Abstract
In Hydra the nervous system is composed of neurons and mechanosensory cells that differentiate from interstitial stem cells (ISCs), which also provide gland cells and germ cells. The adult nervous system is actively maintained through continuous de novo neurogenesis that occurs at two distinct paces, slow in intact animals and fast in regenerating ones. Surprisingly Hydra vulgaris survive the elimination of cycling interstitial cells and the subsequent loss of neurogenesis if force‐fed. By contrast, H. oligactis animals exposed to cold temperature undergo gametogenesis and a concomitant progressive loss of neurogenesis. In the cold‐sensitive strain Ho_CS, this loss irreversibly leads to aging and animal death. Within four weeks, Ho_CS animals lose their contractility, feeding response, and reaction to light. Meanwhile, two positive regulators of neurogenesis, the homeoprotein prdl‐a and the neuropeptide Hym‐355, are no longer expressed, while the “old” RFamide‐expressing neurons persist. A comparative transcriptomic analysis performed in cold‐sensitive and cold‐resistant strains confirms the downregulation of classical neuronal markers during aging but also shows the upregulation of putative regulators of neurotransmission and neurogenesis such as AHR, FGFR, FoxJ3, Fral2, Jagged, Meis1, Notch, Otx1, and TCF15. The switch of Fral2 expression from neurons to germ cells suggests that in aging animals, the neurogenic program active in ISCs is re‐routed to germ cells, preventing de novo neurogenesis and impacting animal survival.
Keywords: Hydra nervous system, interstitial stem cells, adult de novo neurogenesis, gametogenesis, aging, homeoprotein prdl‐a, neuropeptide Hym‐355, evolution of neurogenesis
Introduction
The freshwater hydrozoan Hydra polyp (Fig. 1A) belongs to Cnidaria, a phylum that includes anthozoans (sea anemones and corals) and medusozoans (jellyfish) (Collins et al., 2006). Their common ancestor arose prior to the common ancestor of bilaterians (Fig. 1B) and Cnidaria together with Bilateria are eumetazoans, that is, animals equipped with a nervous system and epithelial layers, the internal one forming a gut, and the external one an epidermis (Fig. 1C). Among cnidarians, the hydrozoan Hydra polyp is well known for its amazing capacity to regenerate within few days any missing body part after amputation including its complete nervous system. In fact, the central body column of the animal is populated all along its life with multipotent interstitial stem cells (ISCs) that beside germ cells and gland cells produce all cells of the nervous system, as well as epithelial stem cells in the epidermis (eESCs) and the gastrodermis (gESCs) (Buzgariu et al., 2015). ISCs produce neuronal precursors that migrate toward the extremities of the animal where they differentiate to replace cells that die or get sloughed off (Bode et al., 1973; Fujisawa, 1989; Teragawa and Bode, 1995; Technau and Holstein, 1996). As a consequence, the Hydra nervous system is much denser at the apical and basal poles. This process that takes several weeks in homeostatic conditions is completed within several days when the animals regenerate its apical or basal extremity (Wenger et al., 2016).
Figure 1.
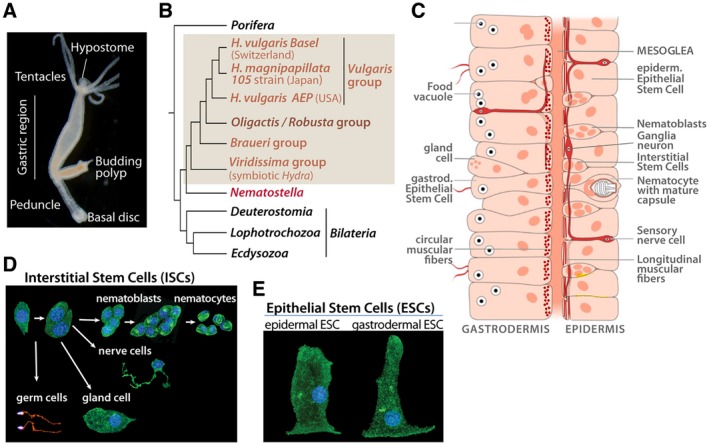
Hydra anatomy, phylogenetic position and cell types. (A) Bright field picture of a budding H. oligactis. Note the typical stalk shape of the peduncle region. (B) Phylogenetic tree showing the respective positions of H. vulgaris and H. oligactis. (C) Schematic view of the bilayered organization of the body column with the gastrodermis (or gut) on the left side and the epidermis on the right one, separated by an acellular matrix named mesoglea. Epithelial cells from both layers contain myofibrils, circular in gESCs and longitudinal in eESCs. Nematoblasts and nematocytes are restricted to the epidermis while nerve cells are present in both layers although more abundant in the outer one. Scheme modified from (Lenhoff and Lenhoff, 1988). (D, E) Identification of the various Hydra cell types, as observed after tissue maceration followed by b‐tubulin immunodetection and DAPI counter staining. Among the derivatives of the multipotent ISCs (D), note the precursors to nematocytes (or cnidocytes) named nematoblasts that divide syncitially before entering differentiation, and the different types of nerve cells: sensory, sensory/motor (bipolar), or ganglion neurons (multipolar). The ESCs (E) are unusual stem cells, that is, self‐renewing and differentiated (Buzgariu et al., 2015). [Colour figure can be viewed at wileyonlinelibrary.com]
Cnidarian neurons that form neurites but no typical axons, therefore, often named “nerve cells,” can be sensory, sensory‐motor bipolar, and multipolar also named ganglia neurons (Fig. 1D). These ganglia neurons function as interneurons that form in some species a well visible nerve ring at the base of the tentacles, often recognized as a simple form of cephalization (Koizumi, 2007). Beside nerve cells, a large pool of mechanosensory cells named cnidocytes or nematocytes contribute to the feeding and defense behaviors, as equipped with a cnidocil that, upon stimulation, triggers the discharge of the cnidocyst, a phylum‐specific organelle that functions as a weapon full of venom (Tardent, 1995). The two main cell lineages that form the Hydra nervous system are quite different at the quantitative level (Bode et al., 1973; David, 1973), the neurons being rather rare (3% of all cell types) and functional for several weeks while the abundant nematocytes (35%) are “single‐usage,” replaced once they have discharged their venom capsule.
Cnidarian and bilaterian nervous systems follow the same principles of synaptic conduction and chemical neurotransmission (Anderson and Spencer, 1989; Kass‐Simon and Pierobon, 2007), even though cnidarian neurons heavily make use of peptides with peptide‐gated ion channels as receptors (Grimmelikhuijzen and Westfall, 1995a; Grunder and Assmann, 2015). The pool of transcription factors involved in bilaterian neurogenesis are largely expressed in cnidarian nervous systems (Grens et al., 1995; Gauchat et al., 1998; 2004; Miljkovic‐Licina et al., 2004; 2007; Galliot et al., 2009; Marlow et al., 2009; Galliot and Quiquand, 2011), suggesting a common origin between cnidarian and bilaterian nervous systems. Still, some neuropeptides impact neuronal differentiation, positively in case of Hym‐355 (Takahashi et al., 2000).
Another amazing feature of Hydra is its ability to survive short antimitotic treatments (hydroxyurea, colchicine) that eliminate the fast cycling ISCs but leave intact the two epithelial stem cell populations (Campbell, 1976; Marcum and Campbell, 1978; Marcum et al., 1980). Upon such treatments, de novo neurogenesis is suppressed and animals progressively loose all their nerve cells. In few weeks, the cell lineages that derive from ISCs are depleted making the animal “nerve‐free” (Sacks and Davis, 1979). Surprisingly, such nerve‐free animals can survive months and years if manually force‐fed and their developmental capacities are not impaired, meaning that animals only equipped with two epithelial layers still regenerate and, if heavily fed, bud. A recent transcriptomic analysis showed that these epithelial cells (Fig. 1E) actually upregulate a large number of genes, including genes normally predominantly expressed in ISCs or their derivatives (Wenger et al., 2016). The functional consequences of these genetic modulations have not been explored yet, but this result highlights an epithelial plasticity at the transcriptional level when neurogenesis fails.
Three main studies performed over the past 70 years indicate that Hv polyps have an extremely long lifespan or might even escape aging completely (Brien, 1953; Martínez, 1998; Schaible et al., 2015). The dynamic maintenance of the three populations of stem cells is necessary for slow aging and more specifically the stock of ISCs for maintaining a continuous neurogenesis in adult animals. How Hydra polyps maintain over long periods of time stocks of adult stem cells without any DNA‐damage or cellular alteration is poorly understood. In a distinct species named H. oligactis (Fig. 1A), aging rapidly follows the induction of gametogenesis, which is obtained by transferring the animals from room temperature (RT) to 10°C (Brien, 1953). After few weeks, the intense production of gametes leads to the depletion of the somatic populations derived from ISCs, and animals cannot maintain their fitness (Yoshida et al., 2006).
More recently, we characterized two distinct H. oligactis strains that exhibit different responses to cold‐induced gametogenesis; one that undergoes aging (Ho_CS) and another that resists to aging (Ho_CR) (Tomczyk et al., 2017). Ho_CS animals exhibit the typical aging phenotype reported by Brien (1953) and Yoshida et al. (2006), we also noted the disorganization of the apical nervous system (Tomczyk et al., 2015). At the epithelial level, we identified two main differences between aging‐sensitive and aging‐resistant animals, (i) the inducibility of the autophagy flux that appears deficient in Ho_CS, and (ii) the self‐renewal of ESCs that progressively and irreversibly decreases in aging animals (Tomczyk et al., 2017). The importance of autophagy for stem cell homeostasis was demonstrated in multiple cell types as recently reviewed by (Boya et al., 2018). Autophagy also plays a key role in the maturation and survival of adult‐generated neurons but the mechanisms remain unknown (Xi et al., 2016). With this study our aim was to characterize at the behavioral, cellular and molecular levels the degeneration of the nervous system in aging H. oligactis.
Materials and Methods
Hydra Culture and Induction of Aging
Strains of Hydra vulgaris (Hv_Basel) or Hydra oligactis, identified as Cold Sensitive (Ho_CS) and Cold Resistant (Ho_CR) were mass‐cultured in Hydra Medium (HM: 1 mM of NaCl, 1 mM of CaCl2, 0.1 mM of KCl, 0.1 mM of MgSO4, 1 mM of Tris‐HCl pH 7.6) and fed twice a week with freshly hatched brine shrimps. To induce aging Ho_CS and Ho_CR animals were transferred from 18°C ± 0.5°C, the standard temperature for Hydra culture, to incubators at 10°C ± 0.3°C. For animals maintained at 10°C all washes were done with pre‐cooled HM (10°C) and transferred to sterile cold culture dishes. For transcriptomic analyses, animals from the Hv_Jussy, Hv_sf1, and Hv_AEP strains were used as reported in Wenger et al. (2016).
Behavioral and Morphometric Analyses
For touch responsiveness, animals were stimulated with tweezers in the peduncle region and observed under the binocular and the time between tweezer stimulation and contraction was noted for each Ho_CS animal maintained either at 18°C or at 10°C for 35 days. For light response, 15 polyps maintained at 18°C or 10°C in dim light were allowed to fully extend in a darkened room, then exposed to a LED light source placed 19 cm above the animals to record body column contractions (at least 25% of their length). The latency period corresponds to the time between initial light exposure and the observed first contraction (5 min at most). For prey capture, 15 polyps per condition were placed in a 24‐well plate pre‐filled with 1 ml HM containing Artemia. After 15 s the animals were transferred to HM‐containing wells, the number of captured Artemia was recorded and normalized by tentacle number. The variations in hypostome size were measured by manually delimiting the hypostome area on the images taken with Leica DM5500. The delimited surface was measured with Fiji.
Immunofluorescence (IF) on Macerated Tissues and on Whole Mounts
Procedures were as described in (Wenger et al., 2016). Briefly, for IF on cells Hydra, tissues were macerated according to (David, 1973). The cell suspension was spread on positively charged Superfrost Plus slides (Thermo Scientific) and dried for two days at RT, then washed in PBS, blocked with PBS, 2% BSA, 0.1% Triton‐X100 for 2 h, and incubated with the anti a‐tubulin antibody overnight at 4°C. After washes in PBSTw (PBS, 0.1% Tween‐20), cells were incubated in the anti‐rabbit AlexaFluor‐488 antibody (1:600, Thermofisher), counterstained with DAPI and imaged on a Leica SP8 confocal microscope. For IF on whole mounts, animals were briefly relaxed in urethane 2% (60 s), then fixed in 4% paraformaldehyde (PFA) prepared in HM when immunodetected with the rabbit polyclonal serum raised against the RFamide neuropeptide (Grimmelikhuijzen, 1985; Wenger et al., 2016). When immunodetected with the rabbit polyclonal serum raised against the prdl‐a homeoprotein (Gauchat et al., 1998), animals were fixed in Lavdowsky fixative (50% of ethanol, 3.7% of formaldehyde, and 4% of acetic acid) for 1 h at 37°C, washed several times in PBS, and incubated in 2N HCl for 30 min for DNA denaturation. Then, whatever the fixation condition, animals were washed in PBS, blocked in PBS, 2% BSA, 0.1% Triton‐X100 for 2 h, incubated overnight at 4°C with the primary antibody (1:1,000), washed in PBSTw and incubated with the appropriate anti‐rabbit AlexaFluor antibody (AlexaFluor‐488 or AlexaFluor‐555, 1:600, Thermofisher). The samples were finally washed in PBSTw, DAPI stained and pictured on a Leica SP8 or Zeiss LSM700 confocal microscope.
BrdU Detection and Whole Mount in situ Hybridization (WM‐ISH)
Animals were incubated in 5 mM 5′‐bromodeoxyuridine (BrdU) for 4 h, washed and maintained in HM for two days, and fixed in 4% PFA/HM for 4 h then in ethanol overnight at −20°C. The transcripts were detected with a digoxygenin (DIG)‐labeled riboprobes specific to Hym355, prdl‐a, TCF15 or Fral2 and the samples were processed for WM‐ISH as in (Bode et al., 2008). In case of H. oligactis animals, the WM‐ISH procedure was shortened at several steps to account for their higher frailty: proteinase K digestion was 8 and 10 min for animals maintained at 10 and 18°C respectively, the 80°C preheat step was 20 min, the probe hybridization step 16 h and the post‐hybridization MAB washes 6 × 15 min. Whatever the species, the hybridized riboprobes were similarly detected by NBT‐BCIP staining, the samples were then post‐fixed in 3.7% FA for 30 min, washed for 15 min in methanol and 3 × 10 min in PBSTr (PBS, 0.1% Triton X‐100). Next, the samples were treated with 2N HCl for 30 min, washed in PBST several times over 15 min, incubated overnight at 4°C with the anti‐BrdU antibody (1:20, Roche), washed 4 × 10 min in PBS and incubated in the anti‐mouse Alexa 488‐coupled secondary antibody (1:500, Molecular Probes) for 4 h at RT. Then animals were labeled with DAPI for 10 min, washed in PBS (4 × 10 min), 5 min in H2O, mounted in Mowiol and imaged on a Leica D5500 microscope equipped with a color DMC2900 and a monochromatic DFC9000 camera.
BrdU and prdl‐a Double Immuno‐Labeling
Intact H. vulgaris were incubated with 5 mM BrdU for 4 h and either amputated at mid‐gastric level and let to regenerate for four days, or washed and maintained intact in HM. At the indicated time, the animals were fixed for 1 h at 37°C in Lavdowsky “minus” (50% ethanol, 3.7% FA), washed several times in PBS, treated with 2N HCl for 30 min and washed again in PBS. After 1 h incubation in 2% BSA, PBS, the samples were incubated overnight with the anti‐BrdU (1:20, Roche) and anti‐prdl_a (1:1,000, Gauchat et al., 1998) antibodies, washed 4 × 10 min in PBS, incubated with the anti‐mouse Alexa 488 and anti‐rabbit Alexa 555 antibodies (1:500, Molecular Probes), washed again 4 × 10 min in PBS, incubated for 10 min in DAPI (1 μg/ml in PBS), washed 2 × 5 min in PBS, briefly in water and mounted in Mowiol. Imaging was performed with a LSM700 Zeiss confocal microscope. For quantification of interstitial cell proliferation in Ho_CS, animals were exposed to BrdU for 24 h at indicated time points of the aging process. At the end of the treatment animals were macerated as in (David, 1973) and immunodetected with the anti‐BrdU antibody as described above. The BrdU + nuclei were counted manually to establish the BrdU‐labeling index.
Transcriptomic Analyses
The quantitative RNA‐sequencing analysis (qRNA‐seq) performed on Ho_CS and Ho_CR animals maintained either at 18°C or at 10°C is detailed in (Tomczyk et al., 2017). The qRNA‐seq performed either on Hv_Jussy animals sliced at five distinct positions along the body axis (Supplementary Figure S1), or on Hv_sf1 animals exposed to drugs (hydroxyurea, colchicine) or to heat‐shock, or on transgenic Hv_AEP animals expressing GFP in one or the other stem cell populations is reported in (Wenger et al., 2016). Values that were obtained in biological triplicates for each condition are available in Table S1 from Wenger et al. (2016). For heatmap representations, each value corresponds to the average number of reads obtained at indicated time points. Orthologous relationships between Hv_sf1, Ho_CR and Ho_CS sequences were assigned manually. GraphPad Prism 7 was used to generate the scatterplots of fold change differences between Ho and Hv RNA‐seq values and the regression analysis. Sequences and RNA‐seq profiles are publicly available, for H. vulgaris at https://Hydratlas.unige.ch, for Ho_CS and Ho_CR at http://129.194.56.90/blast/.
Results
Sustained de novo Neurogenesis in Long Lived H. vulgaris
To demonstrate the de novo production of neurons in homeostatic and regenerating non‐aging animals we used two established markers for neuronal progenitors and nerve cells, the homeoprotein prdl‐a and the neuropeptide Hym‐355, respectively. Both prdl‐a and Hym‐355 are involved in the control of neurogenesis, prdl‐a in the production of apical neuronal progenitors (Gauchat et al., 1998), Hym‐355 in the neuronal differentiation of interstitial progenitors (Takahashi et al., 2000). In homeostatic conditions, both prdl‐a transcripts and the prdl‐a protein are predominantly expressed in apical interstitial progenitors (Fig. 2A,D) while Hym‐355 is expressed in apical and basal subpopulations of neurons, and to a lower extent in the body column (Fig. 2A,B). The animals fixed four days after a short exposure to BrdU do not show any BrdU + cells that express Hym‐355 (Fig. 2B). Under homeostatic conditions, the number of neurons produced each day is rather low as de novo neurogenesis is slow (Hager and David, 1997), explaining why BrdU/Hym‐355 positive neurons cannot be detected in this condition.
Figure 2.
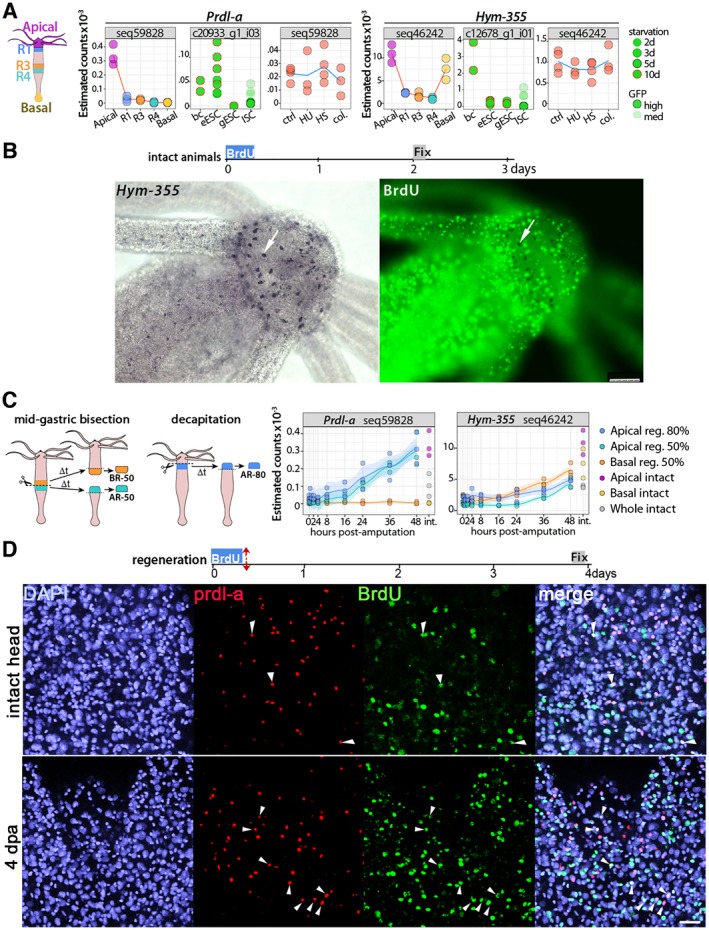
Homeostatic and regeneration‐induced de novo neurogenesis in slow aging Hydra vulgaris. (A) Expression profiles of the neurogenic genes Prdl‐a and Hym‐355 as detected in animals of the heat‐sensitive Hv_sf‐1 strain through three different RNA‐seq approaches performed (a) on tissues from slices of the body column when distinct regions (Head, R1, R3, R4, Foot) were dissected before RNA extraction (see the scheme on the left that indicates the five regions); (b) on distinct cell types, that is, ectodermal epithelial stem cells (eESC), gastrodermal epithelial stem cells (gESC), and interstitial stem cells (ISCs) isolated by flow‐cytometry; (c) on the central body column of animals exposed to drugs (HU and colchicine) or Heat‐shock (HS). All transcriptomic procedures are described in (Wenger et al., 2016). (B) Lack of BrdU+ neurons expressing Hym‐355 in animals exposed to BrdU for 4 h and fixed four days later. Scale bar: 25 μm (left) and 75 μm (right). (C) Expression profiles of Prdl‐a and Hym‐355 transcripts detected by RNA‐seq in regenerating‐tips dissected at various time points of head or foot regeneration. Animals were either bisected at mid‐gastric level (50%) or decapitated (80%). (D) Prdl‐a (red) and BrdU (green) co‐detection in the apex of intact (upper panel) or head‐regenerating (lower panel) H. vulgaris animals. All animals were exposed to a 4 h BrdU pulse, then immediately bisected at mid‐gastric level and fixed three days later. In the apical region shown here prdl‐a expression is nuclear, enhanced in regenerating heads (Gauchat et al., 1998). The prdl‐a+ and prdl‐a+/BrdU+ cells (white arrowheads) are rare in heads of intact animals but numerous in newly regenerated heads. Scale bar: 25 μm. [Colour figure can be viewed at wileyonlinelibrary.com]
During regeneration, the dense nervous systems in the head and peduncle are re‐established, in few days to maintain the animal fitness. Immediately after amputation, neuronal progenitors migrate toward the wound to contribute to the regeneration of the nervous system in the newly formed structure. In the regenerating tips the expression of prdl‐a and Hym‐355 is re‐established with different kinetics: prdl‐a shows a dual regulation with a first immediate but transient expression in gastrodermal cells (Gauchat et al., 1998), followed by an upregulation in interstitial progenitors of the head‐regenerating tip from 16 h post‐amputation (hpa) (Fig. 2C). By contrast, Hym‐355 starts to be upregulated later, detected in both the head‐ and foot‐regenerating tips after 24 hpa (Fig. 2C). To visualize the induction of neurogenesis in regenerating animals, we coupled prdl‐a immunodetection to BrdU labeling: H. vulgaris animals bisected at mid‐gastric position were immediately exposed to BrdU for 4 h, then left to regenerate for four days and fixed for immunodetection. In such conditions, we noticed a high increase in the number of BrdU+/prdl‐a+ nuclei when compared to non‐bisected animals (Fig. 2D), reflecting the anticipated fast neurogenesis process.
These two types of de novo neurogenesis, slow in intact animals and fast in regenerating ones, do not seem to be altered in animals kept for several years as these animals regenerate well the missing structures and maintain their active behaviors, that is, feeding, contracting, and walking (Brien, 1953; Martínez, 1998). By contrast, H. oligactis animals exposed to cold temperature, a condition that induces gametogenesis, rapidly loose their fitness and the ability to maintain a dynamic neurogenesis (Brien, 1953; Yoshida et al., 2006; Tomczyk et al., 2015) (Fig. 3A).
Figure 3.
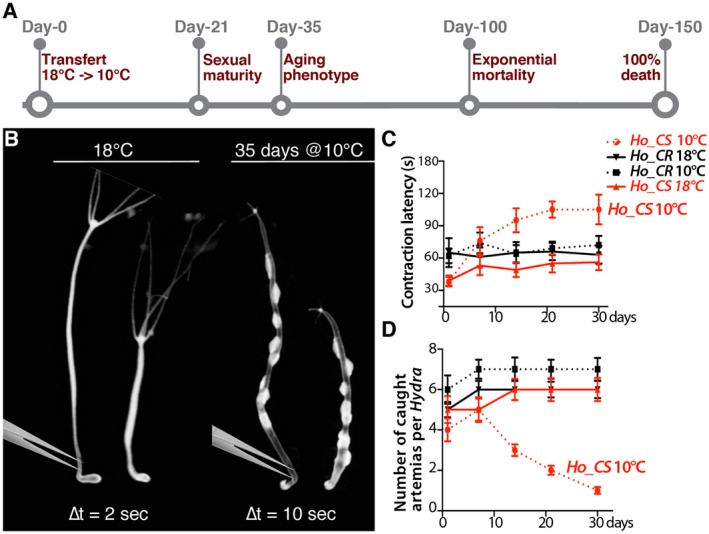
Behavioral alterations in H. oligactis animals undergoing aging. (A) Schematic view of the aging process induced in sexual H. oligactis cold‐sensitive (Ho_CS) animals that undergo gametogenesis upon transfer to cold (10°C). Animals from the closely related H. oligactis cold‐resistant strain (Ho_CR) also undergo gametogenesis within three weeks but do not exhibit any aging phenotype and remain fit over the following months (Tomczyk et al., 2017). (B) Contractility upon mechanical stimulation in Ho_CS animals maintained either at 18°C or at 10°C for 35 days. The contractility in response to tweezer stimulation of the peduncle region was recorded on live animals imaged with an Olympus SZX10 microscope equipped with a DP73 camera (movie). Contraction of the whole animal body takes place within 2 s in animals maintained at 18°C and within 10 s in Ho_CS animals maintained at 10°C for 35 days. (C) Light‐induced contractility of Ho_CS and Ho_CR animals maintained at 10°C (n = 15). The contractility was deduced from the time taken by the animal to elaborate a typical contraction response to a bright, directly focused light source. (D) Prey capture of Ho_CS and Ho_CR animals maintained at 10°C (n = 15) assessed from the number of Artemia fixed on tentacles of each animal normalized by tentacle number. In C and D, the legend codes are the same and the error bars correspond to standard error of the mean (SEM). Note that already two weeks after transfer to 10°C, Ho_CS but not Ho_CR animals show an extended contraction latency after light exposure and lose the ability to capture preys. [Colour figure can be viewed at wileyonlinelibrary.com]
Aging H. oligactis Lose Their Contractility and Feeding Capacity
To test the impact of aging on the various functions of the nervous system, we compared the behaviors of aging and non‐aging animals exposed to mechanical stimulus, light, or food. We first tested the contractility induced by a mechanical stimulus, that is, a pinch in the peduncle region (Fig. 3B). We found that while Ho_CS animals maintained at 18°C contract within 2 s, Ho_CS animals taken 35 days after transfer to 10°C show a much slower reaction time of 10 s (see supplementary movies). We also measured at regular time‐points over a 30 days period after transfer to 10°C, the contraction latency in response to light (Fig. 3C). We recorded an increase in the contraction latency, from 40 to 100 s in Ho_CS animals while the initial value (60 s) remained stable in Ho_CR animals exposed to cold for the same period of time (Fig. 3C). The difference between the two strains was already visible at day‐14 after the temperature switch.
The feeding behavior in Hydra is characterized by the capture of preys on the tentacles, the subsequent opening of the mouth and the coordinated movement of the tentacles to favor the ingestion of the preys through the mouth. This complex behavior is triggered by reduced glutathione release from the captured prey (Loomis, 1955; Grosvenor et al., 1996; Pierobon, 2015). Here animals were individually exposed to preys, that is, swimming Artemias, for 15 s and the number of preys fixed on the tentacles was counted. This test that was performed at various time points after transfer to 10°C, showed a continuous decline in the ability to fix preys in Ho_CS animals, already visible at day‐14 post‐transfer, but not in Ho_CR ones (Fig. 3D). After 30 days, a single prey was fixed on average on tentacles of Ho_CS animals versus six in Ho_CR animals. This decline reflects the inability of the animals to produce functional nematocytes that are normally abundant along the tentacles. After five to six weeks at 10°C, prey capture becomes impossible for Ho_CS animals as their head structures are heavily degenerated. These results show that both the contractility in response to light and the capacity to capture preys start to decline within 14 days after transfer to cold in Ho_CS but not in Ho_CR, progressively decreasing over the following weeks. This indicates that deficiencies in both neurogenesis and nematogenesis precede the morphological changes that appear at day‐30.
Degeneration of the Nervous System in Aging H. oligactis
To investigate the nervous systems of the animals that show a decline of their behavioral functions, we analyzed the nerve cell density along the animal body, first by monitoring the distribution of the Hym‐355 expressing neurons in Ho_CS animals undergoing aging. At 18°C, Hym‐355 neurons distribute in the apical and basal regions as well as along the body column in Ho_CS animals (Fig. 4A). In H. vulgaris Hym‐355 was described as predominantly expressed at the extremities, with only a low number of Hym‐355 neurons present in the body column (Takahashi et al., 2000). In H. oligactis as in H. vulgaris, Hym‐355 expression is restricted to nerve cells however Hym‐355 neurons are also numerous along the body axis, indicating species‐specific variations. After one month at 10°C, we observed a severe loss of Hym‐355 + nerve cells, which are no longer detected after 41 days (Fig. 4A).
Figure 4.
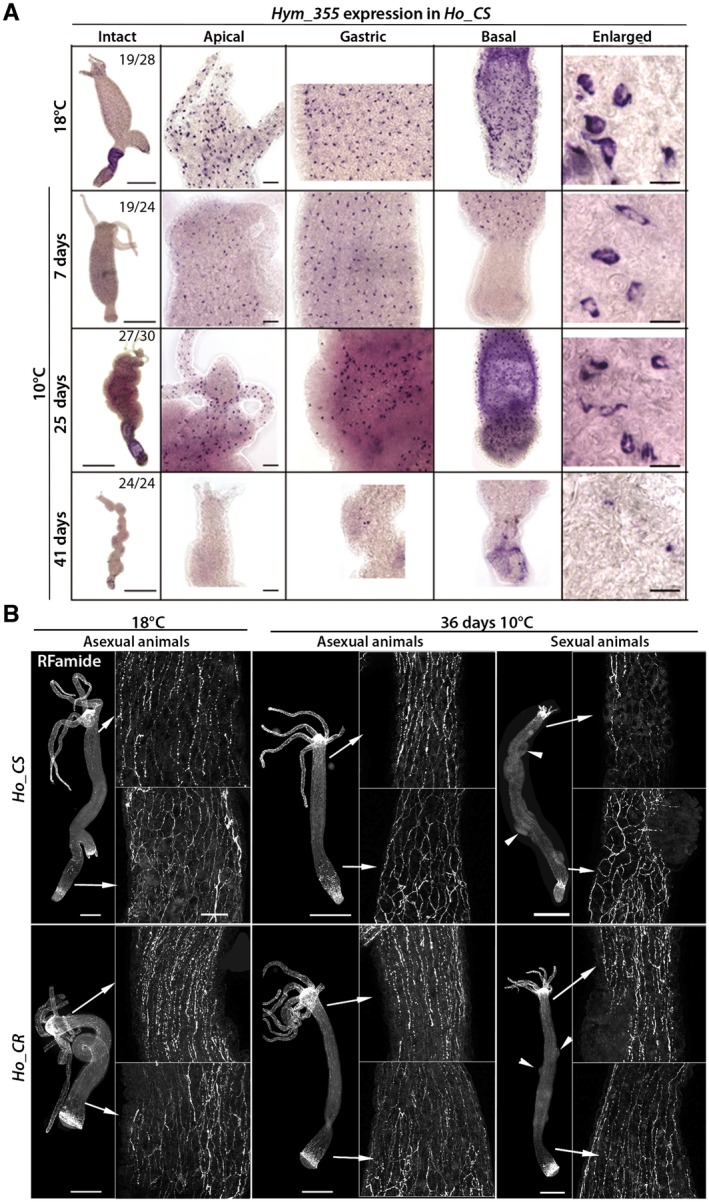
Loss of neuronal density in Ho_CS animals undergoing aging. (A) Hym‐355 expressing nerve cells detected by in situ hybridization along the body column of Ho_CS animals maintained at 18°C or 10°C for 7, 25, or 41 days. Scale bars: 500 µm (intact), 50 µm (apical, gastric, and basal), 10 µm (enlarged). (B) RFamide expressing nerve cells immunodetected in Ho_CS and Ho_CR animals maintained at 18°C or 10°C for 36 days. Arrowheads indicate mature or post‐mature testes. Scale bars: 100 µm. [Colour figure can be viewed at wileyonlinelibrary.com]
In parallel, we detected RFamide expressing neurons in Ho_CS and Ho_CR animals maintained at 18°C or at 10°C (Fig. 4B). RFamides are neuropeptides widely expressed in mature nerve cells along the body axis and at the extremities, highlighting the higher density of the nervous system at the apex and along the peduncle (Grimmelikhuijzen, 1985). As a low fraction of Ho_CS and Ho_CR animals do not undergo gametogenesis after transfer to cold, we used sexual and asexual animals maintained for 36 days at 10°C to discriminate between the effects of cold stress versus gametogenesis and aging on the nervous system. We noted an obvious lower density of RFamide+ nerve cells in the upper part of the body column in sexual Ho_CS animals, well visible in the region directly below the head. We noted the persistence of “old” RFamide+ neurons at the apical extremity as well as in the peduncle region although the nerve net appears disorganized (Fig. 4B). In contrast, Ho_CR animals, sexual, or asexual, maintained at 10°C for 36 days exhibit a nervous system that is not modified in terms of density or organization. Both neuronal markers, Hym‐355 and prdl‐a, point to a clear degeneration of the nervous system in sexual Ho_CS animals maintained for weeks at 10°C.
Loss of de novo Neurogenesis in Aging Hydra Oligactis
The observed loss of RFamide and Hym‐355 neurons could have various causes: (i) a loss of ISC proliferation necessary to replenish the stock of nerve cells, (ii) a disrupted differentiation of neuronal progenitors or (iii) a poor survival of the newly differentiated neurons. After transfer to 10°C both Ho_CS and Ho_CR exhibit a decrease in body size due to the change in the feeding rhythm, from four feedings per week at 18°C to two feedings at 10°C. However in Ho_CS animals this size decrease strongly affects the head structures that become three fold smaller within 35 days (Fig. 5A). As a consequence of the massive production of gametes, the proliferation of somatic interstitial cells measured after a 24 h BrdU pulse drops to reach a minimal value after 32 days at 10°C, leading to a severe depletion of the stock of somatic ISCs as previously reported (Yoshida et al., 2006; Tomczyk et al., 2017). However, the rare somatic ISCs that survive readily cycle at later time‐points (Fig. 5B).
Figure 5.
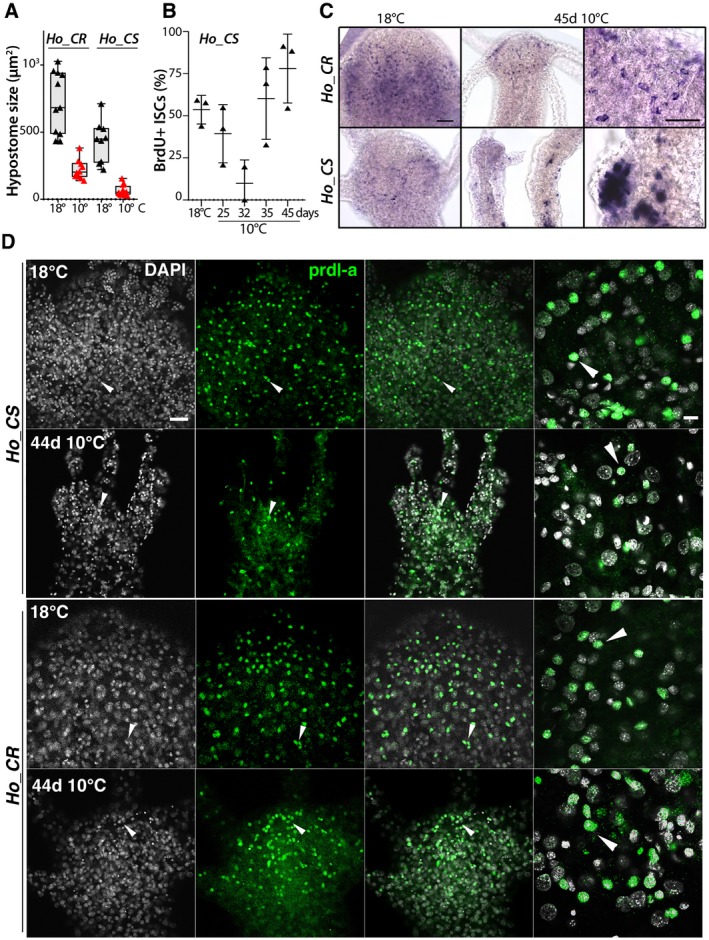
Loss of neuronal progenitors and prdl‐a/prdl‐a expression in aging H. oligactis. (A) Decrease in hypostome size in Ho_CS and Ho_CR animals maintained at 18°C or 10°C for 35 days. (B) Transient decrease in interstitial cycling cells in Ho_CS. (C) Apical expression of prdl‐a in Ho_CS and Ho_CR animals maintained at 18°C or 10°C for 45 days. (D) Immunodetection of the homeoprotein prdl‐a in Ho_CS and Ho_CR animals maintained at 18°C or taken 44 days after transfer to 10°C. Scale bars: 20 µm (left panels) and 10 µm (right panels). Note the progressive decrease in the number of prdl‐a expressing cells in aging Ho_CS when compared to Ho_CR maintained in the same conditions.
To visualize the neuronal progenitors in Ho_CS animals taken at 44 or 45 days after transfer to cold, we detected the cells expressing prdl‐a either by in situ hybridization (Fig. 5C) or by immunodetection with the antibody raised against Hydra prdl‐a (Fig. 5D).
At 18°C prdl‐a is predominantly expressed in neuronal progenitors located in the apical region, a pattern also observed in Ho_CR animals maintained at 10°C for 45 days (Fig. 5C). By contrast, Ho_CS animals maintained at 10°C for 45 days have very few apical cells expressing prdl‐a, while some ectopic patches of non‐neuronal prdl‐a cells can be found in the body column. As anticipated, prdl‐a protein in Ho_CR and Ho_CS animals maintained at 18°C is predominantly apical and nuclear (Fig. 5D, arrowheads), a pattern that remains stable in Ho_CR maintained at 10°C for 44 days. By contrast, in aging Ho_CS we observed a drastic decrease in the number of prdl‐a+ cells, likely reflecting a decrease in the number of neuronal precursors. The loss of apical prdl‐a+ cells suggests that the deterioration of the nervous system in aging Ho_CS is predominantly caused by the lack of de novo homeostatic neurogenesis, responsible for the insufficient renewal of the apical nervous system.
Aging‐Induced Modulations of Genes Potentially Involved in Neurogenesis and Neurotransmission
To further characterize the changes that occur in the nervous system of aging Hydra, we analyzed by quantitative RNA‐seq (qRNA‐seq) the expression of genes potentially associated with neurogenesis (195) and neurotransmission (377), as previously annotated by Wenger et al. (2016). We retrieved the expression levels of these genes in Ho_CS and Ho_CR animals maintained at 10°C for 14, 26, 32, 35, and 45 days (only Ho_CS) as previously reported (Tomczyk et al., 2017). Concerning the three RFamide genes (Hansen et al., 2000), RFamide A and RFamide B show very similar profiles in the two strains except an upregulation of RFamideA in Ho_CS at 45 days (Fig. 6A). The persistence of the RFamideA transcripts in Ho_CS can be linked to their presumptive upregulation in the persisting neurons and/or to their ectopic expression in non‐neuronal cell types. RFamide C, which is no longer expressed after 25 days at 10°C in Ho_CR, exhibits a burst of expression at day‐25 in Ho_CS to become undetectable at day‐45. Concerning prdl‐a, we found its expression seemingly identical between Ho_CS and Ho_CR animals, a result compatible with the patterns detected by in situ hybridization (Fig. 6A).
Figure 6.
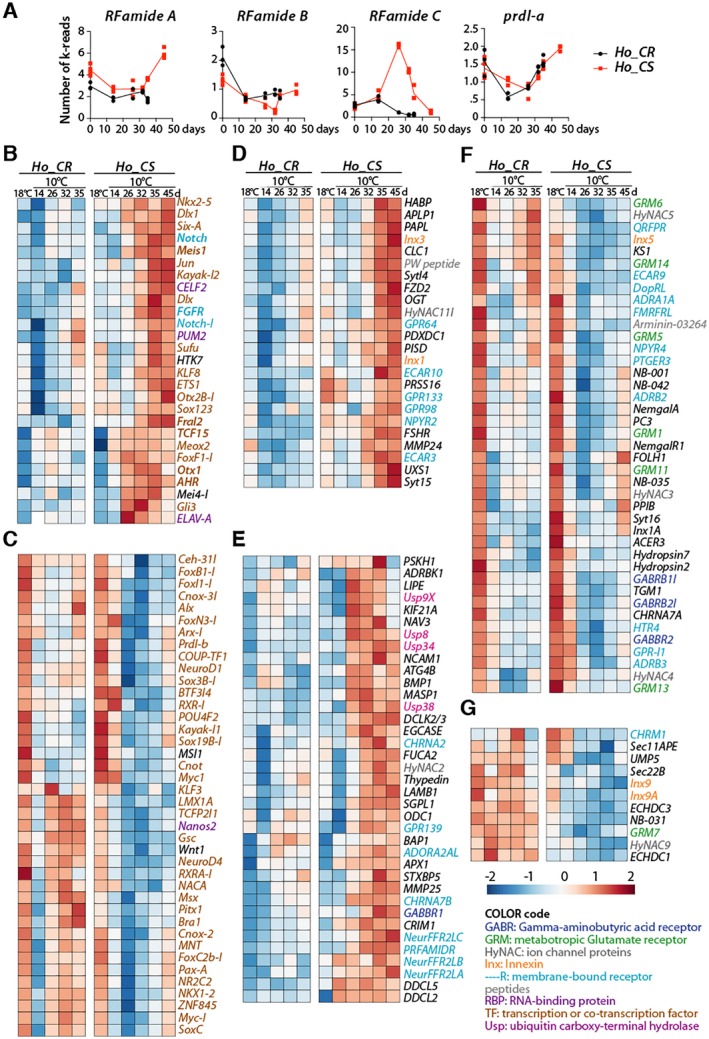
Modulations of neurogenesis and neurotransmission gene expression in Ho_CS and Ho_CR animals maintained at 18°C or 10°C. (A) Expression profiles of the genes encoding the RFamide neuropeptides and the prdl‐a homeoprotein measured by qRNA‐seq in Ho_CR (blue) and Ho_CS (red) animals at indicated time points after transfer to 10°C. (B‐G) Heatmaps showing the relative expression of 67 genes known or expected to regulate neurogenesis (B, C), and 114 genes whose products are predicted to be involved in neurotransmission (D‐G) in Ho_CR and Ho_CS animals transferred to 10°C at day‐0. In panels B, D, E, genes are upregulated in aging Ho_CS animals, while in panels C, F, and G, genes are either upregulated in Ho_CR or downregulated in Ho_CS. Sequences from Hv orthologs can be found in Table S1 from Wenger et al. (2016), expression profiles of neurogenesis genes are shown in Supplementary Figure S1. All expression profiles are accessible at www.hydratlas.unige.ch for Hv sequences, at http://129.194.56.90/blast/ for Ho_CS and Ho_CR ones after tblastn with Hv sequences. [Colour figure can be viewed at wileyonlinelibrary.com]
To get more global view of the modulations affecting the neurogenesis and neurotransmission genes, we produced heatmaps comparing gene expression levels in each strain (Fig. 6B–G). Among the 195 genes potentially associated with neurogenesis, we found 67 genes that are differently modulated in Ho_CS and Ho_CR, with 27 genes upregulated in Ho_CS but not in Ho_CR (Fig. 6B), and 40 genes upregulated in Ho_CR but not in Ho_CS (Fig. 6E). The former group that includes Dlx, Dlx1, ELAV‐A, ETS1, Fral2, Gli3, KLF8, Meis1, Otx1, Otx2B‐1, PUM2, Sox123, and TCF15 (Fig. 6B) might correspond to genes upregulated in the remaining epithelial cells after the loss of the interstitial lineage, and/or to genes linked to the persisting gametogenesis in Ho_CS. The second group likely includes genes involved in the wave of de novo neurogenesis that follows gametogenesis in Ho_CR but not in Ho_CS (Fig. 6C). Among these 40 genes upregulated in Ho_CR, one finds genes associated with interstitial stemness like Myc‐1, Pax‐A, ZNF845, or Nanos2 (Mochizuki et al., 2000; Hobmayer et al., 2012; Wenger et al., 2016). Loss of sustained expression of these genes in Ho_CS likely corresponds to the definitive loss of ISCs and progenitors. Interestingly LMX1A, which is upregulated in epithelial cells of HU‐treated animals (Wenger et al., 2016) is found in this category, suggesting that ESCs in aging Ho_CS are not able to upregulate LMX1A in response to the loss of neurogenesis.
Among the 377 genes associated with neurotransmission, we found 114 genes with expression profiles differently modulated in Ho_CS and Ho_CR. We first identified 24 and 37 genes upregulated at late time points in Ho_CS but not in Ho_CR where a large number show a transient downregulation at day 14 (Fig. 6D, E). This group contains several genes that encode ubiquitin specific peptidases (Usp), which play important roles in the development and the physiology of bilaterian nervous systems as well as in neurodegenerative diseases (Baptista et al., 2012). As an example, Usp8 is implicated in the clearance of protein aggregates (Alexopoulou et al., 2016) and might in Ho_CS be involved in the response of old persisting neurons to the proteomic stress. Another group contains 41 genes similarly downregulated in response to cold exposure in both Ho_CS and Ho_CR, possibly not linked to the aging process (Fig. 6F). Finally, 11 genes are maintained at high levels in Ho_CR but not in Ho_CS (Fig. 6G), likely reflecting the regeneration of a functional nervous system in Ho_CR but not in Ho_CS animals.
Impact of the Loss of Neurogenesis in HU‐Treated H. vulgaris and in Aging Ho_CS
In Hydra, the loss of neurogenesis can easily be achieved with anti‐proliferative treatments, such as hydroxyurea (HU) or colchicine. Such treatment in Ho_CS animals maintained at 18°C, so in the absence of gametogenesis, also leads to an aging phenotype (Tomczyk et al., 2017). This scenario resembles the aging process induced by gametogenesis in Ho_CS where the depletion of somatic interstitial progenitors leads to the disorganization of the nervous system. To investigate a possible similarity on the genetic level we compared the transcriptomic data obtained in Ho_CS and Ho_CR maintained at 10°C (Tomczyk et al., 2017) to those obtained in HU‐treated H. vulgaris (Hv_sf1) (Wenger et al., 2016). In each context we retrieved the number of reads for the neurogenesis and neurotransmission associated genes in two conditions, at 18 and 10°C day‐35 for Ho_CS and Ho_CR animals (Fig. 7A), in untreated and HU‐treated Hv_sf1 animals taken seven days after HU exposure (Fig. 7B). We calculated the log2 fold change (FC) for all selected genes (Fig. 7C) and produced a scatterplot representation of the changes in gene expression between Hv_sf1 post‐HU and Ho_CS or Ho_CR maintained at 10°C.
Figure 7.
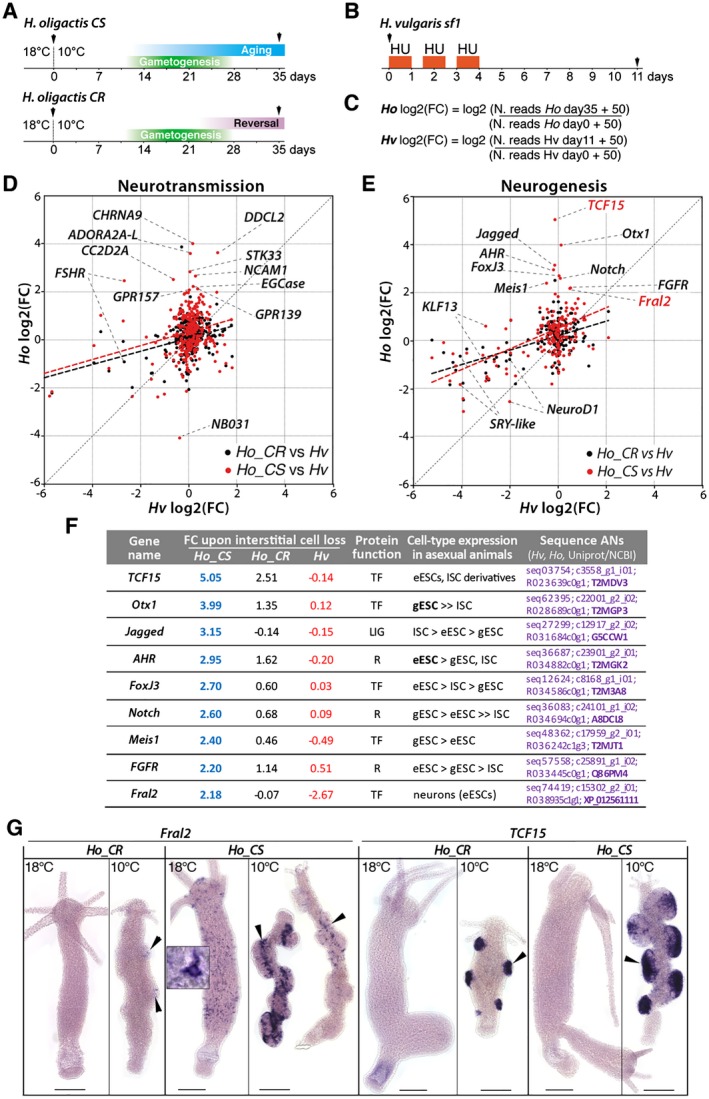
Comparative analysis of gene modulations of neurotransmission and neurogenesis genes in nerve free H. vulgaris, Ho_CS, and Ho_CR. (A, B) Schemes representing the experimental conditions to produce qRNA‐seq data (black arrowheads) from Ho_CS and Ho_CR animals maintained at 10°C for 35 days, or from Hv_sf1 animals exposed to hydroxyurea (HU). (C) Equations used to calculate the log2 fold change (FC) for Ho_CS, Ho_CR animals between day‐0 and day‐35 of cold exposure, and Hv_sf1 animals between day‐0 and day‐11 after HU exposure. (D, E) Scatterplot of FC values from Hv_sf1 (x axis) and Ho_CS or Ho_CR (y axis). The thick dashed lines represent linear regression for the Ho_CS (red) and Ho_CR (black) conditions. (F) Modulations in the expression of nine neurogenic candidate genes in Ho_CS and Ho_CR animals maintained at 10°C for 35 days (blue and black numbers, respectively) and in HU‐treated H. vulgaris (red numbers) as identified in (E). Fold change values (FC) measured in Ho_CS and Ho_CR animals maintained at 10°C for 35 days or in HU‐treated Hv_sf1 animals were normalized over the values measured in Ho animals maintained at 18°C or in untreated Hv animals, respectively. LIG: ligand; R: receptor, TF: transcription factor. Detailed expression profiles are shown in Supplementary Figure S1. (G) Expression patterns of Fral2 and TCF15 in Ho_CS and Ho_CR animals maintained at 18°C or at 10°C for 35 days. Scale bar: 200 µm. [Colour figure can be viewed at wileyonlinelibrary.com]
That way, we identified a series of genes strongly upregulated in Ho_CS but not in Ho_CR. Among the genes upregulated at least fourfold in Ho_CS, we identified ADORA2A‐L, CC2D2A, CHRNA9, DDCL2, EGCase, GPR139, GPR157, NCAM1, and STK33 as neurotransmission genes (Fig. 7D) and AHR, Fral2, FGFR, FoxJ3, Jagged, Meis1, Notch, Otx1, and TCF15 as neurogenesis genes (Fig. 7E,F). These putative neurogenesis genes are upregulated at least twofold in 35 days old Ho_CS animals (blue numbers), much higher than in Ho_CR maintained at 10°C for 35 days (black numbers) where their expression is stable or mildly upregulated (Otx1, FGFR) except in case of TCF15 that is upregulated 2.5 fold. Their upregulation in Ho_CS also contrasts with the stable expression noted in Hv_sf1 having lost their ISCs upon HU treatment (red numbers) except Fral2 that is downregulated.
Expression of two genes Fral2 and TCF15 was verified by in situ hybridization (Fig. 7G). In Hv_AEP maintained at RT, both genes are predominantly expressed in cells from the epidermal layer, epithelial cells but also neurons sticking to these cells (Supplemental Figure S1). Consistently, in Ho_CS animals maintained at 18°C, we found Fral2 expressed at high levels in neurons of the apical region and along the body column, and at low levels in epithelial cells, but we did not detect Fral2‐expressing cells in Ho_CR polyps. After 35 days at 10°C, the testes are already post‐mature in Ho_CR where few Fral2 expressing cells can be detected (arrowheads). In aging Ho_CS where testes are still fully mature, we found Fral2 predominantly expressed at the base, where spermatogonia are found. Fral2 transcripts were also detected in some small cells spread along the body column. This result suggests a role for Fral2 in spermatogenesis as identified in mice (Cohen et al., 1994). Concerning TCF15 expression, it was undetectable in animals maintained at 18°C, but strong in whole testes (Fig. 7G). These modulations of expression patterns indicate that both genes are upregulated upon gametogenesis, Fral2 expression being seemingly shifted from the neuronal lineage to the germ cells in testes.
Discussion
In Hydra neurogenesis is constantly active although at a low rate in intact adult animals, replacing the neurons that get sloughed off at the extremities throughout the animal life. After bisection, neurogenesis becomes activated and rapidly re‐establishes the apical or basal nervous system in the regenerated structure. In this study, we show that the active behaviors are impaired shortly after aging is induced in Ho_CS animals. Indeed, contractility or prey capture behaviors become altered long before the loss of nerve cells or the anatomical disorganization of the nervous system are observed. These alterations, behavioral, anatomical, and cellular, are well supported by the molecular analyses. The transcriptomics analysis detects changes in the expression profile of a number of neurotransmission related genes already two weeks after transfer to cold, such as several GABA and glutamate receptors that are downregulated in both Ho_CS and Ho_CR animals maintained at 10°C. A recent study claims that the neural circuits that mediate the response to glutathione are located in the hypostome and make use of GABA as neurotransmitter (Lauro and Kass‐Simon, 2018). As the downregulation of the GABA receptor genes is more pronounced in Ho_CS than in Ho_CR, this difference might explain the lowered ability of Ho_CS to capture preys.
The homeoprotein Prdl‐a was initially identified as a marker of apical neuronal progenitors with the expected nuclear localization (Gauchat et al., 1998). In aging Hydra, that is, sexual Ho_CS, but not in Ho_CR, we noticed a striking decrease in the number of prdl‐a+ cells while the level of prdl‐a transcripts remained similar between the two strains. After induction of gametogenesis, the number of ISCs rapidly drops in both Ho_CS and Ho_CR (Yoshida et al., 2006; Tomczyk et al., 2017), but goes back to the homeostatic level in Ho_CR while remaining low in Ho_CS. The persistence of prdl‐a+ cells in asexual Ho_CS and Ho_CR animals maintained at 10°C for 44 days suggest that their loss is connected with the aging process. The prdl‐a RNA‐seq expression profile reflects this transient loss of ISCs and the subsequent recovery in Ho_CR. In Ho_CS the prdl‐a RNA‐seq expression profile is similar but the ISCs population remains depleted suggesting an ectopic, possibly epithelial, upregulation of prdl‐a, although not detected at the protein level. Indeed, we were able to detect such ectopic expression in patches of prdl‐a + cells along the body column of aging Ho_CS. This ectopic upregulation might be interpreted as an attempt to reactivate neurogenesis in response to the low number of ISCs and neuronal progenitors, but as the epithelial context is not appropriate, the prdl‐a protein would not be properly translated or stabilized.
Similarly, the comparative transcriptomic analysis between Ho_CR and Ho_CS undergoing gametogenesis reflects the intensity of the loss of neurogenesis on the one hand, and the attempt of the aging animals to rescue neurogenesis on the other hand. Indeed, the loss of mature neurons is well visible in Ho_CS but not in Ho_CR where the number of neuronal progenitors appears unchanged, implying that in Ho_CR, de novo neurogenesis is maintained at low pace to replace the old neurons. This state is reflected in the global qRNA‐seq results with the heatmaps indicating that large sets of neurotransmission and neurogenesis genes are differently downregulated in Ho_CR and Ho_CS, mildly and transiently in Ho_CR, in a sustained fashion in Ho_CS. By contrast, a subset of neurotransmission and neurogenesis genes show the opposite behavior: strongly upregulated in Ho_CS while rather downregulated in Ho_CR. This second behavior suggests two distinct interpretations: (i) an attempt to rescue neurogenesis in Ho_CS by upregulating a series of key genes for neurogenesis; (ii) the capture of neurogenic genes to the benefice of gametogenesis as shown for Fral2.
Among these neurogenesis genes Otx1, Otx2, or Gli3 are involved in brain development in mammals, while Dlx1, Meis1, and PUM2 are involved in neuronal survival and differentiation. Hydra interstitial cells are able to sense their density and respond to a lower density by increasing their proliferation rate (David et al., 1991). As the number of ISCs and neuronal progenitors strongly decreases in aging Ho_CS over the first month, the remaining cells may respond to this low density by over‐expressing genes involved in stem cell proliferation and nerve cell differentiation to re‐establish the nervous system. This scenario could explain how a large set of genes exhibit similar expression levels between Ho_CS and Ho_CR despite a dramatically different number of ISCs recorded in these animals when maintained for 35 days at 10°C.
Here, we should mention that the loss of ISCs and interstitial cells is actually more massive and definitive after three courses of HU treatment than in Ho animals undergoing gametogenesis (Tomczyk et al., 2017; Buzgariu et al., 2018). In fact, in HU‐treated Hv‐sf1 the depletion of ISCs and neuronal progenitors is almost complete and leads to the complete loss of expression of numerous neurogenic genes (Wenger et al., 2016). When we compare the molecular consequences of these two types of neurogenesis loss, we identify nine genes upregulated in aging Ho_CS but not at all or to a lower level in the non‐aging Ho_CR, and at least four‐fold in aging Ho_CS when compared to HU‐treated Hv_sf1. For this latter comparison, the results are only indicative as the Hv and Ho transcriptomes were not generated and processed together, therefore the quantitative approach taken here needs to be confirmed.
Among these genes, we find interestingly FoxJ3, initially identified in the neurectoderm of mouse embryos (Landgren and Carlsson, 2004), but also identified as a regulator of spermatogenesis in adult mice (Ni et al., 2016). These genes are either ubiquitously expressed or predominantly expressed in ESCs, suggesting that their upregulation in aging animals takes place either in epithelial cells or in testes as observed here for Fral2 and TCF15 by in situ hybridization, an experiment that validates the transcriptomic comparative analysis.
Fra‐1 and Fra‐2 are bZIP transcription factors initially identified as stress factors that belong to the Fos‐related family (Franza et al., 1988). In mammals, they are essential for development, involved in tumorigenesis of multiple organs and in fibrotic diseases (Eferl and Wagner, 2003; Wernig et al., 2017), with a key regulation by the ERK pathway (Gillies et al., 2017). In Hydra where five copies of Fra‐like genes are found (Supplementary Figure S2), Fral2 expression switches between asexual and sexual animals, from neurons to male germ cells. As the base of testes contains early spermatogonia that actively proliferate to produce sperm cells, this change in Fral2 expression suggests a recruitment by the germ cells at the expenses of the nerve cells, a competition process possibly leading to aging. Fral2 could potentially play a cytoprotective function in this highly active cell population. The low epidermal expression of TCF15 in standard asexual conditions versus the high gonadic expression in aging animals suggests that in sexual Hydra TCF15 is recruited to play a role in gametogenesis. In mammals TCF15 is a transcription factor implicated in the priming of the pluripotent embryonic stem cells toward differentiation (Davies et al., 2013).
In summary, we demonstrate an aging‐dependent loss of the function, density and organization of nervous system in Hydra. These adverse changes are caused by a partial loss of ISCs and neuronal progenitors that occurs as a consequence of the intense production of gametes from ISCs. We identify some candidate regulators of this somatic to germinal switch as Fral2, pointing to a possible competition for molecular resources between the somatic – neuronal‐ and germ cell fate of ISCs.
The authors thank C. Grimmelikhuijzen (Copenhagen) for kindly providing the anti‐RFamide antibody, Y. Wenger for the assistance with the transcriptomic analyses.
Supporting information
Literature Cited
- Alexopoulou, Z. , Lang, J. , Perrett, R.M. , Elschami, M. , Hurry, M.E.D. , Kim, H.T. , et al. (2016) Deubiquitinase Usp8 regulates α‐synuclein clearance and modifies its toxicity in Lewy body disease. Proceedings of the National Academy of Sciences of the United States of America, 113, E4688–E4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, P.A. and Spencer, A.N. (1989) The importance of cnidarian synapses for neurobiology. Journal of Neurobiology, 20, 435–457. [DOI] [PubMed] [Google Scholar]
- Baptista, M.S. , Duarte, C.B. and Maciel, P. (2012) Role of the ubiquitin‐proteasome system in nervous system function and disease: using C. elegans as a dissecting tool. Cellular and Molecular Life Sciences, 69, 2691–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode, H.R. , Berking, S. , David, C. , Gierer, A. , Schaller, H. and Trenker, E. (1973) Quantitative analysis of cell types during growth and regeneration in hydra. Roux's Archives of Developmental Biology, 171, 269–285. [DOI] [PubMed] [Google Scholar]
- Bode, H. , Lengfeld, T. , Hobmayer, B. and Holstein, T.W. (2008) Detection of expression patterns in Hydra pattern formation. Methods in Molecular Biology, 469, 69–84. [DOI] [PubMed] [Google Scholar]
- Boya, P. , Codogno, P. and Rodriguez‐Muela, N. (2018) Autophagy in stem cells: repair, remodelling and metabolic reprogramming. Development, 145, dev146506. [DOI] [PubMed] [Google Scholar]
- Brien, P. (1953) La pérennité somatique. Biological Reviews, 28, 308–349. [Google Scholar]
- Buzgariu, W. , Al Haddad, S. , Tomczyk, S. , Wenger, Y. and Galliot, B. (2015) Multi‐functionality and plasticity characterize epithelial cells in Hydra. Tissue Barriers, 3, e1068908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzgariu, W. , Wenger, Y. , Tcaciuc, N. , Catunda‐Lemos, A.P. and Galliot, B. (2018) Impact of cycling cells and cell cycle regulation on Hydra regeneration. Developmental Biology, 433, 240–253. [DOI] [PubMed] [Google Scholar]
- Campbell, R.D. (1976) Elimination by Hydra interstitial and nerve cells by means of colchicine. Journal of Cell Science, 21, 1–13. [DOI] [PubMed] [Google Scholar]
- Cohen, D.R. , Sinclair, A.H. and McGovern, J.D. (1994) SRY protein enhances transcription of Fos‐related antigen 1 promoter constructs. Proceedings of the National Academy of Sciences of the United States of America, 91, 4372–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, A.G. , Schuchert, P. , Marques, A.C. , Jankowski, T. , Medina, M. and Schierwater, B. (2006) Medusozoan phylogeny and character evolution clarified by new large and small subunit rDNA data and an assessment of the utility of phylogenetic mixture models. Systematic Biology, 55, 97–115. [DOI] [PubMed] [Google Scholar]
- David, C.N. (1973) A quantitative method for maceration of hydra tissue. Roux's Archives of Developmental Biology, 171, 259–268. [DOI] [PubMed] [Google Scholar]
- David, C.N. , Fujisawa, T. and Bosch, T.C.G. (1991) Interstitial stem cell proliferation in hydra: evidence for strain‐specific regulatory signals. Developmental Biology, 148, 501–507. [DOI] [PubMed] [Google Scholar]
- Davies, O.R. , Lin, C.Y. , Radzisheuskaya, A. , Zhou, X. , Taube, J. , Blin, G. , et al. (2013) Tcf15 primes pluripotent cells for differentiation. Cell Reports, 3, 472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eferl, R. and Wagner, E.F. (2003) AP‐1: a double‐edged sword in tumorigenesis. Nature Reviews Cancer, 3, 859–868. [DOI] [PubMed] [Google Scholar]
- Franza, B.R., Jr., Rauscher, F.J., 3rd., Josephs, S.F. and Curran, T. (1988) The Fos complex and Fos‐related antigens recognize sequence elements that contain AP‐1 binding sites. Science, 239, 1150–1153. [DOI] [PubMed] [Google Scholar]
- Fujisawa, T. (1989) Role of interstitial cell migration in generating position‐dependent patterns of nerve cell differentiation in Hydra. Developmental Biology, 133, 77–82. [DOI] [PubMed] [Google Scholar]
- Galliot, B. and Quiquand, M. (2011) A two‐step process in the emergence of neurogenesis. European Journal of Neuroscience, 34, 847–862. [DOI] [PubMed] [Google Scholar]
- Galliot, B. , Quiquand, M. , Ghila, L. , de Rosa, R. , Miljkovic‐Licina, M. and Chera, S. (2009) Origins of neurogenesis, a cnidarian view. Developmental Biology, 332, 2–24. [DOI] [PubMed] [Google Scholar]
- Gauchat, D. , Escriva, H. , Miljkovic‐Licina, M. , Chera, S. , Langlois, M.C. , Begue, A. , et al. (2004) The orphan COUP‐TF nuclear receptors are markers for neurogenesis from cnidarians to vertebrates. Developmental Biology, 275, 104–123. [DOI] [PubMed] [Google Scholar]
- Gauchat, D. , Kreger, S. , Holstein, T. and Galliot, B. (1998) prdl‐a, a gene marker for hydra apical differentiation related to triploblastic paired‐like head‐specific genes. Development, 125, 1637–1645. [DOI] [PubMed] [Google Scholar]
- Gillies, T.E. , Pargett, M. , Minguet, M. , Davies, A.E. and Albeck, J.G. (2017) Linear integration of ERK activity predominates over persistence detection in Fra‐1 regulation. Cell Systems, 5(549–563), e545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grens, A. , Mason, E. , Marsh, J.L. and Bode, H.R. (1995) Evolutionary conservation of a cell fate specification gene: the Hydra achaete‐scute homolog has proneural activity in Drosophila. Development, 121, 4027–4035. [DOI] [PubMed] [Google Scholar]
- Grimmelikhuijzen, C.J. (1985) Antisera to the sequence Arg‐Phe‐amide visualize neuronal centralization in hydroid polyps. Cell and Tissue Research, 241, 171–182. [Google Scholar]
- Grimmelikhuijzen, C.J. and Westfall, J.A. (1995a) The nervous systems of cnidarians. EXS, 72, 7–24. [DOI] [PubMed] [Google Scholar]
- Grimmelikhuijzen, C.J.P. and Westfall, J.A. (1995b) The nervous systems of Cnidarians In: Breidbach O. and Kutsch W. (Eds.) The Nervous Systems of Invertebrates: An Evolutionary and Comparative Approach. Basel, Switzerland: Birkhaüser Verlag, pp. 7–24. [Google Scholar]
- Grosvenor, W. , Rhoads, D.E. and Kass‐Simon, G. (1996) Chemoreceptive control of feeding processes in hydra. Chemical Senses, 21, 313–321. [DOI] [PubMed] [Google Scholar]
- Grunder, S. and Assmann, M. (2015) Peptide‐gated ion channels and the simple nervous system of Hydra. The Journal of Experimental Biology, 218, 551–561. [DOI] [PubMed] [Google Scholar]
- Hager, G. and David, C.N. (1997) Pattern of differentiated nerve cells in hydra is determined by precursor migration. Development, 124, 569–576. [DOI] [PubMed] [Google Scholar]
- Hansen, G.N. , Williamson, M. and Grimmelikhuijzen, C.J. (2000) Two‐color double‐labeling in situ hybridization of whole‐mount Hydra using RNA probes for five different Hydra neuropeptide preprohormones: evidence for colocalization. Cell and Tissue Research, 301, 245–253. [DOI] [PubMed] [Google Scholar]
- Hobmayer, B. , Jenewein, M. , Eder, D. , Eder, M.K. , Glasauer, S. , Gufler, S. , et al. (2012) Stemness in Hydra – a current perspective. The International Journal of Developmental Biology, 56, 509–517. [DOI] [PubMed] [Google Scholar]
- Lenhoff, H.M. and Lenhoff, S.G. (1988) Trembley‘s polyps. Scientific American, 108–113. [Google Scholar]
- Kass‐Simon, G. and Pierobon, P. (2007) Cnidarian chemical neurotransmission, an updated overview. Comparative Biochemistry and Physiology Part A, 146, 9–25. [DOI] [PubMed] [Google Scholar]
- Koizumi, O. (2007) Nerve ring of the hypostome in hydra: is it an origin of the central nervous system of bilaterian animals? Brain, Behavior and Evolution, 69, 151–159. [DOI] [PubMed] [Google Scholar]
- Landgren, H. and Carlsson, P. (2004) FoxJ3, a novel mammalian forkhead gene expressed in neuroectoderm, neural crest, and myotome. Developmental Dynamics, 231, 396–401. [DOI] [PubMed] [Google Scholar]
- Lauro, B.M. and Kass‐Simon, G. (2018) Hydra's feeding response: effect of GABAB ligands on GSH‐induced electrical activity in the hypostome of H. vulgaris. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology, 225, 83–93. [DOI] [PubMed] [Google Scholar]
- Loomis, W.F. (1955) Glutathione control of the specific feeding reactions of hydra. Annals of the New York Academy of Sciences, 62, 209–228. [Google Scholar]
- Marcum, B.A. and Campbell, R.D. (1978) Development of Hydra lacking nerve and interstitial cells. Journal of Cell Science, 29, 17–33. [DOI] [PubMed] [Google Scholar]
- Marcum, B.A. , Fujisawa, T. and Sugiyama, T. (1980) A mutant hydra strain (sf‐1) containing temperature‐sensitive interstitial cells In: Tardent P. and Tardent R. (Eds.) Developmental and Cellular Biology of Coelenterates. Amsterdam: Elsevier/North Holland, pp. 429–434. [Google Scholar]
- Marlow, H.Q. , Srivastava, M. , Matus, D.Q. , Rokhsar, D. and Martindale, M.Q. (2009) Anatomy and development of the nervous system of Nematostella vectensis, an anthozoan cnidarian. Developmental Neurobiology, 69, 235–254. [DOI] [PubMed] [Google Scholar]
- Martínez, D.E. (1998) Mortality patterns suggest lack of senescence in hydra. Experimental Gerontology, 33, 217–225. [DOI] [PubMed] [Google Scholar]
- Miljkovic‐Licina, M. , Chera, S. , Ghila, L. and Galliot, B. (2007) Head regeneration in wild‐type hydra requires de novo neurogenesis. Development, 134, 1191–1201. [DOI] [PubMed] [Google Scholar]
- Miljkovic‐Licina, M. , Gauchat, D. and Galliot, B. (2004) Neuronal evolution: analysis of regulatory genes in a first‐evolved nervous system, the hydra nervous system. Biosystems, 76, 75–87. [DOI] [PubMed] [Google Scholar]
- Mochizuki, K. , Sano, H. , Kobayashi, S. , Nishimiya‐Fujisawa, C. and Fujisawa, T. (2000) Expression and evolutionary conservation of nanos‐related genes in Hydra. Development Genes and Evolution, 210, 591–602. [DOI] [PubMed] [Google Scholar]
- Ni, L. , Xie, H. and Tan, L. (2016) Multiple roles of FOXJ3 in spermatogenesis: a lesson from Foxj3 conditional knockout mouse models. Molecular Reproduction and Development, 83, 1060–1069. [DOI] [PubMed] [Google Scholar]
- Pierobon, P. (2015) Regional modulation of the response to glutathione in Hydra vulgaris. The Journal of Experimental Biology, 218, 2226–2232. [DOI] [PubMed] [Google Scholar]
- Sacks, P.G. and Davis, L.E. (1979) Production of nerveless Hydra attenuata by hydroxyurea treatments. Journal of Cell Science, 37, 189–203. [DOI] [PubMed] [Google Scholar]
- Schaible, R. , Scheuerlein, A. , Dańko, M.J. , Gampe, J. , Martínez, D.E. and Vaupel, J.W. (2015) Constant mortality and fertility over age in Hydra. Proceedings of the National Academy of Sciences of the United States of America, 112, 15701–15706. 201521002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, T. , Koizumi, O. , Ariura, Y. , Romanovitch, A. , Bosch, T.C. , Kobayakawa, Y. , et al. (2000) A novel neuropeptide, Hym‐355, positively regulates neuron differentiation in Hydra. Development, 127, 997–1005. [DOI] [PubMed] [Google Scholar]
- Tardent, P. (1995) The cnidarian cnidocyte, a high‐tech cellular weaponry. BioEssays, 17, 351–362. [Google Scholar]
- Technau, U. and Holstein, T.W. (1996) Phenotypic maturation of neurons and continuous precursor migration in the formation of the peduncle nerve net in Hydra. Developmental Biology, 177, 599–615. [DOI] [PubMed] [Google Scholar]
- Teragawa, C.K. and Bode, H.R. (1995) Migrating interstitial cells differentiate into neurons in hydra. Developmental Biology, 171, 286–293. [DOI] [PubMed] [Google Scholar]
- Tomczyk, S. , Fischer, K. , Austad, S. and Galliot, B. (2015) Hydra, a powerful model system for aging studies. Invertebrate Reproduction and Development, 59, 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomczyk, S. , Schenkelaars, Q. , Suknovic, N. , Wenger, Y. , Ekundayo, K. , Buzgariu, W. , et al. (2017) Deficient autophagy drives aging in Hydra. bioRxiv. 10.1101/236638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger, Y. , Buzgariu, W. and Galliot, B. (2016) Loss of neurogenesis in Hydra leads to compensatory regulation of neurogenic and neurotransmission genes in epithelial cells. Philosophical Transactions of the Royal Society B: Biological Sciences, 371, 20150040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig, G. , Chen, S.Y. , Cui, L. , Van Neste, C. , Tsai, J.M. , Kambham, N. , et al. (2017) Unifying mechanism for different fibrotic diseases. Proceedings of the National Academy of Sciences of the United States of America, 114, 4757–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi, Y. , Dhaliwal, J.S. , Ceizar, M. , Vaculik, M. , Kumar, K.L. and Lagace, D.C. (2016) Knockout of Atg5 delays the maturation and reduces the survival of adult‐generated neurons in the hippocampus. Cell Death & Disease, 7, e2127–e2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, K. , Fujisawa, T. , Hwang, J.S. , Ikeo, K. and Gojobori, T. (2006) Degeneration after sexual differentiation in hydra and its relevance to the evolution of aging. Gene, 385, 64–70. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


