Abstract
The Cryptococcus pathogenic species complex is a group of opportunistic human fungal pathogens that cause cryptococcal meningoencephalitis, an infection associated with unacceptably high mortality rates. The public health relevance of these pathogens has galvanized extensive research over the past several decades and led to characterization of their sexual cycles. This research has allowed several Cryptococcus species to develop into model fungal organisms for both pathogenesis and basic science studies. Many of these studies require observation of the meiotic process and its associated mating structures, as well as the generation of meiotic progeny with novel phenotypes and genotypes. Herein, we describe how to set up genetic crosses between Cryptococcus strains and observe their mating phenotypes, as well as how to recover progeny from these crosses for further analysis.
Keywords: Cryptococcus, Genetic cross, Mating progeny
INTRODUCTION
The human pathogenic Cryptococcus species complex belongs to the phylum Basidiomycota and is currently composed of seven species, defined based on genetic divergence and reproductive isolation (Sun and Xu 2007; Sun and Xu 2009; Hagen et al. 2015; Kwon-Chung et al. 2017). Most of the species in this complex are haploid opportunistic human pathogens, staying dormant in healthy hosts and only causing disease when the host immune system becomes compromised, such as in AIDS or organ transplant patients. It has been estimated that Cryptococcus was responsible for 223,100 cases of meningitis in 2014, with an associated 20–70% mortality rate for patients in care (Rajasingham et al. 2017). The global impact of this pathogen is magnified by the limited number of antifungal drug treatment options, as well as the recent outbreak of C. gattii infections in the Pacific Northwest of the USA and Canada, which suggests an expansion of the geographic niches of some of these human pathogens (Byrnes et al. 2010).
Pathogenic Cryptococcus species have a bipolar mating system, with two mating types (MATa and MATα) defined by which of the two alternative alleles is present at the single MAT locus (Lengeler et al. 2000; Hull et al. 2002). Close contact between cells of opposite mating types results in the formation of zygotes that are able to undergo filamentous growth and eventually produce basidia and basidiospores. Basidiospores are the products of meiosis followed by multiple rounds of mitosis and are therefore the results of exchange of genetic material between the two parental cells.
Numerous tools and resources are available for Cryptococcus sexual reproduction and genetic studies. Genetic manipulation in Cryptococcus can be achieved by biolistic transformation, Agrobacterium-mediated random insertional mutagenesis, and a novel CRISPR-mediated electroporation method (Toffaletti et al. 1993; Toffaletti and Perfect 1994; Davidson et al. 2000; Esher et al. 2015; Fan and Lin 2018; Jung et al. 2018). Congenic strain pairs (i.e. strains that are genetically identical with the exception of the MAT locus) have been constructed for C. neoformans, C. deneoformans, and C. deuterogattii (Heitman et al. 1999; Nielsen et al. 2003; Zhai et al. 2013). High quality, end-to-end chromosomal level genome assemblies are becoming available for all of the major lineages in the species complex (Loftus et al. 2005; Janbon et al. 2014; Farrer et al. 2015), as well as closely related non-pathogenic sister species (Sun et al. 2017). Additionally, collections of deletion strains of lacking all non-essential genes, transcription factors, and kinases in the C. neoformans genome have been or are being constructed (Liu et al. 2008; Jung et al. 2015; Lee et al. 2016). All of these advances that facilitate genetic manipulation and analyses of specific genes in Cryptococcus have allowed this opportunistic pathogen to become a fungal model organism (Idnurm et al. 2005; Heitman et al. 2011). These resources have aided fungal pathogenesis studies to elucidate fungal virulence factors and host-pathogen interactions, which have allowed significant advancements in our knowledge and treatment of these human pathogens. Furthermore, studies on basic fungal biology in Cryptococcus have contributed to our understanding of sexual reproduction, genome evolution, and evolution of the fungal mating type locus and centromeres (Heitman et al. 2011; Sun et al. 2012; Sun et al. 2017; Yadav et al. 2018).
Sexual reproduction plays important roles in Cryptococcus pathogenesis, as well as in the population dynamics and evolution of natural isolates. Cryptococcal infection begins when desiccated yeast cells or spores, the products of Cryptococcus sexual reproduction, are inhaled into the lungs. Genomic analyses suggest that sexual reproduction readily occurs in populations of Cryptococcus environmental and clinical isolates, providing a route to generate genetic diversity and the potential for increased pathogenicity (Hiremath et al. 2008; Sun et al. 2014; Roth et al. 2018). Therefore, it is crucial to understand the genetic and molecular basis of sexual reproduction in this species. In the following sections, we will illustrate how standard mating assays of Cryptococcus strains are carried out and what one can expect to observe in these assays. Additionally, we will demonstrate how to recover mating products via either spore dissection or screening for phenotypic markers.
STRATEGIC PLANNING
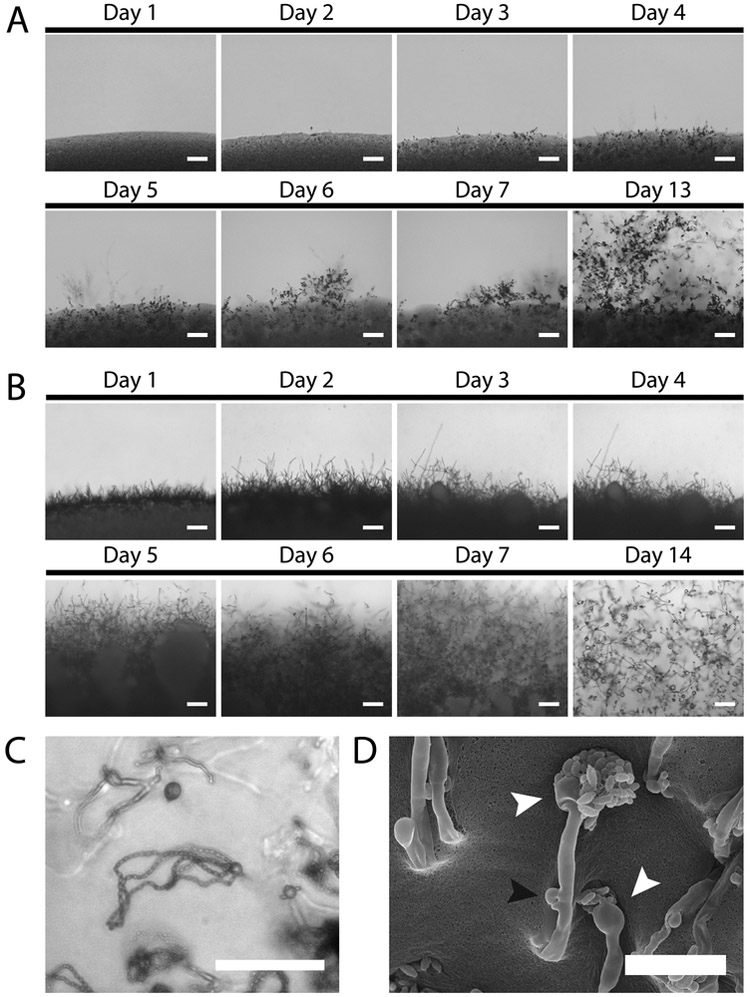
There is a wide range of mating phenotypes and efficiencies among C. neoformans, C. deneoformans, and C. gattii strains. Therefore, consideration should be taken for each genetic cross to determine the appropriate mating medium and to approximate the incubation period that will be required for sufficient mating structure and basidiospore chain production for either imaging or dissection. Murashige and Skoog (MS) agar medium is commonly used for C. neoformans and C. deneoformans dissections, but V8 (pH=5) agar medium can also be used for C. deneoformans dissections. Filamentation and mating structures typically appear within the first 1–4 days of incubation, making this an ideal time for determining mating responses. Figures 1A and 1B show typical mating structure production in a C. neoformans cross on MS and a C. deneoformans cross on V8 (pH=5), respectively. Figures 1C and 1D depict mating structures, including hyphal filaments, fused clamp cells, basidia, and protruding spore chains, from a C. neoformans cross on MS after approximately two weeks of incubation. Dissection from C. neoformans crosses should be done approximately 2 to 3 weeks post inoculation on the mating medium, while basidiospores from C. deneoformans crosses should be dissected later, around 3 to 4 weeks after inoculation.
Figure 1. Mating progression of Cryptococcus reference strains.
A. Filamentation and mating structures produced by a cross between C. neoformans strains H99α and KN99a on MS medium. Hyphal filaments begin appearing after two days of incubation on the MS medium, while basidia appear on day 3. Spore chains are visible by the fifth day, but are robust and far enough from the central mating patch for dissection by day 13. Scale bars represent 100 μM. B.Filamentation and mating structures produced by a C. deneoformans cross between JEC20a and JEC21α on V8 (pH=5) medium. Robust filamentation occurs within the first day and continues with prolonged incubation. After two weeks, basidia with individual spores protruding are visible, although it typically takes 3 to 4 weeks of incubation before enough spore chains will be available for dissection. Scale bars represent 100 μM. C. Spore chains emerging from a basidium produced by a cross between KN99α and KN99a. Basidia producing individual spores and hyphal filaments can be observed in the periphery. Scale bar represents 50 μM. D. Scanning electron microscopy of mating structures produced by a cross between KN99α and KN99a after approximately two weeks of incubation on MS medium. White arrowheads indicate basidia producing spore chains. The black arrowhead indicates a fused clamp cell along a hyphal filament. Scale bar represents 15 μM.
BASIC PROTOCOL 1: Mating Assays
Genetic crosses can be performed between Cryptococcus strains to observe various mating phenotypes and their associated sexual structures, to generate progeny of a desired genotype or phenotype, and to probe species boundaries. Crosses typically consist of two strains of opposite mating types (MATa and MATα), although unisexual reproduction between C. deneoformans parents of the same mating type is also possible. See Figure 2A for appropriate plate setup.
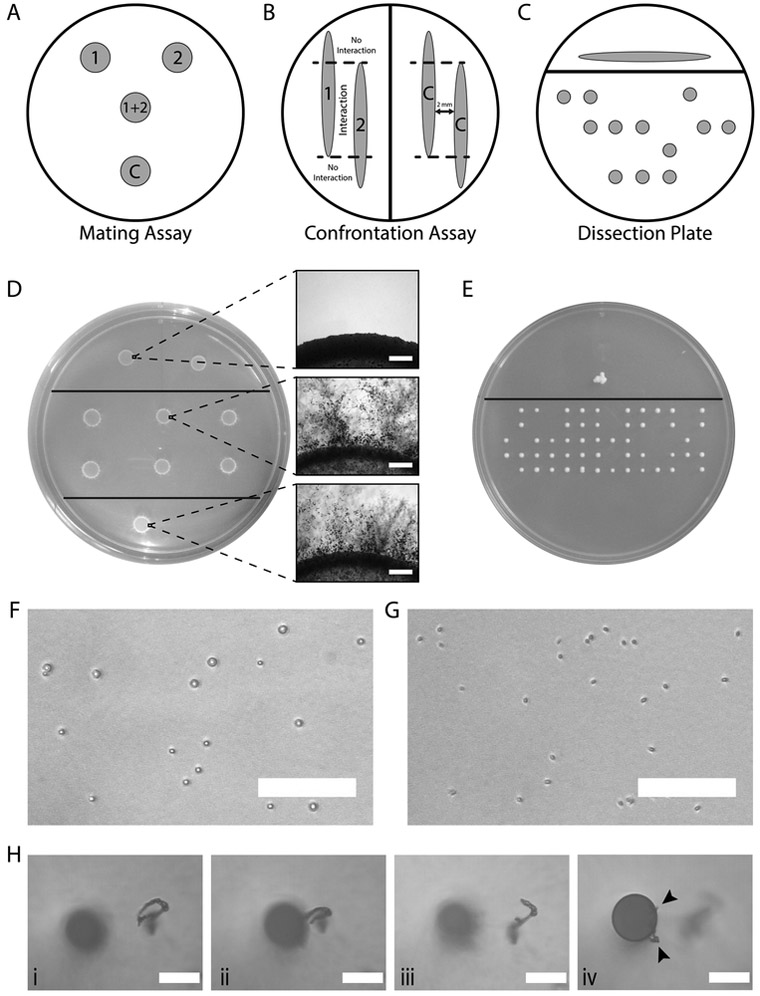
Figure 2. Plate layouts for mating assays, confrontation assays, and spore dissections as well as a comparison of yeast cells and spores.
A. The schematic depicts a basic plate layout that can be used for mating assays as described in Basic Protocol 1. It includes spots of both parental strains alone (1 and 2), to display any possible self-filamentation or confirm that either parent is not contaminated. Below the parents is the combination of parents (1+2), which is the mating patch that will be used for imaging and/or dissection. Finally, the bottom patch (C) should be a control mating with strains known to robustly undergo sexual reproduction together to ensure that the medium supports mating. With appropriate spacing, additional replicates of the mating patch (1+2) can also be included on the plate. B. An example of how to set up confrontation assays to determine if a strain (1) will produce mating structures in response to another strain (2). The approximately 2 mm region between the two strains (area labeled “Interaction”), will be where potential interactions and mating structures can be observed. Outside of the dashed lines (area labeled “No Interaction”), the researcher can determine if either strain is self-filamentous or is responding to the medium alone. The right half of the plate depicts a positive control, which should be between two strains known to strongly interact. C. A representative example of how a dissection plate will appear after incubation. Spores from the mating plate should be placed above the solid black line. Following incubation, this will become a large patch including a mixture of yeast cells and basidiospores. Below the solid black line is where spores from above will be micromanipulated and arranged into a grid. D. A representative mating plate depicting a cross between two KN99α x KN99a mutant strains and appropriate controls on MS medium after a two-week incubation. The top portion of the plate contains either parental strain alone, the middle portion has replicates of the KN99α and KN99a mutant cross, and the bottom portion has a control mating between KN99α and KN99a. Insets show filamentation along the border of each corresponding spot. Scale bars represent 400 μM.E. An example of a dissection plate after four days of incubation with progeny from a KN99α and KN99a cross. Panels F and G are representative images of what will be observed when performing random basidiospore dissections. F. Yeast cells distributed on YPD agar. Note the large size, round shape, and distinct black outline. Scale bar represents 50 μM. G. Spores on YPD agar. Note that spores are smaller and more elongated than the yeast cells. Scale bar represents 50 μM. H. Sequential images of what will be observed during dissection of basidiospores from a single basidium, as the needle i) comes into focus, ii) makes physical contact with a chain of basidiospores, and iii) is retracted, separating spores from the basidium. The final panel (iv) shifts focus to the end of the needle and arrowheads indicate where spores can be seen affixed to the sides of the needle. Scale bars represent 50 μM.
Materials
Cryptococcus strains (overnight culture or cells on agar medium)
Sterile deionized water
1.5 ml microcentrifuge tubes (Eppendorf)
Inoculation picks
Spectrophotometer
Vortex
Mating medium of choice (approximately 1 agar plate for every two unique genetic crosses)
Protocol steps
- Combine Cryptococcus cells for desired cross
- If using cells from agar plates, prepare a microcentrifuge tube with 200 µL of dH2O. Using inoculation picks, add cells of two chosen strains for the cross (an inoculum approximately the size of a matchstick head for each strain). Vortex the cells from the two strains together in the water.
- If using cells from an overnight culture, spin the culture down in a tabletop centrifuge at 3,000 rpm for 5 minutes and wash twice with 5 ml sterile dH2O. Resuspend cells in sterile dH2O to an absorbance of OD600 = 1.0. Combine 100 µL of the two strains to be crossed in a single microcentrifuge tube and vortex until thoroughly mixed.
- Spot 20 µL of the mixed strains onto the mating medium of choice.
- Ensure that each mating spot is approximately 2 cm apart from any other mating spot to minimize interference from other spots on mating. Each agar plate can accommodate up to 9 total spots, including positive and negative controls (Figures 2A and 2D).
- On each mating plate, a positive control involving common laboratory strains should be used to ensure there is no deficiency in the mating medium. For C. neoformans (serotype A) strains, the positive control may be a cross between KN99a x KN99α (Nielsen et al. 2003). For C. deneoformans (serotype D) strains, the positive control may be a cross between JEC20 x JEC21 (Kwon-Chung et al. 1992; Heitman et al. 1999). Negative controls, where each parental strain involved in the experimental cross are spotted alone, should also be included on the mating plate to determine if contamination or self-filamentation may confound results (Figure 2A).
After mating spots have dried (approximately 20 minutes), place mating plates face up in a completely dark area (such as a drawer) at room temperature. Do not place mating plates upside down, parafilm plates, or seal them in any way as this can inhibit mating.
-
Incubate mating spots for up to six weeks, imaging and dissecting spores as necessary.
Note: To observe filamentation, check plates every day; for basidia and basidiospore observation, check every day after day 4. For spore dissection, begin checking mating structures after approximately two weeks of incubation.
ALTERNATE PROTOCOL 1: Liquid-free Mating Assays
High humidity environments are known to decrease the mating efficiency of Cryptococcus strains. If the genetic cross set up following BASIC PROTOCOL 1 fails to produce sufficient mating structures, setting up a cross involving no additional liquid may help improve spore production. This method also produces more dispersed mating structures around the mating patch than the method described in BASIC PROTOCOL 1.
Materials
Cryptococcus strains (on agar medium)
Inoculation picks
Mating medium of choice (approximately 1 agar plate for every two unique genetic crosses)
Protocol steps
Using a sterile inoculation pick, make a small circle of cells of one parental strain on the mating medium of choice.
With a second sterile inoculation pick, make a small circle of the same size with the other parental strain directly on the same spot from step 1.
Include the same positive and negative controls that were described in BASIC PROTOCOL 1.
Place plates in dark, room temperature incubation area as in BASIC PROTOCOL 1 and incubate accordingly.
SUPPORT PROTOCOL 1: Confrontation Assays
Unlike mating assays, confrontation assays are utilized to determine if a strain of interest will produce mating structures in response to pheromone or small molecule production by another strain. This can be determined by placing the two strains in close proximity to each other and observing the production of hyphae in the space separating the two isolates over time. See Figure 2B for appropriate plate setup.
Materials
Cryptococcus strains (on agar medium)
Inoculation picks
V8 (pH=5) or V8 (pH=7) medium plates
Protocol steps
Using a sterile inoculation pick, streak a thin line of one of the two strains on the V8 medium plate.
Using another sterile inoculation pick, streak a thin line of the other strain in parallel with the first strain, approximately 1 to 2 mm apart, beginning approximately 1 cm below where the first line was struck and extending an additional centimeter beyond where the first line ended to establish zones of non-interaction. Similar to BASIC PROTOCOL 1, a positive control should be included on each plate in which two strains known to undergo robust, successful mating are confronted. See plate layout in Figure 2B.
Incubate the plates as in BASIC PROTOCOL 1. Hyphal production should be visible after 1 to 3 days of incubation for mating partners with vigorous interaction. Extended incubation may be needed to observe hyphal production between strains with low-efficiency mating responses.
BASIC PROTOCOL 2: Recover basidiospores by random spore dissection
Isolating C. neoformans basidiospores via microdissection is an important technique for generating progeny with novel genotypes and phenotypes and for measuring germination frequencies to determine species boundaries or meiotic proficiency. To dissect random basidiospores, they must first be isolated from the periphery of a mating patch and moved to nutrient-rich Yeast Extract Peptone Dextrose (YPD) agar medium. Following this isolation, the spores can be individually micromanipulated and spread across the agar plate to ensure each resulting colony is the product of a single progeny. See Figure 2C for how to set up the YPD agar plate that spores will be dissected onto and Figures 2F and 2G for microscopy images of what the experimenter may see during the dissection process.
Materials
Mating spot with sufficient basidiospore chains from BASIC PROTOCOL 1
9” glass Pasteur pipette (VWR cat. no. 14672–380)
Unfolded paperclip
YPD agar plates
Dissection microscope (Zeiss AxioScope.A1 HAL 50, specifically configured for tetrad dissection of yeast)
Dissection needle (Optical Fiber Dissection Kit, 25-micron, Cora Styles Lab Supplies, cat. no. 1025)
Parafilm
Protocol steps
Isolate spores from mating plate and transfer to YPD agar plate
-
1
Observe the periphery of a mating spot under a light microscope and identify an area with robust spore chain production that is as far as possible from the dense mating patch. On the bottom of the mating plate, mark this area for easy identification in step 5.
-
2
Break off the end of a long tip (9 inch) glass Pasteur pipette to obtain a small-diameter piece approximately 2 inches in length.
Note: you will want a paperclip that can fit into the broken Pasteur pipette piece as tightly as possible for the full 1 to 2-inch length. You will also want both ends of the broken pipette piece to be as flat as possible.
-
3
Insert the straightened paperclip fully into the broken pipette piece and sterilize them together over an open flame.
-
4
After cooling, remove the pipette piece from the paper clip and flame the more uneven end of the pipette piece.
-
5
Insert the more uneven end of the pipette piece into the agar at the marked spot from step 1.
Note: Take care to avoid getting any of the dense mating spot itself in the pipette piece, as this area contains many yeast cells.
-
6
Carefully remove the pipette piece with the agar plug remaining inside of the pipette (twist and slowly lift up; angle pipette piece if necessary).
-
7
Briefly flame the other end of the pipette piece (the flatter end) being careful not to let the agar plug with spore chains come too near the flame.
-
8
After cooling, slide the paperclip into the end of the pipette piece where the agar plug is and push the plug to the opposite end of the pipette piece so that the agar plug surface with the spore chains protrudes very slightly.
-
9
Drag the agar plug surface with the spore chains lightly along the surface of a YPD agar plate in a single straight line to deposit spore chains while keeping the agar plug within the pipette piece and avoiding gouging the YPD agar. See Figure 2C for dissection plate layout.
Spore dissection
-
10
Center the dissection needle within the field of view on the dissection microscope and clean the dissection needle by inserting and removing the needle several times in a new, sterile YPD agar plate.
-
11
Place the YPD agar plate from step 9 upside down (so that the agar surface is facing the dissection needle) on the stage of the dissection microscope.
-
12
Move the dissection needle to an area of the plate that does not contain yeast cells and will not be used for dissection later (normally the top most region of the plate, see Figure 2C). Bring the dissection needle into focus so that it is just touching the surface of the agar.
Note: Needle will appear to “jump” slightly as it comes into contact with the agar.
-
13
Move needle away from agar, keeping agar surface within focus.
-
14
Move field to where spores were distributed onto plate and identify spores for dissection.
Note: spores are smaller and slightly elongated in shape (Figure 2G), while yeast cells are typically larger, almost perfectly round, and have a distinct black border around the cell (Figure 2F).
-
15
Slowly bring dissection needle to the agar surface so that the perimeter of the needle comes just into contact with the spore.
Note: Make sure that the needle does not come into contact with any yeast cells.
-
16
After the needle has contacted the agar surface and the needle edge has come into contact with the spore, slowly withdraw the needle and ensure that the spore has remained attached to the needle (it should no longer be in the field of view).
Note: gently tapping the microscope base can help to ensure the spore is stuck to the dissection needle before withdrawing the needle.
-
17
Move the stage to a specific coordinate on the lower region of the plate (see Figure 2C), bring the needle with the spore to the agar surface, lightly tap microscope base to dislodge spore from needle. Continue bringing the needle to the surface (without puncturing agar), lightly tapping, and withdrawing needle as needed until spore has been visibly deposited on agar surface.
-
18
Record coordinates at which the spore was placed.
-
19
Return to area where spores were distributed from mating plate in step 9, identify additional spores for dissection, and repeat steps 14–18.
-
20
After dissecting the appropriate number of spores, remove the YPD agar plate from the dissection scope, parafilm the YPD plate, and leave it to incubate upside down at room temperature.
-
21
Purify germinated spores from the YPD agar dissection plate by streaking them onto the appropriate medium.
Note: spores typically take 3 to 5 days to germinate, but spores from genetically distant crosses may take up to two weeks to germinate. Additionally, if contamination is observed on YPD plate that spores were dissected onto, the progeny from this dissection cannot be used to estimate germination frequencies as contaminating organisms and their secreted compounds could affect germination efficiency.
ALTERNATE PROTOCOL 2: Recover basidiospores by single basidium dissection
The progeny recovered through random spore dissection are, as indicated by the name of the technique, random. That is, there is not information on the “genealogy” of the spores dissected, and they are considered to represent a population sampled from a large number of meiotic events, with the four meiotic products from each meiosis not necessarily all being represented in the progeny dissected. Random dissection is advantageous in cases where a broad sampling of the progeny population is the objective, such as when analyzing genome-wide meiotic recombination, or when isolating a progeny that has a certain combination of the genotypes of the two parents at multiple loci. However, sometimes it is necessary to know which spores were produced from the same meiotic event and in these cases, dissecting spores from a single basidium is required. Because it is known that only one meiotic event occurred within each basidium of Cryptococcus (Idnurm 2010), single basidium dissection in Cryptococcus, to some extent, resembles tetrad dissection in Saccharomyces cerevisiae.
Materials
Mating spot with sufficient basidiospore chains from BASIC PROTOCOL 1
Dissection microscope equipped with dissection needle (see BASIC PROTOCOL 2)
YPD agar plates
Protocol steps
- Collect basidiospores from a single basidium on the mating plate from Basic Protocol 1.
- Place the mating plate upside down (so that the mating spot is facing the dissection needle) on the stage of the dissection microscope.
- Make sure that the needle is away from and not touching the plate surface and mating spot.
- Focus the objective onto the agar surface of the YPD medium plate.
- Locate an area where the basidia and basidiospores are abundant.
- Find one basidium with chains of basidiospores that is well isolated from the other basidia and spores chains.
- Move this basidium into the center of the field.
- Bring the needle closer to the plate surface (this should be done very slowly using the turning knob) until the needle’s shape first appears in the field. (Figure 2H panel i)
- Using the handle, slightly move the needle closer to the targeted basidium and spore chains (now the needle will come into focus).
- Spore chains will be “pulled” onto the needle surface (likely due to electrostatic attraction). (Figure 2H panel ii)
- Using the handle, slightly move the needle away from the plate surface, and in the process, breaking the spore chains, which will result in a portion of the spores from that basidium remaining on the tip of the needle. (Figure 2H panel iii)
- Using the turning knob, move the needle as far as possible from the plate surface.
- Remove the mating plate from the stage of the dissection microscope.
- Transfer the basidiospores onto a fresh YPD plate.
- Place a fresh YPD plate on the stage of the dissection microscope.
- Move the stage, so that the needle is underneath a spot on the plate that is located on the vertical axis of the plate, and about 5–10 cm from the top (upper region of plate in Figure 2C).
- Move the objective to focus on the agar surface of the plate.
- Using the turning knob, very slowly move the needle towards the plate surface until the shape of the needle appears in the field.
- Using the handle, move the needle even closer to the surface until it comes into focus.
- Using the handle, move the needle slightly up and down so that it makes repeated contacts with the agar plate surface. With each contact, a portion of the spores on the tip of the needle will be placed onto the plate surface.
-
Repeat until no additional spores are placed onto the agar plate surface with additional needle-plate contacts.Note: it is helpful to move the stage very slightly following each contact to make sure the spores are relatively well separated from each other while placing the spores onto the plate surface, which will facilitate the following spore separation steps.
- Separate the basidiospores
- Move the field to where spores were distributed onto plate.
- Follow step 15 to step 21 in BASIC PROTOCOL 2.
SUPPORT PROTOCOL 2: Recover mating progeny using dominant drug markers
When sexual reproduction is between two strains that are genetically modified to express different dominant drug resistance markers (e.g. NAT, NEO, or HYG), meiotic progeny can be recovered by simply screening for double-resistant colonies using YPD medium plates supplemented with both drugs.
Materials
Mating spot with sufficient basidiospore chains from BASIC PROTOCOL 1
Sterile scalpel or razor blade
1.5 ml microcentrifuge tubes (Eppendorf)
Sterile deionized water
Vortex
YPD agar plate supplemented with drugs
Parafilm
Protocol steps
- Recover and resuspend mating products.
- Locate areas on the mating plate that contain abundant basidiospores.
- Using sterile scalpel (or razor blade), cut out the regions with spores and transfer them into a microcentrifuge tube.
- Add 1 ml sterile dH2O into the microcentrifuge tube.
- Vortex for 30 seconds.
Plate 100 µl of the cell suspension onto YPD solid medium supplemented with the two drugs to which the parental strains are resistant.
-
Parafilm the plate and incubate the plate upside down at 30 °C for 3 days.
Note: it is recommended that 10x, 100x, and 1000x dilutions of the cell suspension from step 1 should also each be plated onto YPD solid medium supplemented with drugs, so that a plate with a desirable number of colonies (100 – 200) will be obtained.
REAGENTS AND SOLUTIONS
V8 agar medium
50 ml V8 juice
0.5 g KH2PO4
950 ml dH2O
-
40 g Bacto agar (VWR cat. no. 90000–762)
(Note: The pH of the media should be at 5 inherently and can be adjusted to 7 with KCl if necessary)
Autoclave for 40 minutes
Murashige Skoog (MS) agar medium
4.328 g MS mix (entire contents of one foil packet) (Sigma, M5524)
16 g Bacto agar (VWR cat. no. 90000–762)
1000 ml dH2O
Adjust pH to 5.8
Autoclave for 40 minutes
Add 1ml 1000X vitamin mix after autoclaving (Sigma, M7150)
Yeast Extract Peptone Dextrose (YPD) agar medium
65 g YPD agar (VWR cat. no. 90003–278)
5 g Bacto agar (VWR cat. no. 90000–762)
Autoclave 40 minutes
- Potential antibiotic drug modifications:
- If adding antibiotic drugs, allow agar to cool to 50–55°C before addition.
- Add Nourseothricin to a final concentration of 100 µg/ml.
- Add G418 to a final concentration of 200 µg/ml.
- Add Hygromycin to a final concentration of 200 µg/ml.
Synthetic Drop-out (SD) medium
6.7 g YNB without amino acids
20 g Bacto agar
800 ml dH2O
Autoclave for 40 minutes
- After autoclaving, add:
- 100 ml 20% glucose
- 100 ml 10X amino acid stock mix lacking appropriate amino acid
10X amino acid stock mix
Do not add the amino acid for whichever auxotrophy you will be selecting for.
850 ml dH2O
300 mg L-isoleucine
1.5 g L-valine
200 mg adenine sulfate
200 mg L-histidine-HCl
1 g L-leucine
300 mg L-lysine
200 mg L-methionine
500 mg L-phenylalanine
200 mg L-tryptophan
300 mg L-tyrosine
200 mg uracil
4 g L-serine
1 g L-glutamate
- Autoclave 40 minutes, cool to room temperature, then add:
- 50 ml 4% (w/v) L-threonine
- 100 ml 1% (w/v) L-aspartate
YNB + 5-fluoroorotic acid (5-FOA)
20 g Bacto agar
6.7 g YNB without amino acids
50 mg uracil
900 ml dH2O
Autoclave 40 minutes
- While autoclaving, combine:
- 100 ml 10X amino acid stock without uracil
- 1 g 5-FOA
- 20 g glucose
- Heat while stirring to dissolve solutes, making sure not to boil
- Filter sterilize
Add filter-sterilized solution to autoclaved medium (no need to cool)
COMMENTARY
Background Information.
It is thought that the cryptococcal bipolar mating system evolved from a tetrapolar system that is ancestral to Basidiomycetes (Metin et al. 2010; Findley et al. 2012; Sun et al. 2017). The Cryptococcus sexual cycle is well-characterized and was first identified and described in C. neoformans over 40 years ago (Kwon-Chung 1975; Kwon-Chung 1976a; Kwon-Chung 1976b). When mating compatible MATa and MATα cells come into close contact, conjugation between the two cells is initiated, resulting in a dikaryotic zygote. The zygote undergoes a morphological transition and grows as a filament. Fused clamp cells form along the hyphae, bridging over the septa that divide the hyphae into compartments and ensuring accurate nuclear distribution during hyphal development. Eventually, the hyphal tip develops into a basidium, within which the two nuclei fuse and undergo karyogamy and one round of meiosis, producing four meiotic products. These four meiotic products then undergo repeated rounds of mitosis to produce additional nuclei that are packaged into basidiospores, which are located on the surface of the basidium and form four long chains of basidiospores. Thus, Cryptococcus mating bears all of the hallmarks of sexual development in Basidiomycetes, making it an excellent model organism to study fungal sexual reproduction. In addition to mating between cells of opposite mating types, unisexual reproduction can also occur between MATα strains in C. deneoformans (Lin et al. 2005; Feretzaki and Heitman 2013; Sun et al. 2014; Sun and Heitman 2015; Roth et al. 2018)
Controls.
As in all biological experiments, proper controls (positive and negative) should be included in the analysis of sexual reproduction in Cryptococcus for both confrontation and mating assays. For mating assays, a positive control should be a mixture of two strains known to undergo efficient mating. For confrontation assays, a positive control should be a confrontation between two strains with opposite mating types that are known to interact and produce mating structures. The common strain pairs used for positive controls in mating assays are congenic C. neoformans strains KN99a and KN99α or C. deneoformans strains JEC20 (MATa) and JEC21 (MATα). C. neoformans H99 (MATα) crg1Δ and KN99a crg1Δ deletion strains are frequently used in confrontation assays due to their hyper-sensitivity to pheromones (see comment below). Negative controls are typically the solo culture of the strains being tested for mating or mating interactions. Positive and negative controls should be on the same plate as the mating pairs being tested. Figures 2A and 2B depict common layouts of the plates for both mating as well as confrontation assays and include all of the proper controls.
Super-maters.
The mating ability, such as hyphal production, of Cryptococcus isolates has been shown to be a quantitative trait (Lin et al. 2006). Therefore, the absence of signs of successful mating does not necessarily indicate that the strains being tested are sterile. There are several genetically modified Cryptococcus isolates that have been shown to exhibit enhanced mating abilities, such as a crg1Δ strain in which the CRG1 gene that is involved in attenuating the pheromone response has been disrupted (Fraser et al. 2003) and a CPR2OE strain where a constitutively active pheromone receptor-like gene, CPR2, has been overexpressed (Hsueh et al. 2009). These genetically engineered super-maters can be used in cases where mating interaction is absent when standard tester strains are used.
Mating medium.
The most commonly used medium for Cryptococcus mating assay are MS, V8 (pH=5), and V8 (pH=7). It should be noted that if an MS medium plate is used for crosses that involve strains that are auxotrophic for certain amino acids (e.g. adenine or uracil), these amino acids need to be supplemented in the MS medium to ensure proper growth of the auxotrophic strains. Mating agar plates should also be prepared with at least 25 ml of medium to ensure that they do not dry out for up to six weeks of incubation at room temperature. Studies have shown that certain modifications of these standard mating medium plates could enhance mating or induce mating structures, and thus, increase the chance of observing successful sexual reproduction. For example, supplementing media with inositol has been shown to stimulate mating. It has also been indicated that the presence of copper in media has a positive effect on mating. Additionally, glucosamine as the sole carbon source in nutrient-rich media as well as the presence of quorum sensing peptides have also been shown to induce filamentation in Cryptococcus in recent studies (Xue et al. 2007; Kent et al. 2008; Xu et al. 2017; Tian et al. 2018).
Progeny recovery.
When mating is between two strains, each with either a dominant drug resistant marker or an auxotrophic marker, isolation of the mating products based on these phenotypic markers (see SUPPORT PROTOCOL 2) needs to be carried out in two steps because the drugs are ineffective in the minimal medium that is typically used to screen against auxotrophic markers (e.g. SD medium). For example, if a cross is between a nourseothricin-resistant strain and a uracil auxotrophic strain, cells from the mating mixture should first be screened for drug resistant colonies on YPD+nourseothricin medium plates, and then the colonies from the YPD+nourseothricin plates should be replica-plated onto agar plates containing 5-fluoroorotic acid (5-FOA) to screen for uracil auxotrophic isolates. The resulting colonies from the 5-FOA plates will be nourseothricin resistant and uracil auxotrophic, a combination of the phenotypes of the two parental strains. Alternatively, if the nourseothricin resistance marker and the uracil auxotrophic marker are in the same strain and this strain is crossed with an isolate that is wild type regarding these markers, the cells should first be screened on YPD+nourseothricin medium plates and then replica-plated onto SD-uracil solid medium to screen for uracil prototrophic colonies, which represent mating progeny in which the drug marker and uracil auxotrophic marker have segregated. Another method of isolating spores from mating mixtures based on gradient purification using centrifugation was also recently developed, which generates a progeny population resembling that from the random spore dissection method described in BASIC PROTOCOL 2 (Botts et al. 2009).
Troubleshooting.
One of the most common issues that might occur in studies of sexual reproduction in Cryptococcus is that no successful mating and spore production is observed. There are several possible explanations if this happens. First, make sure the medium and controls worked properly (see comments above). If not (e.g. no mating is observed in the positive control), prepare fresh medium plates and repeat. Second, as mentioned in previous comments, mating ability is strain-specific and quantitative, and successful mating could also depend on environmental factors. It might be that the strains being tested have low mating efficiency. If possible, retest the strains with super-maters, or modify the mating media according to previous comments. Third, the absence of successful mating and spore production is a real biological outcome, especially after attempts to induce mating by changing testing strains and modifying mating environments have all failed. Another issue could be low germination rates among the recovered mating progeny. In this case, possible explanations could be that the cross is between strains with highly diverged genomes. This has been observed among the progeny recovered from inter-species crosses in Cryptococcus (Sun and Xu 2007). Genotyping of the strains using MLST markers could help shed light on possible causes of this problem.
Time considerations.
Cryptococcal matings vary greatly in the time they take to produce various mating structures and sufficient basidiospore chains for dissection depending on the species involved in the cross, the mating medium used, and the genetic divergence between the two parental strains. Hyphae protruding from mating mixtures can be visible as early as 1 to 3 days after the mating is set up in efficient crosses and can take up to one week (or even longer) to be visible in crosses where one or both of the strains are less fertile or have mating deficiencies. The most efficient matings will also begin producing basidia and spore chains after approximately 5 to 7 days of incubation, and will continue to produce these structures as incubation is continued. Ideally, C. neoformans spore dissection takes place around two weeks after inoculating the mating medium and C. deneoformans dissections take place 3 to 4 weeks post-inoculation. Spores may be dissected up to 6 weeks after a cross is set up, although germination frequencies tend to decline over time.
ACKNOWLEDGEMENT
This work was funded by the NIH/NIAID R37 MERIT award AI39115–21, R01 grant AI50113–14, and R01 grant AI133654–2 awarded to JH.
LITERATURE CITED
- Botts MR, Giles SS, Gates MA, Kozel TR and Hull CM, 2009. Isolation and characterization of Cryptococcus neoformans spores reveal a critical role for capsule biosynthesis genes in spore biogenesis. Eukaryotic Cell 8: 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes EJ 3rd, Li W, Lewit Y, Ma H, Voelz K et al. , 2010. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog. 6: e1000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RC, Cruz MC, Sia RAL, Allen BM, Alspaugh JA et al. , 2000. Gene disruption by biolistic transformation in serotype D strains of Cryptococcus neoformans. Fung. Genet. Biol. 29: 38–48. [DOI] [PubMed] [Google Scholar]
- Esher SK, Granek JA and Alspaugh JA, 2015. Rapid mapping of insertional mutations to probe cell wall regulation in Cryptococcus neoformans. Fungal Genetics and Biology 82: 9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, and Lin X, 2018. Multiple applications of a transient CRISPR-Cas9 coupled with electroporation (TRACE) system in the Cryptococcus neoformans species complex. Genetics 208: 1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer RA, Desjardins CA, Sakthikumar S, Gujja S, Saif S et al. , 2015. Genome evolution and innovation across the four major lineages of Cryptococcus gattii. mBio 6: e00868–00815. doi:00810.01128/mBio.00868-00815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feretzaki M, and Heitman J, 2013. Genetic circuits that govern bisexual and unisexual reproduction in Cryptococcus neoformans. PLoS Genetics 9: e1003688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findley K, Sun S, Fraser JA, Hsueh Y-P, Averette AF et al. , 2012. Discovery of a modified tetrapolar sexual cycle in Cryptococcus amylolentus and the evolution of MAT in the Cryptococcus species complex. PLoS Genetics 8: e1002528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser JA, Subaran RL, Nichols CB and Heitman J, 2003. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver island, Canada. Eukaryotic Cell 2: 1036–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen F, Khayhan K, Theelen B, Kolecka A, Polacheck I et al. , 2015. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genetics and Biology 78: 16–48. [DOI] [PubMed] [Google Scholar]
- Heitman J, Allen B, Alspaugh JA and Kwon-Chung KJ, 1999. On the origins of congenic MATα and MATa strains of the pathogenic yeast Cryptococcus neoformans. Fungal Genetics and Biology 28: 1–5. [DOI] [PubMed] [Google Scholar]
- Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR and Casadevall A, 2011. Cryptococcus. American Society of Microbiology. [Google Scholar]
- Hiremath SS, Chowdhary A, Kowshik T, Randhawa HS, Sun S et al. , 2008. Long-distance dispersal and recombination in environmental populations of Cryptococcus neoformans var. grubii from India. Microbiology 154: 1513–1524. [DOI] [PubMed] [Google Scholar]
- Hsueh YP, Xue C and Heitman J, 2009. A constitutively active GPCR governs morphogenic transitions in Cryptococcus neoformans. EMBO J 28: 1220–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, Davidson RC and Heitman J, 2002. Cell identity and sexual development in Cryptococcus neoformans are controlled by the mating-type-specific homeodomain protein Sxi1α. Genes Dev. 16: 3046–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, 2010. A tetrad analysis of the basidiomycete fungus Cryptococcus neoformans. Genetics 185: 153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Bahn YS, Nielsen K, Lin X, Fraser JA et al. , 2005. Deciphering the model pathogenic fungus Cryptococcus neoformans. Nat Rev Microbiol 3: 753–764. [DOI] [PubMed] [Google Scholar]
- Janbon G, Ormerod KL, Paulet D, Byrnes EJ III, Yadav V et al. , 2014. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet 10: e1004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K-W, Lee K-T, So Y-S and Bahn Y-S, 2018. Genetic manipulation of Cryptococcus neoformans. Current Protocols in Microbiology 50: e59. [DOI] [PubMed] [Google Scholar]
- Jung K-W, Yang D-H, Maeng S, Lee K-T, So Y-S et al. , 2015. Systematic functional profiling of transcription factor networks in Cryptococcus neoformans. Nature Communications 6: 6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent CR, Ortiz-Bermúdez P, Giles SS and Hull CM, 2008. Formulation of a defined V8 medium for induction of sexual development of Cryptococcus neoformans. Applied and Environmental Microbiology 74: 6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, 1975. A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia 67: 1197–1200. [PubMed] [Google Scholar]
- Kwon-Chung KJ, 1976a. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia 68: 821–833. [PubMed] [Google Scholar]
- Kwon-Chung KJ, 1976b. A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia 68: 943–946. [PubMed] [Google Scholar]
- Kwon-Chung KJ, Bennett JE, Wickes BL, Meyer W, Cuomo CA et al. , 2017. The case for ddopting the “Species Complex” nomenclature for the etiologic agents of Cryptococcosis. mSphere 2: 10.1128/mSphere.00357-00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Edman JC and Wickes BL, 1992. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60: 602–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K-T, So Y-S, Yang D-H, Jung K-W, Choi J et al. , 2016. Systematic functional analysis of kinases in the fungal pathogen Cryptococcus neoformans. Nature Communications 7: 12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler KB, Wang P, Cox GM, Perfect JR and Heitman J, 2000. Identification of the MATa mating-type locus of Cryptococcus neoformans reveals a serotype A MATa strain thought to have been extinct. Proc. Natl. Acad. Sci. USA 97: 14555–14460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Huang JC, Mitchell TG and Heitman J, 2006. Virulence attributes and hyphal growth of C. neoformans are quantitative traits and the MATalpha allele enhances filamentation. PLoS Genetics 2: e187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Hull CM and Heitman J, 2005. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature 434: 1017–1021. [DOI] [PubMed] [Google Scholar]
- Liu OW, Chun CD, Chow ED, Chen C, Madhani HD et al. , 2008. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell 135: 174–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus BJ, Fung E, Roncaglia P, Rowley D, Amedeo P et al. , 2005. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307: 1321–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metin B, Findley K and Heitman J, 2010. The mating type locus (MAT) and sexual reproduction of Cryptococcus heveanensis: insights into the evolution of sex and sex-determining chromosomal regions in fungi. PLoS Genetics 6: e10000961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K, Cox GM, Wang P, Toffaletti DL, Perfect JR et al. , 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect Immun 71: 4831–4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasingham R, Smith RM, Park BJ, Jarvis JN, Govender NP et al. , 2017. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. The Lancet Infectious Diseases 17: 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth C, Sun S, Billmyre RB, Heitman J and Magwene PM, 2018. A high resolution map of meiotic recombination in Cryptococcus deneoformans demonstrates decreased recombination in unisexual reproduction. Genetics 209: 567–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Billmyre RB, Mieczkowski P and Heitman J, 2014. Unisexual reproduction drives meiotic recombination and phenotypic and karyotypic plasticity in Cryptococcus neoformans. PLoS Genetics 10: e1004849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, and Heitman J, 2015. From two to one: unipolar sexual reproduction. Fungal Biology Reviews 29: 118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Hsueh Y-P and Heitman J, 2012. Gene conversion occurs within the mating-type locus of Cryptococcus neoformans during sexual reproduction. PLoS Genet 8: e1002810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, and Xu J, 2007. Genetic analyses of a hybrid cross between serotypes A and D strains of the human pathogenic fungus Cryptococcus neoformans. Genetics 177: 1475–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, and Xu J, 2009. Chromosomal rearrangements between serotype A and D strains in Cryptococcus neoformans. PLoS ONE 4: e5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Yadav V, Billmyre RB, Cuomo CA, Nowrousian M et al. , 2017. Fungal genome and mating system transitions facilitated by chromosomal translocations involving intercentromeric recombination. PLOS Biology 15: e2002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, He G-J, Hu P, Chen L, Tao C et al. , 2018. Cryptococcus neoformans sexual reproduction is controlled by a quorum sensing peptide. Nature Microbiology 3: 698–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toffaletti DL, and Perfect JR, 1994. Biolistic DNA delivery for Cryptococcus neoformans transformation, pp. 303–308 in Molecular Biology of Pathogenic Fungal: A laboratory Manual, edited by Maresca B and Kobayashi GS. Telos Press, New York. [Google Scholar]
- Toffaletti DL, Rude TH, Johnston SA, Durack DT and Perfect JR, 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175: 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Lin J, Zhao Y, Kirkman E, So Y-S et al. , 2017. Glucosamine stimulates pheromone-independent dimorphic transition in Cryptococcus neoformans by promoting Crz1 nuclear translocation. PLOS Genetics 13: e1006982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C, Tada Y, Dong X and Heitman J, 2007. The human fungal pathogen Cryptococcus can complete its sexual cycle during a pathogenic association with plants. Cell Host & Microbe 1: 263–273. [DOI] [PubMed] [Google Scholar]
- Yadav V, Sun S, Billmyre RB, Thimmappa BC, Shea T et al. , 2018. RNAi is a critical determinant of centromere evolution in closely related fungi. PNAS 115: 3108–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai B, Zhu P, Foyle D, Upadhyay S, Idnurm A et al. , 2013. Congenic strains of the filamentous form of Cryptococcus neoformans for studies of fungal morphogenesis and virulence. Infection and Immunity 81: 2626–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]