Abstract
Pre-mRNA splicing, once thought to be a strictly posttranscriptional event in gene expression, is subject to a multitiered network of regulation. Luco et al. now report in Science that this regulation seems to begin with chromatin modifications, suggesting that the histone code may be a prequel to the splicing code.
Intervening sequences (or introns) in most eukaryotic genes must be removed to produce mature mRNA during gene expression. Pre-mRNA is often alternatively spliced to give rise to mRNA isoforms that, in many cases, encode functionally distinct protein products. Although splicing can take place in cell-free extracts, independent of transcription, mounting evidence suggests that the process is largely cotranscriptional, and thus subjected to influence by the transcription process, particularly transcription elongation. In a study recently published in Science, Luco et al. (2010) have now further extended this coupling, opening a new chapter in connecting the histone code to regulated splicing.
First noticed by Ahringer and colleagues, exons show enrichment of the histone H3 lysine 36 trimethyl (H3K36me3) mark relative to introns in the genome of the nematode Caenorhabditis elegans (Kolasinska-Zwierz et al., 2009). Several groups quickly realized, through reanalyzing published nucleosome mapping results (particularly from human T cells), that nucleosomes are depleted in the intron adjacent to the acceptor splice site and enriched within the exons, which appears to largely account for enriched H3K36me3 marks on exons (Andersson et al., 2009; Tilgner et al., 2009; Schwartz et al., 2009). Similarly, strong nucleosome depletion was also observed at the polyA site, followed by enrichment immediately downstream of the polyA site (Spies et al., 2009). Nucleosome position appears to be a transcription-independent chromatin feature because a similar pattern was observed on both actively and poorly transcribed genes, despite the fact that H3K36me3 has previously been linked to transcriptional elongation. While both nucleosome affinity and the level of H3K36me3 are established prior to transcription, highly expressed genes appear to exhibit reduced nucleosomes on exons compared with poorly expressed genes, and the converse is also true for the H3K36 mark (Schwartz et al., 2009), suggesting that they may still be subjected to modulation during transcription.
The ability for the spliceosome to recognize splice sites in the vast abyss of nucleotide sequence has always been something of a conundrum. Now, it seems that nucleosome depletion and enrichment may help delineate where the intron-exon junction lies and facilitate exon definition. Remarkably, the length of a single turn of DNA around the nucleosome is the same average length of a typical human exon preferred by the spliceosome, suggesting an evolutionary conserved preference for nucleosomes to reside in exonic regions.
Nucleosome enrichment is clearly relevant to regulated splicing and is inversely correlated with the strength of splice sites (Tilgner et al., 2009; Spies et al., 2009). This has led to a proposal that nucleosomes may serve as “speed bumps” to elongating RNA polymerase II (Schwartz et al., 2009), similar to a proposed kinetic model suggesting that a reduced transcriptional elongation rate may afford increased time for the recognition of weak splice sites by the splicing machinery during cotranscriptional RNA splicing (Kornblihtt, 2007). Alternatively or in addition, various histone marks may facilitate the recruitment of splicing regulators to recognize cis-acting regulatory elements on emerging nascent RNA to dictate splice-site selection. A number of studies now provide evidence for this extended histone code hypothesis, including a report showing a role for the H3K4me3 binding protein CHD in binding and recruiting U2 snRNP to facilitate efficient splicing (Sims et al., 2007), and most recently, the demonstration of regulated splicing by the H3K36me3 binding protein MRG15 (Luco et al., 2010).
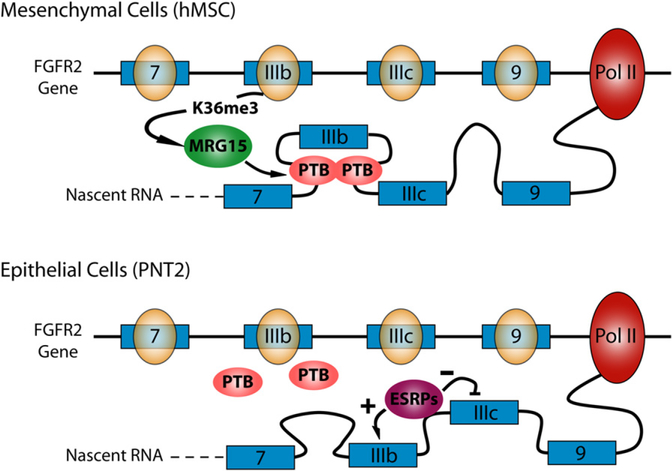
In this pivotal study, Luco et al. (2010) surveyed a number of histone modification events on the model human FGFR2 gene, which undergoes tissue-specific alternative splicing wherein the alternative exons IIIb or IIIc are selectively included in cells of epithelial and mesenchymal lineages, respectively (Figure 1). Elegant studies have revealed that the polypyrimidine tract binding protein PTB serves as a major suppressor of IIIb in mesenchymal cells, whereas several epithelial-specific RNA binding proteins (ESRPs) function as suppressors of IIIc in epithelial cells (Warzecha et al., 2009, and references therein). The selective effect of PTB on IIIb in mesenchyme is not well understood because PTB is abundantly expressed in both mesenchymal and epithelial cells. Luco et al. (2010) now connected an enrichment of the H3K36me3 mark to the action of PTB in suppressing the IIIb choice in human mesenchymal stem cells (hMSCs) (Luco et al., 2010).
Figure 1. A Potential Model for Chromatin Influence on Alternative Splicing of the FGFR2 Gene.
In human mesenchymal cells (hMSCs), the H3K36me3 mark recruits MGR15, which attracts PTB binding at intronic splicing silencer elements near exon IIIb, resulting in IIIb exclusion. Attenuation of this cascade of events by depletion of a H3K36 methyltransferase or MGR15 may derepress IIIb, allowing it to interfere or compete with IIIc inclusion. In epithelial cells, tissue-specific splicing regulators such as ESRPs act to promote IIIb inclusion. Overexpression of the H3K36 methyltransferase or MRG15 may enhance PTB binding, which antagonizes the effect of ESRPs and results in IIIb suppression.
Among the common histone marks they examined, Luco et al. (2010) focused on H3K36me3 and found that its binding protein, MRG15, is able to directly or indirectly interact with PTB to recruit this splicing repressor to the vicinity of exon IIIb, but not exon IIIc. Indeed, overexpression of MRG15 or a methyltransferase for H3K36me3 enhanced PTB recruitment, resulting in reduced IIIb inclusion in FGFR2 splicing in epithelial PNT2 cells. Because IIIb is already fully suppressed in hMSCs, further overexpression of MRG15 or the methyltransferase had little effect. Conversely, downregulation of these factors had no effect on IIIb inclusion in PNT2 cells (likely because of the dominant effect of epithelial-specific factors such as ESRPs), but caused derepressed IIIb to interfere with IIIc inclusion in hMSCs. Luco et al. (2010) next extended the analysis genome-wide, showing that a significant portion of splicing events regulated by PTB were also regulated by MRG15. The authors found that the splicing events regulated by both PTB and MRG15 were similarly affected in response to RNAi against PTB or MRG15. A complete overlap between PTB and MRG15 targets is not expected because many PTB-dependent events may not need additional modulation by MRG15, whereas other MRG15-dependent events might be coupled with different splicing regulators. These questions can now be further pursued by comparing genome-wide MRG15 binding with the recent genome-wide map of PTB binding on RNA (Xue et al., 2009). Interestingly, H3K4me3 distribution across genes exhibits an opposite profile to H3K36me3 between PNT2 and hMSCs, and overexpression of a methyl-transferase for H3K4me3 reduced IIIc inclusion by a mechanism that remains elusive. Furthermore, H3K9me1, a histone mark generally linked to gene repression, is selectively enriched on IIIb in PNT2 cells relative to hMSCs, raising the possibility that RNA polymerase II pauses at IIIb to favor its selection in epithelial cells. These observations leave open a long list of questions to be pursued in future studies.
Collectively, the findings of Luco et al. (2010) demonstrate a clear link between chromatin features and regulated splicing. We may be looking at just the tip of the chromatin modification iceberg, considering the potential combinatory influence of nucleosome positioning on the kinetic coupling between transcription and splicing, appearance of specific cis-acting elements from nascent RNA, and recruitment of splicing regulators that may act in a position- and context-dependent manner. Clearly, complete elucidation of the splicing code must now consider the contribution of the histone code. Indeed, it has been reported that there is increased accuracy in the prediction of splice site usage when information about nucleosome enrichment is added to exon prediction programs (Spies et al., 2009). However, the increase was relatively small, suggesting a long journey ahead of us in predicting the splicing code.
REFERENCES
- Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, and Komorowski J (2009). Genome Res. 19, 1732–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, and Ahringer J (2009). Nat. Genet 41, 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR (2007). Adv. Exp. Med. Biol 623, 175–189. [DOI] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, and Misteli T (2010). Science 327, 996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Meshorer E, and Ast G (2009). Nat. Struct. Mol. Biol 16, 990–995. [DOI] [PubMed] [Google Scholar]
- Sims RJ 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, and Reinberg D (2007). Mol. Cell 28, 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies N, Nielsen CB, Padgett RA, and Burge CB (2009). Mol. Cell 36, 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcárcel J, and Guigó R (2009). Nat. Struct. Mol. Biol 16, 996–1001. [DOI] [PubMed] [Google Scholar]
- Warzecha CC, Sato TK, Nabet B, Hogenesch JB, and Carstens RP (2009). Mol. Cell 33, 591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, et al. (2009). Mol. Cell 36, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]