Abstract
The myeloid translocation gene family member MTG16 is a transcriptional corepressor that relies on the DNA-binding ability of other proteins to determine specificity. One such protein is the ZBTB family member Kaiso, and the MTG16:Kaiso interaction is necessary for repression of Kaiso target genes such as matrix metalloproteinase-7. Using the azoxymethane and dextran sodium sulfate (AOM/DSS) murine model of colitis-associated carcinoma, we previously determined that MTG16 loss accelerates tumorigenesis and inflammation. However, it was unknown whether this effect was modified by Kaiso-dependent transcriptional repression. To test for a genetic interaction between MTG16 and Kaiso in inflammatory carcinogenesis, we subjected single and double knockout (DKO) mice to the AOM/DSS protocol. Mtg16−/− mice demonstrated increased colitis and tumor burden; in contrast, disease severity in Kaiso−/− mice was equivalent to wild type controls. Surprisingly, Kaiso deficiency in the context of MTG16 loss reversed injury and pro-tumorigenic responses in the intestinal epithelium following AOM/DSS treatment, and tumor numbers were returned to near to wild type levels. Transcriptomic analysis of non-tumor colon tissue demonstrated that changes induced by MTG16 loss were widely mitigated by concurrent Kaiso loss, and DKO mice demonstrated downregulation of metabolism and cytokine-associated gene sets with concurrent activation of DNA damage checkpoint pathways as compared with Mtg16−/−. Further, Kaiso knockdown in intestinal enteroids reduced stem- and WNT-associated phenotypes, thus abrogating the induction of these pathways observed in Mtg16−/− samples. Together, these data suggest that Kaiso modifies MTG16-driven inflammation and tumorigenesis and suggests that Kaiso deregulation contributes to MTG16-dependent colitis and CAC phenotypes.
Keywords: Colitis, colitis-associated carcinoma, Kaiso, MTG16, myeloid translocation gene
Introduction
Kaiso (encoded by the ZBTB33 gene) is a transcriptional repressor and member of the BTB/POZ-ZF (broad complex, tramtrack, bric-a-brac/poxvirus and zinc finger) subfamily of transcription factors (1). It was originally determined to bind DNA via a non-methylated consensus Kaiso binding site (KBS, TCCTGCNA), found in genes such as matrix metalloproteinase-7 (MMP7) (1, 2). Additionally, Kaiso has since been determined to bind and suppress methylated CpGs, and has been implicated in repression of several tumor suppressor genes such as CDKN2A, HIC1, and MGMT via this mechanism, indicating Kaiso may broadly function as a tumor promoter (3–5). On the other hand, Kaiso can suppress the pro-tumorigenic WNT signaling pathway by multiple mechanisms, including direct regulation of canonical WNT targets such as MYC, FOS, Siamois, and CCND1, as well as through protein-protein interactions with TCF3/TCF4 which disrupts binding with β-catenin and target promoters (6–9). With both tumor-promoting and tumor-suppressing activities, it is likely that Kaiso dysfunction plays a role in tumorigenesis, yet its exact effects are likely context-specific.
DNA-binding transcription factors such as Kaiso rely on a variety of transcriptional corepressors to form the repression complexes by which they attenuate gene transcription. The myeloid translocation genes (MTGs) are a three-member family consisting of Myeloid Translocation Gene 8 (MTG8, RUNX1T1), Myeloid Translocation Gene 16 (MTG16, CBFA2T3), and Myeloid Translocation Gene Related 1 (MTGR1, CBFA2T2). Each function as a transcriptional corepressor, by acting as a scaffolding protein upon which other transcriptional corepressors, including mSin3a, N-CoR, SMRT, and histone deacetylases (HDACs), assemble (10, 11). Importantly, we have previously determined that Kaiso is an MTG family binding partner, with the zinc finger domain of Kaiso interacting with the nervy homology region 1 (NHR1) domain of MTG16 and thereby recruiting the repressive complex to KBS-containing promoters (12). The functional relevance of the interaction is evident as loss of either protein leads to derepression of targets such as MMP7.
MTG family members have been implicated in both hematologic and solid malignancies including leukemia, colon, and breast cancers (13–15). Indeed, our lab has identified roles for the MTGs in inflammation-driven tumorigenesis, and shown that while MTGR1 is required for tumorigenesis, the absence of MTG16 leads to increased inflammation and tumorigenesis in a murine model of colitis-associated carcinoma (CAC) (16). Kaiso has likewise been implicated in colon tumorigenesis and inflammation, and Kaiso knockout (KO) mice crossed with the tumor-susceptible ApcMin/+ mouse model show resistance to intestinal tumorigenesis by increased tumor latency, while Kaiso overexpression induced intestinal inflammation, activated NF-κB signaling, and promoted tumor formation in the ApcMin/+ model (17–19).
Interestingly, although Kaiso overexpression induced inflammation in the intestine, the impact of Kaiso loss in the setting of inflammatory carcinogenesis has not yet been established. As MTG16 loss greatly exacerbated injury and tumorigenesis in the azoxymethane/dextran sodium sulfate (AOM/DSS) CAC model and modulation of many in vitro target genes was dependent on Kaiso expression, the purpose of this work was twofold: to determine (1) whether Kaiso deficiency modified colitis and/or CAC and (2) whether subsequent loss of Kaiso would modify the Mtg16−/− phenotype. Here, we were surprised to determine that while Kaiso deficiency decreased inflammatory responses at a transcriptional level, it does not lead to altered injury or tumorigenesis in response to the AOM/DSS inflammatory carcinogenesis protocol. Perhaps most surprisingly, while Mtg16−/− mice demonstrated increased injury and tumorigenesis in response to AOM/DSS, concurrent loss of both MTG16 and Kaiso reversed many Mtg16−/−-induced injury responses, in vivo and ex vivo. Taken together, these results indicate that while Kaiso does not modify inflammatory tumorigenesis per se, it likely mediates many of the pro-tumorigenic phenotypes observed with MTG16 loss.
Results
Kaiso and MTG16 are dysregulated in colorectal tumors
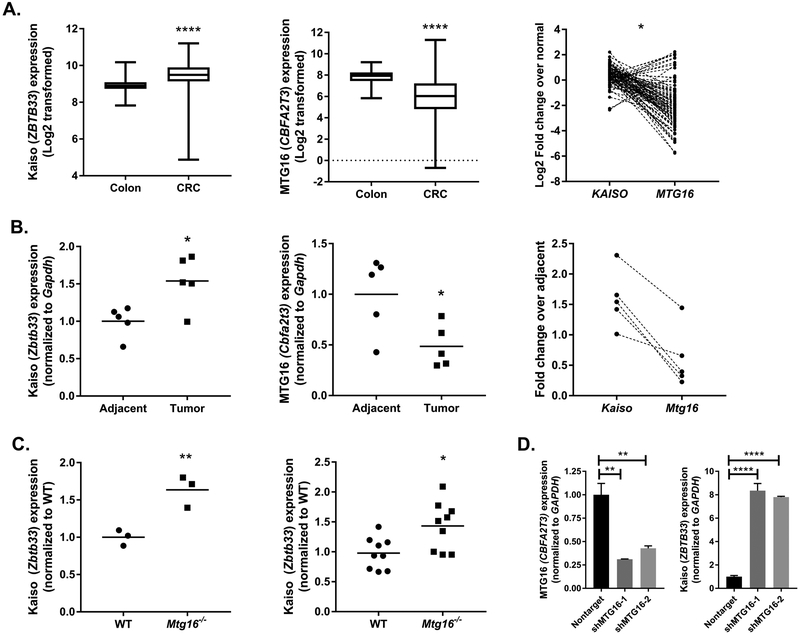
Kaiso is responsible for vectoring MTG16 to a subset of target genes. However, despite in vitro co-regulation of genes such as MMP7, we have previously observed an inverse correlation for MTG16 (CBFA2T3) and KAISO (ZBTB33) expression in human colorectal cancer (CRC) (12). To further extend this observation in a more comprehensive fashion, we queried colorectal cancer cases from The Cancer Genome Atlas (Fig. 1A) (20). As before, we observed KAISO to be upregulated in tumor tissue as compared to adjacent normal tissue (Colon: 8.93±0.05, CRC: 9.45±0.03, P<0.0001, Student’s t test), while MTG16 was downregulated (Colon: 7.8±0.11, CRC: 6.08±0.09, P<0.0001, Student’s t test). Furthermore, this inverse correlation was confirmed by expression analysis of both KAISO and MTG16 within the same tumor (Fig. 1A, right, P=0.03, Spearman correlation).
Figure 1.
Kaiso and MTG16 expression are inversely correlated in colon tumorigenesis. A, Kaiso (ZBTB33), and MTG16 (CBFA2T3) expression were queried from The Cancer Genome Atlas colon adenocarcinoma data set and normalized RSEM expression values were log2 transformed for visualization. Average values for Kaiso (left) and MTG16 (middle) in normal colon (n = 39) and colorectal tumors (CRC; n = 264). Matched expression from 90 random CRC samples shows inverse correlation in individual tumors (right). B, Adjacent non-tumor and tumor tissue was isolated from AOM/DSS treated mice (n = 5). Kaiso (left) and MTG16 (middle) expression was analyzed by qRT-PCR and expression normalized to Gapdh. Data represented as fold change over adjacent non-tumor. Correlation between individual tumor samples is shown on the right. C, Kaiso expression was analyzed in Mtg16−/− intestinal crypt isolates (n = 3, left) and AOM/DSS tumor tissue (n = 9, right) as compared to WT samples. D, MTG16 was knocked down using 2 independent shRNA constructs in HCT116 cells (left) and Kaiso expression (right) was queried by real-time PCR. Expression was normalized to GAPDH and represented as fold change over a nontargeted shRNA control. *P<0.05, **P<0.01, ****P<0.0001; Student’s t test (A & B, left and middle panels, C), Spearman correlation (A & B, right panels), or one-way ANOVA with Tukey’s correction (D).
Previously, we have observed MTG16 downregulation in tumors from the AOM/DSS CAC model, yet Kaiso expression in this model has not yet been reported (21). Therefore, we next assessed Kaiso (Zbtb33) and MTG16 (Cbfa2t3) expression to determine if the trends observed in sporadic CRC were maintained in inflammatory tumorigenesis. Indeed, increased Kaiso (Adjacent: 1±0.09, Tumor: 1.54±0.15, P<0.05, Student’s t test) and decreased MTG16 expression (Adjacent: 0.99±0.16, Tumor: 0.48±0.09, P<0.05, Student’s t test) were seen in AOM/DSS-derived colon tumors as compared to adjacent non-tumor tissue (Fig. 1B). While not statistically significant, a trend toward inverse correlation of Kaiso and MTG16 expression was also observed in the AOM/DSS tumor samples.
As inverse correlation with Kaiso and MTG16 was observed in both human and mouse tumor models, we next investigated whether absence of MTG16 modifies Kaiso expression in the gut. Epithelial crypts were harvested from WT or Mtg16−/− mice and tested for Kaiso expression by quantitative reverse transcription PCR (qRT-PCR). Here, we determined that loss of MTG16 augmented Kaiso expression (WT: 1±0.06, Mtg16−/−: 1.6±0.06, P<0.01 Student’s t test, Fig. 1C). Similarly, Kaiso was upregulated in AOM/DSS-induced tumors from Mtg16−/− mice as compared to WT tumors (WT: 1±0.09, Mtg16−/−: 1.4±0.1, P<0.05, Student’s t test). These results were further confirmed in the HCT116 colorectal cancer cell line, and increased Kaiso expression was observed following MTG16 knockdown (KD) using 2 independent shRNA constructs (Fig. 1D). Conversely, MTG16 expression was not affected by Kaiso loss (data not shown). Taken together, these data suggest that while MTG16 and Kaiso are known to interact at the protein level, there may also be genetic interaction as MTG16 contributes to Kaiso mRNA regulation, which may mediate the inverse expression observed in intestinal tumors.
Kaiso is required for enhanced mucosal injury in Mtg16−/− mice after AOM/DSS treatment
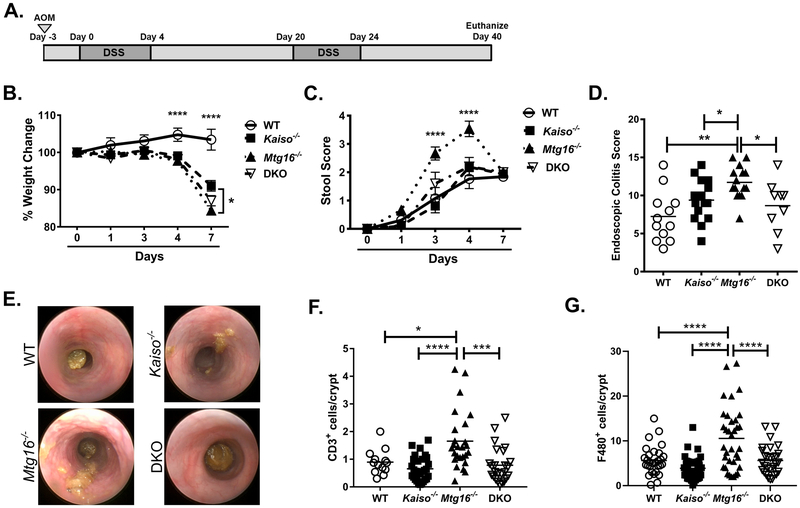
We have recently determined that Mtg16−/− mice demonstrate increased mucosal injury in response to the AOM/DSS protocol, resulting in increased weight loss, stool scores, and morbidity (21). We next aimed to determine whether these phenotypes were dependent on Kaiso expression and whether the inverse MTG16:Kaiso gene expression was relevant to inflammatory tumor formation (12). Cohorts of WT, Mtg16−/−, Kaiso−/−, and double knockout (DKO) mice were treated with 10 mg/kg AOM to initiate DNA damage followed by cyclical administration of 2% DSS for five days, followed by 16 days of recovery (Fig. 2A). Absence of MTG16 resulted in exacerbated injury as evidenced by increased weight loss (P<0.0001 2-way ANOVA vs. WT at day 4 and day 7, Fig. 2B) and increased stool scores, which take into account stool consistency and presence of frank blood (P<0.0001 vs. all groups at day 3 and day 4, 2-way ANOVA, Fig. 2C), both of which were most prominent during the second cycle of DSS administration. In support of these clinical metrics, endoscopy performed at day 29 demonstrated an increase in colitis severity as measured by the murine endoscopic index of colitis severity (MEICS) scoring system (WT: 7.3±1.0, Mtg16−/−: 11.7±0.6, P<0.01, Fig. 2D & 2E) (22). Interestingly, both Kaiso−/− and DKO mice also displayed significantly more weight loss than WT mice in response to DSS, although to a lesser extent than that observed in Mtg16−/− mice. However, neither Kaiso−/− nor DKO mice showed increased stool scores or endoscopic injury indices above that of WT mice (Kaiso−/−: 9.4±0.7, DKO: 8.7±1.1, Fig. 2D, Supplementary Fig. 1).
Figure 2.
Kaiso knockout rescues Mtg16−/− injury phenotypes observed in response to the AOM/DSS protocol. A, Schematic of AOM/DSS inflammatory carcinogenesis protocol. B, Percent weight change during the second DSS cycle. WT weights shown as significance vs. Mtg16−/− at day 4 post-DSS administration (day 4: WT vs. Kaiso−/− **P<0.01; WT vs. DKO ***P<0.001). Significance at day 7 equivalent between WT and remaining genotypes. C, Stool scores during the second DSS cycle as determined using the scoring system described in the “materials and methods” section. Mtg16−/− significance shown against WT and Kaiso−/− at day 3 post-DSS administration (day 3 Mtg16−/− vs. DKO, **P<0.01) and equally significant against all groups at day 4 post-DSS. D, Endoscopic colitis scoring (MEICS scale) determined at day 29 of the AOM/DSS protocol. E, Representative endoscopy images. F, Swiss-rolled colons were stained with antibodies against CD3 to mark T-lymphocytes. Number of positive stromal cells were normalized to the number of crypts per image. G, Swiss-rolled colons were labeled with antibodies against F4/80 to mark macrophages. Number of positive stromal cells were normalized to the number of crypts per image. n = 14 WT, 14 Kaiso−/−, 15 Mtg16−/−, 8 DKO for all in vivo experiments. IHC staining was quantified from 4 fields per mouse, n = 7 mice per genotype. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001; two-way ANOVA with Tukey’s correction (B & C) or one-way ANOVA with Tukey’s correction (D, F, & G).
To complement the stool and endoscopy scores and to determine whether the increased injury is still present post-recovery, we assessed histology of tissue collected at necropsy. Although little crypt damage was observed in any genotype after the 16-day recovery period (data not shown), immunohistochemical (IHC) staining of Mtg16−/− colon tissue demonstrated increased infiltration of T-lymphocytes (WT: 0.9±0.1 CD3+ cells/crypt; Mtg16−/−:1.7±0.2, P<0.05, Fig. 2F) and macrophages (WT: 5.6±0.6 F4/80+ cells/crypt; Mtg16−/−: 10.6±1.2, P<0.0001, Fig. 2G) as compared to WT mice. Infiltration of both immune cell types was equivalent to WT levels in the DKO mice (CD3: 0.8±0.1, P<0.001; F4/80: 5.6±0.6, P<0.0001 vs. Mtg16−/−). Thus, as Kaiso−/−;Mtg16−/− DKO mice demonstrated attenuated injury and inflammatory responses as compared to Mtg16−/− mice, these data suggest that the presence of Kaiso contributes to the heightened injury observed in the Mtg16−/− mice.
Kaiso loss suppresses MTG16-dependent effects in adjacent normal tissue
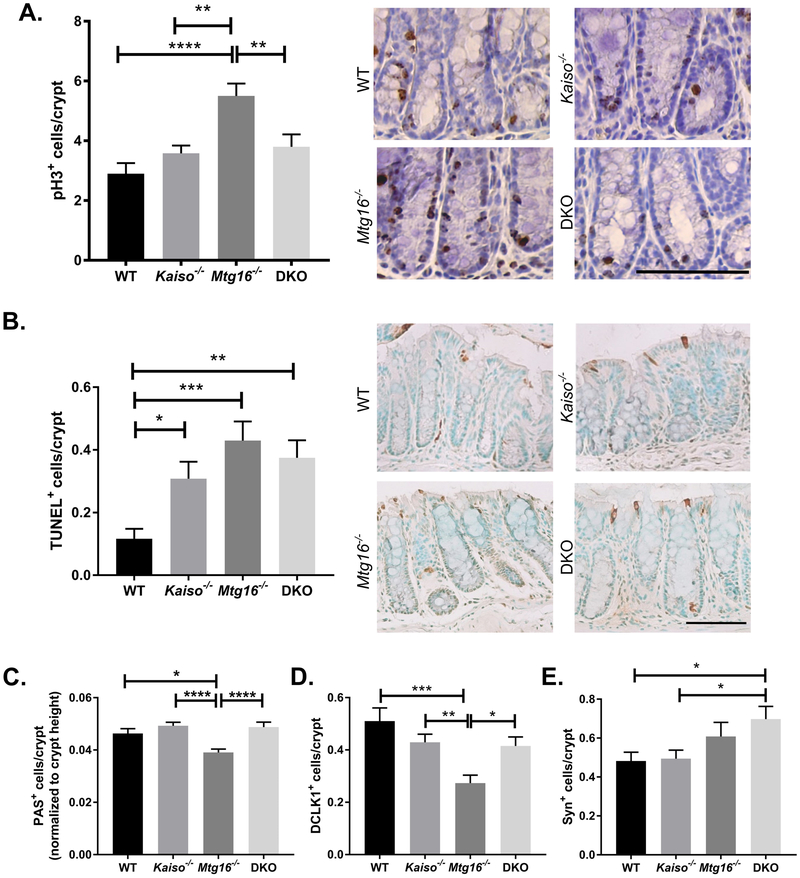
We have previously determined Mtg16−/− intestinal epithelium displays altered proliferation, apoptosis, and secretory lineage allocation at baseline and in the setting of AOM/DSS (21, 23, 24). As these phenotypes likely contribute to the augmented injury response observed in Mtg16−/− AOM/DSS-treated mice, we next analyzed the effect of concurrent MTG16;Kaiso loss in colonic tissue at the conclusion of the AOM/DSS protocol (Fig. 3). In agreement with previous results, Mtg16−/− mice displayed increased proliferation in intestinal epithelial cells as compared to WT mice as measured by IHC staining for phospho-histone H3 (WT: 2.9±0.3 pH3+ cells per crypt, Mtg16−/−: 5.5±0.4, P<0.0001, one-way ANOVA), while the number of proliferative colonocytes in Kaiso−/− tissue was equivalent to that seen in the WT cohort (3.6±0.3, Fig. 3A). However, combined loss of Kaiso and MTG16 in colitic AOM/DSS-treated tissue abrogated the proliferative response observed in Mtg16−/− mice (DKO: 3.8±0.4, P<0.01 vs. Mtg16−/−). This lack of proliferation in DKO mice was not offset by a reduction in apoptosis, as numbers of apoptotic epithelial cells were slightly increased over WT levels in Kaiso−/−, Mtg16−/−, and DKO mice (Fig. 3B).
Figure 3.
Kaiso knockout rescues MTG16-dependent changes in adjacent normal tissue. A, Proliferation was determined in non-tumor tissue at the conclusion of the AOM/DSS protocol by phospho histone-H3 (pH3) immunohistochemistry. Quantification of pH3+ cells/crypt (left) and representative images of each genotype (right). B, TUNEL staining was performed to identify apoptotic cells. TUNEL+ cells/crypt (left) and representative images of each genotype (right). C, Goblet cells were identified by Periodic Acid Schiff (PAS) staining. Crypt height (in pixels) was calculated in Image J software and number of PAS+ cells was normalized to crypt height. D, Tuft cells were identified by DCLK1 staining and quantified as the number of DCLK1+ cells per crypt. E, Enteroendocrine cells were labeled with antibodies against synaptophysin (Syn). Quantification represented as Syn+ cells per crypt. Scale bar = 100μm. n≥10 crypts per mouse, 5 mice per genotype per analysis. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001; one-way ANOVA with Tukey’s correction.
Goblet cells are reduced in the Mtg16−/− small intestine, which secrete mucin to maintain a protective barrier between the intestinal epithelium and gut lumen (24). Importantly, goblet cell loss or dysregulation is believed to both result from intestinal inflammation as well as contribute to the pathology of ulcerative colitis (25–27). To determine whether the goblet cell depletion in the small intestine was also observed in the setting of AOM/DSS-induced colitis in Mtg16−/− mice, we analyzed Periodic acid–Schiff (PAS) staining (Fig. 3C). Indeed, MTG16 loss led to a decrease in PAS positive cells as compared to both WT and Kaiso−/− mice (Mtg16−/−: 0.035±0.001; WT: 0.046±0.002; Kaiso−/−: 0.049±0.001, P<0.0001, one-way ANOVA). As seen with proliferation, goblet cell numbers were also restored to WT levels in the DKO mice (0.045±0.002). We next analyzed additional secretory cell types in the colon. We first used DCLK1 to label tuft cells, a population of chemosensory cells most commonly associated with the secretory lineage and recently determined to protect against intestinal injury (Fig. 3D). Again, DCLK1+ cells were reduced in the Mtg16−/− colons (WT: 0.53±0.05 cells/crypt; Mtg16−/−: 0.29±0.03, P<0.001, one-way ANOVA), yet restored to WT levels in the DKO samples (DKO: 0.42±0.03, P<0.01 vs. Mtg16−/−, one-way ANOVA). Finally, enteroendocrine (EE) cells were assessed by synaptophysin IHC (Fig. 3E). Here, MTG16 loss did not affect EE cell number, in agreement with previous analysis of this cell population in the Mtg16−/− small intestine (24). However, concurrent loss of Kaiso and MTG16 increased EE cell number above both WT and Kaiso−/− cohorts (WT: 0.48±0.04 cells/crypt; Kaiso−/−: 0.49±0.04; DKO: 0.7±0.06, P<0.05, one-way ANOVA). Together, these data suggest Kaiso is required to mediate numerous pro-injury phenotypes induced by MTG16 absence and that Kaiso loss widely promotes differentiation of protective secretory cells in Mtg16−/− mice.
Kaiso loss modifies Mtg16−/−-dependent increases in tumorigenesis
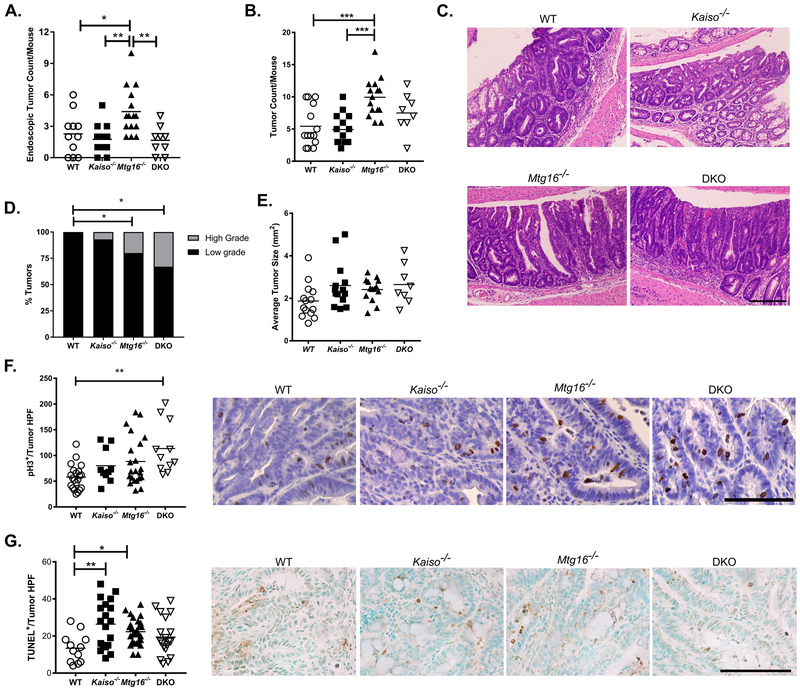
We next evaluated tumor burden induced by the AOM/DSS protocol. As shown in Figure 4A, tumor development tracked very closely with severity of injury when assessed by endoscopy at day 29, with Mtg16−/− mice displaying significantly higher tumor number (4.4±0.6 tumors/mouse) than WT and Kaiso−/− mice (2.3±0.6 and 1.8±0.4 tumors/mouse, respectively). Interestingly, as observed in the injury assessments above, the pro-tumorigenic effect of MTG16 loss was attenuated by concurrent Kaiso deletion (1.7±0.4 tumors/mouse, P<0.01 vs. Mtg16−/−, one-way ANOVA). Consistent with the endoscopy tumor scoring, upon sacrifice, Mtg16−/− mice again showed greater tumor number (9.9±0.8 tumors/mouse, P<0.001, one-way ANOVA) as compared to both the WT (5.4±0.8 tumors/mouse) and Kaiso−/− mice (4.9±0.6 tumors/mouse, Fig. 4B). The tumor number in DKO mice was reduced (7.5±1.1 tumors/mouse), but not statistically different from Mtg16−/− mice, suggesting that concurrent Kaiso-loss partially rescued the pro-tumorigenic phenotype observed in the absence of MTG16. In support of this hypothesis, tumor number in the DKO mice was similar to that observed in WT mice. Together, these data suggest that, while Kaiso has little impact on inflammatory tumorigenesis alone, its expression likely contributes to the increased tumor burden observed in Mtg16−/− mice.
Figure 4.
Concurrent Kaiso loss modifies Mtg16−/− tumor number. A, Tumor number quantified from endoscopy videos at day 29 of the AOM/DSS protocol. B, Number of tumors quantified grossly at sacrifice. C, Representative tumor hematoxylin & eosin images from WT, Mtg16−/−, Kaiso−/−, and DKO mice (20x magnification). Scale bar = 200μm. D, Percentage of mice with high- and low-grade dysplasia as assessed by experienced pathologist. E, Size measurements of WT, Kaiso−/−, Mtg16−/−, or DKO tumors. F, Proliferation was determined by phospho histone-H3 (pH3) immunohistochemistry. Quantification of pH3+ cells/tumor high-powered field (HPF, 40x; left) and representative images of each genotype (right). Scale bar = 100μm. G, TUNEL staining was performed to identify apoptotic cells. TUNEL+ cells/tumor high-powered field (HPF; left) and representative images of each genotype (right). Scale bar = 100μm. n≥10 tumors per genotype per analysis. *P<0.05, **P<0.01, ***P<0.001; one-way ANOVA with Tukey’s correction (A, B, E, F, & G) or Fisher’s exact test (D).
Combined loss of Kaiso and MTG16 increases tumor dysplasia and proliferation
As we observed that the increased proliferation in colitic epithelium and tumor multiplicity in Mtg16−/− mice was partially rescued with concurrent loss of Kaiso, we next analyzed tumor grade and tumor cell proliferation to determine if these metrics were also affected by Kaiso expression. We previously identified significantly higher numbers of tumors with high-grade dysplasia in AOM/DSS treated Mtg16−/− mice, and this was again observed (WT: 0 out of 20 tumors, Mtg16−/−: 5 out of 25 tumors, P<0.05, Fisher’s exact test, Fig. 4C & 4D) (21). Interestingly, a similarly high proportion of tumors with high-grade dysplasia were seen in the DKO cohort (4 out of 16 tumors, P<0.01 vs. WT, Fisher’s exact test). Tumor size, however, was unchanged between the genotypes (WT: 1.8±0.12 mm2, Kaiso−/−: 2.6±0.28, Mtg16−/−: 2.4±1.4, DKO: 2.6±0.33mm2, Fig. 4E). Thus, after tumor establishment, Kaiso loss has little protective effect in the context of MTG16 knock-out (KO).
We next assessed tumors for proliferation via phospho histone-H3 IHC and apoptosis by TUNEL analysis. While proliferation was modestly increased in Mtg16−/− tumors (WT: 58±5.3 pH3+ cells per HPF, Mtg16−/−; 86±10.6, P=0.06, one-way ANOVA, Fig. 4F), the highest proliferation rate was observed in tumors from DKO mice (113±17, P<0.01 vs. WT, one-way ANOVA). Apoptosis, on the other hand, was increased in both Kaiso−/− and Mtg16−/− tumors as compared to those from WT mice (WT: 12.5±2.3 TUNEL+ cells per HPF, Kaiso−/−: 25.7±3.5, Mtg16−/−: 20.9±1.2, Fig. 4G). However, this metric was unchanged in the DKO cohort (15.3±2.7) as compared to WT. Interestingly, as tumors from the DKO mice have the highest proliferation and lowest apoptosis rates without a change in the average tumor size, tumor initiation may be delayed in DKO mice as previously reported for ApcMin/+;Kaiso−/− mice (19). This is also supported by a larger change in tumor number observed between the Mtg16−/− and DKO cohorts at the earlier endoscopic timepoint (Fig. 4A) than at sacrifice (Fig. 4B). Taken together, these data suggest that the reduced tumor burden in DKO mice is likely attributed to decreased proliferation and epithelial damage in the adjacent non-tumor colon tissue, which ultimately attenuate tumor initiation.
Kaiso DKO restores MTG16-dependent transcriptional changes
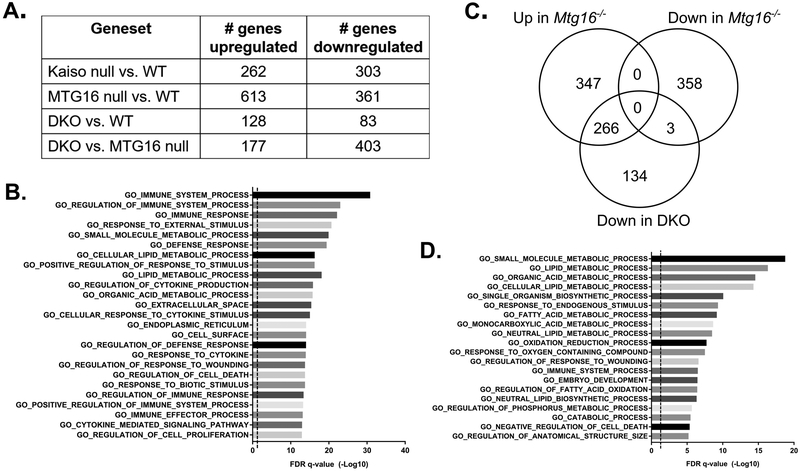
Kaiso and MTG16 both function as transcriptional repressors. Therefore, we determined the transcriptional changes that may decrease intestinal inflammation and inflammatory tumorigenesis in DKO mice. As the functional changes most likely to impact tumor initiation were observed in the colitic non-tumor epithelium (i.e., proliferation, secretory cell number), we analyzed samples of WT, Kaiso−/−, Mtg16−/−, and DKO adjacent non-tumor mucosa by RNA-sequencing. Following analysis, the number of statistically significant genes altered at least 1.5-fold are listed in Figure 5A. Interestingly, while Mtg16−/− samples displayed a large number of upregulated genes (consistent with its role as a transcriptional repressor), DKO mice displayed the lowest number of altered genes as compared to WT (DKO vs. WT: 211 genes). However, robust transcriptional changes were still seen in DKO colons as compared to those from Mtg16−/− mice, with nearly 3 times the number of statistically significant differentially expressed genes as those versus WT (DKO vs. Mtg16−/−: 580).
Figure 5.
MTG16:Kaiso double knockout alters immune and metabolism-associated genes. A, Adjacent normal colon samples were analyzed by RNA-sequencing. Table shows numbers of statistically differentially expressed genes of at least 1.5-fold change between each genotype. B, Gene set overlap against Gene Ontology (GO) curated data sets was performed using the Molecular Signature Database v6.1 software available from the Broad Institute. Graph illustrates 25 of the most statistically enriched datasets in the genes downregulated in DKO mice as compared to Mtg16−/−. Significance is represented as the −Log10 of FDR q-values. Dotted line indicates FDR q-value of 0.05. C, Venn diagram showing the numbers of genes altered in Mtg16−/− samples as compared to WT and whether these genes are downregulated in the DKO samples as compared to Mtg16−/−. D, Gene set overlap showing genes in metabolism and cell growth GO datasets are preferentially downregulated in the 266 genes that were upregulated in Mtg16−/− and downregulated in DKO mice.
We first focused on the genes that were downregulated in the DKO mice as compared to Mtg16−/−. In line with decreased colitis and endoscopy scores (Fig. 2), these 403 genes were found to have significant overlap with several immune- and cytokine-related Gene Ontology (GO) data sets using the Molecular Signatures Database (Fig. 5B) (28, 29). Together, these indicate an attenuated immune and cytokine response and are congruent with reports that Kaiso overexpression induces inflammation in the intestine (17, 18). Indeed, Kaiso loss alone highly downregulated inflammatory pathways, although this downregulation had no apparent effect on colitis severity or CAC tumorigenesis (Supplemental Fig. S2).
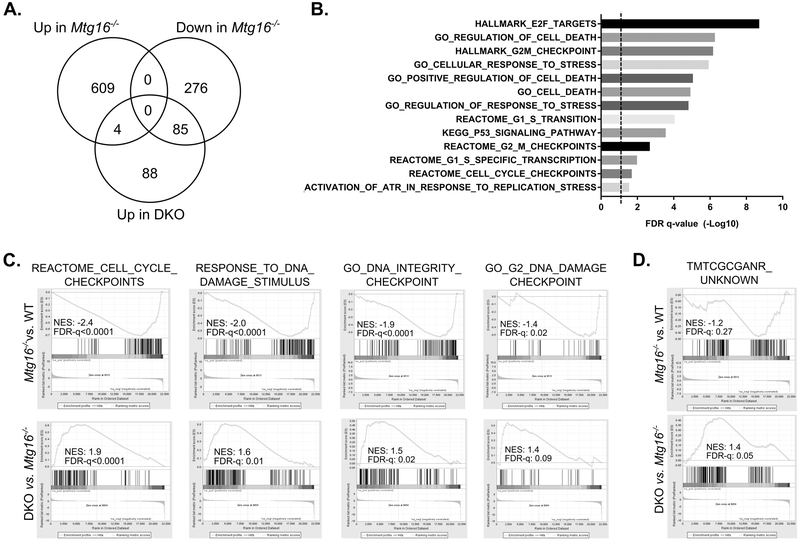
We next investigated whether the specific genes altered by MTG16 KO were modified by concurrent Kaiso loss. Of the 613 genes upregulated in the setting of MTG16 loss, nearly half (43%) were significantly downregulated in the DKO colons as compared to colons from Mtg16−/− mice (Fig. 5C). Analysis of gene set overlaps with these 266 genes determined enrichment for genes regulating various metabolic processes, immune response, cell death, wounding, and tissue development, all of which may help mediate the decreased proliferative response observed in DKO mice (Fig. 5D). Furthermore, the effect of concurrent MTG16 and Kaiso loss was rarely synergistic, and DKO led to further decreases in transcription in less than 1% of the MTG16-downregulated genes. Heighted upregulation of MTG16-regulated genes by DKO was similarly rare, and instead almost half of the genes upregulated in DKO samples (as compared to Mtg16−/−) were downregulated by loss of MTG16 alone (Fig. 6A). Gene set overlap of these 85 genes identified enrichment for cell death and cell cycle checkpoint-related genes (Fig. 6B), suggesting that DKO mice may have an enhanced apoptotic response to cell stress and DNA damage. Inverse expression changes of these gene sets were further confirmed by gene set enrichment analysis (GSEA), which demonstrate a significant downregulation in Mtg16−/− colonic tissue that is restored in DKO mice (Fig. 6C). Importantly, loss of cell cycle checkpoints can favor tumor initiation (e.g., loss of TP53), rendering cells unable to clear initiated/mutated cells (30–32). Together, these data suggest that Kaiso loss, in the setting of MTG16 KO, may attenuate tumor initiation by attenuating inflammation and metabolism while augmenting DNA-damage checkpoint pathways.
Figure 6.
Concurrent Kaiso and MTG16 KO activates cell death and DNA damage checkpoint pathways. A, Venn diagram showing the numbers of genes altered in Mtg16−/− samples as compared to WT and whether these genes are upregulated in the DKO samples as compared to Mtg16−/−. B, Gene set overlap with DNA damage and stress pathway datasets in the 85 genes that were downregulated in Mtg16−/− and upregulated in DKO mice. Significance is represented as the −Log10 of FDR q-values. Dotted line indicates FDR q-value of 0.05. C, Preranked gene set enrichment analysis (GSEA) shows negative enrichment (normalized enrichment score, NES) of cell cycle and DNA damage checkpoint pathways in Mtg16−/− samples (top) and positive enrichment in DKO samples (bottom). D, Preranked GSEA of the “TMTCGCANR” motif gene set coregulated by Kaiso and SMRT. A trend toward negative enrichment was observed in Mtg16−/− samples (top) and positive enrichment in DKO samples (bottom).
Finally, while MTG16 co-represses Kaiso targets containing a KBS motif, Kaiso has been proposed to bind DNA sequences independently of its interaction with MTG16. One such motif, “TMTCGCARN,” was identified as a Kaiso binding site, which it coregulates with the repressor SMRT (33, 34). We therefore determined whether expression of these genes was altered in the setting of Mtg16−/−and/or DKO samples by GSEA (Fig. 6D). Indeed, expression of these genes trended towards downregulation in Mtg16−/− samples, suggesting that loss of the Kaiso:MTG16 interaction frees Kaiso from KBS-containing genes and accentuated repression of these targets. Furthermore, loss of repression was observed with concurrent MTG16 and Kaiso loss. Thus, MTG16 loss appears to deregulate MTG-independent Kaiso transcription, which may also affect colitis and CAC development.
Kaiso loss attenuates Wnt-associated phenotypes in intestinal epithelial cells
In addition to cell-cycle checkpoint genes and TMTCGCARN-associated genes, we have previously determined that increased proliferation and tumorigenesis in Mtg16−/− mice is likely due to increased transcription of WNT target genes (21). We next utilized the intestinal enteroid model to test whether concurrent loss of MTG16 and Kaiso modifies WNT pathway activation in intestinal epithelial cells (21, 23, 35). To directly assess whether Kaiso loss abrogates WNT-associated phenotypes in Mtg16−/− cells, enteroid lines were established from WT and Mtg16−/− mice and Kaiso expression was modified by lentiviral infection of shRNA targeted to murine Zbtb33. Control lines were established by expression of a nontargeted shRNA construct.
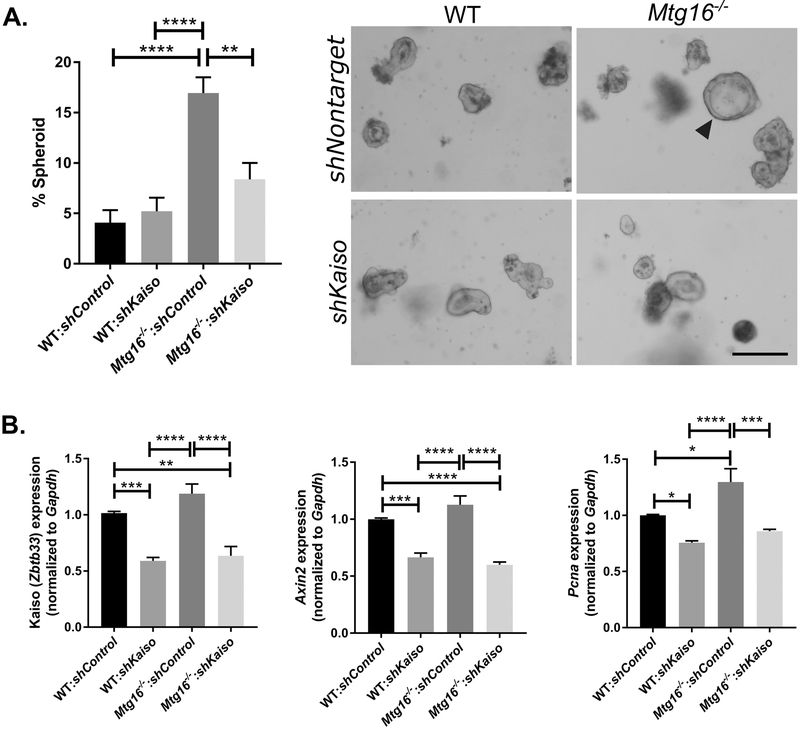
After lentiviral modification, we next assessed WNT-associated phenotypes in the modified enteroid lines. Cystic, spheroid morphology is associated with increased WNT-tone and an undifferentiated state, as enteroids grown in high-WNT conditions can maintain this morphology without progression to crypt budding, evidence for differentiation (36, 37). In line with previous results, loss of MTG16 led to increases in spheroid morphology in enteroids at one day post-split as compared to WT:shcontrol and WT:shKaiso cultures (WT:shcontrol: 4.1±1.2%; WT:shKaiso: 5.2±1.3%; Mtg16−/−:shcontrol: 16.9±1.6%, P<0.0001, one-way ANOVA, Fig. 7A) (24). Interestingly, spheroid numbers were reduced in Mtg16−/− lines following Kaiso KD, and these results were not statistically different than that seen in either WT;shcontrol or WT:shKaiso lines (Mtg16−/−:shKaiso: 8.4±1.6%, P<0.01 vs. Mtg16−/−:shcontrol). This reduction was particularly striking considering that overall levels of Kaiso were only reduced by approximately 50% in both WT and Mtg16−/− enteroid lines (Fig. 7B). In addition to spheroid morphology, qRT-PCR analysis indicated slight increases in the canonical WNT target gene, Axin2, and the proliferation-associated gene, Pcna, in Mtg16−/−:shControl lines. However, Kaiso KD led to robust decreases in these genes in both WT and Mtg16−/− enteroid lines (Fig. 7B). Thus, in line with the decreased proliferation observed in the colon following AOM/DSS treatment, these results indicate that Kaiso expression can modify WNT signaling, which likely decreases injury and tumorigenesis-associated phenotypes in the Mtg16−/− mice.
Figure 7.
Kaiso loss decreases stemness and WNT tone in WT and Mtg16−/− enteroids. A, A nontargeted or Kaiso-targeted shRNA was transduced into enteroids established from WT and Mtg16−/− mice. Spheroid morphology was assessed at one day post passage and normalized to total enteroid number (left). Representative images (right) show spheroid morphology marked by arrowhead. n=4 wells/genotype per 2 independently infected enteroid lines. Scale bar = 200μM. B, Control and knockdown enteroid lines were collected for RNA and analyzed for Kaiso (left), Axin2 (middle), and Pcna (right) expression by qRT-PCR analysis. n= 2 independently infected enteroid lines, samples run in triplicate. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001; one-way ANOVA with Tukey’s correction.
Discussion
MTG16 is a transcriptional co-repressor and serves as a scaffolding protein upon which repressor complexes are nucleated (12). We have previously determined that MTG16 regulates stem cell function in the intestine, and its absence increases proliferation and WNT pathway activation (23, 24). Furthermore, MTG16 protects against intestinal injury in response to DSS and functions as a tumor suppressor in the context of AOM/DSS inflammatory tumorigenesis (21, 23). Like MTG16, Kaiso has been widely considered to be a tumor suppressor and has been shown to inhibit WNT signaling by direct and indirect means (2, 7, 8, 38). However, in vivo studies are instead more consistent with a tumor promoting role, as Kaiso−/− mice display resistance to intestinal polyposis in the ApcMin/+ sporadic tumor model, while Kaiso overexpression in the ApcMin/+ model accelerated tumorigenesis (17, 19).
Kaiso is upregulated in human tumor tissue and recently generated Kaiso overexpressing mice have increased inflammatory signaling both at baseline and in the context of ApcMin/+-induced tumorigenesis (17, 18). Despite these pro-inflammatory and pro-tumorigenic phenotypes, we observed that the absence of Kaiso alone had no effect on colitis severity or development of inflammatory tumorigenesis, and Kaiso−/− tumors were of a similar size, grade, and proliferative rate as those from WT mice. While we did observe widespread downregulation of immune and cytokine-related gene sets in Kaiso−/− colons, as well as robust decreases in WNT-associated genes in the enteroid culture system, this was apparently insufficient to modify tumor formation in the AOM/DSS model. Thus, these data suggest that loss of Kaiso alone has no overt effect on inflammatory tumorigenesis of the colon.
Although Kaiso loss did not attenuate tumorigenesis by itself, Kaiso appeared to modify colitis and tumorigenesis when in the context of MTG16 KO. Interestingly, the DKO tumors were similar to those generated in the Mtg16−/− cohort, with increased rates of high-grade dysplasia, augmented proliferation, and decreased apoptosis. Thus, it is unlikely that the protective effect observed in the DKO mice is mediated by tumoral responses and, instead, Kaiso appeared to preferentially mediate injury responses in response to colitis in the adjacent non-tumor epithelium. Furthermore, transcriptomic analysis indicates activation of cell death and DNA damage checkpoint pathways in response to stress in DKO mice. Taken together, these data suggest concurrent MTG16 and Kaiso loss leads to clearance of initiated tumor cells instead of tumor establishment. Furthermore, the results in intestinal enteroids and decreased proliferation in vivo indicate that the DKO mice have decreased WNT-pathway activation at baseline in comparison to Mtg16−/− mice. Tumor initiation in the intestine largely depends on WNT activation, and data indicates that transformed cells must reach a “threshold” of WNT activity in order to allow for tumor initiation (39–41). This “threshold” is likely more difficult for cells to attain in the setting of Kaiso loss, which may also decrease tumor initiation rates. These results are also consistent with observations using ApcMin/+; Kaiso−/− mice in which Kaiso loss was observed to increase tumor latency and reduce tumor incidence at early timepoints (19). Indeed, the DKO tumors were observed to be a similar size as those from other genotypes despite the heightened proliferation and decreased apoptosis, which would lead to greater tumor size unless latency were increased.
It is worth noting that increased proliferation and WNT-tone is observed in Mtg16−/− mice, which one might expect to accelerate wound healing and decrease overall injury. Indeed, inhibition of the WNT antagonist, Dkk1, leads to faster regeneration following DSS administration (42). However, this inverse effect between proliferation and injury has been consistently observed in Mtg16−/− mice, and similar results are observed following loss of its family member, MTGR1 (21, 23, 43). Further, while stem cells are necessary to repopulate the intestine, proper differentiation is also key to maintaining the intestinal barrier. Indeed, our data demonstrates that loss of MTG16 depletes protective secretory goblet and tuft cells, and previous studies have determined MTG16 loss decreases barrier function in the colon (23). Thus, MTG proteins have multifaceted roles in the intestine, and additional effects on homeostasis and differentiation may offset protection afforded by increased WNT pathway activation.
While these effects were noted in intestinal epithelial cells, both Mtg16−/− and Kaiso−/− lines are global KO models, thus it is possible that alterations in immune cell populations also contribute to the protective effect observed in DKO mice. However, current evidence supports a larger role for epithelial cells and suggest that altered inflammation is likely in response to epithelial-intrinsic changes. For example, transplant of WT bone marrow did not alter response of Mtg16−/− mice to either DSS-induced colitis or AOM/DSS (21, 23). Furthermore, while increased inflammation is observed in the gut following Kaiso overexpression, transgenic Kaiso expression in this model is obtained using the epithelial-specific Villin promoter (17, 18). Thus, the increased inflammation is ultimately an epithelial-cell driven phenotype. Finally, despite large reductions in immune-associated genes, Kaiso loss alone had no apparent effect on intestinal injury or tumorigenesis in the AOM/DSS model. However, the exact contribution of epithelial and immune cell subsets in the DKO model is likely complex, and this interaction remains to be elucidated in future investigations.
Though Kaiso and MTG16 share a subset of targets, both proteins have multiple binding partners and non-overlapping target genes. Kaiso has been shown to bind both specific consensus sequences as well as methylated DNA, yet our previous research demonstrated that Kaiso methylation-dependent binding did not require MTG16 (12). Thus, it is possible that different methylation-specific targets mediate Kaiso’s effect on tumorigenesis in the context of MTG16 deletion. Unfortunately, the data herein do not distinguish whether a subset of Kaiso-exclusive non-overlapping targets mask the MTG16 phenotype upon loss of repression, or whether the Mtg16−/− phenotype is ultimately driven by deregulation of Kaiso target genes, as loss of MTG16 may free Kaiso to bind additional proteins and/or promoters. However, previous research has determined that Kaiso binds a previously “orphan” transcription factor binding motif in complex with SMRT and that, in vitro, Kaiso’s affinity for this methylated palindromic sequence is higher than its affinity for the KBS (33). GSEA identified downregulation of these motif-containing genes in Mtg16−/− samples and upregulation in DKO samples, suggesting 1) that Kaiso regulates these genes independently of MTG16 and 2) that de-repression of these genes may contribute to the protective effect observed in the DKO mice. Furthermore, recent data has also indicated that Kaiso may not solely serve a repressive function, and Kaiso has been reported to associate predominantly with unmethylated, highly active promoters marked with high levels of acetylated histones (44). Thus, one interesting hypothesis is that MTG16 loss may free Kaiso from a repressive function, allowing it to positively regulate genes which may contribute to tumorigenesis. These studies, particularly in the context of the intestine, would be an interesting topic for future research.
In summary, Mtg16−/− and Kaiso−/− mice display very different baseline and, as reported here, AOM/DSS inflammatory carcinogenesis phenotypes. While we have previously determined that absence of MTG16 heightens colitis severity and increases tumor number, these studies are the first to analyze the effect of Kaiso loss in the AOM/DSS system. Interestingly, despite downregulation of cytokine signaling at the transcriptional level, we observed no overt effect on inflammatory tumor development. Further, we were surprised to observe that Kaiso deficiency can rescue the Mtg16−/− phenotype, likely by dampening proliferative responses to injury, modifying the immune response, activating cell death and DNA checkpoint pathways, and attenuated WNT pathway activation. Thus, Kaiso deregulation may be necessary to induce MTG16-dependent changes and suggest that the inverse correlation of KAISO and MTG16 expression observed in human tumors is functionally relevant to tumor initiation and development.
Materials and methods
Murine inflammatory carcinogenesis protocol
Equal numbers of male and female 8–12 week old C57BL/6 wild type (WT) (n=14), Mtg16−/− (n=15), Kaiso−/− (n=14), or Kaiso−/−;Mtg16−/− (DKO, n=8) were used. Cohort numbers were chosen in accordance with previous studies (21). Genotypes were housed individually due to ensure equivalent water/DSS consumption and thus were not randomized. For inflammatory tumorigenesis, mice were injected with 10 mg/kg of azoxymethane (Sigma-Aldrich, St. Louis, MO, USA) intraperitoneally. Three days post-injection, the animals were started on the first of two cycles of 2% DSS ad libitum (see schematic in Fig. 2A). Each cycle lasted 5 days and was followed by a 16-day recovery period. This abbreviated dosing strategy and timepoint were chosen in accordance with previous data indicating Mtg16−/− mice have ~80% mortality in response to an AOM/DSS protocol of 3 cycles of 3% DSS (21). Weights were obtained and reported as the percentage of the original weight one day before DSS administration. In addition, stools were examined for consistency and the presence of blood during the treatment period: normal stools = 0 points, loose stool = 1 point, diarrhea = 2 points, presence of blood = 2 points, and excessive blood = 4 points. During the second cycle of recovery (day 29), colonoscopy (Karl Storz veterinary endoscopy, Goleta, CA, USA) was performed to assess injury and tumor development. Injury and tumor number was evaluated by blinded observers based on the murine endoscopic index of colitis severity (MEICS) that includes 0–3 scoring of thickening of the colon, changes in vasculature, presence of granularity, presence of exudate, and stool consistency (22, 45). Mice were sacrificed on day 40 and colonic tumors were counted macroscopically and measured using calipers. Sections of tumor and normal adjacent colon were collected and stored in RNAlater (Qiagen, Venlo, Netherlands). The remainder of the colon was “Swiss rolled”, formalin fixed overnight, and sectioned for histological analysis. Histological analysis was performed by experienced pathologist blinded to animal genotypes. All in vivo procedures were carried out in accordance with protocols approved by the Vanderbilt Institutional Animal Care and Use Committee.
Cell Culture and shRNA-mediated knockdown
HCT116 cells were purchased from ATCC and authenticated by STR profiling prior to experimentation (ATCC, Manassus, VA, USA). Cells were grown in McCoy’s 5A medium (Gibco, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum (VWR, Radnor, PA, USA), 100 U/ml penicillin, and 100 μg/ml streptomycin and verified to be mycoplasma free (abm, Richmond, BC, Canada). To generate MTG16 knockdown lines, lentivirus was packaged using pMD2.G (a gift from Didier Trono, Addgene plasmid #12259) and psPAX2 (a gift from Didier Trono, Addgene plasmid #12260) vectors. shRNA constructs (clones NM_005187.4–582s1c1 and NM_005187.4–2417s21c1) and a nontargeted scrambled control were purchased in the pLKO.1 lentiviral vector (Sigma-Aldrich, St. Louis, MO, USA).
Enteroids were established as described previously (35, 36, 46). Briefly, 10-cm of the proximal duodenum was harvested from 10-week old littermate WT and Mtg16−/− mice, washed, and minced. Tissue fragments were incubated in chelation buffer (2mM EDTA in PBS) for 30 minutes at 4°C prior to 2 minutes of gentle shaking to free intestinal crypts. Crypts were collected and suspended in growth-factor reduced Matrigel (Corning, Corning, NY, USA) and overlaid with mini-gut culture media (Advanced Dulbecco’s modified Eagle’s medium–F-12 [Gibco], 100 U/ml penicillin, 100 μg/ml streptomycin, 1X N2 [Gibco], 1X B27 [Gibco], 1X Glutamax [Gibco], 1mM HEPES [Gibco], 20% R-spondin conditioned media [generated from R-spondin-expressing cells generously provided by Dr. Jeff Whitsett, Cincinnati Children’s Hospital], 10% Noggin conditioned media [generated from Noggin-expressing cells generously provided by Dr. G.R. van den Brink, described in (47)], and 50ng/ml EGF [R&D Systems, Minneapolis, MN, USA]).
For genetic modification of enteroids, lentivirus was transfected into low-passage 293T cells (ATCC) using the pMD2.G and psPAX2 vectors along with either a nontargeted shRNA control construct or an shRNA construct targeted to murine Kaiso (NM_020256.1–1659s1c1, Sigma-Aldrich). Virus-containing media was collected and concentrated by overnight centrifugation at 8000 rpm. Viral pellets were then resuspended in 250μl complete mini-gut media supplemented with 1μM CHIR-99021 (Tocris, Bristol, UK), 10μM Y-27632 (Tocris), and 8μg/ml polybrene (Millipore, Burlington, MA, USA) and applied to WT and Mtg16−/− enteroid fragments. Virus was spinoculated onto enteroids by centrifugation at 600g for 1 hour, prior to 6-hour incubation at 37°C. Following incubation, infected enteroid fragments were resuspended in Matrigel and overlaid with complete mini-gut media supplemented with 10μM Y-27632 and 1μM CHIR-99021. After antibiotic selection, infected enteroids were split by shearing with a 25-gauge needle and replated. After 24 hours, enteroids were assessed for spheroid morphology and collected for mRNA analysis.
Immunohistochemistry and immunofluorescence staining
Five-micrometer sections of paraffin-embedded colons were cut. Hematoxylin and eosin (H&E) and Periodic Acid Schiff (PAS) staining were performed by the Vanderbilt Translational Pathology Shared Resource according to standard protocols. For IHC analysis, antigen retrieval for pH3, DCLK1, CD3, and F4/80 staining was conducted by 15–20-minute incubation in a 104°C pressure cooker with citrate buffer (pH 6) followed by incubation with serum-free protein block (Dako, Santa Clara, CA, USA). Synaptophysin staining utilized a 20-minute incubation at 97°C with EDTA buffer (pH9). Primary antibodies against synaptophysin (#ab32127, Abcam, Cambridge, UK) and F4/80 (#MCA497R, Bio-Rad) were incubated for 60 minutes at room temperature while primary antibodies against pH3 (#9701, Cell Signaling Technology, Danvers, MA, USA), DCLK1 (#62257, Cell Signaling Technology), and CD3 (#99940, Cell Signaling Technology) were incubated overnight. Staining for all antibodies was visualized with the Dako Envision+ HRP system and DAB. Apoptotic cells were identified with the ApopTag Plus Peroxidase In Situ Apoptosis Kit (Millipore, Burlington, MA, USA) according to the manufacturer’s protocol. Staining indices were generated by counting either the number of positive cells per high-powered field (HPF; 40x objective) within each tumor or the number of positive cells per crypt in at least 10 crypts per mouse by a blinded observer. For the PAS stain, the number of goblet cells counted per crypt was normalized to the crypt height (in pixels) as measured by Image J software. Five-eight mice were used per group per analysis. All imaging and IHC counts were completed in blinded fashion.
RNA analysis
Samples from cell lines, normal mouse intestine, AOM/DSS tumor, and AOM/DSS adjacent normal tissue were homogenized in TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNA was isolated using the RNeasy Mini Kit (Qiagen) and cDNA was prepared using the iScript cDNA synthesis kit (BioRad, Hercules, CA, USA) or qScript XLT cDNA SuperMix (Quntabio, Beverly, MA, USA) with 1–2μg of RNA. qRT-PCR was performed using either PerfeCTa mastermix (Quantabio) and the following primers: Kaiso/Zbtb33 – GAACTCCTTGAATGAACAGCGT, CCCAGCAACTGAGAAGAGC (PrimerBank ID 9937986a1), Axin2 – TGCCCACACTAGGCTGACA, TGACTCTCCTTCCAGATCCCA (PrimerBank ID 31982733a1), Pcna – TTTGAGGCACGCCTGATCC, GGAGACGTGAGACGAGTCCAT (PrimerBank ID 7242171a1), Gapdh – CCGCATCTTCTTGTGCA, CGGCCAAATCCGTTCA, or TaqMan Universal PCR master mix (Thermo Fisher Scientific, Waltham, MA, USA) and probes for MTG16 (mouse Cbfa2t3, Mm00486784_m1; human CBFA2T3, Hs00602520_m1), Kaiso (human ZBTB33, Hs00272725_s1) and Gapdh (mouse, Mm99999915_g1; human, Hs02786624_g1). All qRT-PCR samples were run in triplicate and expression was normalized to Gapdh and represented as fold change over relevant control samples.
For human normal and colorectal cancer RNA expression, Kaiso (ZBTB33) and MTG16 (CBFA2T3) mRNA expression was queried from Illumina HiSeq and Illumina GA RNASeqV2 data in The Cancer Genome Atlas (TCGA) colon adenocarcinoma (COAD) data set (n = 264 CRC, 39 normal colon). Normalized RSEM expression data were log2 transformed for visualization.
RNA-sequencing
RNA from adjacent normal tissue of AOM/DSS treated WT, Kaiso−/−, Mtg16−/−, and DKO mice was isolated using the RNeasy Mini Kit (Qiagen). PE150 RNA-sequencing was performed by Vanderbilt Technologies for Advanced Genomics (VANTAGE) using the NovaSeq6000 (n=3 mice per group). TruSeq adaptors were trimmed by Trimmomatic-0.32 (48). Trimmed paired end reads were aligned to the mouse genome (mm10, downloaded from UCSC) using TopHat (v2.0.11) and differential gene expression was determined by Cuffdiff (v2.1.1) as previously described (49). The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (50) and are accessible through GEO Series accession number GSE12454. Statistical overlap with curated gene sets was determined using the Molecular Signatures Database v6.1, and gene set enrichment analysis (GSEA) of preranked gene sets was performed using GSEA software version 3 maintained by the Broad Institute (28, 29, 34, 51).
Statistical methods
Statistical analysis comparing two or four groups was performed in Graphpad Prism Software using unpaired Student’s t-test (unpaired, two-tailed) or one-way ANOVA and Tukey’s Multiple Comparison post-test, respectively. Samples were excluded if determined to be statistical outliers based on “robust regression and outlier removal” (ROUT) analysis in Graphpad Prism software. Differences in stool scores and percentage weight loss were determined using two-way ANOVA with Tukey’s Multiple Comparison post-test for repeated measurements over time, while dysplasia was analyzed via Fisher’s exact test. Spearman correlation was used to determine correlation. For all studies, center values represent experimental mean, error is represented by standard error of the mean, and P<0.05 is considered significant.
Supplementary Material
Acknowledgements
The authors thank members of the Williams and Reynolds laboratories for thoughtful discussions about this research project. We would also like to thank the Vanderbilt Translational Pathology Shared Resource for aid with histology and the Vanderbilt Technologies for Advanced Genomics for next gen sequencing.
Financial Support: National Institutes of Health (R01DK099204 to CSW; R01AT004821 to KTW; R01CA178030 to SWH; 1F31CA167920 to CWB; F32DK108492 to SPS; P30DK058404 to MKW); the Federal Research Center “Fundamentals of Biotechnology” budget (No 01201371085 to EP); Office of Medical Research, Department of Veterans Affairs (1I01BX001426 to CSW; 1I01BX001453 to KTW; 1IK2BX002126 to LAC)
Competing Interest Statement: The authors declare no competing interest. Financial support includes National Institutes of Health (R01DK099204 to C.W., R01AT004821, to K.W., R01CA178030 to S.H., 1F31CA167920 to C.B., F32DK108492 to S.S.); the Federal Research Center “Fundamentals of Biotechnology” budget (No 01201371085 to E.P.); Office of Medical Research, Department of Veterans Affairs (1I01BX001426 to C.W., 1I01BX001453 to K.W., 1IK2BX002126 to L.C.)
Footnotes
Conflict of Interest Statement: The authors declare no conflict of interest.
References
- 1.Daniel JM, Reynolds AB. The catenin p120(ctn) interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol Cell Biol. 1999;19(5):3614–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spring CM, Kelly KF, O’Kelly I, Graham M, Crawford HC, Daniel JM. The catenin p120ctn inhibits Kaiso-mediated transcriptional repression of the beta-catenin/TCF target gene matrilysin. Exp Cell Res. 2005;305(2):253–65. [DOI] [PubMed] [Google Scholar]
- 3.Lopes EC, Valls E, Figueroa ME, Mazur A, Meng FG, Chiosis G, et al. Kaiso contributes to DNA methylation-dependent silencing of tumor suppressor genes in colon cancer cell lines. Cancer Res. 2008;68(18):7258–63. [DOI] [PubMed] [Google Scholar]
- 4.Filion GJ, Zhenilo S, Salozhin S, Yamada D, Prokhortchouk E, Defossez PA. A family of human zinc finger proteins that bind methylated DNA and repress transcription. Mol Cell Biol. 2006;26(1):169–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buck-Koehntop BA, Martinez-Yamout MA, Dyson HJ, Wright PE. Kaiso uses all three zinc fingers and adjacent sequence motifs for high affinity binding to sequence-specific and methyl-CpG DNA targets. FEBS Lett. 2012;586(6):734–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donaldson IJ, Amin S, Hensman JJ, Kutejova E, Rattray M, Lawrence N, et al. Genome-wide occupancy links Hoxa2 to Wnt-beta-catenin signaling in mouse embryonic development. Nucleic Acids Res. 2012;40(9):3990–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park JI, Kim SW, Lyons JP, Ji H, Nguyen TT, Cho K, et al. Kaiso/p120-catenin and TCF/beta-catenin complexes coordinately regulate canonical Wnt gene targets. Dev Cell. 2005;8(6):843–54. [DOI] [PubMed] [Google Scholar]
- 8.Pozner A, Terooatea TW, Buck-Koehntop BA. Cell-specific Kaiso (ZBTB33) Regulation of Cell Cycle through Cyclin D1 and Cyclin E1. J Biol Chem. 2016;291(47):24538–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruzov A, Savitskaya E, Hackett JA, Reddington JP, Prokhortchouk A, Madej MJ, et al. The non-methylated DNA-binding function of Kaiso is not required in early Xenopus laevis development. Development. 2009;136(5):729–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis JN, McGhee L, Meyers S. The ETO (MTG8) gene family. Gene. 2003;303:1–10. [DOI] [PubMed] [Google Scholar]
- 11.Lutterbach B, Westendorf JJ, Linggi B, Patten A, Moniwa M, Davie JR, et al. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol Cell Biol. 1998;18(12):7176–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrett CW, Smith JJ, Lu LC, Markham N, Stengel KR, Short SP, et al. Kaiso directs the transcriptional corepressor MTG16 to the Kaiso binding site in target promoters. PLoS One. 2012;7(12):e51205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kochetkova M, McKenzie OL, Bais AJ, Martin JM, Secker GA, Seshadri R, et al. CBFA2T3 (MTG16) is a putative breast tumor suppressor gene from the breast cancer loss of heterozygosity region at 16q24.3. Cancer Res. 2002;62(16):4599–604. [PubMed] [Google Scholar]
- 14.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314(5797):268–74. [DOI] [PubMed] [Google Scholar]
- 15.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–13. [DOI] [PubMed] [Google Scholar]
- 16.Barrett CW, Fingleton B, Williams A, Ning W, Fischer MA, Washington MK, et al. MTGR1 is required for tumorigenesis in the murine AOM/DSS colitis-associated carcinoma model. Cancer Res.71(4):1302–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierre CC, Longo J, Mavor M, Milosavljevic SB, Chaudhary R, Gilbreath E, et al. Kaiso overexpression promotes intestinal inflammation and potentiates intestinal tumorigenesis in Apc(Min/+) mice. Biochim Biophys Acta. 2015;1852(9):1846–55. [DOI] [PubMed] [Google Scholar]
- 18.Chaudhary R, Pierre CC, Nanan K, Wojtal D, Morone S, Pinelli C, et al. The POZ-ZF transcription factor Kaiso (ZBTB33) induces inflammation and progenitor cell differentiation in the murine intestine. PLoS One. 2013;8(9):e74160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prokhortchouk A, Sansom O, Selfridge J, Caballero IM, Salozhin S, Aithozhina D, et al. Kaiso-deficient mice show resistance to intestinal cancer. Mol Cell Biol. 2006;26(1):199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDonough EM, Barrett CW, Parang B, Mittal MK, Smith JJ, Bradley AM, et al. MTG16 is a tumor suppressor in colitis-associated carcinoma. JCI Insight. 2017;2(16). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Becker C, Fantini MC, Wirtz S, Nikolaev A, Kiesslich R, Lehr HA, et al. In vivo imaging of colitis and colon cancer development in mice using high resolution chromoendoscopy. Gut. 2005;54(7):950–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams CS, Bradley AM, Chaturvedi R, Singh K, Piazuelo MB, Chen X, et al. MTG16 contributes to colonic epithelial integrity in experimental colitis. Gut. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poindexter SV, Reddy VK, Mittal MK, Williams AM, Washington MK, Harris E, et al. Transcriptional corepressor MTG16 regulates small intestinal crypt proliferation and crypt regeneration after radiation-induced injury. Am J Physiol Gastrointest Liver Physiol. 2015;308(6):G562–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nowarski R, Jackson R, Gagliani N, de Zoete MR, Palm NW, Bailis W, et al. Epithelial IL-18 Equilibrium Controls Barrier Function in Colitis. Cell. 2015;163(6):1444–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergstrom KS, Guttman JA, Rumi M, Ma C, Bouzari S, Khan MA, et al. Modulation of intestinal goblet cell function during infection by an attaching and effacing bacterial pathogen. Infect Immun. 2008;76(2):796–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCormick DA, Horton LW, Mee AS. Mucin depletion in inflammatory bowel disease. J Clin Pathol. 1990;43(2):143–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246(4930):629–34. [DOI] [PubMed] [Google Scholar]
- 31.Syljuasen RG, Krolewski B, Little JB. Loss of normal G1 checkpoint control is an early step in carcinogenesis, independent of p53 status. Cancer Res. 1999;59(5):1008–14. [PubMed] [Google Scholar]
- 32.Rudolph KL, Hartmann D, Opitz OG. Telomere dysfunction and DNA damage checkpoints in diseases and cancer of the gastrointestinal tract. Gastroenterology. 2009;137(3):754–62. [DOI] [PubMed] [Google Scholar]
- 33.Raghav SK, Waszak SM, Krier I, Gubelmann C, Isakova A, Mikkelsen TS, et al. Integrative genomics identifies the corepressor SMRT as a gatekeeper of adipogenesis through the transcription factors C/EBPbeta and KAISO. Mol Cell. 2012;46(3):335–50. [DOI] [PubMed] [Google Scholar]
- 34.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, et al. Systematic discovery of regulatory motifs in human promoters and 3’ UTRs by comparison of several mammals. Nature. 2005;434(7031):338–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–5. [DOI] [PubMed] [Google Scholar]
- 36.Reddy VK, Short SP, Barrett CW, Mittal MK, Keating CE, Thompson JJ, et al. BVES Regulates Intestinal Stem Cell Programs and Intestinal Crypt Viability after Radiation. Stem Cells. 2016;34(6):1626–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyoshi H, Stappenbeck TS. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat Protoc. 2013;8(12):2471–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim SW, Park JI, Spring CM, Sater AK, Ji H, Otchere AA, et al. Non-canonical Wnt signals are modulated by the Kaiso transcriptional repressor and p120-catenin. Nat Cell Biol. 2004;6(12):1212–20. [DOI] [PubMed] [Google Scholar]
- 39.Li Q, Ishikawa TO, Oshima M, Taketo MM. The threshold level of adenomatous polyposis coli protein for mouse intestinal tumorigenesis. Cancer Res. 2005;65(19):8622–7. [DOI] [PubMed] [Google Scholar]
- 40.Buchert M, Athineos D, Abud HE, Burke ZD, Faux MC, Samuel MS, et al. Genetic dissection of differential signaling threshold requirements for the Wnt/beta-catenin pathway in vivo. PLoS Genet. 2010;6(1):e1000816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Christie M, Jorissen RN, Mouradov D, Sakthianandeswaren A, Li S, Day F, et al. Different APC genotypes in proximal and distal sporadic colorectal cancers suggest distinct WNT/beta-catenin signalling thresholds for tumourigenesis. Oncogene. 2013;32(39):4675–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koch S, Nava P, Addis C, Kim W, Denning TL, Li L, et al. The Wnt antagonist Dkk1 regulates intestinal epithelial homeostasis and wound repair. Gastroenterology. 2011;141(1):259–68, 68 e1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez JA, Williams CS, Amann JM, Ellis TC, Moreno-Miralles I, Washington MK, et al. Deletion of Mtgr1 sensitizes the colonic epithelium to dextran sodium sulfate-induced colitis. Gastroenterology. 2006;131(2):579–88. [DOI] [PubMed] [Google Scholar]
- 44.Blattler A, Yao L, Wang Y, Ye Z, Jin VX, Farnham PJ. ZBTB33 binds unmethylated regions of the genome associated with actively expressed genes. Epigenetics Chromatin. 2013;6(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Becker C, Fantini MC, Neurath MF. High resolution colonoscopy in live mice. Nat Protoc. 2006;1(6):2900–4. [DOI] [PubMed] [Google Scholar]
- 46.Brown JJ, Short SP, Stencel-Baerenwald J, Urbanek K, Pruijssers AJ, McAllister N, et al. Reovirus-Induced Apoptosis in the Intestine Limits Establishment of Enteric Infection. J Virol. 2018;92(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heijmans J, van Lidth de Jeude JF, Koo BK, Rosekrans SL, Wielenga MC, van de Wetering M, et al. ER stress causes rapid loss of intestinal epithelial stemness through activation of the unfolded protein response. Cell Rep. 2013;3(4):1128–39. [DOI] [PubMed] [Google Scholar]
- 48.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.