Abstract
Investigation of genetic determinants of Mendelian skin disorders has substantially advanced understanding of epidermal biology. Here we show that mutations in PERP, encoding a crucial component of desmosomes, cause both dominant and recessive human keratoderma. Heterozygosity for a C-terminal truncation, which produces protein that appears to be unstably incorporated into desmosomes, causes Olmsted syndrome with severe periorificial and palmoplantar keratoderma in multiple unrelated kindreds. Homozygosity for an N-terminal truncation ablates expression and causes widespread erythrokeratoderma, with expansion of epidermal differentiation markers. Both exhibit epidermal hyperproliferation, immature desmosomes lacking a dense midline observed via electron microscopy, and impaired intercellular adhesion upon mechanical stress. Localization of other desmosomal components appears normal, in contrast to other conditions caused by mutations in genes encoding desmosomal proteins. These discoveries highlight the essential role of PERP in human desmosomes and epidermal homeostasis, and further expand the heterogeneous spectrum of inherited keratinization disorders.
INTRODUCTION
Disorders of keratinization (DOK) are scaling skin diseases with substantial morbidity and mortality and considerable phenotypic and genotypic heterogeneity. Palmoplantar keratoderma (PPK), or excessive thickening of palms and soles, is a key feature of many DOK including Olmsted syndrome (OS), characterized by palmoplantar and periorificial keratoderma (Duchatelet and Hovnanian, 2015), and many forms of erythrokeratoderma, characterized by widespread erythrokeratotic plaques (Hirano and Harvey, 2011). OS is most often dominant and caused by mutations in TRPV3 (transient receptor potential vanilloid 3, MIM: 607066) (Lin et al., 2012). Most forms of erythrokeratoderma (EK) are also dominant and caused by mutations in GJB3, GJB4, and GJA1 (connexins 31, 30.3, and 43, MIM: 603324, 605425, and 121024) (Boyden et al., 2015, Macari et al., 2000, Richard et al., 1998), LOR (loricrin, MIM: 152445) (Maestrini et al., 1996), or DSP (desmoplakin, MIM: 125647) (Boyden et al., 2016), though recessive forms of EK caused by mutations in ABHD5 (abhydrolase domain containing 5, MIM: 604780) (Lefèvre et al., 2001, Pujol et al., 2005) and KDSR (3-ketodihydrosphingosine reductase, MIM: 136440) (Boyden et al., 2017) have been described.
Other disorders featuring PPK are caused by disruption of desmosomal proteins. Desmosomes are intercellular junctions crucial for cell-cell adhesion, tissue integrity, and epidermal differentiation. They interact with microtubules and mediate localization and function of gap junctions. Genes encoding desmosomal proteins, including cadherins that when mutated cause PPK include DSG1 and DSC2 (cadherins desmoglein 1 and desmocollin 2, MIM: 125670 and 125645) (Gerull et al., 2013, Rickman et al., 1999, Samuelov et al., 2013, Simpson et al., 2009), PKP1 and JUP (armadillo proteins plakophilin 1 and junction plakoglobin, MIM: 601975 and 173325) (McGrath et al., 1997, McKoy et al., 2000), and DSP (Boyden et al., 2016). Hair abnormalities and cardiomyopathy are additional features common among desmosomal disorders.
RESULTS
In ongoing efforts to discover new genetic causes of DOK, including OS and EK, cohorts at INSERM in France and Yale University in the USA were screened for mutations in previously associated genes, followed by exome sequencing of probands without identified pathogenic mutations. Via these analyses, four OS subjects were found to be heterozygous for a nonsense mutation in the last exon of PERP (p.Trp151* or p.Tyr153*), and one EK subject was found to be homozygous for a frameshift mutation in the first exon of PERP (p.Ser38Leufs*52) (Figure 1, Supplementary Table 1, Supplementary Figures S1 and S2).
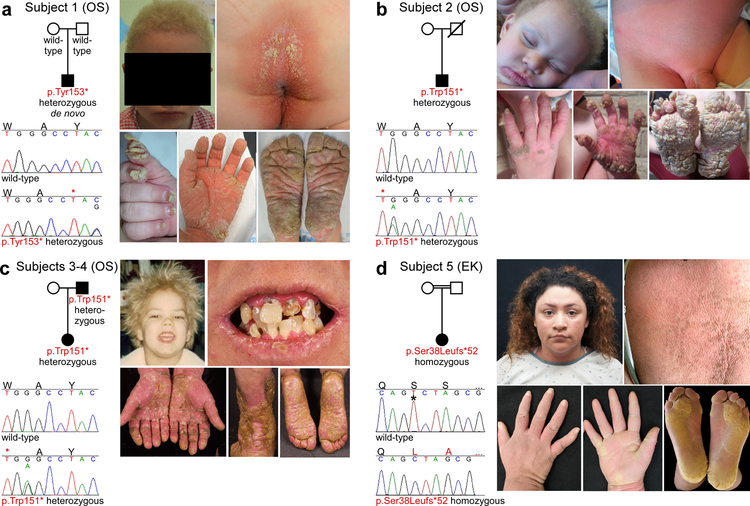
Figure 1. Clinical features and PERP mutations observed in OS and EK subjects.
(a) Subject 1 is heterozygous for PERP p.Tyr153*. He has woolly yellow hair, periorificial keratotic plaques, cheilitis, dystrophic nails, and transgrediens PPK. (b) Subject 2 is heterozygous for PERP p.Trp151*. He has woolly yellow hair, periorificial keratotic plaques, pink-red keratoderma in the diaper area, transgrediens PPK with horny projections, and dystrophic nails. (c) Subjects 3 and 4 (affected father shown) are heterozygous for PERP p.Trp151*. Subject 4 had fine yellow hair and his secondary teeth were prone to caries (shown at age 38), with cheilitis and pink perioral plaques. Cobblestone-like hyperkeratosis over the ankles and transgrediens red keratoderma with yellow scale is present. (d) Subject 5 is homozygous for PERP p.Ser38Leufs*52 (c.112delT, * indicates deleted base). She has woolly hair, generalized erythrokeratoderma, and transgrediens PPK, with thick yellow keratoderma..
Subjects 1–4 presented with OS features. Subject 1 is from the French cohort and is heterozygous for p.Tyr153* (c.459C>G). He is age 6, the second child of unaffected Spanish parents, and presented at age 5 months with cheilitis, periorificial keratotic plaques, keratotic papules on the ear at the junction of the lobule and tragus, hyperkeratotic plaques on the buttocks, transgrediens pink-red PPK which progressed over time to thick and fissured yellow plaques on the fingertips and palms at sites of pressure, hyperkeratotic nails, and woolly yellow hair. Skin in other areas is unaffected, he has no related pain, and teeth, sweating, and heart ultrasound are normal. Subject 2 is from the USA cohort and is heterozygous for p.Trp151* (c.452G>A). He is age 3, born to unaffected Mexican parents, and presented at a walk-in clinic in Arizona with massive, transgrediens PPK extending to the dorsum of wrists and ankles, with yellow horn-like projections present confluently on the feet and on pressure-bearing areas of the fingers, palms, and over the metacarpophalangeal joints. Marked cheilitis, periorificial keratotic plaques, hyperkeratotic plaques on armpits, buttocks, abdomen, groin, and genitals, hyperkeratotic nails, scant eyebrows, and woolly yellow hair strikingly similar to that of Subject 1 are present. There is well-demarcated pink-red keratoderma extending to the lower abdomen, through the inguinal fold to the proximal thighs. His father died in his 20s, preventing assessment of de novo status. Subjects 3 and 4 are a proband and her affected father from the USA cohort. Like Subject 2 they are heterozygous for p.Trp151*, but the nucleotide mutation is different (c.453G>A). Subject 3 is age 4 and presented within the first month of life with pink-red thickening of the palms and soles, which has progressed to confluent palmoplantar keratoderma with thicker scale and fissures developing on pressure-bearing areas. Her hair is yellow, fine, and curly. Her primary teeth are normal and she has chronic cheilitis with small periorificial plaques. Subject 4 is age 40 and had onset of scaling of palms and soles in early childhood, with progression to thick, fissured, cobblestone-like hyperkeratosis on palms and soles which has slowly extended to the ankles, distal shins, and distal calves. He has experienced chronic cheilitis with intermittent periorificial plaques. While his primary teeth were normal he rapidly developed severe caries in his secondary teeth, necessitating extraction of all teeth at age 38. A comprehensive cardiac evaluation at age 39 including echocardiogram showed no evidence of cardiomyopathy. His hair was yellow, fine, and curly as a child, and became darker when he was a teen but remained woolly.
Yellow hair is not a consistent feature of OS, and we know of no cases with both yellow hair and a pathogenic TRPV3 mutation. Genotyping of additional OS subjects for mutations in TRPV3 and PERP will permit determination of whether this is a distinguishing and consistent feature for OS caused by PERP mutation. While there is precedent for dental enamel defects due to mutations in genes encoding desmosomal proteins (Boyden et al., 2016), we have observed only adolescent onset in the secondary teeth of our oldest subject (Subject 4) with a p.Trp151* PERP mutation. Other subjects with dominant mutations are children, and further study will reveal whether this is a defining characteristic of this disorder in adulthood.
In contrast, Subject 5 presented with erythrokeratoderma affecting the entire body. Subject 5 is from the USA cohort and is homozygous for p.Ser38Leufs*52. She was born to consanguineous unaffected Hispanic parents, with woolly hair but normal skin at birth. In the first weeks of life her skin became thickened, red, and rough. When she began to crawl she developed thickening of palms and soles which progressed to thick, yellow hyperkeratosis predominantly on pressure areas of soles, with linear accentuation on the palms. She has normal fingernails but dystrophic toenails. Now age 22, she has recurrent dermatophyte infections, anhidrosis, and normal teeth. A recent echocardiogram was normal. While her PPK and woolly hair bear striking similarities to Subjects 1–4, they are less severe, and she does not have a hair pigmentation abnormality. The erythrokeratoderma affecting virtually all of her skin further distinguishes her EK phenotype from their OS phenotype. Histology of affected skin from Subjects 1 and 5 showed thickening of the epidermis (acanthosis) and a thickened stratum corneum (hyperkeratosis), consistent with diagnoses of OS and EK (Figure 2).
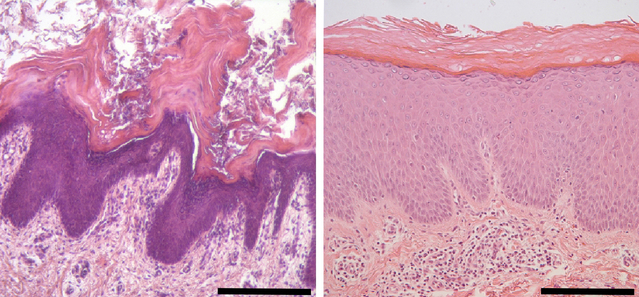
Figure 2. Affected skin of OS and EK subjects with PERP mutations shows marked epidermal hyperplasia.
Hematoxylin-eosin staining of affected skin from Subjects 1 and 5; biopsies used here and throughout were from the buttock at age 1 and the upper thigh at age 22, respectively. Subject 1 (on left) shows hyperplastic epidermis with acanthosis, papillomatosis with elongated rete ridges, hypergranulosis, and orthokeratotic hyperkeratosis. There are some areas of parakeratosis, and no acantholysis. Subject 5 (on right) similarly displays acanthosis, papillomatosis, hypergranulosis, and a compact orthohyperkeratosis. Scale bars are 400 μm.
PERP (p53 effector related to PMP22, MIM: 609301, GenBank: NM_022121 and NP_071404) is a three exon gene encoding a p53/p63 tetraspan membrane protein that is both an apoptosis mediator and a component of desmosomes and other cell junctions (Attardi et al., 2000, Franke et al., 2013, Ihrie et al., 2005, Ihrie et al., 2003). Perp knockout (−/−) mice exhibit hyperproliferative skin and severe blistering symptomatic of compromised adhesion, with postnatal lethality that is partially dependent on strain background (Ihrie et al., 2005). The Perp −/−mice that survive to adulthood display confluent thickened and scaly skin (erythrokeratoderma), PPK, patchy and disorganized fur, and abnormal nails. These features bear striking similarity to those observed in our OS and EK subjects with PERP mutations. They are also reminiscent of the spectrum of skin and dental phenotypes caused by mutations in another essential desmosome component, DSP, which range from severe lethal blistering, to widespread erythrokeratoderma, to PPK alone, depending on the location, type, and zygosity of mutation (Boyden et al., 2016).
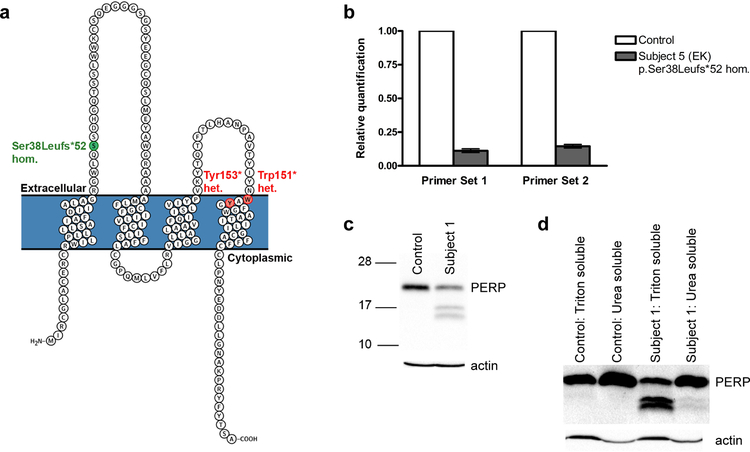
To delineate molecular mechanisms consistent with recessive EK in a subject with a homozygous N-terminal PERP frameshift and dominant OS in two subjects with heterozygous C-terminal PERP terminations (Figure 3a), we considered both in vitro and in vivo data. To conclusively demonstrate that the homozygous p.Ser38Leufs*52 leads to loss of function, we performed qRT-PCR with keratinocyte RNA from EK Subject 5 and observed marked reduction of PERP expression, consistent with nonsense-mediated decay (Figure 3b). In contrast, it is highly unlikely that OS in subjects with the heterozygous p.Trp151* and p.Tyr153* mutations is due to haploinsufficiency, given that Subject 5’s unaffected, consanguineous parents are obligate heterozygotes for p.Ser38Leufs*52, genomic databases contain control subjects heterozygous for PERP truncating mutations (Karczewski et al., 2017), and Perp +/− mice have no discernible phenotype. Given the mutation constraint demonstrated by Subjects 1–4, which suggests that expression of mutant PERP truncated at a specific site is critical to the OS phenotype, we propose that EK and OS caused by PERP mutations have distinct pathobiology.
Figure 3. Effects of a homozygous N-terminal frameshift mutation and heterozygous C- terminal nonsense mutations on PERP mRNA and protein, respectively.
(a) A PERP schematic shows heterozygous (het.) nonsense mutations (Subjects 1–4, OS, red) and the homozygous (hom.) frameshift mutation (Subject 5, EK, green). (b) Quantitative RT-PCR of Subject 5 (p.Ser38Leufs*52 homozygote) keratinocyte RNA shows marked reduction of PERP transcripts, consistent with nonsense-mediated decay. (c-d) Western blots of whole cell (c) and Triton- and urea-soluble (d) Subject 1 and control (age 22, abdominal biopsy) keratinocyte lysates stained for PERP, with actin as loading control. The control displays a band of expected size (~21 kDa), while Subject 1 (p.Tyr153* heterozygote, size prediction ~17 kDa), shows two additional bands (~16 and 17 kDa), enriched in the Triton-soluble fraction.
We further examined the mechanism of dominant OS mutations experimentally, in cells from Subject 1, by examining expression of p.Tyr153* mutant PERP and its subcellular localization, size, and solubility profile. qRT-PCR demonstrated no deficiency in PERP RNA, consistent with terminal exon mutations escaping nonsense-mediated decay (Supplementary Figure S3). PERP immunostaining of keratinocytes from Subject 1 showed normal membrane localization (Supplementary Figure S4). Immunoblotting for PERP demonstrated lower molecular weight bands in Subject 1 keratinocyte lysates, consistent with expression of a truncated form (Figure 3c). These bands are enriched in the Triton soluble fraction (Figure 3d), suggesting that the mutant protein may not be stably incorporated into desmosomes. Dephosphorylation and deglycosylation experiments did not modify the apparent molecular weight of these bands (data not shown). Other desmosomal proteins showed normal solubility profiles consistent with stable incorporation; components of adherens and tight junctions (E-cadherin and claudin 1) also appeared stable (Supplementary Figure S5).
The specific interactions between PERP and desmosomal components, the role of the different PERP protein domains, and the exact function of PERP in desmosomes are yet unknown. Therefore, delineation of the mechanism(s) by which PERP mutations cause OS will require further investigation. PERP mutations in OS could have a gain-of-function, a dominant-negative effect, or a more complex mechanism. There is precedent for C-terminal truncations causing gain-of-function (e.g. epithelial sodium channel mutations that cause hypertension by disrupting removal of the channel from the membrane and thereby increasing renal sodium absorption) (Shimkets et al., 1994, Snyder et al., 1995), and for putative gain-of-function mutations leading to a similar phenotype as definitive loss-of-function mutations in the same gene (Srour et al., 2013). In this case, gain-of-function seems inherently unlikely due to the substantial phenotypic overlap with the recessive loss-of-function disorder. Yet a simple dominant-negative mechanism is not fully satisfactory, given that the OS and EK phenotypes bear phenotypic differences beyond degree of severity. A dominant-negative and neomorphic effect for the heterozygous, tightly clustered C-terminal truncating mutations seems most consistent with these observations. Additional investigation will be necessary to more comprehensively delineate the mechanism of dominant PERP mutations.
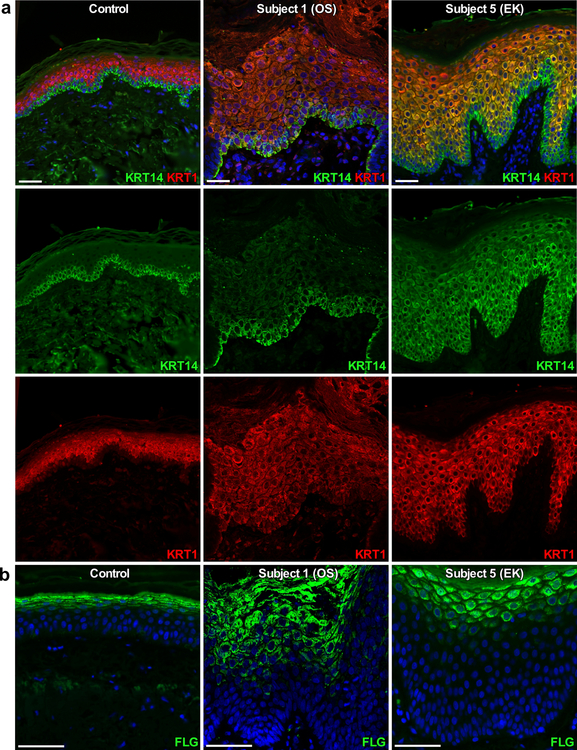
Perp −/− mice display unaltered localization of most desmosomal proteins, though Dsp is more diffuse and intracellul−ar, and Krt14, a marker of basal keratinocytes, is expanded (Ihrie et al., 2005). To further investigate the effects of PERP mutations in our subjects on desmosomes and epidermal development, we immunostained skin biopsies for desmosomal components and markers of epidermal differentiation (Figure 4 and Supplementary Figures S6 and S7). Both OS Subject 1 and EK Subject 5 demonstrate expanded FLG but normal, suprabasal expression of KRT1. KRT1, like KRT10, is an intermediate filament protein and marker of suprabasal keratinocyte differentiation. Furthermore, in contrast to OS Subject 1, EK Subject 5 also shows expansion of KRT14, similar to observations in Perp −/− mice (Ihrie et al., 2005) and other EK disorders, and involucrin staining was present in all suprabasal layers (Boyden et al., 2017). Loricirin staining was localized to the upper granular layers in Subject 1 and Subject 5. However, in Subject 5, increased nuclear LOR localization is observed, similar to previous findings in subjects with keratoderma due to LOR mutation and in mice with keratoderma caused by epidermal-specific suppression of activator protein 1 transcription factor expression. In this mouse model, the keratoderma phenotype is remarkably similar to that of human subjects with LOR mutations and is unchanged when generated in mice lacking LOR, suggesting that nuclear LOR localization, may be a feature of some keratodermas but is not necessary to the pathogenesis (Rorke et al., 2015). Overall, these results suggest a potential defect in terminal differentiation in Subjects 1 and 5. Neither subject demonstrated evidence of abnormal localization of desmosome or other intercellular junction components. Staining for c-Kit and the proliferation marker Ki67 also revealed that both probands exhibit significantly increased mast cell counts in the dermis, and greater percentages of proliferating cells in the basal layer of interfollicular epidermis (Supplementary Figure S8), consistent with hyperproliferation. A similar phenotype is observed in the adult skin of Perp −/− mice, which is hyperproliferative, commonly with infiltrating immune cells, skin inflammation, and susceptibility to infection (Ihrie et al., 2006).
Figure 4. Immunostaining of skin from subjects with PERP mutations for markers of epidermal differentiation.
DAPI nuclear counterstain is in blue; scale bars are 50 μm. Left panels are control tissue (age 32, abdominal biopsy), middle panels are OS Subject 1 (p.Tyr153* heterozygote), and right panels are EK Subject 5 (p.Ser38Leufs*52 homozygote). (a) Positive immunostaining for keratin 14 (KRT14, green) is limited to the epidermal basal layer in the control and Subject 1 but is expanded in Subject 5, while positive immunostaining for keratin 1 (KRT1, red) is limited to suprabasal epidermis in both control and Subjects 1 and 5. (b) Immunostaining for filaggrin (FLG, green) is tightly restricted to the epidermal granular layer in the control, but is expanded in Subjects 1 and 5.
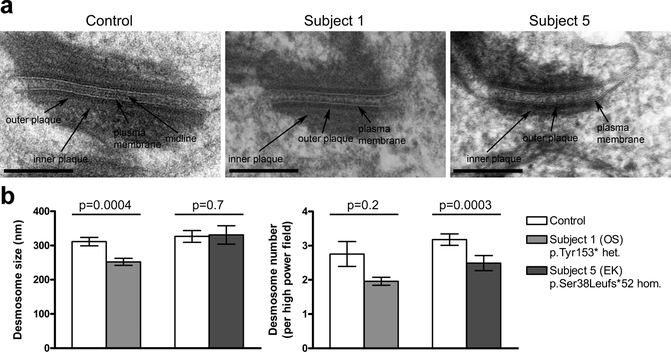
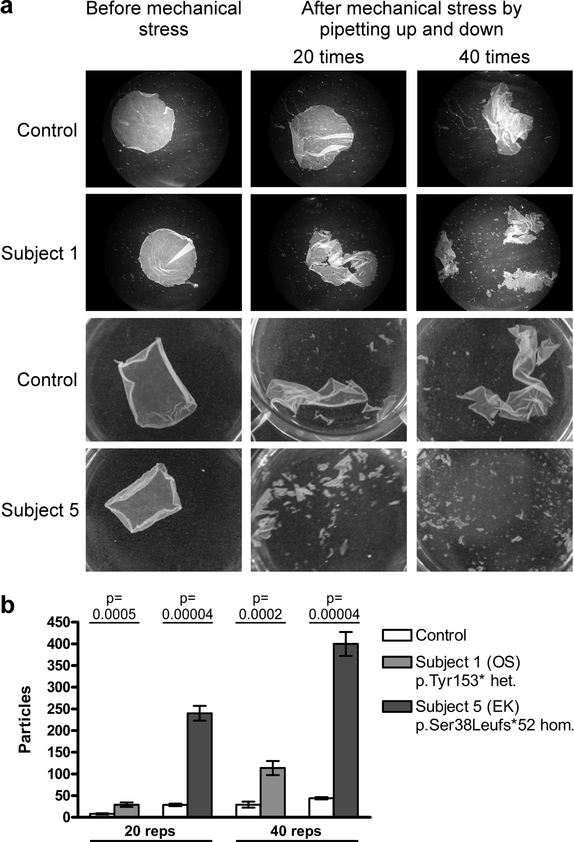
Desmosome abnormalities in Perp −/− mice include greater width, lower electron density, fewer numbers, and aberrant cytoskeletal connections (Ihrie et al., 2005). To characterize desmosomal defects in our subjects with PERP mutations we examined skin from OS Subject 1 and EK Subject 5 by electron microscopy. In both subjects, desmosomes lack the electron-dense midline (EDM) normally present in mature desmosomes (Figure 5a and Supplementary Figure S9a), indicative of an immature state associated with conditions including wound healing, tumor invasion, or keratinocyte mitosis (Brooke et al., 2012). Other desmosomal features, including inner plaques and keratin filament attachments, appeared intact. In the lower suprabasal layer, morphometric analysis demonstrated shorter outer plaque length in Subject 1, and a reduction in numbers of desmosomes in Subject 5 (Figure 5b). In the upper suprabasal layer, desmosome frequency was modestly decreased in Subject 1; there were no significant differences in interplaque width in either layer (Supplementary Figure S9b). Desmosomal adhesive function was investigated by dispase mechanical dissociation assay in keratinocytes cultured from skin biopsies of OS Subject 1 and EK Subject 5. For both subjects increased fragmentation after mechanical stress was observed (Figure 6).
Figure 5. Affected skin from OS and EK subjects with PERP mutations shows desmosomal defects in the lower suprabasal layers.
(a) Electron microscopy shows that desmosomes in skin of both probands lack the dense midline seen in control skin. Scale bars are 200 nm. Additional images are shown in Supplementary Figure 9a. (b) Desmosomes in Subject 1 show a significant reduction in size, while desmosomes in Subject 5 are significantly reduced in number. For controls for Subjects 1 and 5, biopsies were from the dorsal foot of an 8 year old and the breast of a 23 year old, respectively.
Figure 6. Increased mechanical dissociation of keratinocyte monolayers from OS and EK subjects with PERP mutations demonstrates defective intercellular adhesion.
(a) Images of keratinocyte monolayers after repeated pipetting (20 or 40 repetitions) show increased stress- induced fragmentation in both probands. (b) Particle counts quantify the significantly increased dissociation of keratinocyte monolayers in probands. For controls for Subjects 1 and 5, keratinocytes were derived from biopsies from the abdomen of a 22 year old and the shoulder of a 28 year old, respectively.
Interestingly, despite evidence that Perp is a p53/p63-regulated apoptosis mediator (Attardi et al., 2000, Ihrie et al., 2005, Ihrie et al., 2003), Perp −/− mice show no detectable alterations in apoptosis in skin (Ihrie et al., 2005, Marques et al., 2005), are not predisposed to spontaneous tumorigenesis (Ihrie et al., 2006), and are resistant to papilloma development (Marques et al., 2005). However, mice with inducible, epidermis-specific deletion of Perp develop earlier and more numerous squamous cell carcinomas upon chronic UVB irradiation, with defects in UVB- induced apoptosis (Beaudry et al., 2010). Staining of skin from OS Subject 1 for activated caspase 3 revealed no detectable apoptotic cells, similar to control skin (Supplementary Figure S10), and to skin of Perp −/− mice (Ihrie et al., 2005, Marques et al., 2005), but in contrast with OS patients with TRPV3 mutations (Lin et al., 2012).
DISCUSSION
Desmosome abnormalities have been reported in other disorders featuring PPK, including recessive skin fragility syndrome, recessive Naxos disease, and dominant tylosis with esophageal cancer, caused by mutations in PKP1, JUP, and RHBDF2 (rhomboid 5 homolog 2) respectively (Blaydon et al., 2012, McGrath et al., 1997, McKoy et al., 2000). Desmosomes in PKP1-null human skin show altered size and reduced number, with less prominent EDMs (McMillan et al., 2003). Those in Jup-null mouse skin are fewer in number, with structural abnormalities including reduced EDMs (Bierkamp et al., 1999), and those in RHBDF2-mutant human skin lack the EDM (Brooke et al., 2014). Formation of the EDM is central to desmosome adhesive strength, and the importance of Perp to intercellular adhesion has been previously investigated with murine experiments utilizing autoantibodies from the blistering disorder pemphigus vulgaris, which disrupt desmosomal adhesion. When these antibodies are applied to Perp −/− keratinocytes, intercellular adhesion is significantly more impaired than with wild-type cells (Nguyen et al., 2009). Our findings provide further evidence for the role of PERP in desmosomal adhesion. It is possible that other functions of PERP are also affected by the dominant mutations we identify in OS subjects.
The observation of three unrelated OS probands heterozygous for different C-terminal truncating PERP mutations within two residues of each other, one which is de novo and one which is inherited from an affected parent, conclusively demonstrates that these mutations cause OS. An EK subject homozygous for an N-terminal truncating PERP mutation, genetically and phenotypically analogous to the Perp −/− mouse, provides compelling evidence that distinct classes of mutation in PERP cause both dominant and recessive keratinization disorders with partial phenotypic overlap. The consistent features of PPK and woolly hair are hallmarks of desmosomal abnormalities, which display impressively broad phenotypic and genotypic spectrums. Consequently, the demonstration of specific structural abnormalities and adhesion defects in both OS and EK subjects with PERP mutations, despite normal localization of other desmosomal and junctional components, confirms the critical role of desmosomes in these disorders, adds to the wider understanding of skin disease caused by mutations in genes encoding desmosome-associated proteins, and establishes the integral importance of PERP in human skin. These discoveries further highlight the utility of an unbiased genetic approach to DOK.
MATERIALS AND METHODS
Study subjects (Subject 1).
The CPP Ile-de-France Paris approved the study protocol. Subjects provided written informed consent. The proband and his parents provided a blood sample. The proband provided punch biopsies of skin from the buttock. De-identified normal skin from surgical margins was obtained for controls.
Study subjects (Subjects 2–5).
The Yale Human Investigation Committee approved the study protocol. Subjects provided verbal and written informed consent. Each subject provided a blood sample. Subject 5 provided punch biopsies of skin from the hip. De-identified normal skin discarded after skin cancer excisions was obtained for controls.
Exome sequencing and mutation confirmation (Subject 1).
Genomic DNA from the subject and his parents was isolated from peripheral blood using a standard phenol chloroform protocol. DNA purification was performed using Amicon Ultra 30K centrifugal filter units (Millipore). Bar-coded DNA libraries were prepared and exome capture (Agilent 50 Mb SureSelect Human All Exon V3) and sequencing (Life Technologies SOLiD5500) was performed for the trio. Resulting reads (75 bp) were generated with Exact Call Chemistry and aligned to the human reference sequence (hg19) with LifeScope (Life Technologies), and variants were called with GATK and annotated with in-house software (PolyWeb). Mean coverage was 76–92x, with 86− 87% of bases covered >15x. Variants present in databases (dbSNP, 1000 Genomes, Exome Sequencing Project) or previously observed in 1029 in-house exomes were excluded and de novo coding or splicing variants predicted to be damaging by SIFT (Kumar et al., 2009) or PolyPhen-2 (Adzhubei et al., 2013) were verified via Sanger sequencing with standard protocols (Applied Biosystems 3130xl). Additional prediction tools LRT (Chun and Fay, 2009) and MutationTaster (Schwarz et al., 2010) were used.
Exome sequencing and mutation confirmation (Subjects 2–5).
Genomic DNA was isolated from peripheral blood using a standard phenol-chloroform protocol. Bar-coded DNA libraries were prepared and exome capture (Roche EZ Exome 2.0) and sequencing (Illumina HiSeq) was performed by the Yale Center for Genome Analysis. Resulting reads (99 and 74 bp respectively) were aligned to the human reference sequence (hg38) with BWA-MEM (Li, 2013) and variants were called with the Genome Analysis Toolkit (GATK) (McKenna et al., 2010) and annotated with Annovar (Wang et al., 2010) and Variant Effect Predictor (McLaren et al., 2016). For Subjects 2, 3, and 5 mean coverage was 53x, 48x, and 49x, with 89%, 87%, and 74% of bases covered ≥20x, respectively. Frequency of variants was assessed with ExAC (Karczewski et al., 2017), dbSNP (Sherry et al., 2001), 1000 Genomes (Auton et al., 2015), and in-house control exomes. Aligned reads were examined with the Broad Institute Integrative Genomics Viewer (IGV) (Robinson et al., 2011). Verification of mutations was performed via PCR using KAPA2G Fast polymerase (Kapa Biosystems), agarose gel fractionation and visualization, and Sanger sequencing. Primers were designed with ExonPrimer (Rosenbloom et al., 2015, Untergasser et al., 2012) and SNPmasker (Andreson et al., 2006).
Cell culture.
4 mm punch biopsies were explanted to isolate primary keratinocytes, which were maintained in Green medium on a feeder layer of lethally irradiated 3T3 mouse fibroblasts as described previously (Barrandon and Green, 1987). For experiments, keratinocytes were grown in 0.06 mM CaCl2 EpiLife medium (Invitrogen) to 70–80% confluence and then incubated in 1.2 mM CaCl2 EpiLife medium for up to 48 hours to induce formation of desmosomes.
RT-PCR (Subject 1).
Confluent cultured keratinocytes were harvested, RNA was prepared via RNeasy Mini (Qiagen), and cDNA was prepared with random hexamer priming and M-MuLV reverse transcriptase (Fermentas). PCR was performed in triplicate for each sample with qPCR MesaGreen Mastermix (Eurogentec) on a 7500 Sequence Detection or ViiA 7 Real-Time PCR system (Applied Biosystems). The experiment was replicated three times.
RT-PCR (Subject 5).
Keratinocytes grown in one well of a six-well plate were harvested, RNA was prepared via RNA Allprep (Qiagen 80004), and cDNA was prepared with oligo(dT) priming and SuperScript III reverse transcriptase (Invitrogen 18080–093). PCR was performed in triplicate for each sample with Power SYBR Green Master Mix (Applied Biosystems 4367659) on a 7500 Sequence Detection system (Applied Biosystems). The experiment was replicated three times.
RT-PCR primers.
Quantitative PCR was performed with PERP sets 1 and 2 and normalization to PGK. Presence of p.Tyr153* in mRNA was validated with PERP set 3. PERP (5’–3’): F1=CTACGAGGAGGGCTGTCAGA, R1=GCGAAGAAGGAGAGGATGAA; F2=GACCCCAGATGCTTGTCTTC, R2=GCATGAAGGGTGAAGGTCTG; F3=GCCTCTTCGCTTTTGTGG, R3=TCAAAGTCGCCTGGAGAAAC. PGK (5’–3’): F=GTGTGCCCATGCCTGACA, R=TGGGCCTACACAGTCCTTCAA.
Immunoblotting.
Keratinocytes were lysed in buffer (20 mM pH 8 Tris-HCl, 150 mM NaCl, 1% Triton X-100, 0.5% Nonidet P-40, 1 mM pH 8 EDTA pH8, Complete protease inhibitors [Roche]) and rocked for one hour at 4°C. Total protein was isolated by lysis in the above buffer with 9 M urea. Triton-soluble fractions were isolated by centrifugation at 13000 g at 4°C for 20 minutes. Urea-soluble fractions were isolated by resuspension of insoluble material from this preparation in 9 M urea buffer, incubation for 3 hours at room temperature, and centrifugation at 14000 g at room temperature for 30 min. Protein quantification was performed by Bradford assay. 30 μg samples were loaded in Laemmli buffer (62.5 mM pH 6.8 Tris HCl, 5% β- mercaptoethanol, 2% SDS, 10% glycerol, 0.002% bromophenol blue), separated by SDS-PAGE, transferred onto Hybond-ECL membrane (GE Healthcare), and incubated with primary and secondary antibodies, with chemiluminescence detection (ECL Plus Western Blotting Substrate, Pierce). b-actin served as loading control. Experiments were replicated three times.
Immunocytochemistry.
Keratinocytes were cultured on coverslips and fixed in ice-cold methanol for 10 minutes at -20°C or 4% paraformaldehyde for 30 minutes at room temperature. Paraformaldehyde fixed cells were permeabilized with 0.2% TritonX-100 in PBS for 20 minutes. For PERP staining cells were treated with 10 μg/μl Proteinase K for 3 minutes at room temperature. After blocking with 1% fetal bovine serum in PBS, coverslips were incubated with primary antibody overnight at 4°C and Alexa-conjugated secondary antibody for 1 hour at room temperature, mounted on glass slides with Mowiol, and examined with a confocal microscope. Experiments were replicated three times.
Immunohistochemistry (Subject 1).
5 μm FFPE tissue sections were deparaffinized with xylene, and rehydrated with a series of alcohol solutions. Antigen retrieval was performed by boiling in 10 mM pH 6.0 citrate buffer for 20 minutes. Slides were incubated with primary antibody for 1 hour at room temperature and horseradish peroxidase conjugate was detected with an Envision kit (Dako). Counterstain with Mayer’s hematoxylin was performed before mounting with Eukitt quick-hardening mounting medium (Sigma). For immunofluorescent staining slides were incubated with primary antibody overnight at 4°C and with secondary antibody for 60 minutes at room temperature, followed by nuclei staining with 1 μg/ml DAPI and Mowiol mounting.
Immunohistochemistry (Subject 5).
5 μm FFPE tissue sections were deparaffinized with a xylene-ethanol gradient and rinsed in PBS. Antigen retrieval was performed by immersion in modified pH 6.0 citrate buffer for 20 minutes in a steamer. Slides were cooled and rinsed in PBS, and blocked with 10% donkey serum / 1% BSA for 1 hour at room temperature. Slides were incubated with primary antibody overnight at 4°C, washed 3 times with PBS, incubated with secondary antibody for 1 hour at room temperature, and washed 3 times with PBS. Nuclei were stained with 200 ng/ml DAPI before mounting with Mowiol / 1% n-propyl gallate (Sigma- Aldrich 02370).
Antibodies (Subject 1).
Primary antibodies included anti-PERP (N-terminal, Atlas HPA022269; C-terminal, previously reported (Franke et al., 2013)), anti-c-Kit (1:500, Dako A4502), anti-Ki67 (Novocastra RTU-Ki67-MM1), anti-KRT14 (1:2000, Novocastra NCL-L-LL002), anti-KRT1 (1:800, Abcam ab83664), anti-FLG (1:300, Covance PRB-417P), anti-LOR (1:500, Covance PRB-145P) anti-IVL (1:400, Sigma I 9018), anti-DSG1 (immunoblotting, Santa Cruz sc-20114; immunocytochemistry, Abcam ab122913), anti-DSG3 (Life Technologies 32–6300), anti-DSC3 (Progen 6J193), anti-JUP (Santa Cruz sc-8415), anti-DSP (Santa Cruz sc-33555), anti-CDH1 (Cell Signaling 3195), anti-CLDN1 (immunoblotting Santa Cruz sc-166338; immunocytochemistry Abcam ab15098) and anti-cleaved-caspase3 (1:50 Cell Signaling 9661).
Antibodies (Subject 5).
Primary antibodies included rabbit anti-c-Kit (1:500, Dako A4502), rabbit anti-Ki67 (1:300, Abcam ab15580), mouse anti-KRT14 (1:100, Santa Cruz Biotechnology sc-53253), rabbit anti-KRT1 (1:500, Covance PRB-149), goat anti-FLG (1:200, Santa Cruz Biotechnology sc-25897), mouse anti-DSG1 (1:10, Progen 652110), rabbit anti-JUP (1:100, Abcam ab15153), guinea pig anti-DSP (1:50, Progen DP-1), mouse anti-IVL (1:100, ThermoFisher Scientific MA5–11803), and rabbit anti-LOR (1:100, Abcam ab24722). Secondary antibodies were Cy3 donkey anti-rabbit, anti-mouse, and anti-guinea pig IgG and Cy2 donkey anti-goat IgG (1:1000, Jackson Immunoresearch 711-165-152, 715-165-151, 706-165-148, and 705–225-147 respectively).
Cell counting.
For c-Kit, the number of mast cells was counted relative to dermis area. For Ki67, the numbers of labelled cells was counted as a percentage of total basal cells in each 40X field. At least 10 fields were analyzed.
Electron microscopy.
Tissue was fixed by immersion in 2–2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.3–7.4) with rocking for 24–48 hours at 4°C, dehydrated, and embedded in resin. Age-matched controls were used. The analytical methods were described previously (McMillan et al., 2003).
Dispase mechanical dissociation assay.
Keratinocytes were incubated with 2.4 U/ml dispase (BD Biosciences) at 37°C to lift keratinocyte sheets. Released keratinocyte monolayers were subjected to mechanical stress by pipetting as previously described (Ishii et al., 2005).
Statistical analyses.
Statistical significance of observed differences was assessed by two-tailed Mann-Whitney U tests. Means with s.e.m. are shown.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the study subjects and their families for participation in this study. We thank Marina Simon, Laetitia Furio, Irina Tikhonova, Christopher Castaldi, Theodore Zaki, and Kaya Bilguvar for technical assistance, and the Imagine Institute Cell Imaging Core Facility and Professor Franke for the PERP antibody. This work was supported by MEXT KAKENHI (24591620 to A. I.-Y.), Fondation ARC pour la recherche sur le cancer, the Foundation for Ichthyosis and Related Skin Types, the National Institutes of Health (U54 HG006504 to the Yale Center for Mendelian Genomics and R01 AR068392 to K.A.C.) and the Association Ichtyose France (to A.H.).
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
REFERENCES
- Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet 2013;Chapter 7:Unit7.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreson R, Puurand T, Remm M. SNPmasker: automatic masking of SNPs and repeats across eukaryotic genomes. Nucleic Acids Res 2006;34(Web Server issue):W651–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardi LD, Reczek EE, Cosmas C, Demicco EG, McCurrach ME, Lowe SW, et al. PERP, an apoptosis- associated target of p53, is a novel member of the PMP-22/gas3 family. Genes Dev 2000;14(6):704–18. [PMC free article] [PubMed] [Google Scholar]
- Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. A global reference for human genetic variation. Nature 2015;526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrandon Y, Green H. Three clonal types of keratinocyte with different capacities for multiplication. Proc Natl Acad Sci U S A 1987;84(8):2302–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudry VG, Jiang D, Dusek RL, Park EJ, Knezevich S, Ridd K, et al. Loss of the p53/p63 regulated desmosomal protein Perp promotes tumorigenesis. PLoS Genet 2010;6(10):e1001168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierkamp C, Schwarz H, Huber O, Kemler R. Desmosomal localization of beta-catenin in the skin of plakoglobin null-mutant mice. Development 1999;126(2):371–81. [DOI] [PubMed] [Google Scholar]
- Blaydon DC, Etheridge SL, Risk JM, Hennies HC, Gay LJ, Carroll R, et al. RHBDF2 mutations are associated with tylosis, a familial esophageal cancer syndrome. Am J Hum Genet 2012;90(2):340–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden LM, Craiglow BG, Zhou J, Hu R, Loring EC, Morel KD, et al. Dominant De Novo Mutations in GJA1 Cause Erythrokeratodermia Variabilis et Progressiva, without Features of Oculodentodigital Dysplasia. J Invest Dermatol 2015;135(6):1540–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden LM, Kam CY, Hernández-Martín A, Zhou J, Craiglow BG, Sidbury R, et al. Dominant de novo DSP mutations cause erythrokeratodermia-cardiomyopathy syndrome. Hum Mol Genet 2016;25(2):348–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden LM, Vincent NG, Zhou J, Hu R, Craiglow BG, Bayliss SJ, et al. Mutations in KDSR Cause Recessive Progressive Symmetric Erythrokeratoderma. American journal of human genetics 2017;100(6):978–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooke MA, Etheridge SL, Kaplan N, Simpson C, O’Toole EA, Ishida-Yamamoto A, et al. iRHOM2- dependent regulation of ADAM17 in cutaneous disease and epidermal barrier function. Hum Mol Genet 2014;23(15):4064–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooke MA, Nitoiu D, Kelsell DP. Cell-cell connectivity: desmosomes and disease. J Pathol 2012;226(2):158–71. [DOI] [PubMed] [Google Scholar]
- Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res 2009;19(9):1553–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchatelet S, Hovnanian A. Olmsted syndrome: clinical, molecular and therapeutic aspects. Orphanet J Rare Dis 2015;10:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke WW, Heid H, Zimbelmann R, Kuhn C, Winter-Simanowski S, Dörflinger Y, et al. Transmembrane protein PERP is a component of tessellate junctions and of other junctional and non-junctional plasma membrane regions in diverse epithelial and epithelium-derived cells. Cell Tissue Res 2013;353(1):99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerull B, Kirchner F, Chong JX, Tagoe J, Chandrasekharan K, Strohm O, et al. Homozygous founder mutation in desmocollin-2 (DSC2) causes arrhythmogenic cardiomyopathy in the Hutterite population. Circ Cardiovasc Genet 2013;6(4):327–36. [DOI] [PubMed] [Google Scholar]
- Hirano SA, Harvey VM. From progressive symmetric erythrokeratoderma to erythrokeratoderma variabilis progressiva. J Am Acad Dermatol 2011;64(5):e81–2. [DOI] [PubMed] [Google Scholar]
- Ihrie RA, Bronson RT, Attardi LD. Adult mice lacking the p53/p63 target gene Perp are not predisposed to spontaneous tumorigenesis but display features of ectodermal dysplasia syndromes. Cell Death Differ 2006;13(9):1614–8. [DOI] [PubMed] [Google Scholar]
- Ihrie RA, Marques MR, Nguyen BT, Horner JS, Papazoglu C, Bronson RT, et al. Perp is a p63-regulated gene essential for epithelial integrity. Cell 2005;120(6):843–56. [DOI] [PubMed] [Google Scholar]
- Ihrie RA, Reczek E, Horner JS, Khachatrian L, Sage J, Jacks T, et al. Perp is a mediator of p53-dependent apoptosis in diverse cell types. Curr Biol 2003;13(22):1985–90. [DOI] [PubMed] [Google Scholar]
- Ishii K, Harada R, Matsuo I, Shirakata Y, Hashimoto K, Amagai M. In vitro keratinocyte dissociation assay for evaluation of the pathogenicity of anti-desmoglein 3 IgG autoantibodies in pemphigus vulgaris. J Invest Dermatol 2005;124(5):939–46. [DOI] [PubMed] [Google Scholar]
- Karczewski KJ, Weisburd B, Thomas B, Solomonson M, Ruderfer DM, Kavanagh D, et al. The ExAC browser: displaying reference data information from over 60 000 exomes. Nucleic Acids Res 2017;45(D1):D840–D5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009;4(7):1073–81. [DOI] [PubMed] [Google Scholar]
- Lefèvre C, Jobard F, Caux F, Bouadjar B, Karaduman A, Heilig R, et al. Mutations in CGI-58, the gene encoding a new protein of the esterase/lipase/thioesterase subfamily, in Chanarin-Dorfman syndrome. Am J Hum Genet 2001;69(5):1002–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013:13033997v2. [Google Scholar]
- Lin Z, Chen Q, Lee M, Cao X, Zhang J, Ma D, et al. Exome sequencing reveals mutations in TRPV3 as a cause of Olmsted syndrome. Am J Hum Genet 2012;90(3):558–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macari F, Landau M, Cousin P, Mevorah B, Brenner S, Panizzon R, et al. Mutation in the gene for connexin 30.3 in a family with erythrokeratodermia variabilis. American journal of human genetics 2000;67(5):1296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestrini E, Monaco AP, McGrath JA, Ishida-Yamamoto A, Camisa C, Hovnanian A, et al. A molecular defect in loricrin, the major component of the cornified cell envelope, underlies Vohwinkel’s syndrome. Nature genetics 1996;13(1):70–7. [DOI] [PubMed] [Google Scholar]
- Marques MR, Horner JS, Ihrie RA, Bronson RT, Attardi LD. Mice lacking the p53/p63 target gene Perp are resistant to papilloma development. Cancer Res 2005;65(15):6551–6. [DOI] [PubMed] [Google Scholar]
- McGrath JA, McMillan JR, Shemanko CS, Runswick SK, Leigh IM, Lane EB, et al. Mutations in the plakophilin 1 gene result in ectodermal dysplasia/skin fragility syndrome. Nat Genet 1997;17(2):240–4. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20(9):1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKoy G, Protonotarios N, Crosby A, Tsatsopoulou A, Anastasakis A, Coonar A, et al. Identification of a deletion in plakoglobin in arrhythmogenic right ventricular cardiomyopathy with palmoplantar keratoderma and woolly hair (Naxos disease). Lancet 2000;355(9221):2119–24. [DOI] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al. The Ensembl Variant Effect Predictor. Genome Biol 2016;17(1):122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan JR, Haftek M, Akiyama M, South AP, Perrot H, McGrath JA, et al. Alterations in desmosome size and number coincide with the loss of keratinocyte cohesion in skin with homozygous and heterozygous defects in the desmosomal protein plakophilin 1. J Invest Dermatol 2003;121(1):96–103. [DOI] [PubMed] [Google Scholar]
- Nguyen B, Dusek RL, Beaudry VG, Marinkovich MP, Attardi LD. Loss of the desmosomal protein perp enhances the phenotypic effects of pemphigus vulgaris autoantibodies. J Invest Dermatol 2009;129(7):1710–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol RM, Gilaberte M, Toll A, Florensa L, Lloreta J, González-Enseñat MA, et al. Erythrokeratoderma variabilis-like ichthyosis in Chanarin-Dorfman syndrome. Br J Dermatol 2005;153(4):838–41. [DOI] [PubMed] [Google Scholar]
- Richard G, Smith LE, Bailey RA, Itin P, Hohl D, Epstein EH, et al. Mutations in the human connexin gene GJB3 cause erythrokeratodermia variabilis. Nat Genet 1998;20(4):366–9. [DOI] [PubMed] [Google Scholar]
- Rickman L, Simrak D, Stevens HP, Hunt DM, King IA, Bryant SP, et al. N-terminal deletion in a desmosomal cadherin causes the autosomal dominant skin disease striate palmoplantar keratoderma. Hum Mol Genet 1999;8(6):971–6. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol 2011;29(1):24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorke EA, Adhikary G, Young CA, Roop DR, Eckert RL. Suppressing AP1 factor signaling in the suprabasal epidermis produces a keratoderma phenotype. J Invest Dermatol 2015;135(1):170–80. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Rosenbloom KR, Armstrong J, Barber GP, Casper J, Clawson H, Diekhans M, et al. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res 2015;43(Database issue):D670–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelov L, Sarig O, Harmon RM, Rapaport D, Ishida-Yamamoto A, Isakov O, et al. Desmoglein 1 deficiency results in severe dermatitis, multiple allergies and metabolic wasting. Nat Genet 2013;45(10):1244–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 2010;7(8):575–6. [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 2001;29(1):308–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets RA, Warnock DG, Bositis CM, Nelson-Williams C, Hansson JH, Schambelan M, et al. Liddle’s syndrome: heritable human hypertension caused by mutations in the beta subunit of the epithelial sodium channel. Cell 1994;79(3):407–14. [DOI] [PubMed] [Google Scholar]
- Simpson MA, Mansour S, Ahnood D, Kalidas K, Patton MA, McKenna WJ, et al. Homozygous mutation of desmocollin-2 in arrhythmogenic right ventricular cardiomyopathy with mild palmoplantar keratoderma and woolly hair. Cardiology 2009;113(1):28–34. [DOI] [PubMed] [Google Scholar]
- Snyder PM, Price MP, McDonald FJ, Adams CM, Volk KA, Zeiher BG, et al. Mechanism by which Liddle’s syndrome mutations increase activity of a human epithelial Na+ channel. Cell 1995;83(6):969–78. [DOI] [PubMed] [Google Scholar]
- Srour M, Chitayat D, Caron V, Chassaing N, Bitoun P, Patry L, et al. Recessive and dominant mutations in retinoic acid receptor beta in cases with microphthalmia and diaphragmatic hernia. Am J Hum Genet 2013;93(4):765–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3--new capabilities and interfaces. Nucleic Acids Res 2012;40(15):e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38(16):e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.