Summary
Sleep is fundamental to animal survival but is a vulnerable state that also limits how much time can be devoted to critical wake-dependent activities [1]. Although many animals are day-active and sleep at night, they exhibit a midday nap or ‘siesta’ that can vary in intensity and is usually more prominent on warm days. In humans, the balance between maintaining wake or sleeping during the day has important health implications [2], but the mechanisms underlying this dynamic regulation are poorly understood. Using the well-established Drosophila melanogaster animal model to study sleep [3], we identify a new wake-sleep regulator that we term daywake (dyw). Dyw encodes a juvenile hormone binding protein [4] that functions in neurons as a day-specific anti-siesta gene with little effect on sleep levels during the nighttime or in the absence of light. Remarkably, dyw expression is stimulated in-trans via cold-enhanced splicing of the dmpi8 intron [5] from the reverse-oriented but slightly overlapping period (per) clock gene [6]. The functionally integrated dmpi8/dyw genetic unit operates as a ‘behavioral temperate acclimator’ by increasingly counterbalancing siesta-promoting pathways as daily temperatures become cooler and carry reduced risks from daytime heat exposure. While daily patterns of when animals are awake and sleep are largely scheduled by the circadian timing system, dyw implicates a less recognized class of modulatory wake-sleep regulators that primarily function to enhance flexibility in wake-sleep preference, a behavioral plasticity that is commonly observed in animals during the midday, raising the possibility of shared mechanisms.
Keywords: Daywake, sleep-wake behavior, midday siesta, Drosophila, period clock gene, pre-mRNA splicing, dmpi8 intron, juvenile-hormone binding protein, 0.9 gene
eTOC Blurb
Yong and Edery identify an anti-siesta gene in Drosophila, herein termed daywake, that is regulated in an unprecedented manner by splicing of the cold-enhanced dmpi8 intron from the reverse-oriented and slightly overlapping period clock gene. The dmpi8/dyw unit stimulates midday activity on days that carry less risks from heat-exposure.
Results and Discussion
Midday siesta in D. melanogaster is most robust at warmer temperatures and in the presence of light, an adaptive response almost certainly aimed at minimizing exposure to the hot daytime sun [7, 8]. We previously showed that midday siesta levels in D. melanogaster are influenced by the splicing efficiency of a thermosensitive intron (termed dmpi8) in the 3’ untranslated region of the period (per) gene, a central factor in the circadian (≅24 hr) clock [5, 7]. Increases in the splicing efficiency of the dmpi8 intron (D. melanogaster period intron 8) are causally linked to decreases in midday siesta, whereas there is little to no effect on nighttime sleep levels [5, 7, 9, 10]. For example, transgenic flies carrying a construct of the dper gene wherein the 5’ and 3’ splice sites (ss) of the dmpi8 intron were modified to increase its splicing efficiency (herein termed dmpi8UP), exhibit significantly reduced daytime sleep levels compared to their wildtype transgenic controls (herein termed dmpi8WT) [7, 10] (Figure 1B, F).
Figure 1. Effect of dmpi8 splicing efficiency on daytime sleep requires 0.9 protein function but not dPER.
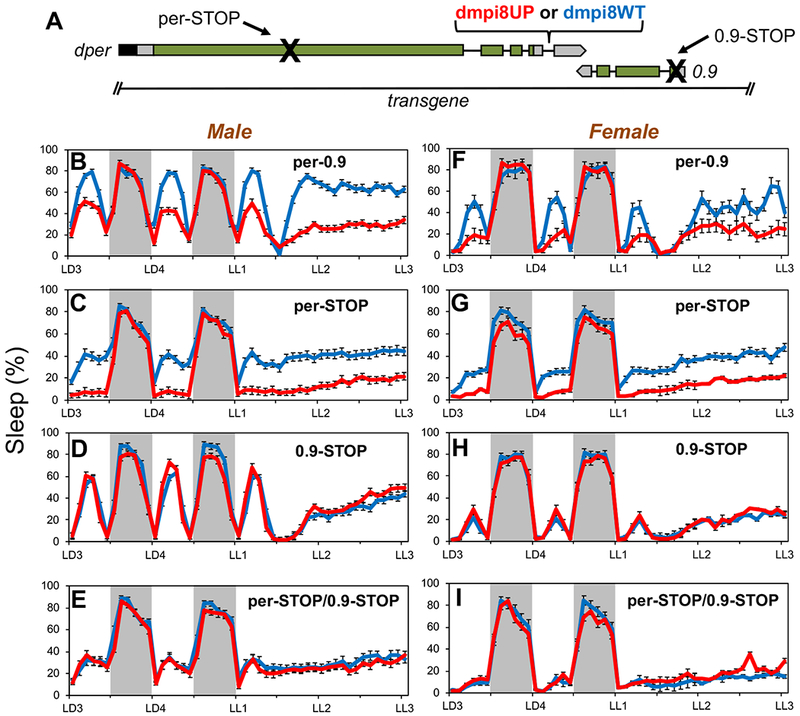
(A) Schematic of the dmpi8WT and dmpi8UP transgenes [10] modified to include a premature stop codon in the open reading frame of dper, 0.9, or both. The transgenes are based on a dper cDNA/genomic hybrid and include approximately 825bp 5’ upstream of the 0.9 transcription unit, which is transcribed in the opposite direction to dper. The 3’ untranslated regions of both dper and 0.9 overlap by 49bp. Rectangles; black (dper circadian regulatory sequence), gray (untranslated regions), green (coding region); black lines (introns). (B-G) Young adult male (B-E) or female (F-I) flies were exposed to 4 days of 12hr light/12 hr dark cycles (LD) followed by three days of continuous light (LL) at 25°C. For each genotype, graphs represent data from 96 individual flies (three independent transgenic lines × 32 individual flies/line) and data pooled to get the population average. Shown are the relative sleep levels during the last 2 days of LD followed by 2 days of LL for transgenic flies carrying the dmpi8UP (red) or dmpi8WT (blue) versions of the wildtype per-0.9 (B, F) per-STOP (C, G), 0.9-STOP (D, H) or double stop (E, I). Grey rectangle, 12 hr dark period. The following lines were used to obtain group averages; dmpi8WT, kx-f4c, m38-k, f9; dmpi8UP, m17, m32, f13; per-STOP[dmpi8WT], m53, f18, f46; per-STOP[dmpi8UP], m55, m131, m138; 0.9-STOP[dmpi8WT], m50, m62, m73; 0.9-STOP[dmpi8UP], m55, f50, f59; per/0.9-STOP[dmpi8WT], m94, m100, f38; per/0.9-STOP[dmpi8UP], m42, m81, f5. Representative examples are shown and similar results were obtained with other independent lines (see STAR Methods and KEY RESORCE TABLE for identity of lines analyzed). See also Figures S1 and S2 for additional results.
Curiously, dmpi8UP flies maintain low sleep levels even when exposed to prolonged periods of constant light (LL) [7] (Figure 1B, F), conditions that abolish circadian rhythmicity and result in constitutively low dPER protein levels [11, 12]. We therefore wondered if the midday sleep effects of dmpi8 splicing might actually be mediated by the small gene 3’ proximal to dper, historically termed ‘0.9kb’ or 0.9 [13] (Figure 1A). The 0.9 gene is transcribed in the reverse orientation to dper, and both transcription units slightly overlap in their 3’ untranslated regions (3’ UTRs). Commonly used dper transgene constructs, including dmpi8UP and dmpi8WT, contain the 0.9 gene (e.g., [14]) (Figure 1A). The 0.9 gene encodes a putative juvenile hormone binding protein (JHBP) [15], but received scant attention once the circadian function of this genomic locus was ascribed to dper [14, 16, 17].
As an initial step to test if the effects of dmpi8 splicing on midday sleep levels are independent of dPER protein activity, we modified the dmpi8UP and dmpi8WT constructs by introducing the classic per01 premature stop codon (Figure 1A), previously shown to eliminate dper function [18, 19]. Multiple independent lines of the per-STOP[dmpi8WT] and per-STOP[dmpi8UP] transgenes were assayed in the per01 genetic background (Figure S1A). We confirmed that the flies are arrhythmic in constant dark conditions (DD; data not shown), although as previously reported they still show day/night fluctuations in wake/sleep levels when maintained in daily 12 hr light/12 dark cycles (LD) [20] (Figure 1C, G). Remarkably, daytime (but not nighttime) sleep levels are significantly lower in per-STOP[dmpi8UP] flies compared to per-STOP[dmpi8WT] flies, a behavioral difference that continues in constant light (Figure 1C, G). This was observed in both males and females (Figure 1C, G), although there is sexual dimorphism whereby females generally exhibit lower siesta levels compared to males [21]. The reduced sleep in per-STOP[dmpi8UP] flies is not due to hyperactivity (Figure S2A), consistent with prior work using dmpi8UP flies [7].
Next, we generated dmpi8UP and dmpi8WT versions where we placed a premature stop codon in the 0.9 reading frame (0.9-STOP[dmpi8UP] and 0.9-STOP[dmpi8WT]), in addition to double stop versions in both dper and 0.9 (per-STOP/0.9-STOP[dmpi8UP] and per-STOP/0.9-STOP[dmpi8WT]) (Figure 1A). In striking fashion, daily sleep levels were virtually identical in LD and LL when comparing the dmpi8UP and dmpi8WT pairs of the 0.9-STOP transgenes, and likewise for the double per/0.9-STOP transgenes (Figure 1D, E, H, I; Figures S1 and S2). As previously noted, baseline sleep levels during the day are lower in arrhythmic mutants (e.g., per01) compared to rhythmic controls [8] (Figure 1). Differences in baseline sleep levels complicates comparisons of absolute sleep levels across different pairs of transgenics (e.g., dmpi8WT/UP vs. per-STOP[dmpi8/UP]), but not the relative effects of dmpi8WT versus dmpi8UP within a pair. Similar results were obtained for each dmpi8WT/UP pair over a wide range of temperatures (18°, 25° and 29°C), even though midday siesta levels are higher on warm days and temperature affects other aspects of daily sleep patterns [5, 7, 8] (Figure S2E–J). Although the effects of our transgenes were assayed in a genetic background containing a wildtype copy of the 0.9 gene, the results clearly show that daytime sleep differences in flies with variant dmpi8 splicing efficiency (i.e., dmpi8WT and dmpi8UP) are mediated via 0.9 function and does not require dPER protein. Other results targeting the endogenous 0.9 gene and ectopic expression are consistent with the results using our transgenic models (see below).
How might dmpi8 splicing link to 0.9 function? Increases in dmpi8 splicing efficiency lead to higher overall daily levels of dper transcripts in adult heads [5, 9, 10], the anatomical location of neural networks regulating daily wake/sleep behavior in D. melanogaster [22]. We wondered if 0.9 transcript levels also follow a parallel relationship with dmpi8 splicing efficiency as that observed for dper. Indeed, total mRNA levels for 0.9 are substantially higher in the heads of dmpi8UP flies compared to the control dmpi8WT flies (Figure 2A, B; Figure S1D, E). The influence of dmpi8 splicing efficiency on 0.9 transcript levels does not require dPER protein function (Figure S2D). We also evaluated norpA (no-receptor-potential A; mutation in phospholipase-C) mutant flies which are visually blind and previously shown to have increased dmpi8 splicing efficiency compared to wildtype controls [23, 24] (Figure 2D). In strong agreement with our transgenic models, 0.9 mRNA levels were markedly increased in the norpA mutant (Figure 2C). Although results with norpA do not necessarily imply a functional relationship with 0.9 levels/activity, they show that the link between increased dmpi8 splicing efficiency and higher 0.9 levels is not just limited to our transgenic models.
Figure 2. Increased dmpi8 splicing efficiency is associated with higher 0.9 transcript levels.
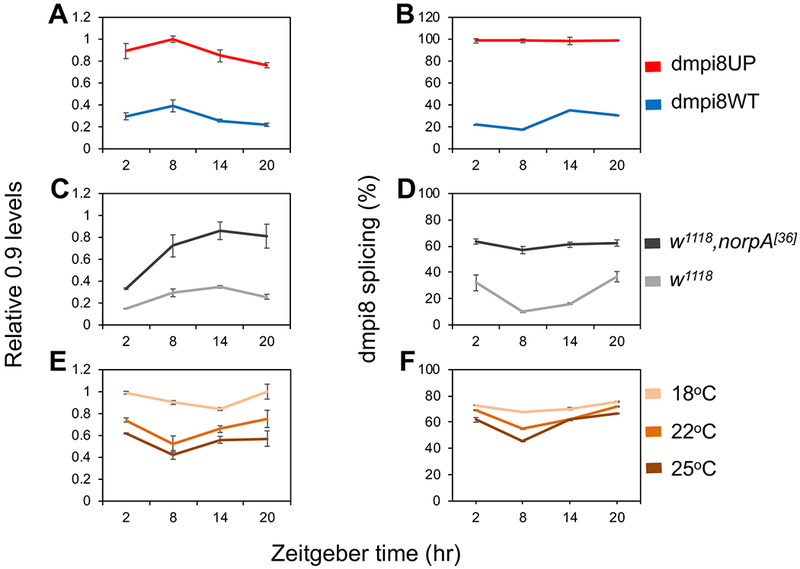
(A-F) Flies were kept at the indicated temperature and entrained by 4 days of LD, followed by collection on the fifth day at the indicated times [where Zeitgeber 0 (ZT0) is defined as lights-on]. RNA was extracted from head extracts and used to measure the relative levels of 0.9 transcripts and dmpi8 splicing efficiency, as indicated. Significant differences were observed for daily 0.9 levels and dmpi8 splicing efficiency, as follows: (A, B) Comparing dmpi8UP and dmpi8WT (two-sided t-test); 0.9 levels; ZT2, 4.3 × 10−3; ZT8, 8.2 × 10−5; ZT14, 2.7 × 10−4; ZT20, 9.9 × 10−6; dmpi8 splicing; ZT2, 2.8 × 10−8; ZT8, 7.1 × 10−9; ZT14, 1.8 × 10−6; ZT20, 5.2 × 10−11. The results are based on pooling data from at least three independent transgenic lines for each genotype; dmpi8WT (f9, m38-k, kx-f4-c), dmpi8UP (m17, f13, m32). (C, D) Comparing w,norpA[36] mutant to its genetic background control (two-sided Student’s t-test); 0.9 levels; ZT2, 7.4 × 10−6; ZT8, 8.0 × 10−3; ZT14, 1.6 × 10−3; ZT20, 3.9 × 10−3; dmpi8 splicing; ZT2, 7.9 × 10−3; ZT8, 8.7 × 10−5; ZT14, 3.4 × 10−5; ZT20, 5.4 × 10−3. (E, F) Comparing three temperatures (ANOVA); 0.9 levels; ZT2, 1.7 × 10−7; ZT8, 5.4 × 10−4; ZT14, 3.6 × 104; ZT20, 1.7 × 10−2; dmpi8 splicing; ZT2, 4.8 × 10−2; ZT8, 8.4 × 10−4; ZT14, 2.4 × 10−2; ZT20, 6.0 × 10−3. For each experiment, approximately 50-100 flies were used for each timepoint. Graphs shown are the average of three independent experiments. See also Figure S1D, E for further results.
Finally, we varied daily temperature to probe for a relationship between dmpi8 splicing efficiency and endogenously expressed 0.9 mRNA levels using wildtype flies. Lower daily temperatures are associated with progressively larger increases in dmpi8 splicing efficiency, consistent with reduced midday siesta [5, 7, 10] (Figure 2F). For temperatures between 18° to 25-26°C, the average daily levels of 0.9 are increasingly elevated as daily temperatures drop (Figure 2E and data not shown), highly consistent with the overall daily increase in dmpi8 splicing efficiency (Figure 2F). However, at the highest temperatures evaluated (e.g., 28° - 29°C) the results with 0.9 levels were inconsistent, sometimes relatively high and other times low, a result obtained with other fly genotypes (data not shown). Although the basis for this apparent high-temperature variability in 0.9 mRNA levels is currently not clear, we note that on warm days (e.g., 29°C) heat strongly suppresses midday wake in a manner that is little influenced by dmpi8 splicing efficiency [7, 9, 10]. Indeed, changes in dmpi8 splicing efficiency are most functionally linked to helping diminish siesta in cool climates as opposed to a mechanism primarily designed to mount a robust midday siesta on hot days [7, 9, 25–27] (see Figure 4). Thus, it is possible that the graded relationship between dmpi8 splicing efficiency and 0.9 levels has thermal limits and mainly functions on cooler days as a mechanism to decrease midday siesta.
We did note low-amplitude variations in the daily levels of 0.9 that varied in profile between genotypes and temperature (Figure 2), but the physiological significance of these fluctuations is presently not clear. Nonetheless, our combined results based on transgenic models, mutants and in wildtype flies over a potentially physiologically-relevant temperature range, reveal a clear connection between increasing dmpi8 splicing efficiency and higher overall levels in 0.9 transcripts. Since increases in dmpi8 splicing efficiency are causally linked to lower midday siesta levels [7], a testable prediction from our findings is that down-regulating 0.9 expression should increase daytime sleep levels.
To this end, we used the Gal4/UAS system [28] in combination with RNA interference (RNAi) [29] against 0.9 (RNAi-0.9) to more directly test a role for 0.9 in wake/sleep regulation. In strong agreement with our hypothesis, driving RNAi-0.9 in dper-expressing cells markedly increased daytime sleep levels relative to both parental control crosses, whereas nighttime sleep levels exhibit little change with this treatment (Figures 3A–E, and S3). These results also point towards dper-expressing cells as key sites for the production of 0.9 in regulating daytime sleep/wake levels—in line with our findings of a causal association between 0.9 levels and dmpi8 splicing efficiency (Figures 2, and S2D). Similar results were obtained with multiple RNAi-0.9 lines (e.g., Figures 3D, E, and S3B), whereas RNAi directed against takeout (the closest homolog to 0.9) [30] and several other jhbs in dper-expressing cells had no effect on daily sleep levels (Figure 3A, B). Increases in daytime sleep levels by driving RNAi-0.9 in dper-expressing cells was observed in both males and females at the two test temperatures of 18° and 25°C (Figures 3C, and S3).
Figure 3. The 0.9 gene suppresses daytime sleep with little to no effect on nighttime sleep levels.
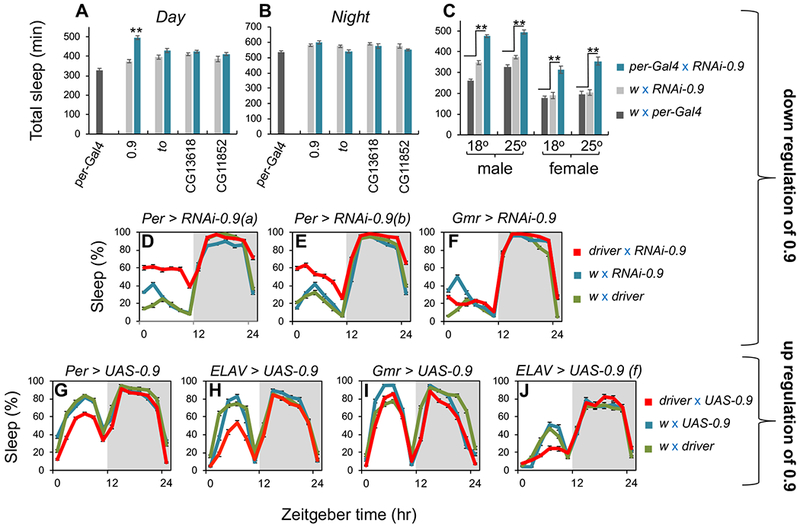
(A, B) Flies were kept for 5 days in LD at 25°C, and shown are total day and night sleep levels (min) for male adult progeny from crosses between per-Gal4 and the indicated UAS-RNAi lines (blue bars); the gene targeted by RNAi is shown at bottom of panels. Contemporaneous control crosses between w1118 (w; the background for both per-Gal4 and the UAS-RNAi lines) and per-Gal4 (black) or the UAS-RNAi lines (gray). (C) Flies were kept for 5 days in LD at either 18°C or 25°C, and shown are total daytime sleep (min) for adult male or female from per-Gal4>RNAi-0.9 (blue); and the parental control crosses, w1118 with per-Gal4 (black) or UAS-RNAi-0.9 (gray). Sleep values are an average from the last 3 days of LD based on 32 individual flies for each cross. **p < 0.001 for experimental group compared to both control crosses. (D-F) Flies were kept for 5 days in LD at 18°C, and shown are the daily sleep levels for female adult progeny for the indicated driver and UAS-RNAi-0.9 (red), and the two parental control crosses between w1118 and the driver (green) or UAS-RNAi-0.9 (blue). RNAi-0.9(a) and RNAi-0.9(b) refers to stock no. 105930 (VDRC) and 56988 (BDSC), respectively. The sleep profiles are an average of the last days of LD based on 32 individual flies for each cross. (G-J) Flies were kept for 5 days in LD at 25°C, and shown are the daily sleep levels for male (except panel J) adult progeny for the indicated driver and UAS-0.9 (red), and the two parental control crosses between w1118 and the driver (green) or UAS-0.9 (blue). For each cross, activity data from 32 individual flies was used, and the sleep profiles shown are an average of the last three days of LD based on pooling results from two independent crossing experiments using a different UAS-0.9 line (f57, f79). Similar results were obtained in other experiments (n = 3). See also Figure S3 and Table S1 for additional results using RNAi-0.9, and see Figure S4 for additional results using UAS-0.9.
We also evaluated several other drivers that express in clock cells, the eyes and sleep centers in the brain. Essentially, expression in dper-cells resulted in the most consistently robust effects on daytime sleep with little change in nighttime sleep. However, other drivers with wide-expression in clock neurons (e.g., Cry16-gal4) also yielded significant increases in daytime sleep levels, whereas no significant effects were observed with several drivers in brain sleep centers or the eyes (Figures 3F, and S3). Although we mainly tested different drivers in males at 25°C (Figures 3A–C, and S3A), similar results with clock and eye drivers were observed in females (Figures 3C–F, and S3B, C). Not surprisingly, females generally provided a more sensitized background to observe manipulations that increase daytime sleep given their lower baseline in midday siesta levels (Figures 3C, and S3) [21, 31].
In stark contrast to RNAi-0.9 treatment, daytime (but not nighttime) sleep levels were decreased when over-expressing 0.9 in dper-expressing cells (Figure 3G), which also shows that 0.9 function on daytime sleep is not dependent on genetic linkage to the dmpi8 intron. Although we only sampled a few drivers besides per-Gal4 in combination with UAS-0.9, similar to the RNAi-0.9 results, expression in Cry-expressing cells but not the eyes (Gmr-Gal4) significantly decreased daytime sleep levels (Figures 3I, and S4A, B). Of the drivers we tested, pan-neural expression (ELAV) yielded the largest decreases in midday siesta levels, a response readily observed in both males and females at 18° and 25°C (Figures 3H, J, and S4C–H). This suggests a direct neuronal function for 0.9 in promoting daytime wake. However, JHBPs can be secreted to target tissues [15], which might also explain why effects were not always observed for RNAi (site-of-production) and overexpression (site-of-action?) in the presence of the same driver. Consistent with our prior findings on dmpi8 splicing efficiency [7], there is no effect of overexpressing 0.9 on daily sleep patterns in constant dark conditions (Figure S4G, H).
In summary, while further studies will be required to better understand how and where 0.9 functions, our findings are remarkably internally consistent and demonstrate that the 0.9 gene is a novel day-specific suppressor of midday siesta that is linked to dmpi8 splicing efficiency. We therefore rename the 0.9 gene as daywake (dyw). The ability of D. melanogaster to mount a robust midday siesta almost certainly reflects its tropical origins [32]. Recent findings implicate a subset of sleep-promoting clock neurons in the fly adult brain termed DN1s (dorsal neurons) as major contributors to the enhanced midday siesta observed at warmer temperatures [33]. We propose that a key adaptive function of the link between dyw and the cold-enhanced splicing of the dmpi8 intron is that it operates in an ‘anti-siesta’ capacity, increasing daytime wake when local weather conditions pose less risks associated with heat exposure (Figure 4). This ability of the dmpi8/dyw unit to attenuate midday siesta might have contributed to the world-wide adaptation of D. melanogaster to temperate regions [34]. Intriguingly, the juvenile hormone signaling pathway was recently implicated in sex-specific sleep patterns [35], suggesting a broader role for this pathway in sleep/wake behavior.
Figure 4. Model for how the dmpi8/daywake genetic unit functions in the thermal adaptation of midday siesta in D. melanogaster.
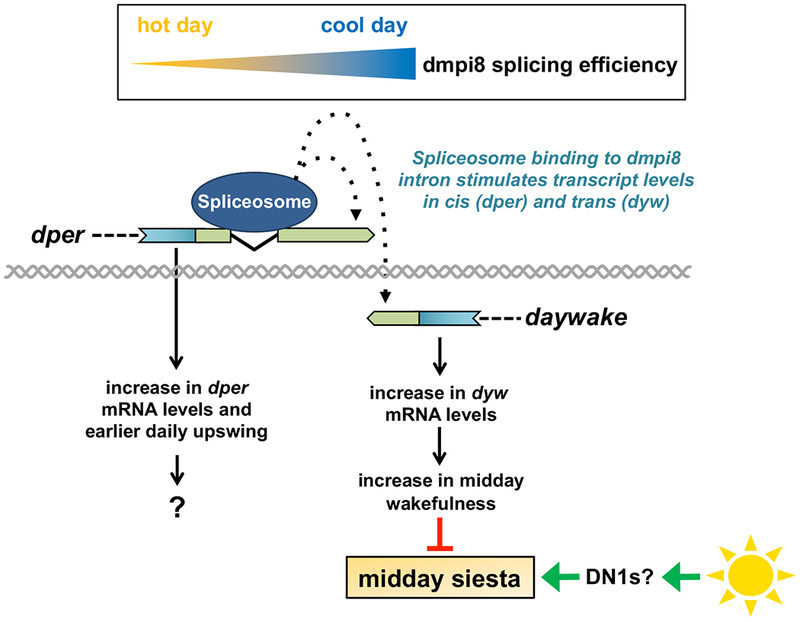
As daily temperatures decline, the splicing efficiency of dmpi8 progressively increases (top), leading to an increase in dper mRNA levels (left). Although it is not clear how dmpi8 splicing regulates dper mRNA levels, active splicing of dmpi8 is required [5]. This suggests that the seeding of splicing factors at the dmpi8 intron stimulates (directly, or indirectly via other interacting factors) the production and/or stability of dper transcripts [5]. The 3’ UTRs of dper and daywake overlap by 49 bp, raising the possibility that this proximity allows the stimulatory mechanism initiated during splicing of the dmpi8 intron to also function over short distances and regulate daywake transcript levels in-trans. Increases in dyw expression promote daytime wakefulness during the midday, leading to a reduction in midday siesta. On warm days, other systems (e.g., heat-activated sleep-promoting DN1s? [33]) become increasingly dominant and evoke a strong midday siesta in a manner that is little influenced by dmpi8 splicing efficiency [7] (bottom, right). This thermal adaptation system provides D. melanogaster the key survival response of mounting a strong siesta on warm days (default state for a tropically originating species), yet the flexibility to increase activity on days when the risks posed by exposure to heat are reduced. For dper and dyw transcripts, blue rectangle (coding region) and green rectangle (3’ UTR); gray double helix (genomic DNA); spliceosome binding is at the dmpi8 intron.
A complex interaction between the circadian timing system and dedicated pathways that promote wake and sleep largely defines the characteristic diurnal or nocturnal wake-sleep patterns observed in animals. D. melanogaster exhibits two prominent clock-controlled bouts of activity (morning and evening) with highly consolidated sleep during most of the night [20, 21]. Much progress has been made in identifying factors and neural circuits that underlie wake/sleep-promoting pathways and how they intertwine with the circadian timing system to generate the characteristic major bouts of activity and nighttime sleep (e.g., [33, 36, 37]). The identification of dyw suggests a less recognized class of wake-sleep regulators that function in a largely circadian-independent manner to increase behavioral plasticity in wake-sleep preference during the midday (and possibly other time periods). In this vein, recent finding indicate that variations in human siesta (e.g., length) is heritable [38, 39]. Moreover, daytime napping has several health benefits, such as improved cognitive functions, whereas excessive daytime sleepiness is correlated with detrimental health [2]. Thus, even though JHBPs are restricted to insects [40], the identification of dyw suggests physiologically similar functions operate in humans. These considerations further reinforce the idea that nighttime sleep and daytime siesta are governed by distinct mechanisms and have different roles.
Active splicing of dmpi8 as opposed to the presence or absence of the intronic sequences is necessary to influence dper mRNA levels [5] (the function of which is still a mystery). Although it is not clear how splicing of the dmpi8 intron modulates dper mRNA levels, spliceosome assembly at 3’ terminal introns can stimulate the binding/activity of factors involved in the process of 3’-end cleavage, leading to increases in mature transcript levels [41, 42]. We speculate that since the 3’ UTR of dyw slightly overlaps with the 3’ UTR of dper, spliceosome binding to the dmpi8 intron can also exert its influence over short-distances to regulate dyw transcript levels (Figure. 4). To the best of our knowledge, co-opting the effects of intron removal at one gene to regulate the transcript levels at an adjoining gene has not been described before. Thus, the study of the dmpi8/dyw unit should not only lead to novel insights on the daytime balance between wakefulness and sleep drive but might reveal new gene regulatory mechanisms.
STAR METHODS text
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Isaac Edery (edery@cabm.rutgers.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Flies were routinely reared at room temperature (22-25°C) and maintained in vials or bottles containing standard agar-cornmeal-sugar-yeast-Tegosept-media (see Key Resources Table for major ingredients). Except for Canton-S wildtype flies, all the other flies used in this study were in the w1118 genetic background (sometimes simplified as w). For behavioral and RNA analysis, young adult flies (3-6 day old) were placed in tubes or vials and then maintained in 12 hr light: 12 hr dark (LD) cycles for at least 4 days at the indicated temperature. Sleep analysis was performed on individual males and females; whereas for measuring RNA levels and dmpi8 splicing efficiency, vials containing both males and females were used. For the dmpi8WT and dmpi8UP flies we used the following independent lines; dmpi8WT, kx-f4c, m38-k, f9; dmpi8UP, m17, m32, f13 [7, 26]. The following flies used in this study were obtained from the Blommington Drosophila Stock Center (BDSC): Canton S wildtype flies (RRID:BDSC_64349), w,norpA[36] (RRID:BDSC_9048), per-Gal4 (RRID:BDSC_7127), cry16-Gal4 (RRID:BDSC_24514), Gmr-Gal4 (RRID:BDSC_1104), ELAV-Gal4 (RRID:BDSC_458), pdf-Gal4 (RRID:BDSC_6899), pdfr(R18H11)-Gal4 (RRID:BDSC_48832), C5-Gal4 (RRID:BDSC_30839), 201Y-Gal4 (RRID:BDSC_4440), RNAi-0.9 (RRID:BDSC_56988), RNAi-to (RRID:BDSC_55982), RNAi-CG13618 (RRID:BDSC_57749), RNAi-CG11852 (RRID:BDSC_57193). RNAi-0.9 (#105930) was obtained from the Vienna Drosophila Resource Center. Mai179-Gal4 [43] was from F. Rouyer; tim(UAS)-Gal4 (TUG) [44] was from J. Blau; and c929-Gal4 [45] was from P. Taghert. For the RNAi experiments, we first crossed the Gal4 driver strains with strains carrying the UAS-dicer2 transgene (BDSC, stocks RRID:BDSC_24651 or RRID:BDSC_24650), which should enhance RNAi-mediated inhibition. For all crosses, we set up contemporaneous control crosses whereby each parental strain was crossed to w1118.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| TRI ReagentR | Sigma-Aldrich | Cat#T9424 |
| AccuPrime™ Taq DNA Polymerase, high fidelity | Invitrogen (ThermoFisher Sci.) | Cat#12346086 |
| SYBR™ Safe DNA Gel Stain | Invitrogen (ThermoFisher Sci.) | Cat#S33102 |
| Drosophila agar (fly food) | LabScientific | FLY-8020 |
| Drosophila yeast (fly food) | LabScientific | FLY-8040 |
| Corn meal, yellow (fly food) | LabScientific | FLY-8010 |
| Light corn syrup (fly food) | LabScientific | FLY-8007 |
| Propionic Acid (fly food) | Frontier Scientific, USA | Cat#JK229213; CAS, 79-09-4 |
| Tegosept (fly food preservative) | Apex (Genesee Scientific, USA) | Cat#20-258; CAS, 99-76-3 |
| Bacto-Agar (behavior tubes) | VWR | Cat#90000-760 |
| Sucrose (behavior tubes) | Sigma-Aldrich | Cat#S7903 |
| KlenTaq1 | AB Bioscience LLC | Cat#1001 |
| dNTP mix | Promega | Cat#U1511 |
| Critical Commercial Assays | ||
| QuikChange II Site-Directed Mutagenesis Kit | Agilent | Cat#200524 |
| HiSpeedR Plasmid Maxi Kit | Qiagen | Cat#12663 |
| RNA to cDNA EcoDry™ Premix (Oligo dT) | TaKaRa | Cat#639543 |
| Experimental Models: Organisms/Strains | ||
| Drosophila melanogaster: p{dmper/8:8}; herein called dmpi8WT | [10] | lines; kx-f4c, m38-k, f9 |
| Drosophila melanogaster: p{dmper/M2M1}; herein called dmpi8UP | [10] | lines; m17, m32, f13 |
| Drosophila melanogaster: per-STOP[dmpi8WT] | This paper | lines; m44, m53, m68, f18, f46 |
| Drosophila melanogaster: per-STOP[dmpi8UP] | This paper | lines; m55, m65, m131, m138, f30 |
| Drosophila melanogaster: 0.9-STOP[dmpi8WT] | This paper | lines; m29, m50, m62, m73, m76 |
| Drosophila melanogaster: 0.9-STOP[dmpi8UP] | This paper | lines; m23, m33, m55, m100, f50, f59 |
| Drosophila melanogaster: per-STOP/0.9-STOP[dmpi8WT] | This paper | lines; m74.1, m94, m100, f38, f55 |
| Drosophila melanogaster: per-STOP/0.9-STOP[dmpi8UP] | This paper | lines: m42, m77, m81, f5 |
| Drosophila melanogaster: Canton-S | Blommington Drosophila Stock Center (BDSC) | RRID:BDSC_64349 |
| Drosophila melanogaster: w1118,norpA[36] | BDSC | RRID:BDSC_9048 |
| Drosophila melanogaster: per-Gal4 | BDSC | RRID:BDSC_7127 |
| Drosophila melanogaster: Cry16-Gal4 | BDSC | RRID:BDSC_24514 |
| Drosophila melanogaster: Gmr-Gal4 | BDSC | RRID:BDSC_1104 |
| Drosophila melanogaster: ELAV-Gal4 | BDSC | RRID:BDSC_458 |
| Drosophila melanogaster: pdf-Gal4 | BDSC | RRID:BDSC_6899 |
| Drosophila melanogaster: pdfr(R18H11)-Gal4 | BDSC | RRID:BDSC_48832 |
| Drosophila melanogaster: C5-Gal4 | BDSC | RRID:BDSC_30839 |
| Drosophila melanogaster: 201Y-Gal4 | BDSC | RRID:BDSC_4440 |
| Drosophila melanogaster: Mai179-Gal4 | Laboratory of F. Rouyer (France) | N/A |
| Drosophila melanogaster: tim(UAS)-Gal4 | Laboratory of J. Blau (USA) | N/A |
| Drosophila melanogaster: c929-Gal4 | Laboratory of P. Taghert (USA) | RRID:BDSC_25373 |
| Drosophila melanogaster: UAS-dicer2 | BDSC | RRID:BDSC_24650 |
| Drosophila melanogaster: UAS-dicer2 | BDSC | RRID:BDSC_24651 |
| Drosophila melanogaster: RNAi-0.9 | BDSC | RRID:BDSC_56988 |
| Drosophila melanogaster: RNAi-0.9 | Vienna Drosophila Resource Center | VDRC ID: 105930; FlyBase ID, FBst0477756 |
| Drosophila melanogaster: RNAi-takeout | BDSC | RRID:BDSC_55982 |
| Drosophila melanogaster: RNAi-CG13618 | BDSC | RRID:BDSC_57749 |
| Drosophila melanogaster: RNAi-CG11852 | BDSC | RRID:BDSC_57193 |
| Drosophila melanogaster: UAS-0.9 | This paper | lines; f57, f58, f79 |
| Oligonucleotides | ||
| See Table S2 | ||
| Recombinant DNA | ||
| plasmid: RH43234 (contains CG2650/0.9 cDNA) | Drosophila Genomics Resource Center (DGRC) | Stock no. 10923; Flybase ID, FBc10263672 |
| plasmid: pUAST | DGRC | Stock no. 1000 |
| plasmid: 8:8; herein renamed dmpi8WT | [10] | hs-CRS-cper-88kx |
| plasmid: M2M1; herein renamed dmpiUP | [10] | hs-CRS-cper-M2M1 |
| plasmid: per-STOP[dmpi8WT] | This paper | per0-88kx #1 |
| plasmid: per-STOP[dmpi8UP] | This paper | per0-M2M1 #2 |
| plasmid: 0.9-STOP[dmpi8WT] | This paper | 88kx-0.9-STOP #3 |
| plasmid: 0.9-STOP[dmpi8UP] | This paper | M2M1-0.9-STOP #5 |
| plasmid: per-STOP/0.9-STOP[dmpi8WT] | This paper | per0–88kx-0.9-STOP #1 |
| plasmid: per-STOP/0.9-STOP[dmpi8UP] | This paper | per0-M2M1-STOP #2 |
| plasmid: UAS-0.9 | This paper | UAS-0.9 #3 |
| Software and Algorithms | ||
| MATLAB | MathWorks | MATLAB_R2017b |
| FaasX Software | Laboratory of F. Rouyer; http://neuro-psi.cnrs.fr/spip.php?article298&lang=en | Kit version: 1.22 |
| DAMSystem3 Data Collection Software | TriKinetics, USA | N/A |
| Fiji Software (ImageJ) | https://fiji.sc | N/A |
METHOD DETAILS
Sleep analysis
Locomotor activity of individual male or female flies (3-6 days old) was continuously monitored and recorded using the Trikinetics system (Waltham, MA, USA), as previously reported [7, 46]. Briefly, individual flies were placed in glass tubes containing 5% sucrose and 2% bacto-agar, followed by loading into DAM2 Activity Monitors (Trikinetics). The activity monitors with flies were placed inside environmental incubators (Percival Scientific) and maintained at the indicated temperature (18°, 25° or 29°C) for at least 5 days of 12 hr light: 12 hr dark cycles [LD; where zeitgeber time 0 (ZT0) is defined as lights-on]. In general, after five days in LD, flies were kept in constant darkness (DD) or constant light (LL) for seven days. Activity counts were collected using the DAMSystem3 Software (Trikinetics), and sleep analysis was done using MATLAB software (MathWorks). Sleep was defined as continuous inactivity lasting five or more min, which is routinely used in the field (e.g., [21]). For each genotype, sex and condition, data from at least 32 individual flies was pooled to generate a group average. In general, data from the last three days of LD were averaged to obtain the sleep plots shown. Free-running periods were calculated based on activity data collected during six consecutive days in DD using the FaasX program (kindly provide by F. Rouyer, France), as previously described [46]. Values for individual flies were pooled to obtain an average value for each genotype.
Generation of the dmpi8WT and dmpi8UP STOP series of transgenic flies (Figure 1A)
All the STOP-series constructs (Figure 1A) were based on the original backbone vectors (hs-CRS-cper-88kx and hs-CRS-cper-M2M1) used to generate dmpi8WT and dmpi8UP flies, originally termed dmper/8:8 and dmper/M2M1, respectively [10]. Essentially, to more clearly reflect the splicing efficiency of the dmpi8 intron, herein we refer to constructs containing the wildtype version of dmpi8 as dmpi8WT (instead of its previously published designation, 8:8), and the enhanced splicing version as dmpi8UP (instead of its previously published designation, M2M1). The dmpi8WT and dmpi8UP transgenes rescue wildtype behavioral and molecular rhythms in per01 flies [10], and contain the entire 0.9 transcription unit (1155 bp) flanked on its 5’ side by about 825 bp of non-transcribed genomic DNA. The 3’ UTRs of 0.9 and dper overlap by 49 bp. Once final transformation vectors were verified (see below), plasmids for injection were prepared using the HiSpeed Plasmid Maxi Ki (Qiagen). Transgenic flies were generated by BestGene, Inc. (Chino Hill, CA, USA) in a w1118 background and subsequently crossed into a w1118, per01 background. The results shown are based on pooling data from multiple independent lines for each transgene (see legend to Figure 1 and Figure S1). A list of all the primers used to generate the different constructs is included in Table S2. More detailed account of how the transgenic constructs were generated are given below:
per-STOP[dmpi8WT] and per-STOP[dmpi8UP]
To generate the per-STOP[dmpi8WT] transgene, we used nested PCR (in total, 4 primers and 3 separate PCR reactions) as follows: In the first PCR, the hs-CRS-cper-88kx plasmid was used to amplify dper genomic sequences from 3177 to 4350bp (all numbering cited herein for dper sequences is according to [14]) using primers P3177F (5’-GCGTCGACGAGCCTAGAGGGCA-3’; bold indicates SgrDI site) and per0 R (5’-AGAAGGACGTAGCAACCGTTCTAGATGAGGAAGCGGTATGGCTTG-3’; bold is per01 mutation) in the presence of AccuPrime Taq DNA Polymerase (Invitrogen). In a second PCR, the hs-CRS-cper-88kx plasmid was used to amplify dper genomic sequences from 4350 to 7312bp using primers P7312R (5’-AACCTTAGGGCTGAGAAGGGTGGT-3’; bold indicates Bsu36I site) and per0 F (5’-CAAGCCATACCGCTTCCTCATCTAGAACGGTTGCTACGTCCTTCT-3’; bold is per01 mutation). The two amplified products have overlapping ends and served as a linear template in the third PCR using primers P3177F and P7312R. The final amplified product (3177 – 7312bp) was sequenced and we verified that it included the per01 mutation and no other changes. Finally, the hs-CRS-cper-88kx plasmid and the final PCR product were digested with SgrDI (cutting site, 3177bp) and Bsu36I (cutting site, 7307bp), and ligated to generate per-STOP[dmpi8WT] (designated in-house as, per0-88kx). The same strategy as above was used to generate per-STOP[dmpi8UP] (in-lab designation, per0-M2M1), except that hs-CRS-cper-M2M1 was used instead of hs-CRS-cper-88kx. The region between 3177-7312 bp was sequenced in the final plasmids prior to usage in fly transformation. The following transgenic lines were obtained and analyzed; per-STOP[dmpi8WT], m44, m53, m68, f18, f46; per-STOP[dmpi8UP], m55, m65, m131, m138, f30.
0.9-STOP[dmpi8WT] and 0.9-STOP[dmpi8UP]
We used the QuikChange II Site-Directed Mutagenesis kit (Agilent) in the presence of hs-CRS-cper-88kx to convert codons for amino acids 21 and 22 (TGCAGA) in the 0.9 open reading frame into two sequential translation stop codons (TGATGA) using primers STOP-CG2650F (5’-CAGCTGGGTTTCCTGATGAGTGGACGCCTCCG-3’; bold, point mutations create stop codons) and STOP-CG2650R (5’-CGGAGGCGTCCACTCATCAGGAAACCCAGCTG-3’; bold, point mutations create stop codons). Sequencing analysis confirmed the presence of the two stop codons and no other changes between 7300-9305 bp. Next, to ensure that the mutagenesis did not introduce unwanted changes to the hs-CRS-cper-88kx plasmid, we subjected the mutagenized plasmid to a round of PCR to amplify the region between 7307- 9305 bp with primers Bsu36I.R1 (5’-GCCCTAAGGTTTATATATCCG-3’; bold, indicates natural Bsu36I site at position 7307 bp) and EcoRI.9300.F (5’-TTGAATTCAATGTAAAATGGTT-3’; bold indicates the natural EcoRI site at position 9305 bp, which marks the 3’ end of the ‘dper’ genomic insert). The PCR product was digested with Bsu36I and Agel (cutting site, 8405 bp) followed by sub-cloning into doubly digested (Bsu36I/AgeI) hs-CRS-cper-88kx and hs-CRS-cper-M2M1, generating 0.9-STOP[dmpi8WT] (in-lab designation, 88-kx-0.9-STOP) and 0.9-STOP[dmpi8UP] (inlab designation, M2M1-STOP). Before using the final transformation vectors, we confirmed that sequences from 7307-8420 bp have both stop codons in the 0.9 reading frame and no other changes (data not shown). The following transgenic lines were obtained and analyzed; 0.9-STOP[dmpi8WT], m29, m50, m62, m73, m76; 0.9-STOP[dmpi8UP], m23, m33, m55, m100, f50, f59.
per-STOP/0.9-STOP[dmpi8WT] and per-STOP/0.9-STOP[dmpi8UP]
The double STOP constructs were generated by replacing the region from Bsu36I (7307 bp) to AgeI (8405 bp) in the per-STOP[dmpi8WT] and per-STOP[dmpi8UP] vectors with the corresponding regions from 0.9-STOP[dmpi8] and 0.9-STOP[dmpi8UP]. This generates the final transformation vectors, per/0.9-STOP[dmpi8WT] (in-lab designation, per0-88-kx-0,9-STOP) and per/0.9-STOP[dmpi8UP] (in-lab designation, per0-M2M1-STOP). We confirmed that the desired mutations were present in the final constructs. The following transgenic lines were obtained and analyzed; per-STOP/0.9-STOP[dmpi8WT], m74.1, m94, m100, f38, f55; per-STOP/0.9-STOP[dmpi8UP], m42, m77, m81, f5.
Generation of UAS-0.9 flies
To generate UAS-0.9 flies we first obtained the plasmid RH43234, which contains a cDNA version of the entire 0.9 open reading frame, from the Drosophila Genomics Resource Center (DGRC). RH43234 was subjected to PCR in the presence of primers CG2650.EcoRI.F (5’-TAAGAATTCATGCAGCTAACCGGTGCC-3’; bold, introduces an EcoRI site just upstream to the natural ATG translation start signal) and CG2650.XbaI.R (5’-TATATCTAGATCATTCCTTTTCGAAGAACTCG-3’; bold, introduces an XbaI site just downstream of the 0.9 stop codon). The 0.9 PCR product and the pUAST plasmid (stock no. 1000, DGRC) were digested with EcoRI and Xba I, followed by ligation to generate the UAS-0.9 transformation vector. Transgenic flies in the w1118 genetic background were generated by BestGene Inc. (Chino Hill, CA, USA) and multiple independent lines were obtained and analyzed (f57, f58, f79).
Measuring dmpi8 splicing efficiency and 0.9 levels
The splicing efficiency of dmpi8 in flies was measured as previously described [10, 25], with some minor modifications. Briefly, vials containing ~50 young (2- to 6-day-old) adult flies were placed in environmental chambers (Percival, USA) at the indicated temperature and exposed to at least five 12hr light/12 hr dark cycles (LD). At selected times during LD (routinely 3rd-5th day), flies were collected by freezing and heads isolated. Total RNA was extracted from heads using TRI ReagentR (Sigma-Aldrich) and a portion reverse transcribed using the RNA to cDNA EcoDry Premix (Oligo dT) kit (TaKaRa), followed by PCR in the presence of KlenTaq1 (AB Bioscience LLC) to measure dmpi8 splicing efficiency as previously described [10, 25]. In order to differentiate between the transgenic derived dper mRNA transcripts from the endogenously derived per01 transcripts we used the forward primers P6851-StulF (5’-ACACAGCACGGGGATGGGAGGC-3’) and P6851 (5’ ACACAGCACGGGGATGGGTAGT 3’), respectively. The P6851 primer will only amplify endogenously derived dper mRNA (e.g., per01 mRNA), whereas the P6851-StulF primer will only amplify transgenic derived dper transcripts that contain the engineered Stu1 site upstream of the dper translation stop codon. All RT-PCR reactions included the dper P7184r reverse primer (5’-GGCTTGAGATCTACATTATCCTC-3’); and the forward primer CBP294F (5’-TGATTGTGATGGGCCTGGACAAGT-3’) and reverse primer CBP536R (5’-GTCCAAGCGAGTGCCATTCACAAA-3’) to target transcripts from the non-cycling Cap Binding Protein 20 (CBP20) gene as an internal control, as previously described [10, 25]. PCR products were separated and visualized by electrophoresis on 2% agarose gels containing SYBR Safe DNA Gel Stain (Invitrogen., USA), and the bands quantified using a Multiimage III Imager (Alpha Innotech) and Fiji software (ImasgeJ). The values of dper-containing amplified products were normalized relative to CBP20. A similar strategy was also used to measure the relative levels of 0.9 transcripts, we used the primers CG2650-F1 (5’-CCAACTCGATGATGGTCAAGAG-3’) and CG2650-R1 (5’-GTCGTTGAACAGATTCGACAGG-3’), in addition to CBP294F and CBP536R to measure CBP20 transcript levels as an internal control (see above). Values for 0.9 mRNA were normalized relative to CBP20. Each experiment contained controls for specificity (e.g., no RT or absence of primers) and samples were taken at different PCR cycle lengths to ensure that the amplified products were in the linear range for quantification. For each experiment, the data for each timepoint represent ~50-60 flies and the data shown are an average of at least three independent experiments.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis of data sets mainly involved Student’s t-test (two-tailed, unpaired) and one-way analysis of variance (ANOVA). Samples were not randomized and we did not predetermine sample size. However, each data point shown for sleep analysis is based on data collected from at least 32 individual flies (usually two sets, males and females separately). In addition, for the transgenic lines we generated (i.e., STOP-series and UAS-0.9) we used multiple independent lines and pooled results to obtain group averages. The data are presented as mean values for group averages with error bars representing s.e.m. Experiments were repeated multiple times (usually 3 or more) and representative examples are shown. In the Figure legends are stipulated the fly lines used, the number of flies and p values (also see Table S1).
Supplementary Material
Highlights.
Control of Drosophila siesta by the per dmpi8 intron is independent of PER protein
Dmpi8 splicing modulates siesta in-trans via the adjoining daywake (dyw) gene
Dyw functions as a potent anti-siesta gene without affecting nighttime sleep levels
The dmpi8/dyw unit reduces siesta on days that carry less risks from heat-exposure
Acknowledgments
We are very grateful to Zhicao Zhang, who was instrumental in suggesting a possible role for 0.9 in mediating the effects of dmpi8 splicing on daytime sleep, and performing some of the initial experiments. This work was supported by funding from the National Institutes of Health to I.E (R01NS42088 and R01NS105780). Fly stocks obtained from the Bloomington Drosophila Stock Center (NIH P400d018537) and the Vienna Drosophila Resource Center (VDRC) were used in this study. We also thank the Drosophila Genomics Resource Center (supported by NIH grant 2P40OD010949) for plasmids.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare that there are no competing interests.
References
- 1.Keene AC, and Duboue ER (2018). The origins and evolution of sleep. J Exp Biol 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mantua J, and Spencer RMC (2017). Exploring the nap paradox: are mid-day sleep bouts a friend or foe? Sleep Med 37, 88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sehgal A, and Mignot E (2011). Genetics of sleep and sleep disorders. Cell 146, 194–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prestwich GD, Wojtasek H, Lentz AJ, and Rabinovich JM (1996). Biochemistry of proteins that bind and metabolize juvenile hormones. Arch Insect Biochem Physiol 32, 407–419. [DOI] [PubMed] [Google Scholar]
- 5.Majercak J, Sidote D, Hardin PE, and Edery I (1999). How a circadian clock adapts to seasonal decreases in temperature and day length [see comments]. Neuron 24, 219–230. [DOI] [PubMed] [Google Scholar]
- 6.Zehring WA, Wheeler DA, Reddy P, Konopka RJ, Kyriacou CP, Rosbash M, and Hall JC (1984). P-element transformation with period locus DNA restores rhythmicity to mutant, arrhythmic Drosophila melanogaster. Cell 39, 369–376. [DOI] [PubMed] [Google Scholar]
- 7.Cao W, and Edery I (2015). A novel pathway for sensory-mediated arousal involves splicing of an intron in the period clock gene. Sleep 38, 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parisky KM, Agosto Rivera JL, Donelson NC, Kotecha S, and Griffith LC (2016). Reorganization of Sleep by Temperature in Drosophila Requires Light, the Homeostat, and the Circadian Clock. Curr Biol 26, 882–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Low KH, Chen WF, Yildirim E, and Edery I (2012). Natural variation in the Drosophila melanogaster clock gene period modulates splicing of its 3’-terminal intron and mid-day siesta. PloS one 7, e49536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Low KH, Lim C, Ko HW, and Edery I (2008). Natural variation in the splice site strength of a clock gene and species-specific thermal adaptation. Neuron 60, 1054–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price JL, Dembinska ME, Young MW, and Rosbash M (1995). Suppression of PERIOD protein abundance and circadian cycling by the Drosophila clock mutation timeless. The EMBO journal 14, 4044–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zerr DM, Hall JC, Rosbash M, and Siwicki KK (1990). Circadian fluctuations of period protein immunoreactivity in the CNS and the visual system of Drosophila. J Neurosci 10, 2749–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reddy P, Zehring WA, Wheeler DA, Pirrotta V, Hadfield C, Hall JC, and Rosbash M (1984). Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms. Cell 38, 701–710. [DOI] [PubMed] [Google Scholar]
- 14.Citri Y, Colot HV, Jacquier AC, Yu Q, Hall JC, Baltimore D, and Rosbash M (1987). A family of unusually spliced biologically active transcripts encoded by a Drosophila clock gene. Nature 326, 42–47. [DOI] [PubMed] [Google Scholar]
- 15.Gilbert LI, Granger NA, and Roe RM (2000). The juvenile hormones: historical facts and speculations on future research directions. Insect Biochem Mol Biol 30, 617–644. [DOI] [PubMed] [Google Scholar]
- 16.Hamblen M, Zehring WA, Kyriacou CP, Reddy P, Yu Q, Wheeler DA, Zwiebel LJ, Konopka RJ, Rosbash M, and Hall JC (1986). Germ-line transformation involving DNA from the period locus in Drosophila melanogaster: overlapping genomic fragments that restore circadian and ultradian rhythmicity to per0 and per- mutants. Journal of neurogenetics 3, 249–291. [DOI] [PubMed] [Google Scholar]
- 17.Lorenz LJ, Hall JC, and Rosbash M (1989). Expression of a Drosophila mRNA is under circadian clock control during pupation. Development 107, 869–880. [DOI] [PubMed] [Google Scholar]
- 18.Konopka RJ, and Benzer S (1971). Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A 68, 2112–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu Q, Jacquier AC, Citri Y, Hamblen M, Hall JC, and Rosbash M (1987). Molecular mapping of point mutations in the period gene that stop or speed up biological clocks in Drosophila melanogaster. Proc Natl Acad Sci U S A 84, 784–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wheeler DA, Hamblen-Coyle MJ, Dushay MS, and Hall JC (1993). Behavior in light-dark cycles of Drosophila mutants that are arrhythmic, blind, or both. J Biol Rhythms 8, 67–94. [DOI] [PubMed] [Google Scholar]
- 21.Cirelli C (2003). Searching for sleep mutants of Drosophila melanogaster. Bioessays 25, 940–949. [DOI] [PubMed] [Google Scholar]
- 22.Dubowy C, and Sehgal A (2017). Circadian Rhythms and Sleep in Drosophila melanogaster. Genetics 205, 1373–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collins BH, Rosato E, and Kyriacou CP (2004). Seasonal behavior in Drosophila melanogaster requires the photoreceptors, the circadian clock, and phospholipase C. Proc Natl Acad Sci U S A 101, 1945–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Majercak J, Chen WF, and Edery I (2004). Splicing of the period gene 3′-terminal intron is regulated by light, circadian clock factors, and phospholipase C. Mol Cell Biol 24, 3359–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao W, and Edery I (2017). Mid-day siesta in natural populations of D. melanogaster from Africa exhibits an altitudinal cline and is regulated by splicing of a thermosensitive intron in the period clock gene. BMC Evol Biol 17, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z, Cao W, and Edery I (2018). The SR protein B52/SRp55 regulates splicing of the period thermosensitive intron and mid-day siesta in Drosophila. Sci Rep 8, 1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Y, and Edery I (2018). Parallel clinal variation in the mid-day siesta of Drosophila melanogaster implicates continent-specific targets of natural selection. PLoS Genet 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGuire SE, Roman G, and Davis RL (2004). Gene expression systems in Drosophila: a synthesis of time and space. Trends Genet 20, 384–391. [DOI] [PubMed] [Google Scholar]
- 29.Mohr SE (2014). RNAi screening in Drosophila cells and in vivo. Methods 68, 82–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarov-Blat L, So WV, Liu L, and Rosbash M (2000). The Drosophila takeout gene is a novel molecular link between circadian rhythms and feeding behavior. Cell 101, 647–656. [DOI] [PubMed] [Google Scholar]
- 31.Ishimoto H, Lark A, and Kitamoto T (2012). Factors that Differentially Affect Daytime and Nighttime Sleep in Drosophila melanogaster. Frontiers in neurology 3, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pool JE, Corbett-Detig RB, Sugino RP, Stevens KA, Cardeno CM, Crepeau MW, Duchen P, Emerson JJ, Saelao P, Begun DJ, et al. (2012). Population Genomics of sub-saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genet 8, e1003080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo F, Yu J, Jung HJ, Abruzzi KC, Luo W, Griffith LC, and Rosbash M (2016). Circadian neuron feedback controls the Drosophila sleep--activity profile. Nature 536, 292–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keller A (2007). Drosophila melanogaster’s history as a human commensal. Curr Biol 17, R77–81. [DOI] [PubMed] [Google Scholar]
- 35.Wu B, Ma L, Zhang E, Du J, Liu S, Price J, Li S, and Zhao Z (2018). Sexual dimorphism of sleep regulated by juvenile hormone signaling in Drosophila. PLoS Genet 14, e1007318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Artiushin G, and Sehgal A (2017). The Drosophila circuitry of sleep-wake regulation. Curr Opin Neurobiol 44, 243–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ly S, Pack AI, and Naidoo N (2018). The neurobiological basis of sleep: Insights from Drosophila. Neurosci Biobehav Rev 87, 67–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lane JM, Liang J, Vlasac I, Anderson SG, Bechtold DA, Bowden J, Emsley R, Gill S, Little MA, Luik AI, et al. (2017). Genome-wide association analyses of sleep disturbance traits identify new loci and highlight shared genetics with neuropsychiatric and metabolic traits. Nat Genet 49, 274–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopez-Minguez J, Morosoli JJ, Madrid JA, Garaulet M, and Ordonana JR (2017). Heritability of siesta and night-time sleep as continuously assessed by a circadian-related integrated measure. Sci Rep 7, 12340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vanaphan N, Dauwalder B, and Zufall RA (2012). Diversification of takeout, a male-biased gene family in Drosophila. Gene 491, 142–148. [DOI] [PubMed] [Google Scholar]
- 41.McCracken S, Lambermon M, and Blencowe BJ (2002). SRm160 splicing coactivator promotes transcript 3’-end cleavage. Mol Cell Biol 22, 148–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nesic D, and Maquat LE (1994). Upstream introns influence the efficiency of final intron removal and RNA 3’-end formation. Genes Dev 8, 363–375. [DOI] [PubMed] [Google Scholar]
- 43.Picot M, Cusumano P, Klarsfeld A, Ueda R, and Rouyer F (2007). Light activates output from evening neurons and inhibits output from morning neurons in the Drosophila circadian clock. PLoS Biol 5, e315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blau J, and Young MW (1999). Cycling vrille expression is required for a functional Drosophila clock. Cell 99, 661–671. [DOI] [PubMed] [Google Scholar]
- 45.Taghert PH, Hewes RS, Park JH, O’Brien MA, Han M, and Peck ME (2001). Multiple amidated neuropeptides are required for normal circadian locomotor rhythms in Drosophila. J Neurosci 21, 6673–6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chiu JC, Low KH, Pike DH, Yildirim E, and Edery I (2010). Assaying locomotor activity to study circadian rhythms and sleep parameters in Drosophila. Journal of visualized experiments : JoVE [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


