Abstract
Increased expression of the full-length androgen receptor (AR-FL) and AR splice variants (AR-Vs) drives the progression of castration-resistant prostate cancer (CRPC). The levels of AR-FL and AR-V transcripts are often tightly correlated in individual CRPC samples, yet our understanding of how their expression is co-regulated is limited. Here, we report a role of c-Myc in accounting for coordinated AR-FL and AR-V expression. Analysis of gene expression data from 159 metastatic CRPC samples and 2142 primary prostate tumors showed that the level of c-Myc is positively correlated with that of individual AR isoforms. A striking positive correlation also exists between the activity of the c-Myc pathway and the level of individual AR isoforms, between the level of c-Myc and the activity of the AR pathway, and between the activities of the two pathways. Moreover, the c-Myc signature is highly enriched in tumors expressing high levels of AR, as is the AR signature in c-Myc-high-expressing tumors. Using shRNA knockdown, we confirmed c-Myc regulation of expression and activity of AR-FL and AR-Vs in cell models and a patient-derived xenograft model. Mechanistically, c-Myc promotes the transcription of the AR gene and enhances the stability of the AR-FL and AR-V proteins without altering AR RNA splicing. Importantly, inhibiting c-Myc sensitizes enzalutamide-resistant cells to growth inhibition by enzalutamide. Overall, this study highlights a critical role of c-Myc in regulating the coordinated expression of AR-FL and AR-Vs that is commonly observed in CRPC and suggests the utility of targeting c-Myc as an adjuvant to AR-directed therapy.
Keywords: c-Myc, androgen receptor, splice variant, castration-resistant prostate cancer
Introduction
The androgen receptor (AR) is the main therapeutic target in prostate cancer. Androgen deprivation therapy, the first-line treatment for advanced prostate cancer, disrupts AR signaling by decreasing androgen levels or by inhibiting AR activity with AR antagonists. However, almost all patients experience progression to the presently incurable stage, termed castration-resistant prostate cancer (CRPC) (reviewed in [1]). AR signaling is sustained and remains critical in CRPC, and several new and more potent AR-directed drugs have been developed to target the sustained AR activity in CRPC (reviewed in [2]). Among these, the potent AR antagonists apalutamide [3] and enzalutamide [4] as well as the CYP17A1 inhibitor abiraterone [5] have been approved by the FDA for treatment of CRPC. However, the development of therapy-resistant disease is an inevitable outcome (reviewed in [2]).
Increased expression of AR splice variants (AR-Vs) has been ascribed as an important mechanism of resistance to AR-directed therapies, including enzalutamide and abiraterone [6–10]. AR-Vs are truncated AR isoforms that lack the functional ligand-binding domain, but most AR-Vs retain the N-terminal transactivation domain and the DNA-binding domain (reviewed in [11]). As a result, many AR-Vs display constitutive transcriptional activation properties [12–18] and accordingly, high levels of AR-Vs, specifically, AR-V7, ARv567es, and AR-V9, have been linked to poor prognosis and short survival of CRPC patients [6, 14, 15, 19–22]. The critical involvement of AR-Vs in the progression of CRPC underscores the need to understand how they are generated so that effective therapeutic strategies can be designed to curb AR-V production.
Several mechanisms have been proposed to mediate AR-V expression. One is the rearrangement of the AR gene [23, 24]. Modeling AR-gene rearrangement using the TALEN technology led to the expression of the ARv567es variant without the full-length AR (AR-FL) in clonally selected cells [24]. Thus, AR gene rearrangement might be an important mechanism of AR-V production in the subset of prostate cancers in which AR-Vs are the major form of AR expressed, whereas other mechanisms might underlie the co-expression of AR-FL and different AR-Vs that is commonly observed in the same cells from CRPC patients [25]. Several splicing factors, such as U2AF65 [26, 27], ASF/SF2 [26], hnRNPA1 [28, 29], and hnRNPF [30], along with the RNA-binding protein Sam68 [31], the transcription factor YB-1 [32], and the molecular chaperone HSP90 [33] have been shown to selectively regulate AR-V7 splicing without affecting AR-FL splicing. However, the levels of AR-FL and AR-V transcripts are often tightly correlated in individual clinical specimens and xenograft models [18, 19, 34, 35]. This study was set out to investigate mechanisms leading to coordinated expression of AR-FL and AR-Vs.
Androgen deprivation has been shown to enhance the rate of AR-gene transcription to produce more AR pre-mRNA [26]. This indicates that, in addition to alternative splicing, a transcriptional mechanism may drive the expression of AR-FL and AR-Vs after AR-directed therapies. Here, we focused on investigating the role of c-Myc in this transcriptional mechanism of regulation because: 1) c-Myc is one of the most overexpressed genes in prostate cancer [36] and is critically involved in prostate cancer progression (reviewed in [37]), 2) c-Myc binds to regulatory regions of the AR gene to induce AR-gene transcription [38, 39], and 3) frequent upregulation of c-Myc in CRPC and a positive correlation between c-Myc and AR mRNA levels were identified in a study of 140 metastatic CRPC samples [40]. We confirmed this positive correlation in 159 metastatic CRPC and 2142 primary prostate cancer samples. We further identified a striking positive correlation between the activity of the c-Myc pathway and the levels of AR-FL and different AR-Vs, between the level of c-Myc and the activity of the AR pathway, and between the activities of the two pathways. Using cell models and a patient-derived xenograft model, we demonstrated the importance of c-Myc in regulating the expression of AR-FL and AR-Vs. Mechanistically, this is mediated through transcriptional regulation coupled with modulation of protein stability without impacting AR RNA splicing. Together, our results provide a rationale for targeting c-Myc to curb AR-FL and AR-V expression for more effective treatment of prostate cancer.
Results
c-Myc expression and activity positively correlate with AR expression in human prostate cancer specimens
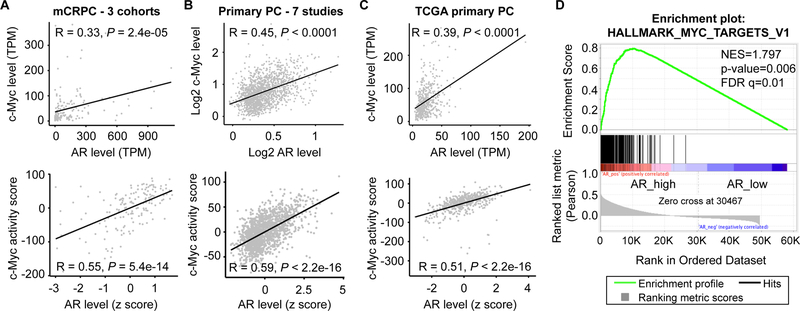
The levels of c-Myc and AR transcripts were reported previously to be positively correlated in metastatic CRPC samples [40]. To validate this observation in other cohorts, we analyzed the RNA-seq data of 159 metastatic CRPC samples from the “Stand Up To Cancer East Coast Prostate Cancer Research Group” (SU2C) project (n = 51) [41], the Prostate Cancer Medically Optimized Genome-Enhanced Therapy (PROMOTE) study (n = 74) [42], and the Beltran cohort (n = 34) [43]. Consistent with the previous report, the level of c-Myc RNA was positively correlated with that of AR in these samples (Fig. 1A, top panel). We also evaluated the activity of the c-Myc pathway in these samples using two established c-Myc gene expression signatures, the Schuhmacher [44] and the Jung [45] signatures. Both signatures showed a strong positive correlation with AR transcript level (Fig. 1A, bottom panel; Supplementary Fig. S1A). To assess whether these correlations exist in primary prostate tumors, we analyzed gene expression data from 1642 prostatectomy samples from 7 different studies on a clinical-grade microarray platform [46–53] and the RNA-seq data from 500 primary tumors in TCGA cohort. The data from analysis of primary tumors were very similar to those from analysis of CRPC samples, showing a positive correlation between c-Myc and AR expression levels and between the activity of the c-Myc pathway and the level of AR (Fig. 1B–1C and Supplementary Fig. S1B–S1C). Moreover, unbiased Gene Set Enrichment Analysis (GSEA) demonstrated that the transcriptional profiles of the tumors that express a high level of AR were highly enriched for the two hallmark c-Myc pathway gene sets and that the enrichment was comparable to that of the AR pathway (Fig. 1D and Supplementary Figs. S2 & S3).
Figure 1. c-Myc level and activity positively correlate with AR level in human prostate cancer samples.
A-C, Pearson’s correlation coefficient analysis showing positive correlation between c-Myc and AR levels and between c-Myc activity score and AR level in 159 metastatic CRPC (mCRPC) samples (A), a meta dataset of 1642 primary prostate cancer (PC) samples (B), and the 500 TCGA primary prostate cancer samples (C). The c-Myc activity score was computed as the sum of z-scores for the Schuhmacher c-Myc-target signature. For analysis of correlation between expression levels and activity scores, the expression levels were computed as the z-scores for AR levels. D, GSEA showing enrichment of the hallmark Myc_targets_v1 pathway in mCRPC samples that express a high level of AR. TPM, transcripts per million.
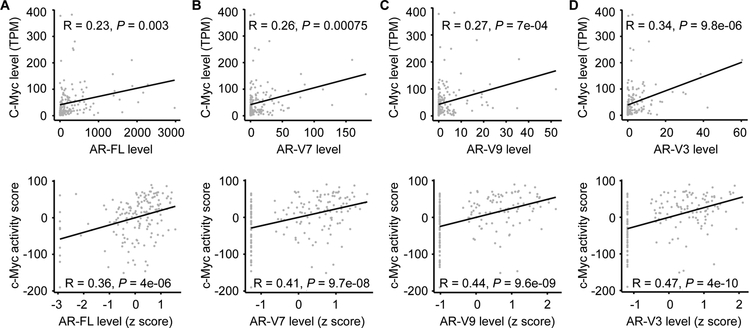
Next, we analyzed the CRPC samples for expression of individual AR isoforms, including AR-FL and three of the more abundantly expressed AR-Vs, AR-V7, -V9, and -V3 [41]. Concordant with our data from overall AR expression, the levels of individual AR isoforms were all positively correlated with c-Myc level and activity (Fig. 2 and Supplementary Fig. S4). Thus, the gene expression data from this large collection of clinical samples from different cohorts support our hypothesis that c-Myc positively regulates the co-expression of AR-FL and AR-V transcripts. We did not include the primary tumors in this analysis because they express minimal amounts of AR-Vs.
Figure 2. c-Myc level and activity positively correlate with AR-FL and AR-V levels in metastatic CRPC samples.
Pearson’s correlation coefficient analysis showing that c-Myc level (top panels) and activity (bottom panels) positively correlate with the levels of AR-FL (A), -V7 (B), -V9 (C), and -V3 (D) in 159 metastatic CRPC samples. The AR-FL, -V7, -V9, and -V3 levels were quantified as normalized number of RNA-seq reads spanning AR exons 7–8, 3-CE3, 3-CE5, and 2-CE4 splice junctions, respectively. The c-Myc activity score was computed as the sum of z-scores for the Schuhmacher c-Myc-target signature. TPM, transcripts per million.
c-Myc knockdown blocks AR-V7 induction by AR-directed therapies
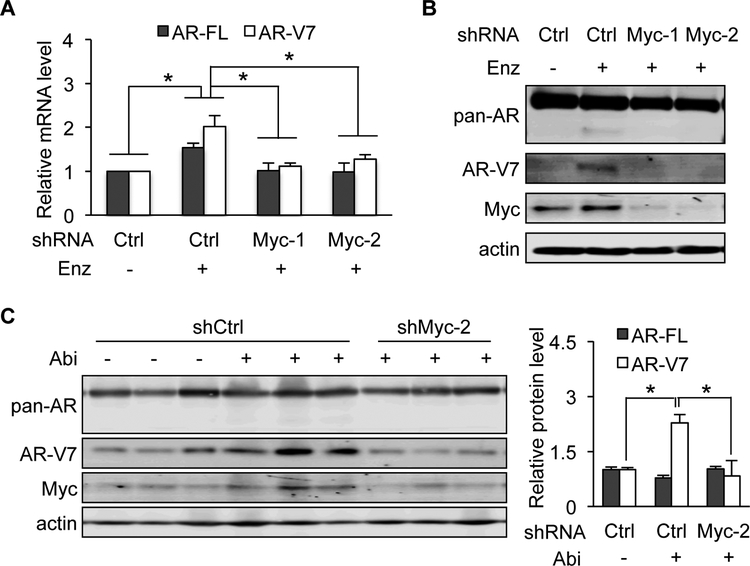
To validate the role of c-Myc in AR-FL and AR-V expression in response to treatment with enzalutamide or abiraterone, we knocked down the expression of c-Myc in VCaP prostate cancer cells and the castration-resistant LuCaP 35CR patient-derived xenograft tumors [54]. As expected [26], enzalutamide treatment led to an upregulation of AR-FL mRNA as well as AR-V7 mRNA and protein in VCaP cells (Figs. 3A and 3B). Unlike AR-V7, AR-FL was upregulated only at the RNA level, not at the protein level. This was related to stabilization of AR-FL protein upon androgen binding (reviewed in [55]) that is blocked by enzalutamide, whereas the stability of the ligand-binding-domain-truncated AR-V7 protein is independent of androgen. c-Myc knockdown using lentiviruses expressing either of the two c-Myc shRNAs abolished enzalutamide induction of AR-FL and AR-V7 expression (Fig. 3A and 3B). Similarly, knockdown of c-Myc blocked abiraterone upregulation of AR-V7 protein in LuCaP 35CR xenograft tumors in vivo (Fig. 3C). Together, these cell culture and xenograft studies provide experimental support to the role of c-Myc in regulating AR-FL and AR-V7 expression in response to AR-directed therapies.
Figure 3. Knockdown of c-Myc blocks enzalutamide/abiraterone upregulation of AR-FL/AR-V7.
A & B, qRT-PCR (A) and Western blotting with a pan-AR or AR-V7 antibody (B) showing that c-Myc knockdown blocked enzalutamide (Enz) induction of AR-FL mRNA as well as AR-V7 mRNA and protein expression in VCaP cells. Cells were treated with 10 μM Enz at 24 h after shCtrl- or shMyc-lentivirus transduction. C, Western blot analysis showing loss of ability of abiraterone (Abi) to induce AR-V7 expression after c-Myc knockdown in LuCaP 35CR xenograft tumors. Right panel, quantitation of AR-FL and -V7 protein levels. *, P < 0.05.
c-Myc knockdown attenuates basal AR-FL and AR-V expression
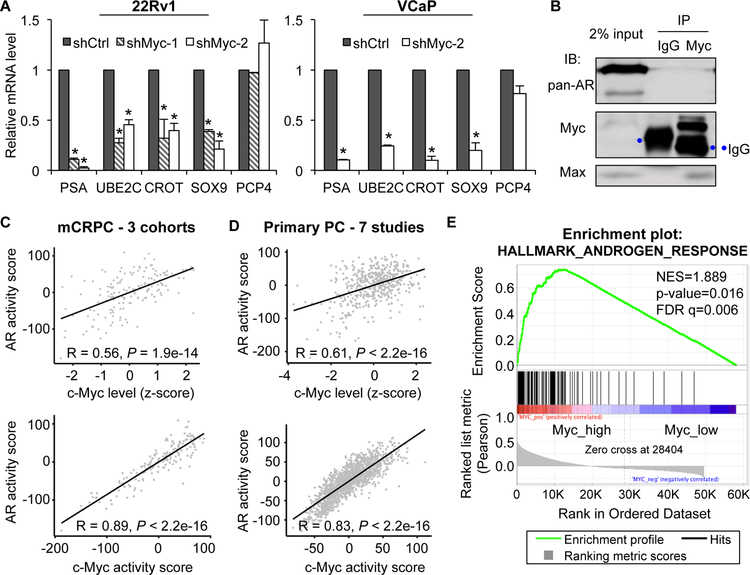
We next assessed the role of c-Myc in supporting basal expression of AR-FL and AR-Vs. The levels of AR-FL and AR-V transcripts (Fig. 4B–D) and proteins (Fig. 4A) were significantly reduced after c-Myc knockdown in all of the AR-V-expressing human prostate cancer cell models tested, 22Rv1, LNCaP95, and VCaP. Importantly, the effect was not limited to AR-V7. Other AR-Vs were similarly downregulated after c-Myc knockdown (Fig. 4B). These results provide direct evidence of the role of c-Myc in regulating the expression of AR-FL and different AR-Vs.
Figure 4. Knockdown of c-Myc decreases basal expression of AR-FL and AR-Vs.
Western blotting with a pan-AR or AR-V7 antibody (A) and qRT-PCR analyses (B - D) showing a reduced expression of AR-FL and AR-Vs in shMyc-lentivirus-transduced cells compared to the control cells. *, P < 0.05 from the shCtrl group.
c-Myc knockdown mitigates AR-FL and AR-V target-gene expression
In concordance with decreased levels of AR-FL and AR-Vs, the expression of AR-FL and AR-V targets, prostatic-specific antigen (PSA), ubiquitin conjugating enzyme E2C (UBE2C) [56], carnitine O-octanoyltransferase (CROT) [57], and sex-determining region Y-box 9 (SOX9) [57], was greatly diminished after c-Myc knockdown in both 22Rv1 and VCaP cells (Fig. 5A; the non-AR target, PCP4, was included to show selectivity). This was unlikely to be a result of direct interaction between c-Myc and AR-FL or c-Myc and AR-Vs since co-immunoprecipitation experiment failed to detect c-Myc/AR-FL or c-Myc/AR-V interaction (Fig. 5B). We then analyzed the 159 metastatic CRPCs, 1642 meta-set of primary tumors, and 500 TCGA primary tumors for their individual AR activity using the Nelson [58] and the Bluemn [59] AR gene expression signatures and assessed the correlation of AR activity with c-Myc level and with c-Myc activity. The AR activity calculated with both signatures displayed a strong positive correlation with c-Myc level (Figs. 5C, 5D, Supplementary Fig. S5, top panels) and with c-Myc activity (Figs. 5C & 5D, Supplementary Fig. S5, bottom panels) in all 3 sets of samples. Additionally, unbiased GSEA showed a striking enrichment of the AR pathway in the tumors that express a high level of c-Myc, and the enrichment was analogous to that of the c-Myc pathway (Figs. 5E, Supplementary Figs. S6 & S7). Together, the knockdown experiment and the human gene expression data support a positive regulation of AR signaling by c-Myc.
Figure 5. c-Myc positively regulates AR activity.
A, qRT-PCR showing a downregulation of AR-FL and AR-V targets, PSA, UBE2C, CROT, and SOX9, but not the non-AR target PCP4 in shMyc-lentivirus-transduced cells compared to the control cells. *, P < 0.05 from the shCtrl group. B, Co-immunoprecipitation with a c-Myc antibody showing no direct association between c-Myc and AR-FL or c-Myc and AR-V7 in VCaP cells. Immunoblotting with a Max antibody was included as a positive control. C & D, Pearson’s correlation coefficient analysis showing a positive correlation between c-Myc level and AR activity and between c-Myc and AR activities in 159 mCRPC (C) and 1642 primary prostate cancer samples (D). The c-Myc and AR activity scores were computed as the sum of z-scores for the Schuhmacher c-Myc signature and the Nelson AR signature, respectively. E, GSEA showing enrichment of the hallmark androgen activity pathway in mCRPC samples that express a high level of c-Myc.
c-Myc promotes AR-gene transcription and enhances AR-FL and AR-V protein stability
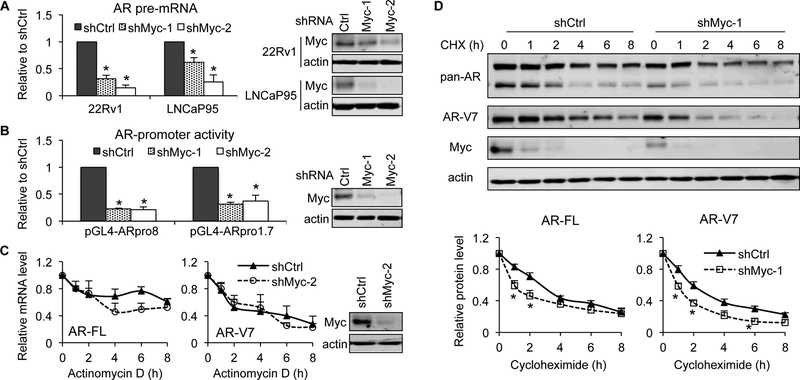
c-Myc is known to induce AR-gene transcription through binding to regulatory regions of the AR gene [38, 39]. Consistently, after knocking down c-Myc expression in 22Rv1 and LNCaP95 cells, we observed a marked decrease in the levels of AR pre-mRNA (Fig. 6A) and the activities of both the 1.7-kb (−600 to +1,115) and the 8-kb (−6,885 to +1,115) AR promoters that contain promoter/enhancer regions and the entire 5’-untranslated region (Fig. 6B). We also measured AR-FL and AR-V7 mRNA stability in response to c-Myc knockdown but did not detect any significant changes (Fig. 6C). In contrast, c-Myc knockdown decreased the stability of both AR-FL and AR-V7 proteins (Fig. 6D). This could explain the more significant decrease of AR proteins, especially the AR-FL, compared to AR mRNAs, in response to c-Myc knockdown (Fig. 4). Collectively, these findings indicate that c-Myc regulates the expression of AR-FL and AR-Vs by promoting the transcription of the AR gene and enhancing the stability of the AR-FL and AR-V7 proteins.
Figure 6. c-Myc knockdown inhibits AR-gene transcription and destabilizes AR-FL and AR-V7 proteins.
c-Myc knockdown was achieved by shMyc-lentivirus transduction. A, qRT-PCR showing a reduction of AR pre-mRNA after c-Myc knockdown. B, Luciferase assay showing decreased activities of AR promoters after c-Myc knockdown in 22Rv1 cells. Cells were transfected with the pGL4-ARpro8 or pGL4-ARpro1.7 construct in bulk and reseeded in triplicate 24 h post transfection for luciferase assay. C, mRNA stability assay showing no significant change in AR-FL or -V7 mRNA stability after c-Myc knockdown in 22Rv1 cells. Cells were treated with 10 μM actinomycin D for the indicated duration and collected for qRT-PCR. Right panels in A-C, Western blotting confirmation of c-Myc knockdown. D, Protein stability assay showing accelerated AR-FL or -V7 protein degradation after c-Myc knockdown in 22Rv1 cells. Cells were treated with 10 μg/ml cycloheximide (CHX) for the indicated duration and collected for Western blotting. Bottom panels, quantitation of AR-FL and -V7 protein levels. *, P < 0.05 from the shCtrl group.
c-Myc does not appear to regulate AR RNA splicing
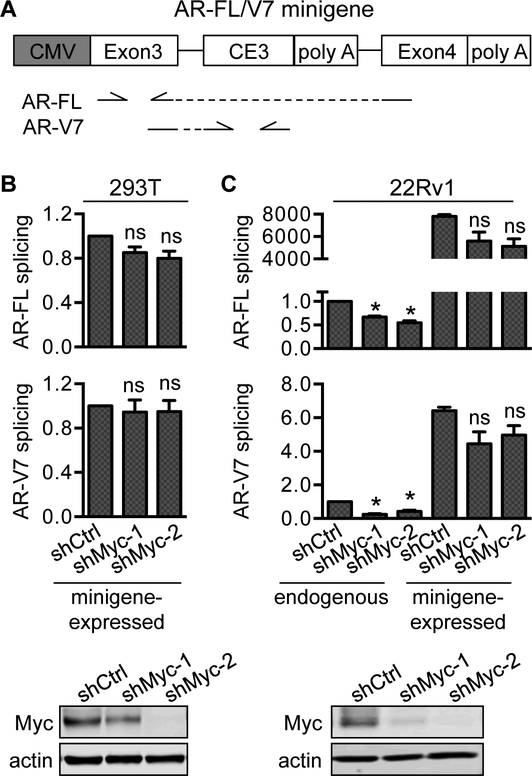
Because the levels of AR-FL and the AR-Vs that we analyzed correlated positively with c-Myc level and activity in metastatic CRPC samples (Fig. 2 and Supplementary Fig. S4) and they were similarly downregulated after c-Myc knockdown (Fig. 4B), we reasoned that c-Myc might not regulate AR RNA splicing to produce a specific AR isoform. To test this hypothesis, we transfected control or c-Myc-knockdown AR-null 293T cells or AR-FL/V7-expressing 22Rv1 cells with a CMV-promoter-driven AR-FL/V7 minigene construct [26], which has the AR-V7-specific cryptic exon 3 (CE3) and its ~400-bp flanking sequence inserted between exon 3 and exon 4 of the human AR gene (Fig. 7A). We then measured the levels of exon3-exon4-spliced and exon3-CE3-spliced transcripts that are indicative of AR-FL and -V7 splicing, respectively. Because the expression from the CMV promoter is not regulated by c-Myc (Supplementary Fig. S8), changes in the levels of minigene-expressed exon3-exon4 and exon3-CE3 transcripts would be suggestive of altered AR-FL or -V7 splicing. Our results did not show any statistically significant change in minigene-expressed exon3-exon4 or exon3-CE3 transcript after c-Myc knockdown (Fig. 7B & C), supporting our hypothesis of c-Myc not altering AR RNA splicing.
Figure 7. Knockdown of c-Myc does not affect AR-V7 alternative splicing.
A, Schematic diagram of the AR-FL/V7 minigene construct. The locations of the qPCR primers used to amplify exon3-exon4 and exon3-CE3 transcripts are depicted below the diagram. B, qRT-PCR analysis of minigene-expressed exon3-exon4 and exon3-CE3 transcripts showing no alteration of AR-FL or -V7 RNA splicing after c-Myc knockdown. At 24 h after shCtrl- or shMyc-lentivirus transduction, 293T or 22Rv1 cells were transfected with mock vector or the AR-FL/V7 minigene construct and collected for qRT-PCR analyses 48 h post transfection. *, P < 0.05 from the shCtrl group. ns, not statistically significant from the shCtrl group. Bottom panels in B and C, Western blotting confirmation of c-Myc knockdown.
c-Myc inhibition alleviates enzalutamide resistance
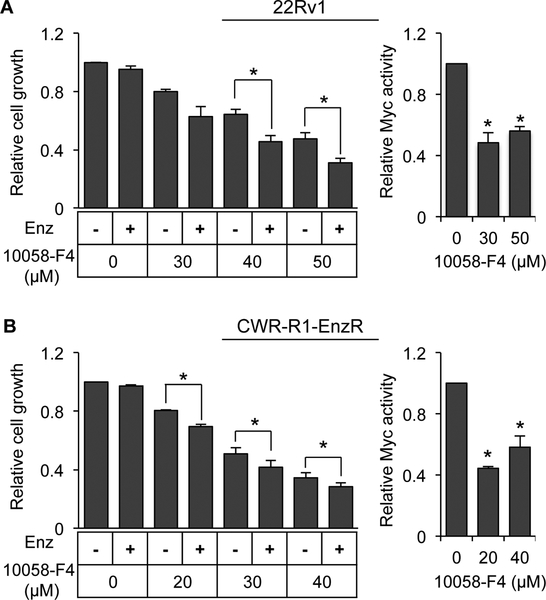
Increased expression of AR-Vs is known to contribute significantly to resistance of CRPC to enzalutamide [6–8, 10, 20, 21]. To test whether c-Myc inhibition could be a viable approach to alleviate enzalutamide resistance, we measured the growth response of 22Rv1 and CWR-R1-EnzR cells to combination treatment with enzalutamide and the 10058-F4 c-Myc inhibitor. 10058-F4 inhibits c-Myc activity by disrupting the heterodimerization between c-Myc and its activation partner Max [60]. 22Rv1 cells are intrinsically resistant to enzalutamide [8, 61], and CWR-R1-EnzR cells are enzalutamide-resistant subline of CWR-R1 cells [62]. Enzalutamide treatment did not affect the growth of these cells, whereas the c-Myc inhibitor caused a dose-dependent suppression of cell growth in both models (Fig. 8). Importantly, enzalutamide became effective in suppressing cell growth in the presence of the c-Myc inhibitor (Fig. 8). Consistent with c-Myc knockdown, treatment with 10058-F4 led to a downregulated expression of AR and AR-V7 as well as their target genes (Supplementary Fig. S9). These data suggested that inhibiting c-Myc could sensitize enzalutamide-resistant cells to growth inhibition by enzalutamide.
Figure 8. c-Myc inhibition sensitizes enzalutamide-resistant prostate cancer cells to growth inhibition by enzalutamide.
Left panels, SRB assay of the growth of 22Rv1 (A) and enzalutamide-resistant CWR-R1 (CWR-R1-enzR) (B) cells treated with 10 μM enzalutamide (Enz) with or without the 10058-F4 c-Myc inhibitor for 72 h. Right panels, luciferase assay with a luciferase construct driven by the E-box c-Myc-binding motif confirming c-Myc inhibition by 10058-F4.
Discussion
Despite the commonly observed tight correlation between the levels of AR-FL and AR-V transcripts in individual clinical CRPC samples and xenograft models, our understanding of the mechanism underlying their coordinated expression is limited. Here, we report an important role of c-Myc in accounting for this coordinated expression. Our analysis of RNA-seq data from 159 metastatic CRPC samples from three different cohorts showed positive correlations between the level of c-Myc and the level of individual AR isoforms (AR-FL, -V7, -V9, -V3) and between the activity of the c-Myc pathway and the level of each AR isoform. The correlations between c-Myc level/activity and AR level were also observed in a meta-dataset of 1642 primary prostate tumors and in 500 TCGA primary prostate tumors. Significantly, in unbiased GSEA, the c-Myc pathway is a top-enriched pathway in the tumors that express a high level of AR. Collectively, these gene expression results support a possible role of c-Myc in regulating AR-FL/AR-V expression. Experimentally, through the use of an shRNA knockdown approach in cell models and in a patient-derived xenograft model, we confirmed c-Myc regulation of the expression and activity of AR-FL and AR-Vs. We found that c-Myc promotes the transcription of the AR gene and enhances the stability of the AR-FL and AR-V proteins without altering AR RNA splicing or AR transcript stability. We further show that inhibition of c-Myc could sensitize enzalutamide-resistant cells to growth inhibition by enzalutamide. Together, the findings reported here reveal a mechanism underlying coordinated expression of AR-FL and AR-Vs in CRPC. The tight correlation between the levels of AR-FL and AR-V transcripts in individual clinical samples underscores the clinical relevance of this mechanism.
Notably, our gene expression analyses extend beyond the correlation between c-Myc level/activity and AR expression level. We identified a strong positive correlation between the level/activity of c-Myc and the activity of the AR pathway and the very high enrichment of the AR gene expression signature in tumors that express a high level of c-Myc, indicating that high AR levels in these tumors are associated with high AR activities. Together with our experimental results showing that c-Myc knockdown leads to a decreased expression of AR-FL and AR-Vs and a concomitant downregulation of their target genes, these lines of evidence provide compelling support for c-Myc as a positive regulator of AR signaling, as opposed to the antagonistic effect of c-Myc on AR signaling that was reported in a previous study [63]. The discrepancy between our data and theirs could be due to the difference in experimental approaches. In their study, most of the work was conducted in LNCaP prostate cancer cells using inducible c-Myc overexpression [63]. It is notable that the endogenous c-Myc gene is already amplified and overexpressed in LNCaP cells [64] and that an excessive amount of c-Myc is known to induce oncogenic stress and apoptosis [65, 66]. In fact, we found that ectopic expression of c-Myc could induce cell death even in cells without c-Myc amplification. As such, we only used the knockdown approach in our study. The antagonistic effect of c-Myc on AR signaling that they observed might be a response to oncogenic stress imposed by overexpressed c-Myc.
Analyzing the number of RNA-seq reads spanning the splice junctions unique to each AR isoform allowed us to quantify the levels of AR-FL and different AR-Vs. To our knowledge, this is the first report on the positive correlations between the level/activity of c-Myc and the level of individual AR isoforms in CRPC samples. Significantly, knockdown of c-Myc leads to a similar decrease in the expression of AR-FL and different AR-Vs both in vitro and in vivo. These data suggest a role of c-Myc in regulating the co-expression of AR-FL and AR-Vs. In support of this contention, our AR-FL/AR-V7 minigene assay showed that c-Myc does not regulate AR RNA splicing to selectively produce AR-FL or -V7. This is in contrast to the results from a previous study showing c-Myc promoting AR alternative splicing to generate AR-V7 by upregulating the expression of the hnRNPA1 splicing factor [28]. Interestingly, similar to our results, when they knocked down c-Myc in VCaP and 22Rv1 cells, AR-FL and AR-V7 levels were equally downregulated [28]. However, overexpression of hnRNPA1 restored the expression of AR-V7 but not AR-FL, leading to their conclusion that c-Myc, via hnRNPA1, promotes AR alternative splicing to generate AR-V7 [28]. In our study, we specifically assessed AR-FL and AR-V7 splicing by measuring the levels of exon3-exon4-spliced and exon3-CE3-spliced transcripts expressed from an AR-FL/V7 minigene construct, and we did not detect alteration in AR-FL or -V7 splicing after c-Myc knockdown. The rescuing effect that they observed with hnRNPA1 overexpression perhaps is due to overexpressed hnRNPA1 stimulating AR alternative splicing to produce AR-V7. It would be important to assess the contribution of the endogenous hnRNPA1 to c-Myc-mediated AR-FL and -V7 expression.
One mechanism that accounts for c-Myc regulation of coordinated expression of AR-FL and AR-Vs is through promoting the transcription of the AR gene. c-Myc knockdown leads to a decrease in the activities of two AR promoters, the level of the AR pre-mRNA, and thereby the levels of AR-FL and AR-V mRNAs. Since the activities of the 1.7-kb (−600 to +1,115) and the 8-kb (−6,885 to +1,115) AR promoters are similarly downregulated by c-Myc knockdown, the c-Myc-regulatory site is very likely to reside in the 1.7-kb region. This is consistent with the presence of a previously identified c-Myc-binding site in this region, nucleotides −571 to −451 [39]. In addition to promoting AR-FL and AR-V expression at the transcriptional level, we found that c-Myc could extend the half-lives of AR-FL and AR-V7 proteins. Notably, this is unlikely to be a result of c-Myc physically interacting with AR-FL or -V7 protein as our co-immunoprecipitation assay failed to detect a c-Myc-AR-FL complex or a c-Myc-AR-V complex. To our knowledge, the ability of c-Myc to increase AR-FL or AR-V protein stability has not been reported before. In fact, our understanding of AR-V protein stability is rather limited. Elucidating the underlying mechanism may help us identify additional therapeutic targets to block AR-V signaling.
With the use of the 10058-F4 c-Myc inhibitor, which disrupts c-Myc and Max dimerization [60], we provide the proof of concept for the potential of inhibiting c-Myc to alleviate enzalutamide resistance. However, the poor bioavailability and rapid metabolism of 10058-F4 have limited its in vivo applicability [67]. New 10058-F4 analogs are being developed, and they have displayed improved pharmacokinetic properties (reviewed in [68]). Additionally, with novel computer-aided virtual screening approach to maximize the throughput of structure-based drug discovery, the development of potent, specific, and clinically viable inhibitors of c-Myc/Max dimerization is foreseeable [69]. Moreover, indirect targeting of c-Myc by inhibiting the transcription, translation, stability, and transcriptional activity of Myc as well as by synthetic lethality has proven tractable, with many molecules having progressed to clinical trials (reviewed in [68]). The potentials of different c-Myc-inhibiting strategies to improve the efficacy of AR-directed therapy, especially in the in vivo setting, warrant exploration.
In summary, our study highlights a critical role of c-Myc in regulating the coordinated expression of AR-FL and AR-Vs that is commonly observed in CRPC and suggests the utility of targeting c-Myc as an adjuvant to AR-directed therapy.
Materials and Methods
Cell lines and reagents
22Rv1, VCaP, and 293T cells were obtained from ATCC. LNCaP95 and CWR-R1-EnzR cells were provided by Drs. Alan Meeker and Donald Vander Griend, respectively. Cells used in all experiments were within 3 months of resuscitation of frozen cell stocks established within 3 passages after receipt of the cells. Cell authentication was performed at the Genetica DNA Laboratories, and cells were regularly evaluated for mycoplasma contamination. Enzalutamide, abiraterone acetate, and 10058-F4 were purchased from Selleck Chemicals.
Correlation coefficient analysis and GSEA
The Affymetrix Human Exon 1.0 ST microarray data from 7 studies of 1642 radical prostatectomy samples were downloaded from Gene Expression Omnibus with accession numbers GSE46691 [46, 47], GSE62116 [48], GSE62667 [49, 50], GSE72291 [51], GSE79915 [52], GSE79956 [53], and GSE79957 [53]. After normalization using the Single Channel Array Normalization algorithm [70], the data were pooled together for downstream analysis as previously reported [71]. The TCGA RNA-seq data of 500 primary prostate cancer samples were downloaded from Genomic Data Commons. The RNA-seq data of 3 cohorts of metastatic CRPC adenocarcinoma samples were downloaded from dbGaP, including 51 samples from the SU2C project (dbGaP accession pht004946.v1.p1) [41], 74 samples from the PROMOTE study (dbGaP accession phs001141.v1.p1) [42], and 34 samples from the Beltran study (dbGaP accession phs000909.v1.p1) [43]. RNA-seq gene expression analysis was performed using RSEM [72]. The transcripts per million (TPM) values from the RSEM output were used for downstream analysis. To determine AR-FL and AR-V expression, RNA-seq reads were aligned to the human hg38 genome using STAR (--clip5pNbases 6 --outFilterMultimapNmax 20 --outSAMtype BAM SortedByCoordinate --outWigType wiggle --outWigNorm None). Junction read numbers were generated from .SJ.out.tab splice junction files, and the numbers of reads spanning the splice junctions unique to each AR isoform were quantified and normalized by total splice junction reads [73].
For correlation analysis, two Myc-target signatures, Schuhmacher [44] and Jung [45], were used to evaluate c-Myc activity. Two AR-target signatures, the Nelson signature [58] and the Bluemn signature [59], were used to gauge AR activity. The activity scores were computed as the sum of z-scores for these c-Myc or AR signature genes. For analysis of correlation between expression levels and activity scores, the expression levels were computed as the z-scores for c-Myc or AR levels. Pearson’s correlation coefficient analysis was performed using R 3.4.3.
GSEA was performed with 1000 permutations. The genes were ranked using the Pearson’s correlation matrix with continuous phenotype labels for c-Myc or AR, and the datasets were run against the hallmark gene sets in the Molecular Signatures Database.
Western blotting
The procedure was described previously [74]. The following antibodies were used: anti-β-actin (Catalog #3700S; Cell Signaling Technology), anti-AR (N-20 (Catalog #sc-816) and 441 (Catalog #sc-7305), Santa Cruz Biotechnology), anti-AR-V7 (Catalog #31-1109-00; RevMAb Biosciences), and anti-c-Myc (Catalog #5605S; Cell Signaling Technology). The experiments were performed three times, and the levels of AR and c-Myc were normalized by that of β-actin.
Quantitative reverse transcription-PCR (qRT-PCR) and cell growth assay
qRT-PCR was performed as described [75], and the qPCR primer-probe sets and SYBR Green qPCR primers were from IDT. The sequences of the customer-designed primer-probe sets and primer pairs are listed in Supplementary Table S1. The primer sequences for the AR-minigene assay were described before [26]. qRT-PCR was conducted three times in triplicate. Cell growth was determined by the sulforhodamine (SRB) assay as described [76]. SRB was performed three times in six replicates.
Lentivirus Packaging and Transduction
The lentiviral c-Myc shRNA constructs were purchased from Sigma. Lentivirus was prepared in 293T cells in a 10-cm plate using a standard second-generation packaging system. Briefly, 293T cells were co-transfected with a lentivirus vector and the pCMV-dR8.2 dvpr and pCMV-VSV-G packaging vectors [77] (gifts from Dr. Bob Weinberg; Addgene plasmid # 8454 & 8455) at a ratio of 6:5:1 using Polyethylenimine (Polysciences). Pooled medium containing lentivirus collected each day from Day 2 to Day 4 post-transfection was concentrated using a Lenti-X Concentrator (Clontech). 22Rv1, LNCaP95, or VCaP cells were seeded in 6-well plates at 4 × 105 cells per well. The next day, the cells in each well were transduced with 1/15 of the concentrated lentivirus in normal growth media. The transduction was conducted in triplicate and repeated for the following 2 days.
DNA transfection and reporter-gene assay
22Rv1 and 293T cells were transfected by using the Lipofectamine 3000 (Invitrogen) and the TurboFect (Thermo) reagent, respectively, per instruction of the manufacturer. The AR-minigene construct was described previously [26]. Six luciferase reporter plasmids were used: PSA-luc (driven by the promoter of the PSA gene [78]), UBE2C-luc (driven by a minimal promoter and three repeats of an AR-V-specific promoter element in the UBE2C gene [79]), pGL4-ARpro8 and pGL4-ARpro1.7 (driven by an 8 or 1.7 kb fragment of the 5’-flanking region of the AR gene, respectively [80]), E2F2-Luc (driven by a minimal CMV promoter and tandem repeats of the E-box sequence that is responsive to c-Myc [81]), and CMV-LUC2CP (driven by the CMV promoter [82]; a gift from Dr. Gideon Dreyfuss; Addgene #62857). To ensure an even transfection efficiency, we conducted the transfection in bulk and then split the transfected cells for luciferase assay [83]. The transfection and reporter gene assays were performed three times in triplicate.
mRNA and protein stability assays
Actinomycin D (10 μM) or cycloheximide (10 μg/ml) was added to the cultures to stop new RNA or protein synthesis, respectively, at 24 h after shCtrl- or shMyc-lentivirus transduction. AR-FL or AR-V7 mRNA levels were measured by qRT-PCR, and protein levels were measured by Western blotting at indicated time points. The mRNA and protein stability assays were performed three times in triplicate.
Co-immunoprecipitation assay
Cells were pelleted, washed twice with cold PBS, and lysed in 500 μl of lysis buffer (1% NP-40, 20 mmol/L Tris, pH 7.4, 140 mmol/L NaCl, and 2 mmol/L EDTA) containing a protease inhibitor cocktail (Sigma). The suspension was left on ice for 30 min, followed by centrifugation at 12,000 rpm for 10 min. The lysate was then used for immunoprecipitation with a c-Myc antibody (Sigma) or an IgG control. The immune complex was precipitated using Protein G Dynabeads (Thermo Fisher Scientific) and washed three times with lysis buffer. The precipitate was resuspended in the LDS sample buffer (Thermo Fisher Scientific), boiled for 10 min, and subjected to Western blot analysis. The co-immunoprecipitation assay was conducted three times.
In vivo study with LuCaP 35CR patient-derived xenograft
Male CB17 SCID mice were obtained from Charles River at 4–6 weeks of age. After 1 week of adaptation, mice were castrated via a scrotal approach. At two days after castration, mice were inoculated subcutaneously with LuCaP 35CR tumor bits as described [54]. When the tumors reached ~200 mm3, they were randomized to receive 1/15 of the concentrated lentivirus encoding either the control or the c-Myc shRNA (n = 6 for control shRNA; n = 3 for c-Myc shRNA), and the lentivirus was injected daily into each tumor for 6 days. The mice received control shRNA lentivirus were then randomized and treated with vehicle control (5% benzyl alcohol and 95% safflower oil, n = 3) or abiraterone (0.5 mmol/kg/d in vehicle, n = 3) daily through intraperitoneal injection for 14 days [9], and all the mice received c-Myc shRNA lentivirus were treated with abiraterone in the same fashion. At the termination of the experiment, the mice were sacrificed, and the tumors were collected for molecular analysis. Investigators were not blinded to the experimental groups. All animal procedures were approved by Tulane University Institutional Animal Care and Use Committee.
Statistical analysis
Sample size for cell culture and animal studies was selected based on the ability to achieve an overall significance level of P = 0.05 and 80% power. The Student two-tailed t test was used to determine the mean differences between two groups. P < 0.05 is considered significant. Data are presented as mean ± SEM.
Supplementary Material
Acknowledgements
We are grateful to Dr. Alan Meeker at Johns Hopkins University for providing LNCaP95 cells, to Dr. Donald Vander Griend at the University of Illinois at Chicago for providing CWR-R1-EnzR cells, and to Dr. Robert Matusik at Vanderbilt School of Medicine for providing the ARR3-luc construct. We appreciate the support from the Tulane Cancer Next Generation Sequence Analysis core for utilization of resources and expertise for this work.
Footnotes
Competing interests
This work was supported by the following grants: the National Institutes of Health grants R01CA188609, R01AI101046, R01AI106676, P01CA214091, RCMI 2G12MD007595, and P20GM103518; Department of Defense grants W81XWH-15-1-0439, W81XWH-16-1-0317, W81XWH-16-1-0318, and W81XWH-14-1-0485; National Natural Science Foundation of China Project 81430087. The Richard M Lucas Foundation supported the development of the LuCaP 35CR model. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
References
- 1.Knudsen KE, Scher HI. Starving the addiction: new opportunities for durable suppression of AR signaling in prostate cancer. Clin Cancer Res 2009; 15: 4792–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nuhn P, De Bono JS, Fizazi K, Freedland SJ, Grilli M, Kantoff PW et al. Update on Systemic Prostate Cancer Therapies: Management of Metastatic Castration-resistant Prostate Cancer in the Era of Precision Oncology. Eur Urol 2019; 75: 88–99. [DOI] [PubMed] [Google Scholar]
- 3.Smith MR, Saad F, Chowdhury S, Oudard S, Hadaschik BA, Graff JN et al. Apalutamide Treatment and Metastasis-free Survival in Prostate Cancer. N Engl J Med 2018; 378: 1408–1418. [DOI] [PubMed] [Google Scholar]
- 4.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med 2012; 367: 1187–1197. [DOI] [PubMed] [Google Scholar]
- 5.de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L et al. Abiraterone and Increased Survival in Metastatic Prostate Cancer. N Engl J Med 2011; 364: 1995–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 2014; 371: 1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao B, Qi Y, Zhang G, Xu D, Zhan Y, Alvarez X et al. Androgen receptor splice variants activating the full-length receptor in mediating resistance to androgen-directed therapy. Oncotarget (1802 pii) 2014; 5: 1646–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Chan SC, Brand LJ, Hwang TH, Silverstein KA, Dehm SM. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res 2013; 73: 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mostaghel EA, Marck BT, Plymate SR, Vessella RL, Balk S, Matsumoto AM et al. Resistance to CYP17A1 Inhibition with Abiraterone in Castration-Resistant Prostate Cancer: Induction of Steroidogenesis and Androgen Receptor Splice Variants. Clinical Cancer Res 2011; 17: 5913–5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nadiminty N, Tummala R, Liu C, Yang J, Lou W, Evans CP et al. NF-kappaB2/p52 Induces Resistance to Enzalutamide in Prostate Cancer: Role of Androgen Receptor and Its Variants. Mol Cancer Ther 2013; 12: 1629–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao S, Zhan Y, Dong Y. Emerging data on androgen receptor splice variants in prostate cancer. Endocr Relat Cancer 2016; 23: T199–T210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan SC, Li Y, Dehm SM. Androgen receptor splice variants activate androgen receptor target genes and support aberrant prostate cancer cell growth independent of canonical androgen receptor nuclear localization signal. Journal of Biological Chemistry 2012; 287: 19736–19749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res 2008; 68: 5469–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo Z, Yang X, Sun F, Jiang R, Linn DE, Chen H et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res 2009; 69: 2305–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu R, Dunn TA, Wei S, Isharwal S, Veltri RW, Humphreys E et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res 2009; 69: 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu R, Isaacs WB, Luo J. A snapshot of the expression signature of androgen receptor splicing variants and their distinctive transcriptional activities. The Prostate 2011; 71: 1656–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun S, Sprenger CC, Vessella RL, Haugk K, Soriano K, Mostaghel EA et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J Clin Invest 2010; 120: 2715–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watson PA, Chen YF, Balbas MD, Wongvipat J, Socci ND, Viale A et al. Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc Natl Acad Sci USA 2010; 107: 16759–16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hornberg E, Ylitalo EB, Crnalic S, Antti H, Stattin P, Widmark A et al. Expression of androgen receptor splice variants in prostate cancer bone metastases is associated with castration-resistance and short survival. PLoSOne 2011; 6: e19059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scher HI, Lu D, Schreiber NA, Louw J, Graf RP, Vargas HA et al. Association of AR-V7 on Circulating Tumor Cells as a Treatment-Specific Biomarker With Outcomes and Survival in Castration-Resistant Prostate Cancer. JAMA Oncol 2016; 2: 1441–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welti J, Rodrigues DN, Sharp A, Sun S, Lorente D, Riisnaes R et al. Analytical Validation and Clinical Qualification of a New Immunohistochemical Assay for Androgen Receptor Splice Variant-7 Protein Expression in Metastatic Castration-resistant Prostate Cancer. European Urol 2016; 70: 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Todenhofer T, Azad A, Stewart C, Gao J, Eigl BJ, Gleave ME et al. AR-V7 Transcripts in Whole Blood RNA of Patients with Metastatic Castration Resistant Prostate Cancer Correlate with Response to Abiraterone Acetate. J Urol 2017; 197: 135–142. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Alsagabi M, Fan D, Bova GS, Tewfik AH, Dehm SM. Intragenic rearrangement and altered RNA splicing of the androgen receptor in a cell-based model of prostate cancer progression. Cancer Res 2011; 71: 2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nyquist MD, Li Y, Hwang TH, Manlove LS, Vessella RL, Silverstein KA et al. TALEN-engineered AR gene rearrangements reveal endocrine uncoupling of androgen receptor in prostate cancer. Proc Natl Acad Sci USA 2013; 110: 17492–17497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science 2015; 349: 1351–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu LL, Xie N, Sun S, Plymate S, Mostaghel E, Dong X. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene 2014; 33: 3140–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z, Zhou N, Huang J, Ho TT, Zhu Z, Qiu Z et al. Regulation of androgen receptor splice variant AR3 by PCGEM1. Oncotarget 2016; 7: 15481–15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nadiminty N, Tummala R, Liu C, Lou W, Evans CP, Gao AC. NF-kappaB2/p52:c-Myc:hnRNPA1 Pathway Regulates Expression of Androgen Receptor Splice Variants and Enzalutamide Sensitivity in Prostate Cancer. Mol Cancer Ther 2015; 14: 1884–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tummala R, Nadiminty N, Lou W, Evans CP, Gao AC. Lin28 induces resistance to anti-androgens via promotion of AR splice variant generation. Prostate 2016; 76: 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan L, Zhang F, Xu S, Cui X, Hussain A, Fazli L et al. Histone demethylase JMJD1A promotes alternative splicing of AR variant 7 (AR-V7) in prostate cancer cells. Proc Natl Acad Sci U S A 2018; 115: E4584–E4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stockley J, Markert E, Zhou Y, Robson CN, Elliott DJ, Lindberg J et al. The RNA-binding protein Sam68 regulates expression and transcription function of the androgen receptor splice variant AR-V7. Sci Rep 2015; 5: 13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shiota M, Fujimoto N, Imada K, Yokomizo A, Itsumi M, Takeuchi A et al. Potential Role for YB-1 in castration-resistant prostate cancer and resistance to enzalutamide through the androgen receptor V7. J Natl Cancer Inst 2016; 108:djw005. [DOI] [PubMed] [Google Scholar]
- 33.Ferraldeschi R, Welti J, Powers MV, Yuan W, Smyth T, Seed G et al. Second-Generation HSP90 Inhibitor Onalespib Blocks mRNA Splicing of Androgen Receptor Variant 7 in Prostate Cancer Cells. Cancer Res 2016; 76: 2731–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu Z, Chen S, Sowalsky AG, Voznesensky OS, Mostaghel EA, Nelson PS et al. Rapid induction of androgen receptor splice variants by androgen deprivation in prostate cancer. Clin Cancer Res 2014; 20: 1590–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharp A, Coleman I, Yuan W, Sprenger C, Dolling D, Nava Rodrigues D et al. Androgen receptor splice variant-7 expression emerges with castration resistance in prostate cancer. J Clin Invest 2018;129:192–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet 2007; 39: 41–51. [DOI] [PubMed] [Google Scholar]
- 37.Koh CM, Bieberich CJ, Dang CV, Nelson WG, Yegnasubramanian S, De Marzo AM. MYC and Prostate Cancer. Genes Cancer 2010; 1: 617–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grad JM, Dai JL, Wu S, Burnstein KL. Multiple androgen response elements and a Myc consensus site in the androgen receptor (AR) coding region are involved in androgen-mediated up-regulation of AR messenger RNA. Mol Endocrinol 1999; 13: 1896–1911. [DOI] [PubMed] [Google Scholar]
- 39.Nadiminty N, Tummala R, Lou W, Zhu Y, Zhang J, Chen X et al. MicroRNA let-7c suppresses androgen receptor expression and activity via regulation of Myc expression in prostate cancer cells. J Biol Chem 2012; 287: 1527–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao L, Schwartzman J, Gibbs A, Lisac R, Kleinschmidt R, Wilmot B et al. Androgen receptor promotes ligand-independent prostate cancer progression through c-Myc upregulation. PLoS One 2013; 8: e63563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015; 161: 1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L, Dehm SM, Hillman DW, Sicotte H, Tan W, Gormley M et al. A prospective genome-wide study of prostate cancer metastases reveals association of wnt pathway activation and increased cell cycle proliferation with primary resistance to abiraterone acetate-prednisone. Ann Oncol 2018; 29: 352–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 2016; 22: 298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schuhmacher M, Kohlhuber F, Holzel M, Kaiser C, Burtscher H, Jarsch M et al. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res 2001; 29: 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jung M, Russell AJ, Liu B, George J, Liu PY, Liu T et al. A Myc Activity Signature Predicts Poor Clinical Outcomes in Myc-Associated Cancers. Cancer Res 2017; 77: 971–981. [DOI] [PubMed] [Google Scholar]
- 46.Erho N, Crisan A, Vergara IA, Mitra AP, Ghadessi M, Buerki C et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One 2013; 8: e66855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakagawa T, Kollmeyer TM, Morlan BW, Anderson SK, Bergstralh EJ, Davis BJ et al. A tissue biomarker panel predicting systemic progression after PSA recurrence post-definitive prostate cancer therapy. PLoS One 2008; 3: e2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karnes RJ, Bergstralh EJ, Davicioni E, Ghadessi M, Buerki C, Mitra AP et al. Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at risk patient population. J Urol 2013; 190: 2047–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klein EA, Yousefi K, Haddad Z, Choeurng V, Buerki C, Stephenson AJ et al. A genomic classifier improves prediction of metastatic disease within 5 years after surgery in node-negative high-risk prostate cancer patients managed by radical prostatectomy without adjuvant therapy. Eur Urol 2015; 67: 778–786. [DOI] [PubMed] [Google Scholar]
- 50.Prensner JR, Zhao S, Erho N, Schipper M, Iyer MK, Dhanasekaran SM et al. RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol 2014; 15: 1469–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Den RB, Feng FY, Showalter TN, Mishra MV, Trabulsi EJ, Lallas CD et al. Genomic prostate cancer classifier predicts biochemical failure and metastases in patients after postoperative radiation therapy. Int J Radiat Oncol Biol Phys 2014; 89: 1038–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Freedland SJ, Choeurng V, Howard L, De Hoedt A, du Plessis M, Yousefi K et al. Utilization of a Genomic Classifier for Prediction of Metastasis Following Salvage Radiation Therapy after Radical Prostatectomy. Eur Urol 2016; 70: 588–596. [DOI] [PubMed] [Google Scholar]
- 53.Ross AE, Johnson MH, Yousefi K, Davicioni E, Netto GJ, Marchionni L et al. Tissue-based Genomics Augments Post-prostatectomy Risk Stratification in a Natural History Cohort of Intermediate- and High-Risk Men. Eur Urol 2016; 69: 157–165. [DOI] [PubMed] [Google Scholar]
- 54.Corey E, Quinn JE, Buhler KR, Nelson PS, Macoska JA, True LD et al. LuCaP 35: a new model of prostate cancer progression to androgen independence. The Prostate 2003; 55: 239–246. [DOI] [PubMed] [Google Scholar]
- 55.Burnstein KL. Regulation of androgen receptor levels: implications for prostate cancer progression and therapy. J Cell Biochem 2005; 95: 657–669. [DOI] [PubMed] [Google Scholar]
- 56.Hu R, Lu C, Mostaghel EA, Yegnasubramanian S, Gurel M, Tannahill C et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res 2012; 72: 3457–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Z, Wu D, Thomas-Ahner JM, Lu C, Zhao P, Zhang Q et al. Diverse AR-V7 cistromes in castration-resistant prostate cancer are governed by HoxB13. Proc Natl Acad Sci U S A 2018; 115: 6810–6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nelson PS, Clegg N, Arnold H, Ferguson C, Bonham M, White J et al. The program of androgen-responsive genes in neoplastic prostate epithelium. Proc Natl Acad Sci U S A 2002; 99: 11890–11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bluemn EG, Coleman IM, Lucas JM, Coleman RT, Hernandez-Lopez S, Tharakan R et al. Androgen Receptor Pathway-Independent Prostate Cancer Is Sustained through FGF Signaling. Cancer Cell 2017; 32: 474–489 e476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yin X, Giap C, Lazo JS, Prochownik EV. Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene 2003; 22: 6151–6159. [DOI] [PubMed] [Google Scholar]
- 61.Cao B, Qi Y, Yang Y, Liu X, Xu D, Guo W et al. 20(S)-protopanaxadiol inhibition of progression and growth of castration-resistant prostate cancer. PLoS One 2014; 9: e111201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kregel S, Chen JL, Tom W, Krishnan V, Kach J, Brechka H et al. Acquired resistance to the second-generation androgen receptor antagonist enzalutamide in castration-resistant prostate cancer. Oncotarget 2016; 7: 26259–26274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barfeld SJ, Urbanucci A, Itkonen HM, Fazli L, Hicks JL, Thiede B et al. c-Myc Antagonises the Transcriptional Activity of the Androgen Receptor in Prostate Cancer Affecting Key Gene Networks. EBioMedicine 2017; 18: 83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nag A, Smith RG. Amplification, rearrangement, and elevated expression of c-myc in the human prostatic carcinoma cell line LNCaP. Prostate 1989; 15: 115–122. [DOI] [PubMed] [Google Scholar]
- 65.Chen CH, Zhang J, Ling CC. Transfected c-myc and c-Ha-ras modulate radiation-induced apoptosis in rat embryo cells. Radiat Res 1994; 139: 307–315. [PubMed] [Google Scholar]
- 66.Askew DS, Ashmun RA, Simmons BC, Cleveland JL. Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene 1991; 6: 1915–1922. [PubMed] [Google Scholar]
- 67.Guo J, Parise RA, Joseph E, Egorin MJ, Lazo JS, Prochownik EV et al. Efficacy, pharmacokinetics, tisssue distribution, and metabolism of the Myc-Max disruptor, 10058-F4 [Z,E]-5-[4-ethylbenzylidine]-2-thioxothiazolidin-4-one, in mice. Cancer Chemother Pharmacol 2009; 63: 615–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Whitfield JR, Beaulieu ME, Soucek L. Strategies to Inhibit Myc and Their Clinical Applicability. Front Cell Dev Biol 2017; 5: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carabet LA, Lallous N, Leblanc E, Ban F, Morin H, Lawn S et al. Computer-aided drug discovery of Myc-Max inhibitors as potential therapeutics for prostate cancer. Eur J Med Chem 2018; 160: 108–119. [DOI] [PubMed] [Google Scholar]
- 70.Piccolo SR, Sun Y, Campbell JD, Lenburg ME, Bild AH, Johnson WE. A single-sample microarray normalization method to facilitate personalized-medicine workflows. Genomics 2012; 100: 337–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao SG, Chang SL, Erho N, Yu M, Lehrer J, Alshalalfa M et al. Associations of Luminal and Basal Subtyping of Prostate Cancer With Prognosis and Response to Androgen Deprivation Therapy. JAMA Oncol 2017; 3: 1663–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011; 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ungerleider N, Concha M, Lin Z, Roberts C, Wang X, Cao S et al. The Epstein Barr virus circRNAome. PLoS Pathog 2018; 14: e1007206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dong Y, Zhang H, Hawthorn L, Ganther HE, Ip C. Delineation of the Molecular Basis for Selenium-induced Growth Arrest in Human Prostate Cancer Cells by Oligonucleotide Array. Cancer Res 2003; 63: 52–59. [PubMed] [Google Scholar]
- 75.Dong Y, Lee SO, Zhang H, Marshall J, Gao AC, Ip C. Prostate specific antigen expression is down-regulated by selenium through disruption of androgen receptor signaling. Cancer Res 2004; 64: 19–22. [DOI] [PubMed] [Google Scholar]
- 76.Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc 2006; 1: 1112–1116. [DOI] [PubMed] [Google Scholar]
- 77.Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 2003; 9: 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pang S, Dannull J, Kaboo R, Xie Y, Tso CL, Michel K et al. Identification of a positive regulatory element responsible for tissue-specific expression of prostate-specific antigen. Cancer Res 1997; 57: 495–499. [PubMed] [Google Scholar]
- 79.Xu D, Zhan Y, Qi Y, Cao B, Bai S, Xu W et al. Androgen Receptor Splice Variants Dimerize to Transactivate Target Genes. Cancer Res 2015; 75: 3663–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cao B, Liu X, Li J, Liu S, Qi Y, Xiong Z et al. 20(S)-protopanaxadiol-aglycone downregulation of the full-length and splice variants of androgen receptor. Int J Cancer 2013; 132: 1277–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sears R, Ohtani K, Nevins JR. Identification of positively and negatively acting elements regulating expression of the E2F2 gene in response to cell growth signals. Mol Cell Biol 1997; 17: 5227–5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Younis I, Berg M, Kaida D, Dittmar K, Wang C, Dreyfuss G. Rapid-response splicing reporter screens identify differential regulators of constitutive and alternative splicing. Mol Cell Biol 2010; 30: 1718–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dong Y, Zhang H, Gao AC, Marshall JR, Ip C. Androgen receptor signaling intensity is a key factor in determining the sensitivity of prostate cancer cells to selenium inhibition of growth and cancer-specific biomarkers. Mol Cancer Ther 2005; 4: 1047–1055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.