Abstract
It has been documented that the airway microbiome differs between asthmatics and healthy individuals. However, the microbial signature associated with asthma remains unclear. One of the major limitations is that the case-control study design used in existing studies may not adequately control for baseline variation in bacterial abundance between individuals. In order to further identify asthma-associated airway microbes, we performed a family-based pilot study. In this study, we investigated the microbiome of induced sputum samples from 14 participants from three two-generational pedigrees. Each pedigree has both asthmatic and non-asthmatic offspring. We compared the differences in the relative abundance of bacteria between asthmatic and non-asthmatic siblings within each family and identified bacterial genus with the same trend across the three families. Our results, for the first time, linked asthma with decreased abundance of sputum Granulicatella and verified the previous finding that Veillonella is increased in the airway of asthmatic subjects. This demonstrates the promise of a family-based study design in the search for a bacterial signature for asthma.
Keywords: Asthma, family-based study design, induced sputum, bacteria, the airway microbiome, 16S ribosomal RNA, Granulicatella, Veillonella
To the Editor,
Introduction
The search for a bacterial signature in asthma is ongoing (1, 2). One potential limitation of existing studies is the case-control study design may not adequately control for baseline variation in the bacterial composition between individuals, which may lead to attenuated association results. Family members generally have relatively similar bacteria composition (3, 4). It is, therefore, advantageous to adopt a family-based study design to reduce inter-familial variation and identify novel asthma-associated microbes.
Microbial exposure in early life determines bacteria colonization (5, 6). Since Firmicutes and Actinobacteria dominate in the indoor environment, a more similar pattern of airway bacteria from these two phyla is expected among family members, especially siblings, compared to non-family members (7). Compositional discrepancies between siblings with and without asthma may indicate a disease-specific microbe.
Here we conducted a preliminary family-based study. The bacterial phyla Firmicutes and Actinobacteria from induced sputum were evaluated using 16S ribosomal RNA (rRNA) sequencing to compare the relative abundance of different bacteria genera between asthmatic and non-asthmatic sibling(s). Our study identified that the relative abundance of Granulicatella is decreased in asthmatic siblings compared to non-asthmatic siblings across all three families.
Methods
This study has been approved by Yale University Human Research Protection Program. Fourteen subjects from three two-generation pedigrees were recruited (Figure E1). Each participating family had both asthmatic and non-asthmatic offspring. Enrolled subjects were invited to the Yale-New Haven Hospital Research Unit for sputum induction and collection. The induced sputum sample was incubated with storage reagent, and then extracted and subjected to16S rRNA sequencing. Sequence data were trimmed, filtered for quality, and clustered at 97% identity into operational taxonomic units. Only bacterial sequencing from phyla Firmicutes and Actinobacteria (>98% in total sequencing reads) were retained for analysis. The bacteria profiling is shown in Figure E2. The α-diversity of each microbial community was calculated and compared by family and by asthma status. UniFrac distance between the sputum bacterial communities of each subject pair was generated. The comparison of genus percentage between asthmatic and healthy siblings within each family were analyzed. The significance across the three families was then evaluated. Detailed methods are in supporting information.
Results
The characteristics of subjects are summarized in Table E1. Compared with non-asthmatics, asthmatics have higher BMI and lower percentage of predicted forced expiratory volume in 1 second (FEV1). As shown in Figure 1A and B, the α-diversity (Shannon’s index) did not significantly differ by family or asthma status. However, the α-diversity tended to have higher variation and lower values, in general, for asthmatic compared to non-asthmatic subjects. This observation indicates that the sputum microbial community in asthmatics have been disturbed and are prone to harbor less diverse gram-positive bacteria. UniFrac distances between the bacterial communities of participating subjects were calculated. The mean distance of each individual with all subjects within the same family and the mean distance with all subjects outside that family were plotted in Figure 1C. In two of the three families (Family 1 and 3), family members have more similar sputum bacterial composition than non-family members. Asthmatics exhibits the same trend with other non-asthmatic family members.
Figure 1.
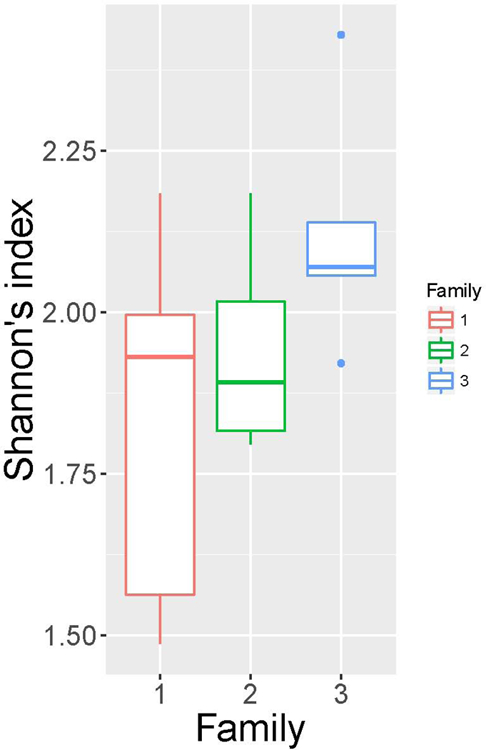
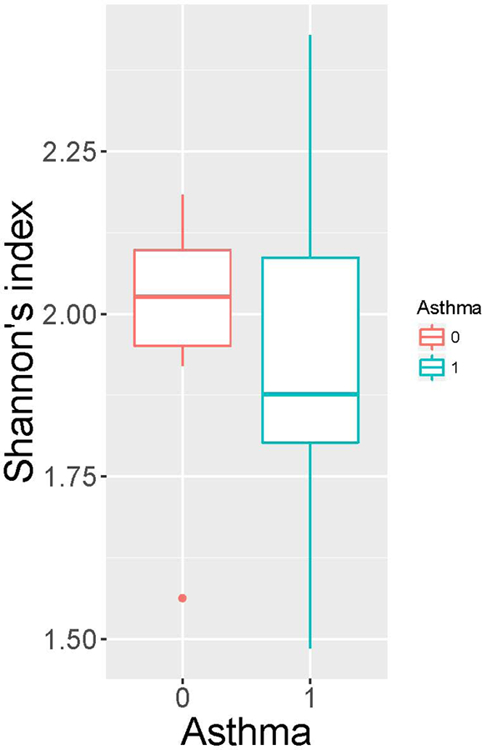
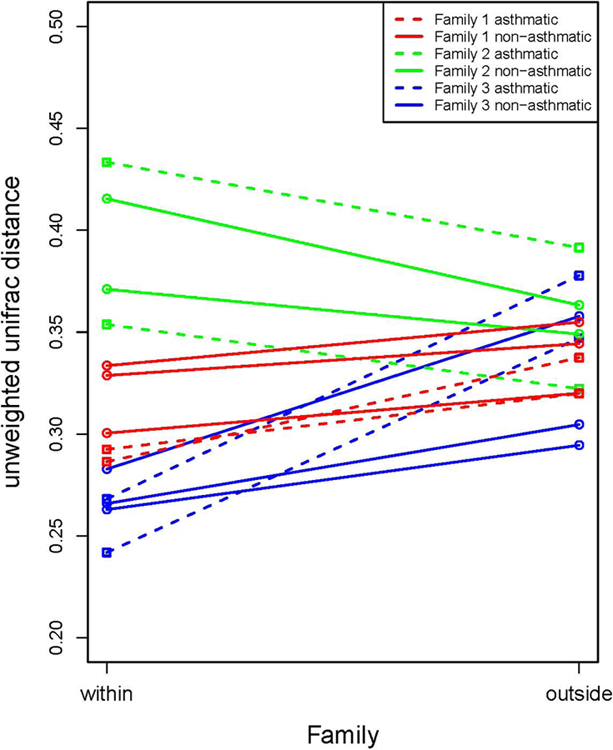
α-diversity (Shannon’s index) among families (A), and between people with asthma (indicated as 1) and not with asthma (indicated as 0) (B). No significant differences were observed by student t-test. Mean distance of an individual with subjects within his/her family and mean distance of the individual with subjects outside his/her family (unweighted UniFrac) (C). Family is indicated by colors; asthma status is indicated by line types.
Next, we focused on the discordant siblings in each household and conducted a genus-level analysis to identify bacteria that displayed differences in abundance between the asthmatic and non-asthmatic sibling(s). We identified eight genera whose average relative abundance is greater than 1% among offspring. As shown in Figure 2 and Table E2, two genera, Granulicatella and Veillonella, have concordant direction of association with asthma across the three families. The genus, Granulicatella, exhibited a significantly reduced abundance in asthmatics, compared to non-asthmatics (p-value=0.043, paired t-test) and genus Veillonella exhibited an increased abundance, however non-significant, in asthmatics compared to non-asthmatics (p-value=0.078, paired t-test).
Figure 2.
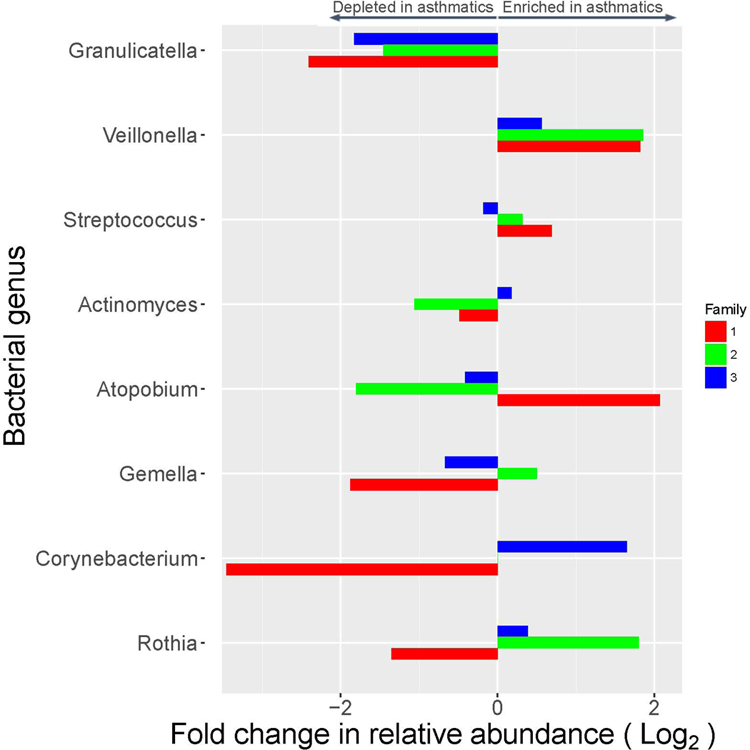
Bacterial genera that were enriched or depleted in relative abundance in asthmatic versus non-asthmatic sibling(s) across three families.
Discussion
To our knowledge, this is the first attempt to apply a family-based design to study the sputum microbiome for asthma. Our results suggest that family members share more similar bacterial composition in the induced sputum, compared to non-family members. A family- based study design is, therefore, promising to diminish baseline variation caused by different household environmental exposures and enhance the association signals between asthma and the sputum microbes.
We applied a two-step strategy that leveraged the family-based nature of our study: we first compared the differences in the relative abundance of bacteria between asthmatic sibling and non-asthmatic siblings within each household. Next, we horizontally compared the abundance of each bacterial genus across the three families to identify those with the same trend. For the first time, our study associates asthma with Granulicatella in the sputum microbiome. Granulicatella are commensal microbes in the respiratory tract (8). It shows a wide range of relative abundance in the sputum microbiome in our data (1%−14.3%), suggesting the abundance of Granulicatella is highly influenced by environmental exposure, which has been reported before in a home-based study (9). Individuals from different household harbor very different relative abundance of Granulicatella in the sputum microbiome, making it difficult for case-control studies to capture this association. However, in our family-based study we were able to compare the relative abundance between case and control siblings to reveal this pattern. We also found the Genus Veillonella was enriched in asthmatic siblings in all three families, which supports previous finding that increased Veillonella is associated with enhanced lung inflammation (10). This is also concordant with the finding that Veillonella is enriched in the bronchial microbiome of asthmatic subjects (1).
Due to lack of information on diet, we made the assumption that siblings within the same household were exposed to similar diet and the eating habits were initiated and nurtured within the family settings. Future study should gather information on diet to define its role in influencing airway bacterial community within each household.
We have included a limited number of families for this study. A large-scale family-based study is necessary to further investigate the impact of shared indoor environment on the host microbiome and improve our understanding of asthma associated microbes in the airway.
Sincerely,
Supplementary Material
Acknowledgements
We thank all participants in this study. We thank Keli Sorrentino, Hannah Strickland, Julie Kezik and Kathleen Clark for their effort in patient recruitment and sputum induction procedures. We thank Dr. Andrew Kau for critical comments on this manuscript.
Funding:
This work was supported by the National Institutes of Health [grant numbers: 1R01HL116742, F32AI114097].
Abbreviations:
- FEV1
forced expiratory volume in 1 second
- rRNA
ribosomal RNA
Footnotes
Conflict of Interests
The authors declare that they have no conflicts of interest
References
- 1.Durack J, Lynch SV, Nariya S, Bhakta NR, Beigelman A, Castro M, et al. Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. J Allergy Clin Immunol 2017;140(1):63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chung KF. Airway microbial dysbiosis in asthmatic patients: A target for prevention and treatment? J Allergy Clin Immunol 2017;139(4):1071–81. [DOI] [PubMed] [Google Scholar]
- 3.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature 2012;486(7402):222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw L, Ribeiro ALR, Levine AP, Pontikos N, Balloux F, Segal AW, et al. The Human Salivary Microbiome Is Shaped by Shared Environment Rather than Genetics: Evidence from a Large Family of Closely Related Individuals. MBio 2017;8(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jatzlauk G, Bartel S, Heine H, Schloter M, Krauss-Etschmann S. Influences of environmental bacteria and their metabolites on allergies, asthma, and host microbiota. Allergy 2017;72(12):1859–67. [DOI] [PubMed] [Google Scholar]
- 6.von Mutius E The microbial environment and its influence on asthma prevention in early life. J Allergy Clin Immunol 2016;137(3):680–9. [DOI] [PubMed] [Google Scholar]
- 7.Hanson B, Zhou Y, Bautista EJ, Urch B, Speck M, Silverman F, et al. Characterization of the bacterial and fungal microbiome in indoor dust and outdoor air samples: a pilot study. Environ Sci Process Impacts 2016;18(6):713–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beck JM, Schloss PD, Venkataraman A, Twigg H 3rd, Jablonski KA, Bushman FD, et al. Multicenter Comparison of Lung and Oral Microbiomes of HIV-infected and HIV-uninfected Individuals. Am J Respir Crit Care Med 2015;192(11):1335–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hosgood HD 3rd, Sapkota AR, Rothman N, Rohan T, Hu W, Xu J, et al. The potential role of lung microbiota in lung cancer attributed to household coal burning exposures. Environ Mol Mutagen 2014;55(8):643–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segal LN, Alekseyenko AV, Clemente JC, Kulkarni R, Wu B, Gao Z, et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome 2013;1(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


