation-specific PCR (MSP).
Fig. 2.
a The process for bisulfite conversion is comprised of four main chemical reactions with three intermediate washing steps to prevent carryover of reagents. b The chemistry is detailed for each step. Deamination and sulphonation convert the unmethylated cytosine into uracil-sulphonate (1). The converted DNA is then bound onto the beads for transportation(2). The uracil-sulfonate is desulfonated resulting in the genomic base uracil (3). The final step is to reverse the binding process to recover the DNA from the beads (4). c Photograph of aqueous reagents loaded onto a single lane of droplet chip. Each reagent is contained in a round well that holds the droplet within it. The wells are connected either by a single open channel (i.e., between I and II) to merge the droplets or a narrow sieve (II through VII) to separate the beads from the droplet by surface tension
Critically, the use of SSBs enables the MOB workflow in the microfluidic droplet format because the beads facilitate a robust transport medium for the surface-bound nucleic acids through a lane of isolated droplets, each of which contains a required MOB reagent. An image of the chip with three parallel lanes is shown in Fig. 2b. Each lane consists of one wide and five circular reservoir chambers, each containing aqueous MOB reagent droplets that are isolated within topographic walls. A hydrophobic coating on the bottom surface of the device and mineral oil pre-loaded in each lane of chip to ensure that the aqueous reagents are maintained in droplet form.
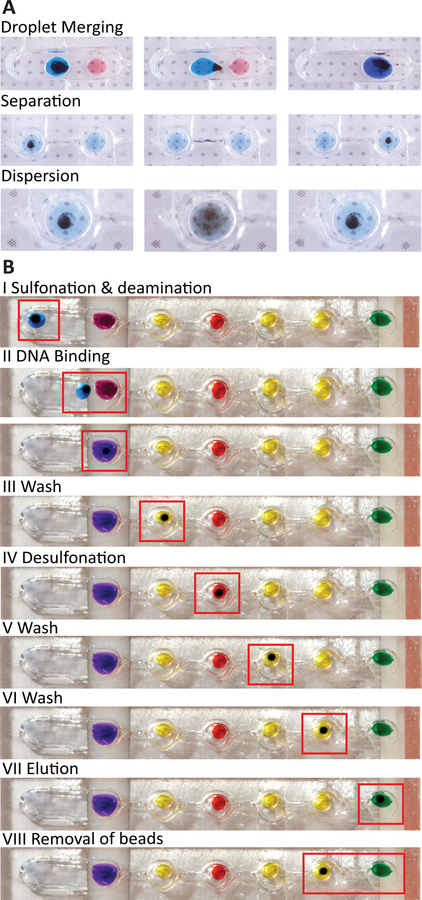
To facilitate a droplet-based MOB workflow, we have designed the device to allow facile manipulation of SSBs to merge droplets, separate from droplets, and disperse within droplets (Fig. 3a). For example, in the chamber containing the bisulfite reagent droplet and the binding buffer (i.e., Fig. 3b, i and ii), the first droplet can be transported across the wide chamber by the SSB cluster due to the high surface tension, resulting in merging of both droplets. In chambers connected by narrow sieve structures (i.e., Fig. 3b, ii through vii), SSBs can be effectively separated from reagent droplets and transported into downstream reagent droplets. Such separation is possible because the narrow sieve creates a surface energy barrier to keep reagent droplets in the chamber, while the SSB cluster can escape due to the significantly higher magnetic force relative to capillary force at the interface. This design provides an efficient and robust means of sequential buffer exchange necessary for bisulfite conversion. Finally, SSBs within droplets can readily disperse by simply removing the magnet; this dispersion thus allows mixing with-in droplets.
Fig. 3.
a The steps required for bisulfite conversion are demonstrated in their corresponding reagent droplets. As the beads and DNA are transported across the chip surface, the DNA is exposed to each subsequent reagent of the BSC process. b There are three main modes of transportation for the beads. Droplet merging is done across an open channel, where the lower surface energy allows the droplet to be transported with the bead cluster. The separation process is achieved by moving the beads across a sieve where the droplet is retained while the SSB cluster can move. Dispersion is performed by removing the magnetic force and allowing the magnetic beads to resuspend via Brownian motion
On-chip sample processing begins by first mixing 2 μl of extracted genomic DNA (gDNA) with 13 μl of the Lightning Conversion reagent (Zymo) and 5 μl of SSB (Promega Magnesil KF MD) to form a droplet that is then loaded into the first reservoir (Fig. 3b, step i). The sample is subsequently heated to 95 °C for 8 min in order to denature the sample into single-stranded DNA (ssDNA). The temperature is then lowered to 54 °C for 1 h to allow the bisulfite to complete deamination of unmethylated cytosine and sulfonation to yield uracil-sulfonate. After this process, the droplet is mixed together with a 60 μl droplet of M-binding buffer (step ii) and briefly incubated to allow the DNA to adsorb onto the silica surface of the beads. The SSB are then washed with 40 μl ethanol (step iii) to eliminate any remaining bisulfite reagent before being transported into a 20 μl M-desulphonation buffer droplet (step iv). This alkali solution desulfonates the uracil-sulfonate into stable uracil bases. The SSB are washed twice in 40 μl M-wash buffer droplets (steps v and vii) in order to remove any residual sodium hydroxide from the solution. Finally, the remaining DNA is eluted from the SSB into Tris-EDTA buffer (step vii) and the SSB are decanted into the previous well to allow the droplet containing purified, bisulfite-converted DNA to be retrieved from the chip for downstream analysis (step viii).
2.2. Chip design and manufacturing
The device consists of a linear array of cylindrical chambers (5 mm diameter), interconnected by sieve structures 1 mm wide and 4 mm long. The total spacing from well center to center is 9 mm, to allow for compatibility with multi-channel pipettors. A typical device has a depth of 5 mm. The device is fabricated using only a single piece of polydimethylsiloxane (PDMS) bonded to a thin coverslip, followed by hydrophobic coating. The PDMS pattern was designed using Solidworks 3D CAD (Solidworks Corp.) and translated into a negative acrylonitrile butadiene styrene (ABS) plastic mold generated using a 3D printer (Stratasys Dimension 1200es). PDMS prepared at 1:10 crosslinker-to-base ratio (Sylgard 184, Dow Corning Corp.) is subsequently poured into this mold and incubated at 80 °C for >1 h. The resulting PDMS structure is bonded to a 2′′ × 3′′ coverslip using an oxygen-plasma treatment of the surface. Finally, the chip is coated in Teflon to provide hydrophobic properties to the surface to prevent contact between the reagents and the glass. Heating and cooling of the reagents are performed by regulating the chip temperature via two independent thermoelectric elements, as shown in Fig. S1.
2.3. Performance evaluation protocol
In order to evaluate the efficiency and reliability of on-chip conversion, the converted products were assessed using a standard Methylight assay. This assay relies upon carefully designed primers and Taqman probes to quantify bisulfite-converted target loci. The primers are designed to exclusively amplify only fully-converted target sequences, resulting in amplification only when complementary template is present.
In the initial assessment of bisulfite conversion on chip, we compared conversion of an unmethylated human gDNA sample using the BSC chip with the standard in-tube MOB process. Converted DNA from both methods were analyzed via Methylight assay using primers targeting a locus within the β-Actin gene (Mori et al. 2008). An unconverted gDNA sample was used as a negative control. The qPCR master mix was prepared using 300 nM for each primer, 200 nM dNTPs, 500 nM hybridization probe, and 1 unit of Platinum Taq (Invitrogen 10966), 16.7 mM ammonium sulfate, 67 mM TRIS (pH 8.8), 6.7 mM magnesium chloride and 10 mM mercaptoethanol. The relative DNA quantity for both proceses was obtained from the CT values, using the chip process as the reference value.
Subsequently, we assessed the performance of BST chip using the average and variability of cycle threshold (CT) values (Bustin et al. 2009; Svec et al. 2015). In order to assess the chip’s ability to process a wide range of input DNA in a linear fashion, we used an input DNA ranging from 1 to 100 ng. In order to demonstrate linearity in DNA output from the device for both converted and unconverted DNA, we used a validated set of methylated and unmethylated primers targeting a locus within the promoter region of ADAMTS1 using the same qPCR conditions as described previously (Yi et al. 2013). The estimated quantity was normalized using the CT value for 1 ng as a reference.
3. Results
3.1. Simplified, faster protocol
The bisulfite conversion on droplet platform significantly reduces both the number of pipetting steps required for the conversion process and the total time required, particularly when processing multiple samples. Under the droplet microfluidic approach, repetitive and operator-dependent pipetting steps required for reagent aspiration and loading are replaced by particle transport across reagent chambers. As a result, pipetting steps are reduced to the two steps required for initial loading and final retrieval of DNA sample from the chip. This reduces the number of steps per sample (Fig. 4), but more importantly, enables the use of multichannel-pipettors for one-step reagent loading.
Fig. 4.
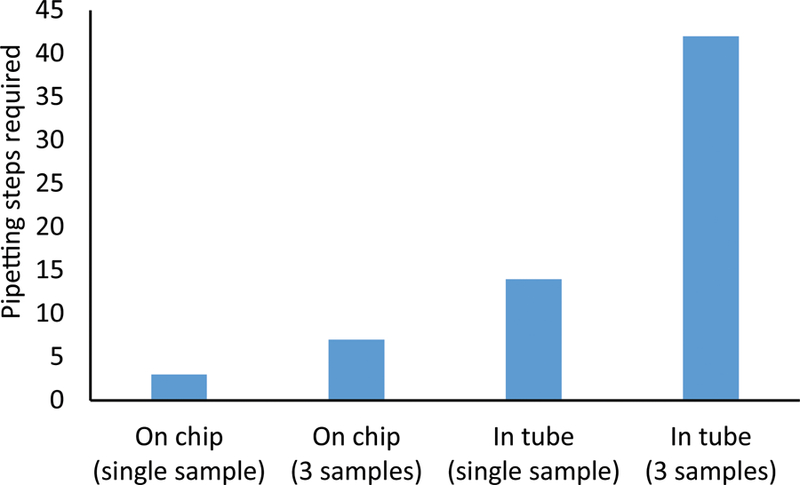
The BSC chip greatly reduces the number of manual pipetting steps, particularly when processing multiple samples in parallel. Tube-based benchtop process require numerous liquid transfer steps into and out of the tube, while the BSC chip liquid transfers are limited to sample loading and retrieval
The selection of faster conversion reagents from the Zymo Lightning Kit, namely the bisulfite conversion reagent allow for even further reduction in processing time. The overall amount of time saved per sample is even more substantial when three samples are processed simultaneously using a multichannel-pipettor.
3.2. Performance in downstream analysis
The converted DNA from the BSC chip platform exhibits comparable performance to the benchtop tube process when analyzed using qPCR. We initially tested the converted DNA using a conversion - specific PCR primer set which does not take into account methylation status (B-actin). We found the CT values for the BSC chip process and the tube based process to be 28.34 and 28.45 with a standard deviation of 0.14 and 0.25, respectively (Fig. 5a). Similar conversion efficiency was observed between the two processes, suggested by the comparable mean CT value. As expected, bisulfite conversion is required for amplification as indicated by the lack of amplification in the unconverted negative control.
Fig. 5.
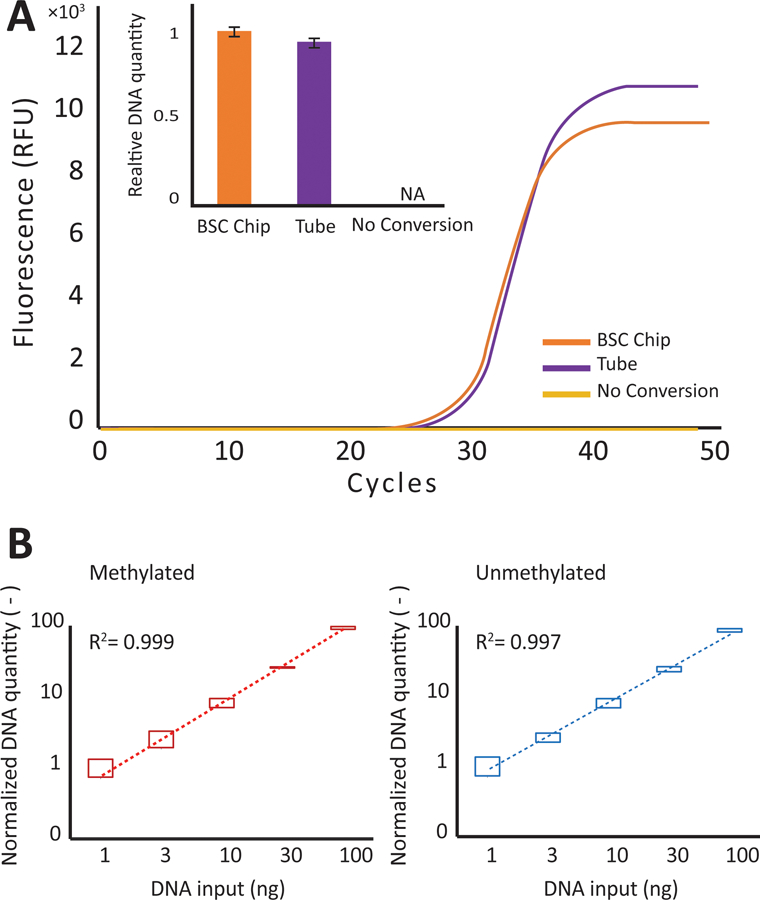
Comparison between chip and tube performance. A) The qPCR amplification curve for the tube process (shown in blue) is compared to the chip process (shown in red). B) The calculated DNA recovery (based on CT value and normalized to 1 ng) for varying quantities of DNA are compared across methylated and unmethylated sequences. The top and bottom bars reflect the variability across multiple devices and different channels
To further assess the device’s ability to process DNA without bias towards DNA input quantity or methylation status, we compared the converted output of unmethylated and methylated DNA controls at various concentrations using methylation - specific primer sets targeting the ADAMTS1 promoter region. The processed DNA concentration (Fig. 5b) shows a linear relationship with the initial DNA concentration and demonstrates that the BSC chip can operate at high yield across a wide range of sample input quantity. We were able to perform simultaneous conversion on a BSC device with all three lanes to demonstrate low bias and high reproducibility across replicates (Fig. 5b). Importantly, we established that qPCR linearity is preserved across replicate features, indicative of high process fidelity using the droplet device.
3.3. Reliability and reproducibility
The bisulfite conversion process performed on chip exhibits high reproducibility across runs. The low variability (Fig 5b error bars) is translated into accurate quantification when used in conjunction with qPCR. This variability is comparable regardless of methylation status. Additionally, this variability is also comparable to that exhibited by the Zymo Lightning Kit when performed in tube.
The reduced processing time required for the conversion reagents from the Zymo Lightning Kit not only significantly reduces the total processing time, but also minimizes evaporation (Fig. S2). Protection from significant evaporation helps to maintain stable buffer conditions for highly reproducible conversion results. Additionally, there was no visible precipitation of reagents in the droplets due to saturation, which could lead to co-precipitation of DNA and lower yields.
4. Discussion
In this report, we presented a droplet microfluidic device for performing BSC with a significant reduction in reagent volumes, as well as complexity, when compared to the benchtop in-tube process. This was achieved while maintaining equivalent performance in all conversion metrics tested. All necessary steps were performed via droplet merging, droplet separation and mixing through magnetic actuation of SSBs. The reduction in sample volume is especially convenient for the analysis of low-volume samples that are likely to suffer from reduced yield when diluted for processing with the tube-based benchtop protocol.
The modular nature of the device also suggests multiple paths through which the functionality of the BSC chip can be augmented and enhanced. The simplicity of an open-surface droplet design is amenable to facile integration with automation equipment for operator-free sample injection and retrieval, while future iterations of the device may be parallelized at a larger scale for high-throughput sample processing. The chip can potentially interface with other surface droplet solutions. For example, DNA extraction can be performed on the chip prior to conversion (Lehmann et al. 2006). Likewise, the chip can incorporate downstream analysis such as whole genome (Spits et al. 2006), or locus-specific (Ohashi et al. 2007) methylation assessment.
In summary, the robustness of the on-chip BSC, in addition to the simplified sample processing workflow and volume reduction demonstrate substantial benefit over existing bench-top processes. Furthermore, the platform’s potential for full automation and scalability lays the groundwork for integrating sample pre-processing in high-throughput DNA methylation analyses.
Supplementary Material
Acknowledgments
The authors would also like to thank funding sources from National Institutes of Health (R01CA155305, U54CA151838, R21CA186809).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10544-015-0029-8) contains supplementary material, which is available to authorized users.
References
- Bailey VJ, Zhang Y, Keeley BP, Yin C, Pelosky KL, Brock MV, Baylin SB, Herman JG, Wang T-H, Clin. Chem 56, 1022 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AP, Nature 321, 209 (1985) [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT, Clin. Chem 55, 611 (2009) [DOI] [PubMed] [Google Scholar]
- Chiou CH, Shin DJ, Zhang Y, Wang T-H, Biosens. Bioelectron 50, 91 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das PM, Singal R, J. Clin. Oncol 22, 4632 (2004) [DOI] [PubMed] [Google Scholar]
- Dedeurwaerder S, Defrance M, Calonne E, Denis H, Sotiriou C, Fuks F, Epigenomics 3, 771 (2011) [DOI] [PubMed] [Google Scholar]
- Dupont J, Tost J, Jammes H, Glynne I, Anal. Biochem 333, 119 (2004) [DOI] [PubMed] [Google Scholar]
- Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW, Nucleic Acids Res 28, e32 (2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Oncogene 21, 5400 (2002) [DOI] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL, Proc. Natl. Acad. Sci. U. S. A 89, 1827 (1992) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijs MAM, Microfluid. Nanofluid 1, 22 (2004) [Google Scholar]
- Herman JG, Latif F, Weng Y, Lerman MI, Zbar B, Liu S, Samid D, Duan DS, Gnarra JR, Linehan WM, Proc. Natl. Acad. Sci. U. S. A 91, 9700 (1994) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB, Proc. Natl. Acad. Sci. U. S. A 93, 9821 (1996) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Baylin SB, N. Engl, J. Med 349, 2042 (2003) [DOI] [PubMed] [Google Scholar]
- Keeley B, Stark A, Pisanic TR, Kwak R, Zhang Y, Wrangle J, Baylin SB, Herman JG, Ahuja N, Brock MV, Wang T-H, Clin. Chim. Acta 425, 169 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Jebrail MJ, Sinha A, Bent ZW, Solberg OD, Williams KP, Langevin SA, Renzi RF, Van De Vreugde JL, Meagher RJ, Schoeniger JS, Lane TW, Branda SS, Bartsch MS, Patel KD, PLoS One 8, e68988 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-G, Cheong KH, Huh N, Kim S, Choi J-W, Ko C, Lab Chip 6, 886 (2006) [DOI] [PubMed] [Google Scholar]
- Lehmann U, Vandevyver C, Parashar VK, Gijs MAM, Angew. Chem. Int. Ed 45, 3062 (2006) [DOI] [PubMed] [Google Scholar]
- Mariella R Jr., Biomed. Microdevices 10, 777 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin SA, Sidransky D, Nat. Med 1, 686 (1995) [DOI] [PubMed] [Google Scholar]
- Miller EM, Wheeler AR, Anal. Chem 80, 1614 (2008) [DOI] [PubMed] [Google Scholar]
- Mitchell P, Nat. Biotechnol 19, 717 (2001) [DOI] [PubMed] [Google Scholar]
- Mori R, Wang Q, Kd D, Jk P, Pv D, Prostate 380, 1 (2008) [Google Scholar]
- Ohashi T, Kuyama H, Hanafusa N, Togawa Y, Biomed. Microdevices 9, 695 (2007) [DOI] [PubMed] [Google Scholar]
- Park S, Zhang Y, Lin S, Wang T-H, Yang S, Biotechnol. Adv 29, 830 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte TH, Bardell RL, Weigl BH, Clin. Chim. Acta 321, 1 (2002) [DOI] [PubMed] [Google Scholar]
- Shin DJ, Zhang Y, Wang T-H, Microfluid. Nanofluid 17, 425 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spits C, Le Caignec C, De Rycke M, Van Haute L, Van Steirteghem A, Liebaers I, Sermon K, Nat. Protoc 1, 1965 (2006) [DOI] [PubMed] [Google Scholar]
- Srinivasan V, Pamula VK, Fair RB, Lab Chip 4, 310 (2004) [DOI] [PubMed] [Google Scholar]
- Svec D, Tichopad A, Novosadova V, Pfaffl MW, Kubista M, Biomol. Detect. Quantif 3, 9 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velev OD, Prevo BG, Bhatt KH, Nature 426, 515 (2003) [DOI] [PubMed] [Google Scholar]
- Verpoorte E, Lab Chip 3, 60N (2003) [DOI] [PubMed] [Google Scholar]
- Vossen RHAM, Aten E, Roos A, den Dunnen JT, Hum. Mutat 30, 860 (2009) [DOI] [PubMed] [Google Scholar]
- Wojdacz TK, Dobrovic A, Nucleic Acids Res 35, e41 (2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu AR, Kawahara TLA, Rapicavoli NA, Van Riggelen J, Shroff EH, Xu L, Felsher DW, Chang HY, Quake SR, Lab Chip 12, 2190 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z, Laird PW, Nucleic Acids Res 25, 2532 (1997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JM, Guzzetta AA, Bailey VJ, Downing SR, Van Neste L, Chiappinelli KB, Keeley BP, Stark A, Herrera A, Wolfgang C, Pappou EP, Iacobuzio-Donahue CA, Goggins MG, Herman JG, Wang T-H, Baylin SB, Ahuja N, Clin. Cancer Res 19, 6544 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang T-H, Adv. Mater 25, 2903 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Park S, Liu KJ, Tsuan J, Yang S, Wang T-H, Lab Chip 11, 398 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.