SUMMARY
While rods in the mammalian retina regenerate rhodopsin through a well-characterized pathway in cells of the retinal pigment epithelium (RPE), cone visual pigments are thought to regenerate in part through an additional pathway in Müller cells of the neural retina. The proteins comprising this intrinsic retinal visual cycle are unknown. Here, we show that RGR opsin and retinol dehydrogenase-10 (Rdh10) convert all-trans-retinol to 11-cis-retinol during exposure to visible light. Isolated retinas from Rgr+/+ and Rgr−/− mice were exposed to continuous light, and cone photoresponses were recorded. Cones in Rgr−/− retinas lost sensitivity at a faster rate than cones in Rgr+/+ retinas. A similar effect was seen in Rgr+/+ retinas following treatment with the glial-cell toxin, α-aminoadipic acid. These results show that RGR opsin is a critical component of the Müller-cell visual cycle, and that regeneration of cone visual pigment can be driven by light.
In Brief
Morshedian et al. report that RGR opsin in Müller cells functions to regenerate cone visual pigments during light exposure, and is the likely isomerase of the intrinsic retinal visual cycle. RGR opsin is required to maintain cone sensitivity during sustained light exposure.
INTRODUCTION
Light perception in the retina begins with the absorption of a photon by an opsin visual pigment. The light-absorbing molecule in most animals is 11-cis-retinaldehyde (11cRAL), which is coupled to the opsin protein as a Schiff base and converted by light to the lower-energy isomer, all-trans-retinaldehyde (atRAL). The resulting change in opsin conformation activates its associated G protein and thereby the visual transduction cascade in both vertebrate and invertebrate photoreceptors. Rhabdomeric photoreceptors in the eyes of insects and other invertebrates contain bistable opsins where the atRAL remains covalently coupled to the protein following its activation (Fain et al., 2010). Absorption of a second photon converts the atRAL back to 11cRAL, which restores light sensitivity without the need for enzymatic synthesis of 11cRAL. In contrast, ciliary photoreceptors, such as rods and cones in the retinas of mammals contain bleaching opsins that dissociate following light activation to yield free atRAL and unliganded apo-opsin. Light sensitivity is restored when apo-opsin combines with another 11cRAL to form a new visual pigment. To sustain light sensitivity, ciliary photoreceptors must therefore be supplied with chromophore at a rate that matches its rate of consumption through photoisomerization. Under dim light, this conversion is carried out in cells of the retinal pigment epithelium (RPE) by an enzyme pathway called the visual cycle. Under daylight conditions however, photoisomerization of visual opsins in rods and cones far outstrips the synthesis of 11cRAL by RPE cells (Mata et al., 2002). The mechanism whereby mammalian photoreceptors maintain light sensitivity under daylight conditions is unknown.
Accumulating evidence suggests the existence of a second visual cycle that regenerates cone visual pigments in the neural retina. Cones, but not rods, were shown to recover photosensitivity following light exposure in isolated retinas from multiple species including humans and mice (Goldstein, 1970; Hood and Hock, 1973; Wang and Kefalov, 2009). Müller cells have been implicated in this process by several previous observations: (i) Müller cells express multiple retinoid-processing proteins including cellular retinaldehyde binding protein (CRALBP, Bunt-Milam and Saari, 1983), cellular retinol binding protein-1 (CRBP1, Huang et al., 2009), retinol dehydrogenase-10 (Rdh10, Wu et al., 2004), retinol dehydrogenase-11 (Rdh11, Haeseleer et al., 2002), and retinol dehydrogenase-14 (Rdh14, Haeseleer et al., 2002); (ii) cultured Müller cells take up all-trans-retinol (atROL) and release 11-cis-retinol (11cROL) into the medium (Betts-Obregon et al., 2014; Das et al., 1992); and (iii) treatment of isolated retinas with the glial-cell toxin, α-aminoadipic acid (α-AAA) (Jablonski and lannaccone, 2000) abolished recovery of cone sensitivity in isolated retina (Wang and Kefalov, 2009). Since only 11cRAL can regenerate bleached opsin, these observations suggest further that cones, but not rods, contain an 11-cis-retinol dehydrogenase (11cRDH) activity that converts 11cROL to 11cRAL (Mata et al., 2002; Sato and Kefalov, 2016). Hence, the proposed Müller-cell visual cycle provides a ‘private pipeline’ of chromophore precursor to regenerate cone opsin.
Cones are responsible for vision in bright light and operate at high rates of opsin photoisomerization. Recovery of cone sensitivity was shown to be limited by chromophore supply (Wang et al., 2014), emphasizing the importance of the retinal visual cycle to cone function. Two proteins were tentatively identified as components of the Müller-cell visual cycle: dihydroceramide desaturase-1 (Des1) (Kaylor et al., 2013) and multifunctional O-acyltransferase (MfAt) (Kaylor et al., 2014). When co-expressed in cultured cells, Des1 and MFAT converted atROL to 11-cis-retinyl esters (11cRE’s), a lipid-soluble storage form of 11cROL. This ‘isomerosynthase’ activity was also observed in homogenates of cone-dominant chicken and ground squirrel retinas, but was undetectable in homogenates of rod-dominant mouse or cow retinas (Mata et al., 2002). Retinas from cone-dominant species contain 11cRE’s, possibly due to Des1—MFAT activity, while 11cRE’s were undetectable in rod-dominant retinas (Mata et al., 2002). Recent studies have shown that mice with a conditional null mutation of the Des1 gene in Müller cells recover cone sensitivity normally (Kiser et al., 2019). For these reasons, Des1 probably plays no role in the regeneration of mouse cone opsin. RPE cells contain a retinoid isomerase (retinal pigment epithelium-specific 65-kDa protein or Rpe65) coupled to a retinylester synthase (lecithin retinol acyltransferase or LRAT); however, neither Rpe65 nor LRAT is expressed in the neural retina (Kiser et al., 2019; Mata et al., 2005). Hence, the LRAT—Rpe65 isomerase system does not contribute to cone opsin regeneration in isolated retinas. The proteins responsible for 11cROL synthesis by Müller cells and the recovery of cone sensitivity in isolated retinas following a photobleach are hence unknown.
‘Retinal G protein-coupled receptor’ (RGR) opsin is a non-visual opsin in intracellular membranes of RPE and Müller cells (Pandey et al., 1994). In contrast to the visual opsins in photoreceptors, RGR opsin covalently binds atRAL in the dark, which is isomerized to 11cRAL upon exposure to light (Hao and Fong, 1999). Despite its name, RGR opsin lacks the conserved (E/D)R(Y/W/F) and NPxxY(x)5,6F motifs required for interaction of a receptor with its G protein (Franke et al., 1992; Fritze et al., 2003). Thus, RGR is probably not a signaling molecule, consistent with its proposed role as a photoisomerase (Hao and Fong, 1999). Point mutations in the human Rgr gene are responsible for the inherited blinding disease, retinitis pigmentosa (RP) in a small subset of cases (Morimura et al., 1999). Mice with a knockout mutation in the Rgr gene showed lower levels of rhodopsin and diminished rod b-wave amplitudes by in vivo electroretinography after exposure to bright light, suggesting that RGR plays a role in chromophore synthesis (Chen et al., 2001a). However, other studies on Rgr−/− mice found no light-dependent effects of RGR on rod photopigment regeneration (Maeda et al., 2003; Wenzel et al., 2005).
In this study, we present the first evidence for a role of RGR opsin in the regeneration of cone visual pigment. We show that RGR opsin functionally pairs with Rdh10 to carry out the light-dependent conversion of atROL to 11cROL. This activity was found in cultured cells expressing both RGR and Rdh10, and in retinal fractions from normal, but not Rgr−/− mutant mice lacking RGR opsin (Chen et al., 2001a). These findings suggest that RGR opsin and Rdh10 serve together as a light-dependent 11cROl generator. To test this possibility, we measured the sensitivity of cones in isolated retinas from normal and Rgr−/− mice during exposure to bright light. Cones in Rgr−/− retinas lost sensitivity at a significantly faster rate than cones in Rgr+/+ retinas. A similar effect was seen in Rgr+/+ retinas following treatment with α-AAA. These results indicate that maintenance and recovery of cone sensitivity in isolated mouse retinas requires a light-driven visual cycle that depends on RGR opsin. Thus, ciliary photoreceptors of vertebrates, like the rhabdomeric photoreceptors of invertebrates, can use light itself to regenerate visual pigment.
RESULTS
Coupled photoisomerization and oxidoreduction of vitamin A by RGR opsin and Rdh10
Müller cells take up atROL discharged by rods and cones during light exposure and release 11 cROL to regenerate cone visual pigments (Betts-Obregon et al., 2014; Das et al., 1992; Mata et al., 2002). This activity could be carried by RGR opsin in conjunction with a reversible retinol dehydrogenase with dual-substrate specificity. RGR opsin from bovine RPE was found to co-purify with retinol dehydrogenase-5 (Rdh5) (Simon et al., 1995), suggesting an interaction between these proteins (Chen et al., 2001b). However, Rdh5 is not expressed in the retina (Huang et al., 2009) and hence is an unlikely component of the retina visual cycle. In contrast, Rdh10 is present in both RPE and Müller cells (Wu et al., 2004). Further, Rdh10 exhibits dual-substrate specificity, favoring oxidation of atROL (Wu et al., 2002) and reduction of 11cRAL (Farjo et al., 2009). These properties suggest that RGR opsin and Rdh10 may cooperate in the presence of light to convert atROL to 11cROL. To test this possibility, we expressed bovine RGR opsin and Rdh10 alone and together in HEK-293T cells. Cells were placed in assay media containing atROL and were maintained in darkness or exposed to UV-filtered white light (400-nm cutoff) for 30 minutes. Media and cells from each culture dish were extracted into hexane and analyzed for retinoid content by normal-phase high performance liquid chromatography (HPLC). Low levels of 11cROL were observed in media from all cells maintained in darkness (Fig. 1A). Similarly, media from light-exposed cells expressing RGR alone, Rdh10 alone, or neither protein also contained low 11cROL. However, media from cells co-expressing RGR opsin and Rdh10 and exposed to light contained dramatically higher 11cROL (Fig. 1A). Chromatograms are shown in Fig. S1. At the same time, levels of 11cRAL were not increased in media from light-exposed cells expressing RGR opsin and Rdh10 (Fig. S2). These findings suggest close cooperativity between RGR opsin and Rdh10 such that 11cRAL produced by RGR photoisomerization is quantitatively reduced to 11cROL before it can escape into the media.
Figure 1. Production of 11cROL from atROL by RGR opsin and Rdh10 in cells exposed to light.

(A) HEK-293T cells were transfected with equal amounts of non-recombinant plasmid (pcDNA3.1) or plasmids containing the coding regions for Rdh10 (pcDNA3.1-bRdh10), RGR opsin (pcDNA3.1-bRGR) or both. After two days in culture to allow for protein expression, the culture media were supplemented with 5.0 μM atROL and 2% BSA under dim red light. The cell cultures (~80% confluent) were then incubated at 37°C for 30 minutes in the dark or exp osed to monochromatic light (470 nm ± 10 nm at 0.2 W/m2). Cell culture media were collected, extracted, and analyzed for retinoid content by normal-phase HPLC. Note the production of 11cROL only in media from light-exposed cells expressing both Rdh10 and RGR opsin (p < 0.001) relative to the other transfection and light-exposure conditions. (B) HEK cells were transfected with non-recombinant plasmid (pcDNA3.1) alone, plasmid containing the coding region for RGR opsin plus pcDNA3.1, or plasmid containing the coding region for RGR opsin plus plasmid containing the coding region for Rdh5 (human), Rdh8 (human), Rdh10 (human, mouse, or chicken), Rdh11 (human), or Rdh14 (human), as indicated. After two days in culture, the culture media were supplemented with 5.0 μM atROL and 2% BSA under dim red light. All cell cultures were then incubated for 30 minutes during exposure to monochromatic light (470 nm ± 10 nm at 0.2 W/m2). Culture media were collected, extracted, and analyzed by normal-phase HPLC for retinoid contents, expressed as pmoles per culture dish. Note the production of 11cROL (p < 0.001) in medium from cells expressing Rdh10 and RGR opsin, but not from cells expressing the other retinol dehydrogenases plus RGR opsin. (C) Action spectrum of RGR and Rdh10. HEK-293T cells were transfected with equal amounts of non-recombinant plasmid (pcDNA3.1) or plasmids containing the coding regions for Rdh10 (pcDNA3.1-bRdh10), RGR opsin (pcDNA3.1-bRGR), or both. After two days in culture, the media were supplemented with 5 μM atROL and 2% BSA under dim red light. The cell plates (~80% confluent) were exposed to monochromatic light (20-nm bandwidth) of wavelengths 425—575 nm for 30 minutes, or maintained in darkness. The light intensities were adjusted to deliver the same photon flux at each wavelength (0.35 W/m2 at 425 nm to 0.26 W/m2 at 575 nm). The media above these plates were collected, extracted and analyzed for retinoid contents, which are expressed as pmoles per mg total protein. Cells exposed to full-spectrum (FS) light (4.0 W/m2) showed the greatest production of 11cROL (p < 0.001) versus the other light conditions. Wavelengths between 425 and 525 nm showed increased production of 11-cis-retinol (p < 0.01) relative to cells maintained in darkness or the other monochromatic light wavelengths. Each bar represents n = 3 plates of transfected cells. Data represent mean ± SD; n.s., not significant, * p < 0.05, ** p < 0.01, *** p < 0.001, one-way ANOVA with Tukey’s post hoc analysis.
RGR opsin and Rdh10 interact specifically
To test whether the interaction between RGR and Rdh10 is specific, we co-expressed RGR opsin in 293T cells with other retinol dehydrogenases from RPE and Müller cells. As before, we incubated these expressing 293T cells in media containing atROL and exposed the cells to UV-filtered white light. Negative controls included non-recombinant plasmid, RGR opsin alone, or Rdh10 alone. Cells expressing RGR opsin and Rdh10 from human, mouse or chicken produced significant amounts of 11cROL (Fig. 1B). In contrast, cells expressing RGR opsin alone or RGR plus Rdh5, Rdh8, Rdh11 or Rdh14 produced only background levels of 11cROL. These data indicate that Rdh10 interacts selectively with RGR opsin to convert atROL into 11cROL, although Rdh11—Rdh14 also act on 11c-and at-retinoids (Haeseleer et al., 2002). Similar rates of 11cROL formation were observed with Rdh10’s from mouse, human or chicken (Fig. 1B), indicating similar activities of the three homologs. The redox photoisomerase activity of RGR opsin is therefore specific to Rdh10.
The action spectrum of RGR photoisomerase activity corresponds to its absorption spectrum
RGR opsin combined with atRAL exhibits absorption maxima (λmax) at 375 and 466 nm, corresponding to non-protonated and protonated forms of the retinylidene Schiff base (Hao and Fong, 1996). Since the ocular medium blocks transmittance of light below 400 nm in humans (Boettner and Wolter, 1962), only protonated RGR opsin may function in vivo as a retinaldehyde photoisomerase. To confirm that RGR opsin is responsible for the light-dependent conversion of atROL to 11cROL, we repeated the assays on cells expressing RGR and Rdh10 with monochromatic light (20-nm bandwidth) of wavelengths 425 to 575 nm. Light intensities were adjusted to yield equal photon fluxes at each wavelength. Other plates of expressing cells were exposed to UV-filtered full-spectrum light or were maintained in darkness, as positive and negative controls. After incubating the live cells for 30 minutes in the presence of atROL, we extracted the culture media and determined the retinoid content by HPLC. Synthesis of 11cROL by these cells varied strongly with light conditions (Fig. 1C). Again, we observed only background 11cROL in media from expressing cells incubated in darkness, and dramatic synthesis of 11cROL by cells exposed to full-spectrum visible light (Fig. 1C). Of cultures exposed to narrow-band light, the highest synthesis of 11cROL occurred at 470 nm, with reduced synthesis at longer and shorter wavelengths (Fig. 1C). This action spectrum for 11cROL synthesis by RGR-expressing cells overlaps the absorption spectrum of protonated RGR opsin (Hao and Fong, 1996), providing further evidence that all-trans (at) to 11-cis (11c) retinoid photoisomerization was carried out by RGR opsin in these cells.
Redox-photoisomerase activity in mouse retina and RPE microsomes
If light-dependent conversion of atROL to 11cROL by expressing HEK cells mirrors the in vivo activity of Rdh10 and RGR, similar redox-photoisomerase activity should be present in retinas. To test this possibility, we prepared microsomal membranes from wild-type (Rgr+/+) and Rgr−/− mouse retinas and RPE-containing eyecups (see STAR Methods). Microsomes from Rgr+/+ mouse retinas produced 2.7-fold greater 11cROL during light exposure than did Rgr−/− retina microsomes (Fig. 2A). In contrast, we observed no significant difference in levels of 11cRAL between Rgr+/+ and Rgr−/− retina microsomes. The endogenous 11cRAL in Rgr+/+ and Rgr−/− retina microsomes is from unbleached rhodopsin and cone-opsin pigments in these membrane samples (Fig. 2A). The increased 11cRAL following light exposure, which occurred independent of RGR opsin, probably reflects at- to 11c- photoisomerization of N-ret-PE (Kaylor et al., 2017). These results corroborate our observations that light and RGR can mediate production of 11cROL by HEK cells expressing RGR and Rdh10 (Fig. 1A).
Figure 2. Conversion of atROL to 11cROL and 11cRAL by light-exposed microsomes from wild-type and Rgr−/− retinas and RPE.

Retinas and RPE-containing eyecups were collected from two-month-old Rgr+/+ (129/Sv) and Rgr−/− mice. Microsomes were prepared from each tissue. The samples were supplemented with 2% BSA and 5 μM atROL. One set of samples was extracted to determine the endogenous retinoid content (endo). The remaining samples were placed in cuvettes and agitated during exposure to 470-nm monochromatic light (20-nm bandwidth) at 0.2 W/m2 for 30 minutes at 37°C (light). Retinoids were extracted and analyzed by normal-phase HPLC. (A) 11cROL and 11cRAL synthesized by retina microsomes from wild-type and Rgr−/− mice is expressed as pmoles per mg total protein. Note the several-fold reduction in 11cROL (p < 0.001) and unchanged 11cRAL produced by Rgr−/− versus wild-type retina microsomes in light. (B) 11cROL and 11cRAL synthesized by RPE microsomes from Rgr+/+ and Rgr−/− mice expressed as pmoles per mg total protein. Note the severalfold reduction in both 11cROL and 11cRAL produced by Rgr−/− versus wild-type RPE microsomes in light (p < 0.001). Each bar represents n = 3 samples of microsomes. Data represent mean ± SD; n.s., not significant, * p < 0.05, ** p < 0.01, *** p < 0.001, one-way ANOVA with T ukey’s post hoc analysis.
Since RGR opsin and Rdh10 are also both present in RPE internal membranes (Chen et al., 2001b; Pandey et al., 1994), we tested for redox photoisomerase activity in microsomes from Rgr+/+ and Rgr−/− mouse RPE. Here, we observed nearly five-fold greater production of 11cROL by light-exposed Rgr+/+ versus Rgr−/− RPE microsomes (Fig. 2B), suggesting that RGR opsin and Rdh10 in the RPE also exhibit redox-photoisomerase activity. In contrast to retina microsomes, we observed several-fold lower 11cRAL production in dark- and light-exposed RPE microsomes from Rgr−/− versus Rgr+/+ mice (Fig. 2B). This can be explained by the three-fold lower Rpe65-isomerase activity in RPE homogenates from Rgr−/− versus Rgr+/+ mice (Radu et al., 2008; Wenzel et al., 2005).
The conversion of atROL to 11cROL carried out by mouse retina and RPE microsomes (Figs. 2A and 2B) was observed with no dinucleotide cofactor added to the assay mixtures. Since microsomal membranes are depleted of cytoplasmic contents, these findings suggest that NADPH/NADP+ cofactor may remain in association with Rdh10, switching between reduced and oxidized forms with oxidation of atROL and reduction of 11cRAL.
RGR opsin is expressed in Müller cells of the mouse retina
Although RGR opsin has been shown to be expressed in Müller cells of human (Trifunovic et al., 2008), bovine (Pandey et al., 1994) and chicken retinas (Diaz et al., 2017), it has not been detected in cells of the mouse retina (Tao et al., 1998; Trifunovic et al., 2008). To establish that RGR opsin is expressed in mouse retinas, we collected neural retinas and RPE-containing eyecups from Rgr+/+ (129/Sv) and Rgr−/− mice, prepared protein homogenates from these tissues, and analyzed by immunoblotting with an antibody against mouse RGR opsin. Immunoreactive bands corresponding to RGR opsin were present in lanes containing isolated neural retinas and RPE-containing eyecups (Fig. 3A). To establish RGR opsin expression in mouse Müller cells, we performed immunofluorescence analysis on thick (18-μm) retina sections from Rgr+/+ and Rgr−/− mice. As a control, we used antibodies against CRALBP, which is also expressed in RPE and Müller cells. RGR opsin and CRALBP co-localized in Müller-cell endfeet, the inner nuclear layer containing Müller-cell nuclei, and in the apical microvilli of Müller cells above the outer limiting membrane (Fig. 3B). Importantly, we observed no RGR immunoreactivity in retinas from Rgr−/− mice (Fig. 3B). These data confirm that RGR opsin is expressed in mouse Müller cells.
Figure 3. RGR opsin is expressed in mouse Müller cells.

(A) Representative immunoblot containing homogenates of neural retinas (retina) and RPE-containing eyecups (RPE) from Rgr+/+ and Rgr−/− mice. The blot was probed with the anti-RGR opsin (Pin 2) antibody. Lanes were loaded with 90 or 50 μg total protein as indicated. (B) Immunofluorescence analysis of RGR opsin and CRALBP in 18-μm retina sections from Rgr+/+ and Rgr−/− mice. Sections were probed with rabbit polyclonal anti-RGR Pin3 (red) and mouse monoclonal anti-RLBP1 clone 1H7 (CRALBP, green). Images were acquired on an Olympus FluoView FV1000 confocal microscope under a 40x oil objective. Note the co-localization of RGR opsin and CRALBP in Müller-cell endfeet, the inner nuclear layer (INL) and in the apical microvilli of Müller cells above the outer limiting membrane (OLM).
Contribution of RGR opsin to the cone photoresponse under continuous illumination
If the atROL to 11cROL redox-photoisomerase activity of Rdh10 and RGR opsin plays a role in vision, loss of RGR opsin should affect cone function during light exposure. To explore this possibility, we crossed the Rgr−/− mutation onto the Gnat1−/− background to yield Rgr−/− Gnat1−/− mice with absent rod but normal cone photoresponses (Calvert et al., 2000; Chen et al., 2001a). Isolated retinas (without the RPE) were collected and placed individually into a specially designed recording chamber that has been shown to support cone photoresponses for several hours (Vinberg et al., 2014). The retinas were perfused with a medium containing synaptic inhibitors to suppress responses from retinal interneurons and glial cells (Vinberg et al., 2014; Wang and Kefalov, 2009), and 10 μM atROL, to maintain constant substrate concentration during the recordings. Cone photoresponses were elicited with test flashes at 565 nm, which photoisomerizes M opsin (λmax = 508 nm) with 107-fold greater efficiency than S opsin (λmax = 357 nm) (Govardovskii et al., 2000). Recordings were made in dark-adapted retinas immediately before and at various times during continuous exposure to 505-nm background light at an intensity of 9.1 × 106 photons (ϕ) μm−2 s−1, estimated to bleach 106 M-opsin pigment molecules per second (106 P* s−1) (see STAR Methods).
Representative recordings from Rgr+/+ Gnat1−/− retinas are shown in Fig. 4A. Under continuous light, cone responses gradually diminished in sensitivity and maximum amplitude during the one-hour recording period. In the absence of any pigment regeneration, the background light can be calculated to bleach essentially all of the M-opsin pigment during the one-hour exposure, leaving less than a single unbleached pigment molecule per cone. A bleach of this magnitude would be expected to produce dramatically lower response amplitudes than those observed (Fig. 4A), suggesting concurrent regeneration of M opsin in Rgr+/+ Gnat1−/− retinas during light exposure. To test whether RGR opsin contributes to cone regeneration, we repeated the experiment using retinas from Rgr−/− Gnat1−/− mice. Here, we observed more rapid loss of cone response amplitude (Fig. 4B). These results suggest that, under continuous illumination, retinas lacking RGR undergo faster net bleaching of cone opsins. Next, we pre-incubated Rgr+/+ Gnat1−/− retinas in a medium containing 10 mM α-AAA. This potent gliotoxin acts by inhibiting the glutamate transporter and glutamine synthetase in Müller cells (Jablonski and Iannaccone, 2000; McBean, 1994). Pre-incubation with α-AAA was shown to block recovery of cones in isolated mouse retinas (Wang and Kefalov, 2009). We recorded cone photoresponses in α-AAA-treated Rgr+/+ Gnat1−/− retinas during light exposure. These retinas exhibited rapid loss of cone response amplitude (Fig. 4C), similar to untreated Rgr−/− Gnat1−/− retinas (Fig. 4B). Thus, loss of RGR opsin, due to a mutation in its gene or pharmacological ablation of Müller cells, results in diminished cone response amplitude and sensitivity during light exposure.
Figure 4. Representative transretinal responses of (A) Rgr+/+ Gnat1−/− mice, (B) Rgr−/− Gnatl−/− mice, and (C) Rgr+/+ Gnat1−/− mice incubated with α-AAA.
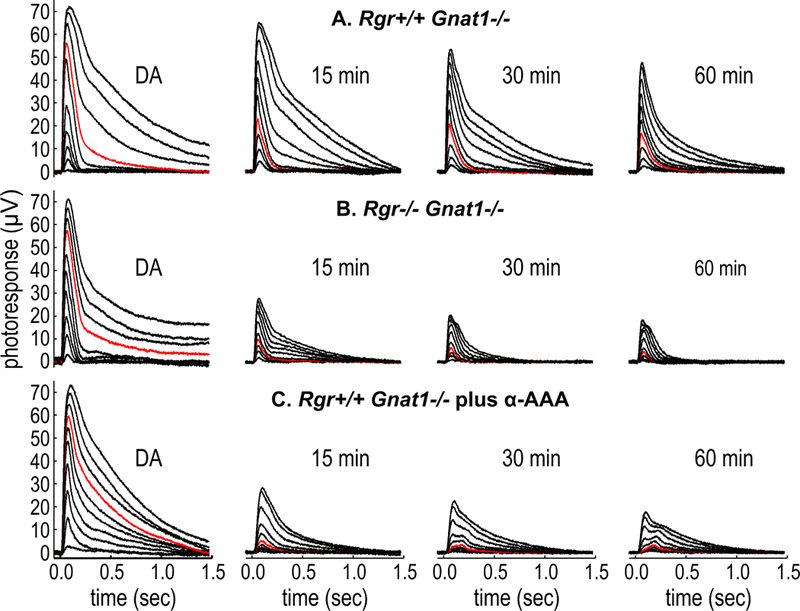
M-Cone isolated photoreceptor recordings were made in dark-adapted (DA) retinas and after 15, 30, and 60 minutes of exposure to a continuous 505-nm background light (9.1 × 106 ϕ μm−2 s−1). The stimulating light flashes were produced by a 565-nm LED (flashes ranging from 65 ϕ μm−2 to 5.4 × 107 ϕ μm−2 effective at the λmax of the M cone pigment). Red traces show responses to a constant flash of an intensity of 3.7 × 105 ϕ μm−2. Note the lower response amplitudes in Rgr−/− versus Rgr+/+ retinas, or when isolated Rgr+/+ retinas were incubated in α-AAA.
Effect of RGR opsin on cone sensitivity
The effect of RGR opsin on cone sensitivity were quantified by plotting response amplitudes in Fig. 4 against the number of photons (ϕ) contained in each of the flashes. Figs. 5A–5C show response-intensity curves as a function of duration of background exposure for the three conditions in Fig. 4. The curves have been fitted with the Hill equation (1),
| (1) |
where r is the amplitude of the response, rmax is the maximum response amplitude, I is the number of incident 565-nm photons in the flash (ϕ μm−2), and n is an exponent. Best-fitting values of rmax and n are given in the figure legend. The curves were shifted to the right and downward from the decrease in sensitivity and maximum amplitude, initially as a result of adaptation of the cones to the background light (see Fig. S3 and Burkhardt, 1994). The curves then continued to shift and decrease in maximum amplitude, presumably from bleaching of pigment and gradual loss of chromophore during perfusion of the retina despite the addition of atROL to the medium. In the absence of RGR, or after exposure of the retina to α-AAA, the curves were dramatically shifted further downward and to the right (Figs. 5B and 5C). These changes in sensitivity and response amplitude are compared in Figs. 5D and 5E. No significant changes in sensitivity or response amplitude were observed over the 60-min recording duration when retinas were maintained in darkness. Sensitivities were significantly lower at 30, 45, and 60 minutes in Rgr−/− Gnat1−/− retinas and Rgr+/+ Gnat1−/− retinas treated with α-AAA compared to Rgr+/+ Gnat1−/− control retinas (see figure legend). No significant differences were detected between untreated Rgr−/− Gnat1−/− and α-AAA-treated Rgr+/+ Gnat1−/− retinas.
Figure 5. The effect of RGR and Müller cells on intensity-response, photosensitivity and maximum response amplitude of M-cones in Rgr+/+ Gnatl−/−, Rgr−/− Gnat1−/− and Rgr+/+ Gnat1−/− retinas incubated in α-AAA in the presence of a continuous light.

Changes in the response intensities in dark-adapted retinas from mice of the indicated genotypes at 15 min, 30 min, 45 min and 60 min in the presence of a continuous light (9.1 × 106 ϕ μm2 s−1), plotted as a function of incident photons from mouse cones in (A) Rgr+/+ Gnat1−/− (n = 9), (B) Rgr−/− Gnat1−/− (n = 8), and (C) Rgr+/+ Gnat1−/− incubated with α-AAA (n = 5). Curves are best fits to the Hill equation (Eqn. 1) with the following parameter values: (A) Vmax = 55.1, I1/2 = 4.4 × 104, n = 0.58 (DA); Vmax = 50.0 and I1/2 = 1.0 × 106, n = 0.66 (15 min); Vmax = 48.0, I1/2 = 1.7 × 106, n = 0.66 (30 min); Vmax = 45.3 I1/2 = 2.2 × 106, n = 0.67 (45 min) and ; Vmax = 40.0, I1/2 = 2.1 ×106, n = 0.81 (60 min). (B) Vmax = 53.0, I1/2 = 4.9 × 104, n = 0.52 (DA); Vmax = 30.2 and I1/2 = 1.4 × 106, n = 0.79 (15 min); Vmax = 23.9, I1/2 = 2.3 × 106, 0.88 (30 min); Vmax = 20.5, I1/2 = 3.9 × 106, n = 0.80 (45 min) and Vmax = 17.3, I1/2 = 3.7 × 106 n = 0.79 (60 min). (C) Vmax = 55.1, I1/2 = 4.7 × 104, n = 0.57 (DA); Vmax = 32.4 and I1/2 = 3.3 × 106, 0.59 (15 min); Vmax = 26.6, I1/2 = 4.0 × 106, n = 0.74 (30 min); Vmax = 24.6, I1/2 = 5.4 × 106, n = 0.73 (45 min) and ; Vmax = 18.2, I1/2 = 5.6 × 106, n = 0.79 (60 min). Half-maximal response amplitude of the Rgr+/+ Gnat1−/− at 60 min is shown with a dashed line in (A). The same flash intensity generates significantly smaller responses at 60 min in Rgr−/− Gnat1−/−and Rgr+/+ Gnat1−/− incubated with α-AAA. (D) Mean maximum response amplitude and (E) relative dim flash sensitivity from recordings of Fig. 4 were calculated and plotted (with S.E.) as a function of time. Maximum amplitudes were significantly smaller (see Star Methods) at 15, 30, 45 and 60 min in Rgr−/− Gnat1−/− retinas than in Rgr+/+ Gnat1−/− retinas (p <0.01, 0.002, 0.002, and 0.001). That was also true for Rgr+/+ Gnat1−/− retinas and retinas treated with α-AAA acid (p <0.05, 0.04, 0.01, and 0.002). There was no significant difference between Rgr−/− Gnat1−/− or Rgr+/+ Gnat1−/− α-AAA- treated retinas (p uniformly > 0.6). No significant change in response amplitude could be detected when either Rgr+/+ Gnat1−/− or Rgr−/− Gnat1−/− retinas were kept in darkness. (E) Sensitivities were significantly lower at 30, 45, and 60 min in Rgr−/− Gnat1−/− retinas (p < 0.046, 0.034, and 0.02) or retinas treated with α-AAA acid (p < 0.05, 0.034, and 0.02). No significant difference could be detected between Rgr−/− Gnat1−/− and Rgr+/+ Gnat1−/− α-AAA-treated retinas (p uniformly > 0.8).
Post-bleach recovery of cone sensitivity in the dark
Conversion of atROL to 11 cROL by cells expressing Rdh10 and RGR was only observed following exposure to light (Figs. 1A and 1C), suggesting that RGR opsin has no isomerase activity in the dark. An earlier study however showed dark recovery of cones in isolated Gnat1−/− mouse retinas following a deep photobleach, which was interpreted to support the existence of a ‘dark’ isomerase in Müller cells (Wang and Kefalov, 2009). To understand this apparent discrepancy, we repeated the published experiment (Wang and Kefalov, 2009) using an identical 30-second illumination calculated to bleach ~90% of mouse M-opsin pigment in isolated Rgr+/+ Gnat1−/− and Rgr−/− Gnat1−/− retinas, followed by incubation in darkness. Substantially lower recovery of cone sensitivity was seen in Rgr−/− Gnat1−/− retinas (Fig. 6). Both retinas showed an initial, approximately three-fold gain in sensitivity immediately after the bleach, probably due to dark adaptation of the visual transduction cascade (Fig. S3) versus regeneration of cone opsin. After 60 minutes, however, the Rgr−/− Gnat1−/− retinas exhibited ~10-fold lower cone sensitivity than the Rgr+/+ Gnat1−/− retinas.
Figure 6. The role of RGR in dark adaptation.

Time dependence of normalized M-cone sensitivity following a 565nm light exposure calculated to bleach 90% of the cone photopigment. The following animals were used for bleach exposures with the following durations (the intensity of the bleaching light was adjusted to give the same calculated bleach): Rgr+/+ Gnat1−/− with 30 second stimulus (n = 8) Rgr−/− Gnat1−/− with 30 second stimulus (n = 5), Rgr+/+ Gnat1−/− with 350 ms stimulus (n = 5), and Rgr+/+ Gnat1−/− with 5 min stimulus (n = 5). Post bleach recovery of sensitivity was significantly different between Rgr+/+ Gnat1−/− bleached in 30 seconds and Rgr−/− Gnat1−/− bleached in 30 seconds (p < 0.025, 0.048, 0.024, and 0.03 at 5, 15, 30 and 60 min after the bleach). The duration of the bleach also affected the recovery of sensitivity. Rgr+/+ Gnat1−/− bleached with 350 ms stimulus showed significantly smaller recovery compared to Rgr+/+ Gnat1−/− bleached with the 30 second stimulus (p < 0.045, 0.049, 0.017 and 0.03 at 5, 15, 30 and 60 min after the bleach). There was no statistically significant difference between Rgr+/+ Gnat1−/− bleached in 30s and Rgr+/+ Gnat1−/− bleached in 5 min.
Regeneration of a cone opsin pigment by RGR opsin and Rdh10 in Müller cells requires multiple steps: (i) dissociation of bleached cone opsin; (ii) reduction of atRAL to atROL by Rdh8; (iii) transit of atROL from cone to Müller cell; (iv) oxidation of atROL to atRAL by Rdh10; (v) photoisomerization of atRAL to 11cRAL by RGR opsin; (vi) reduction of 11cRAL to 11cROL by Rdh10; (vii) transit of 11cROL from Müller cell to cone; (viii) oxidation of 11cROL to 11cRAL by the unidentified cone 11cRDH; and (ix) conversion of apo-opsin to cone opsin pigment by recombination with 11cRAL. Since these events take time to complete, short exposure of a retina to bright light might have a different effect than long exposure to dim light conveying similar total photons. To test this possibility, we repeated the experiment of Fig. 6 on Rgr+/+ Gnat1−/− retinas with a much brighter light of shorter duration calculated to bleach the same 90% of M-opsin in 350 ms instead of in 30 seconds. Here, we observed much less RGR-dependent recovery of cone sensitivity (blue squares, Fig. 6). We also repeated the experiment on Rgr+/+ Gnat1−/− retinas with a dimmer light calculated to bleach 90% of M-opsins in five minutes. This dimmer, longer-duration light exposure yielded much greater cone recovery in comparison to the 30-second bleach, exemplified by the ~10-fold higher sensitivity already at five minutes after the bleach (green versus black squares, Fig. 6). This difference may reflect in part greater dark adaptation after exposure to the dimmer bleaching light, but it also suggests that there may have been significant RGR- dependent M-opsin regeneration even during the five-minute light exposure.
Effect of N-ret-PE photoisomerization on the cone photoresponse under continuous illumination
N-retinylidene phosphatidylethanolamine (N-ret-PE) is a conjugate of retinaldehyde and PE in OS disks. N-ret-PE occurs in the same stereoisomeric configurations as retinaldehyde. Recently, at-N-ret-PE was shown to undergo specific photoisomerization to 11c-N-ret-PE upon exposure to blue light (Kaylor et al., 2017). Further, 11c-N-ret-PE efficiently donated its 11cRAL to bleached opsin, regenerating visual pigment in rods and cones (Kaylor et al., 2017). To compare the contributions of N-ret-PE- and RGR-photoisomerization to pigment regeneration, we took advantage of the difference in the visible absorption spectra between N-ret-PE (λmax = 450 nm) and M-opsin (λmax = 508 nm). While the photosensitivity of M-opsin is similar at 450 and 560 nm (Govardovskii et al., 2000), the photosensitivity of N-ret-PE is nearly 30-fold higher at 450 versus 560 nm (Kaylor et al., 2017). Accordingly, we measured cone photoresponses to flash families in retinas from Rgr+/+ Gnat1−/− and Rgr−/− Gnat1−/− mice during exposure to monochromatic background light at 450 or 560 nm. Cone responses in Rgr+/+ Gnat1−/− retinas exposed to 450-nm background light declined slowly over 60 minutes (Fig. 7A), reflecting the contributions of both RGR opsin and N-ret-PE photoisomerization to pigment regeneration. Cone responses in Rgr−/− Gnat1−/− retinas exposed to 450-nm light showed a faster decline (Fig. 7B). The responses here reflect M-opsin regeneration through photoisomerization of N-ret-PE, but not RGR opsin. Finally, we measured cone photoresponses in Rgr−/− Gnat1−/− retinas during exposure to 560-nm background light (Fig. 7C). The responses here reflect cone pigment regeneration with minimal contributions from RGR opsin or N-ret-PE. The maximum cone response amplitudes at various times during the 60-minute light exposures are shown in Fig. 7D, while cone sensitivities calculated from the same recordings are shown in Fig. 7E. Both the cone sensitivity and maximum amplitude were significantly lower in retinas illuminated with the 560-nm light (figure legend). Mean intensity-response curves for these experiments are shown in Supplementary Fig. S4. These results suggest that photoisomerization of rGr opsin and N-ret-PE both contribute to cone recovery.
Figure 7. Relative contributions of RGR and N-ret-PE photoisomerization to sensitivity and maximal response amplitude in M-cones.

Representative transretinal ERG responses, (A) through (C); changes in maximum response amplitude (D); and relative photosensitivity, (E) of M-cones of Rgr+/+ Gnat1−/− in continuous 450-nm light (n = 7), Rgr−/− Gnat1−/− in continuous 450-nm light (n = 7) and Rgr−/− Gnat1−/− in continuous 560-nm light (n = 7). M-Cone isolated photoreceptor recordings were made in dark-adapted retinas and after 15, 30, 45 and 60 minutes of exposure to a continuous background light. The intensities of the 450-nm or 560-nm backgrounds were set to the same value of 9.1 × 106 photons (ϕ) μm−2 s−1 effective at the λmax of the M cone pigment. The stimulating light flashes were produced by a 565-nm LED (flashes ranging from 65 ϕ μm−2 to 5.4 × 107 ϕ μm−2 effective at the λmax of the M cone pigment). The red traces show responses to a constant flash of 3.7 × 105 ϕ μm−2. Note the lower response amplitudes in Rgr−/− Gnat1−/− in continuous 450-nm light and even lower amplitudes of Rgr−/− Gnat1−/− responses in continuous 560-nm light. Mean maximum response amplitude (D) and relative dim flash sensitivity (E) of individual recordings in (A), (B) and (C) are calculated and plotted (with S.E.) as a function of time. The maximum amplitudes were significantly lower at 15, 30, 45, and 60 min in Rgr−/− Gnat1−/− retinas in 450-nm light (p < 0.047, 0.045, 0.039 and 0.031) and Rgr−/− Gnat1−/− at 560-nm (p < 0.004, 0.008, 0.006 and 0.005) compared to Rgr+/+ Gnat1−/− in 450-nm. The maximum amplitudes were also significantly lower at 45, and 60 min in Rgr−/− Gnat1−/− retinas in 450-nm light compared to Rgr−/− Gnat1−/− retinas in 560-nm light (p < 0.048 and 0.042). The sensitivities were significantly lower at 30, 45, and 60 min in Rgr−/− Gnat1−/− retinas in 450-nm light (p < 0.019, 0.033, and 0.003) and at 15, 30, 45 and 60 min in Rgr−/− Gnat1−/− at 560-nm (p < 0.022, 013, 0.026, and 0.002) compared to Rgr+/+ Gnat1−/− in 450-nm. The sensitivities were also significantly lower at 15, 45, and 60 min in Rgr−/− Gnat1−/− retinas in 560-nm light compared to Rgr−/− Gnat1−/− retinas in 450-nm light (p < 0.019, 0.033, and 0.003).
DISCUSSION
This work describes a new mechanism for the regeneration of cone visual pigment. Illumination of RGR opsin in cells that also express Rdh10 results in the conversion of atROL to 11cROL. Since bleached cones, but not rods, can recover light sensitivity upon addition of 11cROL (Jones et al., 1989), production of 11cROL by RGR—Rdh10 allows cones to escape competition from rods for limited chromophore in bright light. If RGR opsin affects light-dependent regeneration of cone visual pigment, cone function should be impaired in light-exposed retinas from Rgr−/− mice. To test this possibility, we generated Rgr−/− Gnat1−/− double-mutant mice that lack both RGR opsin and rod α-transducin. Cone responses were similar in dark-adapted retinas of the two genotypes (Fig. 4). However, striking differences were observed between retinas that express or lack RGR opsin when recordings were made under continuous background illumination. While Rgr+/+ retinas exhibited only gradual loss of cone sensitivity under constant background light, Rgr−/− retinas lost cone sensitivity much more rapidly (Fig. 5). These observations indicate that RGR opsin contributes to sustained cone vision under daylight conditions.
Co-expression of RGR opsin and Rdh10 yielded a novel catalytic activity that converts atROL to 11cROL upon exposure to visible light (Fig. 1A). Following photoisomerization, the 11cRAL product remains bound to the RGR opsin protein (Chen et al., 2001b). This restriction on RGR turnover is overcome by mass action through reduction of 11cRAL to 11cROL by Rdh10. We observed similar redox-photoisomerase activity in microsomes from normal mouse retinas, which was much reduced in microsomes from Rgr−/− retinas (Fig. 2A). Cultured Müller cells were previously shown to take up atROL and release 11cROL into the medium through an unknown mechanism (Betts-Obregon et al., 2014; Das et al., 1992). Also, bleached cones, but not rods, have been shown to recover light sensitivity upon addition of 11cROL (Goldstein, 1970; Hood and Hock, 1973; Jones et al., 1989). The results presented here suggest that RGR opsin and Rdh10 are the proteins responsible for 11cROL production during light exposure by Müller cells, as depicted in Fig. 8.
Figure 8. Hypothesized Müller-cell visual cycle.
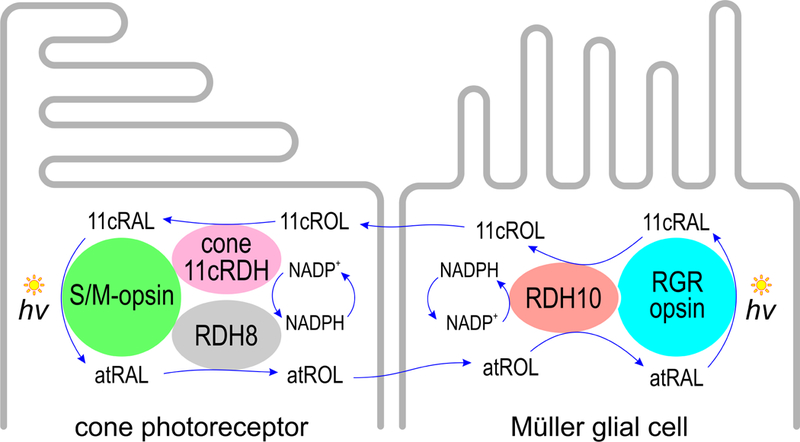
During light exposure, Müller cells take up atROL released by cones and rods (not shown). Rdh10 oxidizes atROL to atRAL, which forms a visual pigment with RGR opsin and is isomerized to 11cRAL upon absorption of a photon (hv). The 11cRAL is reduced to 11cROL by Rdh10, balancing the redox reaction in the Müller cell with no net consumption of NADPH or NADP+. The 11cROL is taken up by a cone cell, which contains an as-yet unidentified 11cRDH that oxidizes 11cROL to 11cRAL. The 11cRAL combines with apo-opsin to regenerate the M-opsin pigment. Absorption of a photon (hv) by M-opsin activates the visual transduction cascade as the first step in visual perception (not shown). The bleached M-opsin releases atRAL, which is reduced by Rdh8 to atROL, completing the visual cycle.
Since RGR opsin and Rdh10 are also both expressed in RPE cells, we analyzed microsomes from Rgr+/+ and Rgr−/− RPE for redox-photoisomerase activity. Similar to wild-type retina microsomes, RPE microsomes also showed light-dependent conversion of atROL to 11cROL (Figs. 2A and 2B). These results suggest that RPE cells, in addition to Müller cells, provide 11cROL for light-dependent regeneration of cone pigments. Thus, RPE cells may contain two visual cycles: the LRAT—Rpe65 pathway (Saari, 2016), and the new pathway defined here by RGR—Rdh10. While RGR opsin is abundant in Müller cells of human (Trifunovic et al., 2008), bovine (Jiang et al., 1993; Pandey et al., 1994) and chicken retinas (Diaz et al., 2017), it is much less abundant in mouse retinas (Tao et al., 1998; Trifunovic et al., 2008) (Fig. 3A). Mice are nocturnal animals and thereby have less need for a photoisomerase to maintain cone sensitivity under daylight conditions. Despite its low abundance in mouse retinas, we observed robust biochemical (Fig. 2) and physiological (Figs. 4–6) phenotypes in isolated retinas from Rgr+/+ versus Rgr−/− mice. These results suggest that RGR opsin contributes more to cone pigment regeneration in diurnal animals, such as humans, cows and chickens, where RGR opsin is more abundant in Müller cells (Diaz et al., 2017; Pandey et al., 1994; Trifunovic et al., 2008).
If the retinoid-isomerase activity of RGR opsin is light dependent, as shown previously for retinaldehyde (Hao and Fong, 1999) and here for retinol (Figs. 1A and1C), why do cones exhibit RGR-dependent recovery in the dark following a photobleach (Fig. 6)? The initial phase of post-bleach recovery observed in both Rgr+/+ and Rgr−/− retinas probably reflects dark adaptation of the visual transduction cascade (Fig. 6). The slower, RGR-dependent recovery may reflect regeneration of M-opsin from a storage pool of 11cROL or 11cRAL previously produced by RGR during light exposure. This storage pool may correspond to CRALBP in Müller cells (Bunt-Milam and Saari, 1983), which binds 11cRAL and 11cROL with high affinity but has low affinity for other retinoid isomers (Saari and Bredberg, 1987). In agreement with this notion, cone recovery after light exposure was shown to depend critically on CRALBP in isolated mouse retinas (Xue et al., 2015).
We are used to thinking that visual pigments in vertebrate rods and cones regenerate pigment through a biochemical mechanism involving the enzymatic visual cycle in RPE cells, whereas rhabdomeric photoreceptors of invertebrates use light to regenerate opsin (Fain et al., 2010). This strict dichotomy is no longer valid. Here we show that cones in mice recover light sensitivity through a photic mechanism involving RGR opsin in Müller cells, in addition to the LRAT—Rpe65 visual cycle in RPE cells. Although this RGR-dependent photic mechanism is different from photoregeneration of bistable opsins in rhabdomeric photoreceptors, it offers the same advantages. With photic regeneration, the rate of chromophore synthesis scales with light intensity, while metabolic regeneration of chromophore is limited by enzyme turnover. Also, conversion of atROL to 11cROL is endergonic (ΔG = +4.1 kcal/mol) (Rando and Chang, 1983). For the LRAT—Rpe65 visual cycle, retinol isomerization comes at the metabolic cost of an activated fatty acid for each absorbed photon (Rando, 1991). Similar to insects and other invertebrates, vertebrates can now claim use of solar energy to power regeneration of cone visual pigment.
STAR ★ METHODS OUTLINE
Detailed methods are provided in the online version of this paper and include the following:
KEY RESOURES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-RGR | Andreas Wenzel | Pin 3 |
| Guinea pig polyclonal anti-RGR | Andreas Wenzel | Pin 2 |
| IRDye 800CW donkey anti-guinea pig antibody | LI-COR | 926–32411 |
| Mouse Monoclonal ANTI-FLAG® M2 antibody | Sigma-Aldrich | F3165 |
| Mouse anti-Myc Tag Antibody, clone 4A6 | Millipore Sigma | 05–724 |
| Donkey anti-Mouse 800 secondary antibody IgG (H+L) | Li-COR | 926–32212 |
| Mouse monoclonal RLBP-1 (clone 1H7) antibody | Sigma-Aldrich | WH0006017M1 |
| Goat anti-Rabbit IgG (H+L) secondary antibody, Alexa Fluor 488 | Thermo Fisher | A-11008 |
| Goat anti-mouse IgG (H+L) secondary antibody, Alexa Fluor 568 | Thermo Fisher | A-11004 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| All-trans-retinol | Sigma-Aldrich | 95144; CAS# 68–26-8 |
| Bovine serum albumin | Sigma-Aldrich | A6003; CAS# 9048–46-8 |
| Polyfect™ Transfection Reagent | Qiagen | 301107 |
| Hydroxylamine hydrochloride | Sigma-Aldrich | 255580; CAS# 5470–11-1 |
| Hexanes | Thermo Fisher | H303–4; CAS# 110–54-3 |
| 1,4-dioxane | Sigma-Aldrich | 34857; CAS# 123–91-1 |
| Ames’ medium | Sigma-Aldrich | A1420 |
| Sodium bicarbonate | Sigma-Aldrich | S6014; CAS# 144–55-8 |
| L-aspartic acid | Sigma-Aldrich | A9256; CAS# 56–84-8 |
| DL-2-amino-4-phosphonobutyric acid | Tocris Bioscience | 0101; CAS# 20263–07-4 |
| Sodium L-lactate | Sigma-Aldrich | 71718; CAS# 867–56-1 |
| Benzonase nuclease | Sigma-Aldrich | E1014–25KU; CAS# 9025–65-4 |
| Halt protease inhibitor cocktail (100×) | Thermo Scientific | 78429 |
| Micro BCA protein assay kit | Thermo Scientific | 23235 |
| NuPAGE LDS sample buffer (4×) | Novex (Life Technologies) | NP0007 |
| NuPAGE sample reducing agent (10×) | Novex (Life Technologies) | NP0009 |
| NuPAGE 12% Bis-Tris gel | Novex (Life Technologies) | NP0342BOX |
| Immobilon-FL transfer membrane | Merck Millipore | IPFL00010 |
| Odyssey blocking buffer™ (PBS) | LI-COR | 927–40000 |
| Donkey serum | Sigma-Aldrich | D9663–10ML |
| Anased xylazine injection solution | Akorn | NADA# 139–236 |
| Ketamine hydrochloride | Putney | NADA# 26637–731-51 |
| DMEM (phenol free) | Thermo Fisher | 21063–029 |
| Penicillin-streptomycin | Thermo Fisher | 15070–063 |
| BCA Protein Assay Kit | Thermo Fisher | 23225 |
| Sodium dodecyl sulfate (SDS) | Sigma-Aldrich | L3771; CAS# 151–21-3 |
| Fetal bovine serum | Thermo Fisher | 16140–071 |
| Nicotinamide adenine dinucleotide phosphate (reduced) | Sigma-Aldrich | N7505; CAS# 2646–71-1 |
| Protein G Dynabeads™ immunoprecipitation kit | Thermo Fisher | 10007D |
| TRIS base | Thermo Fisher | BP152; CAS# 77–86-1 |
| Sodium chloride | Thermo Fisher | S271–500; CAS# 7647–14-5 |
| Glycerol | Thermo Fisher | G33–500; CAS# 56–81-5 |
| NP-40 detergent Surfact-Amps™ solution | Thermo Fisher | 28324: CAS# 9016–45-9 |
| Phosphate buffered saline (PBS) | Thermo Fisher | 20012–050 |
| Tween 20 | Thermo Fisher | BP337–500; CAS# 9005–64-5 |
| Sodium borohydride | Sigma-Aldrich | 213462; CAS# 16940–66-2 |
| Triton X-100 | Sigma-Aldrich | T-9284; CAS# 9002–93-1 |
| normal goat serum | Sigma-Aldrich | G9023 |
| Prolong Gold antifade reagent with DAPI mounting solution | Thermo Fisher | P36935 |
| Experimental Models: Cell Lines | ||
| HEK-293T Cell Line | ATCC | CRL-11268 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Rgr −/− | Henry Fong Lab | University of Southern California |
| Mouse: Gnat1 −/− | Janis Lem | Tufts University, Boston |
| Mouse: Rgr −/− / Gnat1 −/− double knock-out | This paper | |
| Mouse: wild-type 129S6/SvEv Tac | Taconic Biosciences | TAC 129SVE |
| Oligonucleotides | ||
| (Rd8): forward-5′GGTGACCAATCTGTTGACAATCC | PMID: 22447858 | |
| (Rd8): reverse-5′GCCCCATTTGCACACTGATGAC | PMID: 22447858 | |
| (Rpe65 codon 450): forward- 5′CCTTTGAATTTCCTCAAATCAATTA | PMID: 18474598 | |
| (Rpe65 codon 450): reverse- 5′TTCCAGAGCATCTGGTTGAG | PMID: 18474598 | |
| (Gnat1−/−): WT forward- 5′GTTCATTGCCATCATCTACGG | PMID: 11095744 | |
| (Gnat1−/−): WT reverse- 5′GCATTGTGCCTTCCTCAATAG | PMID: 11095744 | |
| (Gnat1−/−): KO forward- 5′AGCACAGCTTTCCTTTCAGG | PMID: 11095744 | |
| (Gnat1−/−): KO reverse- 5′CAGAAAGCGAAGGAGCAAAG | PMID: 11095744 | |
| (RGR−/−): forward (‘RGR-Oligo1’)- 5′TGCATTTTCCTGTGAGATGG | PMID: 18474598 | |
| (RGR−/−): reverse (‘RGR-Oligo2’)- 5′GCTCAGTACCAGCAGGTTGC | PMID: 18474598 | |
| (RGR−/−): reverse (‘RGR-Oligo3’)- 5′GGGGAACTTCCTGACTAGGG | PMID: 18474598 | |
| Recombinant DNA | ||
| Plasmid: pcDNA 3.1+ | Thermo Fisher | V79020 (vector for all constructs) |
| Human RDH5 | Genscript | NM_001199771.1 |
| Human RDH8 | Genscript | NM_015725.2 |
| Human RDH10 | Genscript | NM_172037.5 |
| Human RDH14 | Genscript | NM_020905.4 |
| Chicken RDH10 | Genscript | NM_001199459.1 |
| Human RDH11 | Krzysztof Palczewski | NM_016026.4 |
| Mouse RDH10 | Origene | NM_133832.3 |
| Bovine RDH10 | Genscript | NM_174734.2 |
| Bovine RGR | Genscript | NM_175775.2 |
| Chicken RGR | Genscript | NM_001031216.1 |
| Software and Algorithms | ||
| SigmaPlot (biochemistry data) | Systat Software Inc. | http://www.sigmaplot.co.uk/products/sigmaplot/sigmaplot-details.php |
| OriginPro (physiology data) | OriginLab | http://www.originlab.com/Origin |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Gabriel H. Travis (travis@jsei.ucla.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animal use and care statement
This study was carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, and the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research. The animal use protocol was approved by the University of California, Los Angeles Animal Research Committee (Permit Number: A3196–01). Euthanasia was performed by cervical dislocation in deeply anesthetized mice by intraperitoneal injections (xylazine 10 mg/kg and ketamine 100 mg/kg). All efforts were made to minimize pain and discomfort in mice used in this study.
Mice and Genotyping
All mice were reared under 12-hour cyclic light. Rgr−/− mice were generously provided by Henry Fong. The genotyping protocol was as previously reported (Radu et al., 2008). Wild-type (129/Sv) mice were purchased from Taconic Biosciences, Inc. All mice used were tested to exclude the spontaneous Rd8 and Rpe65-M450 mutations using the primers (Rd8): forward-5’GGTGACCAATCTGTTGACAATCC and reverse-5’GCCCCATTTGCACACTGATGAC; and (Rpe65): forward- 5’CCTTTGAATTTCCTCAAATCAATTA and reverse-5’TTCCAGAGCATCTGGTTGAG. To determine the genotype at Gnat1, we used the wild-type primer set: forward-5’GTTCATTGCCATCATCTACGG and reverse-5’GCATTGTGCCTTCCTCAATAG; and the knockout primer set: forward-5’AGCACAGCTTTCCTTTCAGG and reverse-5’CAGAAAGCGAAGGAGCAAAG. To determine the genotype at Rgr, we used the multiplexed wild-type and knockout primer set forward- 5’TGCATTTTCCTGTGAGATGG (‘RGR-oligo1’), reverse-5’GCTCAGTACCAGCAGGTTGC (‘RGR- oligo2’), and reverse-5’GGGGAACTTCCTGACTAGGG (‘RGR-oligo3’). For enzymatic assays, retina and RPE-containing eyecups were isolated from the eyes of two-month-old wild-type (129/Sv) and Rgr−/− mice of both sexes.
HEK-293T Cells
Authenticated human female embryonic kidney epithelial cells were purchased from ATCC (HEK 293T/17; CRL-11268). Cells were grown and maintained in DMEM (Gibco/Thermofisher) supplemented with 10% heat-inactivated fetal bovine serum and antibiotics (100 u/mL of penicillin G and 100 μg/mL of streptomycin) at 37°C in 5% CO2. These cells constitutively express the simian virus 40 (SV40) large T antigen and clone 17 was selected specifically for its high transfectability.
METHOD DETAILS
General enzyme assay conditions
All experimental manipulations involving retinoids were performed in a darkroom under dim red light. Protein samples and solutions were kept on ice until use. Stocks of atROL were freshly dissolved in ethanol and stored on ice. Stock concentration was determined by UV-VIS spectroscopy using the reported extinction coefficient (ϵ) for all-trans-ROL (λmax = 325 nm, ϵ = 52,770 M−1cm−1)(Leenheer et al., 2000). Unless otherwise stated, all chemicals and solvents were purchased from Sigma-Aldrich. Protein concentrations were measured using the Micro BCA Protein Assay Kit (Pierce).
Normal-phase HPLC analysis of retinoids
Retinoids were extracted from assay mixtures after quenching the reactions with methanol (2 mL) followed by addition of 25 μL 5% SDS (0.2% SDS final concentration) and brine (50 μL). To protect retinaldehydes, retinal oximes were generated by addition of hydroxylamine hydrochloride (500 μL of 1.0 M solution) (Sigma), vortexing, and incubation at room temperature for 15 minutes. The samples were then twice extracted by addition of two mL aliquots of hexane followed by brief vortexing and centrifugation at 3000 × g for five minutes to separate phases. Pooled hexane extracts were added to 13 × 100 mm borosilicate test tubes and evaporated to dryness under a stream of nitrogen. Dried samples were then dissolved in 115 μL hexane and analyzed by normal-phase liquid chromatography in an Agilent 1100 series chromatograph equipped with a photodiode-array detector using an Agilent Zorbax RX-SIL column (4.6 × 100 mm, 1.8 μM) using a 0.24–10% dioxane gradient in hexane, at a flow rate of 0.9 mL per minute. Spectra (190–550 nm) were acquired for all eluted peaks. The identity of each eluted peak was established by comparing its spectrum and elution time with those of authentic retinoid standards. Sample peaks were quantitated by comparing peak areas to calibration curves established with retinoid standards.
Activity of RGR and RDH10 in dark versus light
Bovine RDH10 (NM_174734.2) and bovine RGR (NM_175775.2) cDNA’s were synthesized by GenScript and subcloned into the mammalian expression vector, pcDNA 3.1 (ThermoFisher). HEK-293T cells were grown in DMEM (Gibco/Thermofisher) supplemented with 10% heat-inactivated fetal bovine serum and antibiotics (100 U/mL of penicillin G and 100 μg/mL of streptomycin) at 37°C in 5% CO2. HEK-293T cells were transfected (PolyFect, Qiagen) with non-recombinant pcDNA3.1, pcDNA3.1- RDH10, and/or pcDNA3.1-RGR. When more than one type of clone were used, the transfection were done with equal amounts (50/50) of plasmid. After approximately 40 hours in culture, the medium above the cells was removed and replaced with phenol red-free DMEM (Gibco/ThermoFisher: 21063–029) supplemented with 5 μM atRoL, 2% bovine serum albumin (BSA), and 250 μM NADPH. Plates were placed in a 37°C incubator and exposed for 30 minutes to monochromatic light (470nm +/− 10nm at 0.4 W/m2) or kept in the dark. The light wavelength and intensity was measured with a spectroradiometer (Black-comet CXR-SR-50, StellarNet Inc.) The monochromatic light was generated by a custom monochromator (Newport Instruments) with a xenon arc lamp. After each assay, the media was separated from the cells by centrifugation (five minutes at 1,000 rpm in a Sorvall Legend RT). The media were extracted as described above for retinoid analysis.
Immunoblot analysis
The immunoblot analysis was performed following similar methods previously published (Kaylor et al., 2013; Lenis et al., 2017). Retinas and RPE-containing eyecups were dissected from euthanized 129/Sv (Rgr+/+) and Rgr−/− mice (about 12–14 weeks old), and homogenized in 1X PBS (pH 7.2) with Halt protease inhibitor cocktail on ice. The homogenates were treated with benzonase nuclease for one hour at room temperature followed by 1% (final concentration) SDS at 4 °C for 20 minutes. The treated homogenates were centrifuged at 3,000 × g for 5 minutes and the supernatants were collected and stored at −80 °C for further analysis. The total protein concentration of each sample was determined by a micro BCA protein assay (Thermo Scientific) according to the protocol suggested by the manufacturer. Protein samples were heat-denatured in NuPAGE LDS sample buffer and NuPAGE sample reducing agent, and then separated by a NuPAGE 12% Bis-Tris gel (Novex by Life Technologies). Proteins were transferred to an Immobilon-FL PVDF transfer membrane (Merck Millipore) using a Trans-Blot SD semi-dry transfer cell (BIO-RAD). The blot/membrane was blocked in Odyssey blocking buffer and probed with a guinea pig anti-RGR (‘Pin2’) primary antibody (courtesy of Andreas Wenzel) at 1:1000 dilution followed by an IRDye 800CW donkey anti-guinea pig (LI-COR) secondary antibody at 1:15000 dilution in Odyssey blocking buffer with 0.1% Tween 20 and 0.5% donkey serum. The blot/membrane was imaged by an Odyssey CLx Infrared Imaging System (LICOR).
Immunocytochemistry of mouse retina sections
Eyes were enucleated and fixed in 2% PFA in 0.1M sodium phosphate buffer for one hour. Eyecups were prepared by removal of anterior segments then infiltrated with 10–30% sucrose, embedded in OCT, and cut into 18-μm sections. Sections were reduced in 0.1 M NaBH4, washed in PBS, permeabilized with 0.1% Triton x-100, and blocked with 1% BSA and 5% normal goat serum. Slides were probed overnight at 4°C with rabbit polyclonal anti-RGR ‘Pin3’ (red) and mouse monoclonal anti- RLBP-1 clone 1H7 (green). All sections were washed and incubated with Alexa Fluor secondary antibodies for two hours at room temperature. Slides were mounted with ProLong Gold antifade reagent containing DAPI. Images were obtained using Olympus FluoView FV1000 confocal microscope under a 40x oil objective.
Activity of RGR with different RDH’s in the retina
Human RDH5 (NM_001199771.1), RDH8 (NM_015725.2), RDH10 (NM_172037.4), and RDH14 (NM_020905.3), as well as chicken RDH10 (NM_001199459.1), were synthesized by GenScript and placed in mammalian expression vector pcDNA 3.1 (ThermoFisher). Mouse RDH10 (NM_133832.3; Origene) and human RDH11 (NM_016026.3; generously provided by Krzysztof Palczewski) were also placed in the mammalian expression vector pcDNA 3.1. Retinol dehydrogenase clones were transfected as described above into HEK 293T cells with bovine RGR (50/50 plasmid mix). Background controls were transfected by replacing the RDH clone with non-recombinant pcDNA3.1 (RGR only control) or by transfecting the cells only with pcDNA 3.1 (cell background control). After culturing forapproximately 40 hours to express proteins, the media were changed as described above. All plates were exposed to 30 minutes of monochromatic light (470nm +/− 10nm at 0.4 W/m2) and treated as described above for retinoid analysis.
Action Spectrum of Bovine RGR/RDH10 Activity
HEK-293T cells were transfected (PolyFect, Qiagen) with five μg each of pcDNA3.1-bRDH10 and pcDNA3.1-bRGR plasmid. After ~ 40 hours the media were replaced with phenol red-free DMEM supplemented with five μM atROL and 2% bovine serum albumin (BSA) (Sigma). The plates were either kept in the dark or illuminated with monochromatic light at wavelengths of 425 to 575 nm with 20–30 nm increments at 37°C for 30 minutes. The monoch romatic light was generated by monochromator and measured by spectroradiometer as described above. The light intensities were adjusted (from 0.35 W/m2 at 425 nm to 0.26 W/m2 at 575 nm) such that each wavelength delivered the same photon flux. The media were extracted as described above and analyzed for retinoid content.
Retinoid Photoisomerization in Mouse Retina and RPE Microsomes
Retina and RPE-containing eyecups were isolated from the eyes of two-month-old wild-type (129/Sv) and Rgr−/− mice. Identical tissues were combined and homogenized in 500 μl mL of pH 7.0 phosphate citrate (PC) buffer (Sigma) for each mouse strain using glass-glass homogenizers (Kontes). The bulk homogenates were pelleted at 15,000 × g (Eppendorf 5424 centrifuge) for five minutes at 4°c. The S1- supernatants were collected and re-spun at 100,000g (Sorvall M-150 ultra-centrifuge) for 60 minutes at 4°C. The S2-supernatants were discarded and the P2- pellets were resuspended in 3.5 mL of PC buffer. Samples of each microsome preparation were used for protein determinations. Triplicate aliquots containing 500 μL of microsomes plus 2% BSA and 5 μM atROL were used in the assays. One set of samples was immediately extracted to determine the content of retinoids before light exposure in order to ascertain the endogenous retinoid profile. The other samples were placed in cuvettes and agitated, exposed to monochromatic light (470 nm +/− 10nm at 0.4 W/m2) for 30 minutes at 37°C. Retinoids were extracted and analyzed by normal-phase HPLC as described above. The remaining 11cROL and 11cRAL levels reflect retinoids synthesized during light exposure in microsomes from wild-type and Rgr−/− retinas.
Electrophysiology
Eyes from mice were enucleated under dim red light or in darkness by means of infrared goggles (American Technologies Network Corporation, San Francisco, CA, USA). The anterior portion of the eye was cut and the lens and cornea were removed in darkness with a dissection microscope. The retina was isolated from the eyecup, and the retinal pigment epithelium was removed with fine tweezers. The retina was then mounted on filter paper (Millipore, 0.45 μm), on the bottom compartment of a perfusion chamber (Vinberg et al., 2014), with the photoreceptor side up in complete darkness. One Ag/AgCl pellet electrode was placed in contact with electrode solution on ganglion cell side of the retina, and another was placed in the solution bathing the photoreceptors. The electrodes were connected to a differential amplifier (Warner instruments DP-311).
During recording, the photoreceptors were continuously perfused with Ames’ medium (Sigma Chemical, St Louis, MO, USA), containing an additional 1.9 g/l NaHCO3 and equilibrated with 95% O2 / 5% CO2. This solution was supplemented with 2 mM aspartic acid, 40 μM DL-2-amino-4-phosphonobutyric acid (AP4, Tocris Bioscience, Bristol, UK), 4 mM L-lactate, and 10 μM atROL in 0.05% bovine serum albumin (Sigma). The osmolarity of the medium was adjusted to 284 mOsm with a vapor-pressure osmometer (Wescore, Logan, UT). Temperature was maintained at 36–38C with an automatic temperature controller (Warner instruments, Hamden, CT).
Illumination was delivered with an OptoLED optical system (Cairn Research, Faversham, UK) coupled to an inverted microscope. The 565-nm test flashes and 505-nm background were produced by monochromatic LEDs at the appropriate wavelengths, but the 450-nm and 560-nm illuminations were provided by a white LED coupled to 10-nm-bandwidth interference filters (Andover Corp, Salem, NH), as in previous experiments (Kaylor et al., 2017). The intensities of the test and bleaching lights were calibrated with a photodiode (OSI Optoelectronics, Hawthorne, CA), and the intensities of the 450-nm and 560-nm background lights were set to the same number of photons by adjusting the current of the white LED. To calculate the number of pigment molecules bleached by this illumination, we compared the number of photons per μm2 required to produce a half-maximal response (I½) of single dark-adapted mouse M cones in retinal slices (Kaylor et al., 2017) with the I½ of the dark-adapted M-cone response of whole retina (Fig. 2). The ratio was then multiplied by the collecting area of the cones, referenced to the collecting area of rods obtained from single-photon responses in slice recordings. Our value of 0.105 is close to that obtained by Vinberg and coworkers (Vinberg et al., 2014). Recordings were filtered and sampled at 1 kHz. Data were displayed and analyzed with PCLAMP (Molecular Devices, Sunnyvale, CA) and Origin Plotting software (OriginLabs, Cambridge, MA).
Pigment bleaching was achieved by illuminating the isolated retina inside the recording chamber. The fraction of pigment bleached is independent of the photoreceptor collecting area and could be estimated from: F = 1 – exp (– IPt ), where F is the fraction bleached, I is the intensity of the bleaching light, t is the time of exposure of the bleaching light, and P is the in situ photosensitivity of vertebrate photopigment (5.7 × 10–9 μm 2), (Nymark et al., 2012; Woodruff et al., 2004). It is important to note that the use of this equation assumes that there was no pigment regeneration.
QUANTIFICATION AND STATISTICAL ANALYSIS
Production of 11cROL and 11cRAL from atROL by RGR and RDH10 co-expressing cells in 470 nm light was analyzed by one-way ANOVA with Tukey’s post hoc analysis. Each bar represents n = 3 plates of transfected cells. Data represent mean ± sD; n.s., not significant, * p < 0.05, ** p < 0.01, *** p < 0.001.
The action spectrum of bovine RGR with bovine RHD10 was analyzed by one-way ANOVA with Tukey’s post hoc analysis. Each bar represents n = 3 plates of transfected cells. Data represent mean ± SD; n.s., not significant, * p < 0.05, ** p < 0.01, *** p < 0.001.
RGR knockout mouse microsome photoisomerase activity was analyzed by one-way ANOVA with Tukey’s post hoc analysis. Each bar represents n = 3 samples of microsomes. Data represent mean ± SD; n.s., not significant, * p < 0.05, ** p < 0.01, *** p < 0.001.
Differences in sensitivity and maximum amplitude of physiological recordings were tested with the Student’s T test assuming a two-tailed distribution and unequal variance. The values of the reported probabilities p are given in the figure legends. We deemed significant any differences for which p was less than 0.05.
Supplementary Material
Highlights.
RGR opsin and Rdh10 convert atROL to 11cROL upon exposure to visible light
Normal mouse retinas maintain cone sensitivity during exposure to background light
Rgr−/− mouse retinas progressively lose cone sensitivity during light exposure
Treatment of normal mouse retinas with a Müller-cell toxin replicates the Rgr−/− phenotype
ACKNOWLEDGEMENTS
We gratefully acknowledge Henry Fong for providing Rgr−/− mice, Janis Lem for the Gnat1−/− mice, Andreas Wenzel for the Pin2 and Pin3 anti-RGR Ab’s, and Rosalie Crouch for providing 11cRAL. This work was supported by NEI, National Institutes of Health Grants R01-EY024379 (to G.H.T.); R01-EY025002 (to R.A.R.), R01-EY029817 (A.P.S.); and R01-EY001844 (G.L.F.); NEI, National Institutes of Health Core Grant P30-EY000331; and a Research to Prevent Blindness unrestricted grant (to the Jules Stein Eye Institute). G.H.T. is the Charles Kenneth Feldman Professor of Ophthalmology at UCLA.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Betts-Obregon BS., Gonzalez-Fernandez F., and Tsin AT. (2014). Interphotoreceptor retinoid-binding protein (IRBP) promotes retinol uptake and release by rat Muller cells (rMC-1) in vitro: implications for the cone visual cycle. Invest Ophthalmol Vis Sci 55, 6265–6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner EA., and Wolter JR. (1962). Transmission of the Ocular Media. Invest Ophthalmol Vis Sci 1, 776–783. [Google Scholar]
- Bunt-Milam AH., and Saari JC. (1983). Immunocytochemical localization of two retinoid-binding proteins in vertebrate retina. Journal of Cell Biology 97, 703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt DA. (1994). Light adaptation and photopigment bleaching in cone photoreceptors in situ in the retina of the turtle. J Neurosci 14, 1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert PD., Krasnoperova NV., Lyubarsky AL., Isayama T., Nicolo M., Kosaras B., Wong G., Gannon KS., Margolskee RF., Sidman RL., et al. (2000). Phototransduction in transgenic mice after targeted deletion of the rod transducin alpha-subunit. Proc Natl Acad Sci U S A 97, 13913–13918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Hao WS., Rife L., Wang XP., Shen DW., Chen J., Ogden T., Van Boemel GB., Wu LY., Yang M., and Fong HKW. (2001a). A photic visual cycle of rhodopsin regeneration is dependent on Rgr. Nat Genet 28, 256–260. [DOI] [PubMed] [Google Scholar]
- Chen P., Lee TD., and Fong HKW. (2001b). Interaction of 11-cis-retinol dehydrogenase with the chromophore of retinal G protein-coupled receptor opsin. Journal of Biological Chemistry 276, 21098–21104. [DOI] [PubMed] [Google Scholar]
- Das SR., Bhardwaj N., Kjeldbye H., and Gouras P. (1992). Muller cells of chicken retina synthesize 11-cis-retinol. Biochemical Journal 285, 907–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz NM., Morera LP., Tempesti T., and Guido ME. (2017). The Visual Cycle in the Inner Retina of Chicken and the Involvement of Retinal G-Protein-Coupled Receptor (RGR). Molecular neurobiology 54, 2507–2517. [DOI] [PubMed] [Google Scholar]
- Fain GL., Hardie R., and Laughlin SB. (2010). Phototransduction and the evolution of photoreceptors. Current biology : CB 20, R114–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farjo KM., Moiseyev G., Takahashi Y., Crouch RK., and Ma JX. (2009). The 11-cis-retinol dehydrogenase activity of RDH10 and its interaction with visual cycle proteins. Invest Ophthalmol Vis Sci 50, 5089–5097. [DOI] [PubMed] [Google Scholar]
- Franke RR., Sakmar TP., Graham RM., and Khorana HG. (1992). Structure and function in rhodopsin. Studies of the interaction between the rhodopsin cytoplasmic domain and transducin. Journal of Biological Chemistry 267, 14767–14774. [PubMed] [Google Scholar]
- Fritze O., Filipek S., Kuksa V., Palczewski K., Hofmann KP., and Ernst OP. (2003). Role of the conserved NPxxY(x)5,6F motif in the rhodopsin ground state and during activation. Proc Natl Acad Sci U S A 100, 2290–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein EB. (1970). Cone pigment regeneration in the isolated frog retina. Vision research 10, 1065–1068. [DOI] [PubMed] [Google Scholar]
- Govardovskii VI., Fyhrquist N., Reuter T., Kuzmin DG., and Donner K. (2000). In search of the visual pigment template. Visual neuroscience 17, 509–528. [DOI] [PubMed] [Google Scholar]
- Haeseleer F., Jang GF., Imanishi Y., Driessen CA., Matsumura M., Nelson PS., and Palczewski K. (2002). Dual-substrate specificity short chain retinol dehydrogenases from the vertebrate retina. J Biol Chem 277, 45537–45546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao W., and Fong HK. (1996). Blue and ultraviolet light-absorbing opsin from the retinal pigment epithelium. Biochemistry 35, 6251–6256. [DOI] [PubMed] [Google Scholar]
- Hao WS., and Fong HKW. (1999). The endogenous chromophore of retinal G protein-coupled receptor opsin from the pigment epithelium. Journal of Biological Chemistry 274, 6085–6090. [DOI] [PubMed] [Google Scholar]
- Hood DC., and Hock PA. (1973). Recovery of cone receptor activity in the frog’s isolated retina. Vision research 13, 1943–1951. [DOI] [PubMed] [Google Scholar]
- Huang J., Possin DE., and Saari JC. (2009). Localizations of visual cycle components in retinal pigment epithelium. Mol Vis 15, 223–234. [PMC free article] [PubMed] [Google Scholar]
- Jablonski MM., and Iannaccone A. (2000). Targeted disruption of Muller cell metabolism induces photoreceptor dysmorphogenesis. Glia 32, 192–204. [DOI] [PubMed] [Google Scholar]
- Jiang M., Pandey S., and Fong HK. (1993). An opsin homologue in the retina and pigment epithelium. Invest Ophthalmol Vis Sci 34, 3669–3678. [PubMed] [Google Scholar]
- Jones GJ., Crouch RK., Wiggert B., Cornwall MC., and Chader GJ. (1989). Retinoid requirements for recovery of sensitivity after visual-pigment bleaching in isolated photoreceptors. Proc Natl Acad Sci U S A 86, 9606–9610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaylor JJ., Cook JD., Makshanoff J., Bischoff N., Yong J., and Travis GH. (2014). Identification of the 11-cis-specific retinyl-ester synthase in retinal Muller cells as multifunctional O-acyltransferase (MFAT). Proc Natl Acad Sci U S A 111, 7302–7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaylor JJ., Xu T., Ingram NT., Tsan A., Hakobyan H., Fain GL., and Travis GH. (2017). Blue light regenerates functional visual pigments in mammals through a retinyl-phospholipid intermediate. Nature communications 8, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaylor JJ., Yuan Q., Cook J., Sarfare S., Makshanoff J., Miu A., Kim A., Kim P., Habib S., Roybal CN., et al. (2013). Identification of DES1 as a vitamin A isomerase in Muller glial cells of the retina. Nature chemical biology 9, 30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiser PD., Kolesnikov AV., Kiser JZ., Dong Z., Chaurasia B., Wang L., Summers SA., Hoang T., Blackshaw S., Peachey NS., et al. (2019). Conditional deletion of Des1 in the mouse retina does not impair the visual cycle in cones. Faseb J, fj201802493R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenheer A.P.d., Lambert WE., and Van Bocxlaer JF. (2000). Modern chromatographic analysis of vitamins, 3rd edn (New York: Marcel Dekker; ). [Google Scholar]
- Lenis TL., Sarfare S., Jiang Z., Lloyd MB., Bok D., and Radu RA. (2017). Complement modulation in the retinal pigment epithelium rescues photoreceptor degeneration in a mouse model of Stargardt disease. Proc Natl Acad Sci U S A 114, 3987–3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T., Van Hooser JP., Driessen CA., Filipek S., Janssen JJ., and Palczewski K. (2003). Evaluation of the role of the retinal G protein-coupled receptor (RGR) in the vertebrate retina in vivo. Journal of neurochemistry 85, 944–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata NL., Radu RA., Clemmons R., and Travis GH. (2002). Isomerization and oxidation of vitamin a in cone-dominant retinas. A novel pathway for visual-pigment regeneration in daylight. Neuron 36, 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata NL., Ruiz A., Radu RA., Bui TV., and Travis GH. (2005). Chicken retinas contain a retinoid isomerase activity that catalyzes the direct conversion of all-trans-retinol to 11 -cis-retinol. Biochemistry 44, 11715–11721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBean GJ. (1994). Inhibition of the glutamate transporter and glial enzymes in rat striatum by the gliotoxin, alpha aminoadipate. Br J Pharmacol 113, 536–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimura H., Saindelle-Ribeaudeau F., Berson EL., and Dryja TP. (1999). Mutations in RGR, encoding a light-sensitive opsin homologue, in patients with retinitis pigmentosa. Nat Genet 23, 393–394. [DOI] [PubMed] [Google Scholar]
- Nymark S., Frederiksen R., Woodruff ML., Cornwall MC., and Fain GL. (2012). Bleaching of mouse rods: microspectrophotometry and suction-electrode recording. The Journal of physiology 590, 2353–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S., Blanks JC., Spee C., Jiang M., and Fong HK. (1994). Cytoplasmic retinal localization of an evolutionary homolog of the visual pigments. Exp Eye Res 58, 605–613. [DOI] [PubMed] [Google Scholar]
- Radu RA., Hu J., Peng J., Bok D., Mata NL., and Travis GH. (2008). Retinal Pigment Epithelium-Retinal G Protein Receptor-Opsin Mediates Light-dependent Translocation of All-trans-retinyl Esters for Synthesis of Visual Chromophore in Retinal Pigment Epithelial Cells. J Biol Chem 283, 19730–19738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando RR. (1991). Membrane phospholipids as an energy source in the operation of the visual cycle. Biochemistry 30, 595–602. [DOI] [PubMed] [Google Scholar]
- Rando RR., and Chang A. (1983). Studies on the catalyzed interconversion of vitamin A derivatives. J Am Chem Soc 105, 2879–2882. [Google Scholar]
- Saari JC. (2016). Vitamin A and Vision. Subcell Biochem 81, 231–259. [DOI] [PubMed] [Google Scholar]
- Saari JC., and Bredberg DL. (1987). Photochemistry and stereoselectivity of cellular retinaldehyde-binding protein from bovine retina. J Biol Chem 262, 7618–7622. [PubMed] [Google Scholar]
- Sato S., and Kefalov VJ. (2016). cis Retinol oxidation regulates photoreceptor access to the retina visual cycle and cone pigment regeneration. The Journal of physiology 594, 6753–6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon A., Hellman U., Wernstedt C., and Eriksson U. (1995). The retinal pigment epithelial-specific 11-cis retinol dehydrogenase belongs to the family of short chain alcohol dehydrogenases. Journal of Biological Chemistry 270, 1107–1112. [PubMed] [Google Scholar]
- Tao L., Shen D., Pandey S., Hao W., Rich KA., and Fong HK. (1998). Structure and developmental expression of the mouse RGR opsin gene. Mol Vis 4, 25. [PubMed] [Google Scholar]
- Trifunovic D., Karali M., Camposampiero D., Ponzin D., Banfi S., and Marigo V. (2008). A highresolution RNA expression atlas of retinitis pigmentosa genes in human and mouse retinas. Invest Ophthalmol Vis Sci 49, 2330–2336. [DOI] [PubMed] [Google Scholar]
- Vinberg F., Kolesnikov AV., and Kefalov VJ. (2014). Ex vivo ERG analysis of photoreceptors using an in vivo ERG system. Vision research 101, 108–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JS., and Kefalov VJ. (2009). An alternative pathway mediates the mouse and human cone visual cycle. Current biology : CB 19, 1665–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JS., Nymark S., Frederiksen R., Estevez ME., Shen SQ., Corbo JC., Cornwall MC., and Kefalov VJ. (2014). Chromophore supply rate-limits Mammalian photoreceptor dark adaptation. J Neurosci 34, 11212–11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel A., Oberhauser V., Pugh EN Jr., Lamb TD., Grimm C., Samardzija M., Fahl E., Seeliger MW., Reme CE., and von Lintig J. (2005). The retinal G protein-coupled receptor (RGR) enhances isomerohydrolase activity independent of light. J Biol Chem 280, 29874–29884. [DOI] [PubMed] [Google Scholar]
- Woodruff ML., Lem J., and Fain GL. (2004). Early receptor current of wild-type and transducin knockout mice: photosensitivity and light-induced Ca2+ release. The Journal of physiology 557, 821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu BX., Chen Y., Chen Y., Fan J., Rohrer B., Crouch RK., and Ma JX. (2002). Cloning and characterization of a novel all-trans retinol short-chain dehydrogenase/reductase from the RPE. Invest Ophthalmol Vis Sci 43, 3365–3372. [PubMed] [Google Scholar]
- Wu BX., Moiseyev G., Chen Y., Rohrer B., Crouch RK., and Ma JX. (2004). Identification of RDH10, an All-trans Retinol Dehydrogenase, in Retinal Muller Cells. Invest Ophthalmol Vis Sci 45, 3857–3862. [DOI] [PubMed] [Google Scholar]
- Xue Y., Shen SQ., Jui J., Rupp AC., Byrne LC., Hattar S., Flannery JG., Corbo JC., and Kefalov VJ. (2015). CRALBP supports the mammalian retinal visual cycle and cone vision. J Clin Invest 125, 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


