Abstract
Background
Cystic fibrosis (CF) lung disease commences in infancy, and understanding the role of the microbiota in disease pathogenesis is critical. This study examined and compared the lower airway microbiota of infants with and without CF and its relationship to airway inflammation in the first months of life.
Methods
Infants newly-diagnosed with CF were recruited into a single-centre study in Melbourne, Australia from 1992–2001. Bronchoalveolar lavage was performed at study entry. Healthy infants undergoing bronchoscopy to investigate chronic stridor acted as controls. Quantitative microbiological culture was performed and inflammatory markers were measured contemporaneously. 16S ribosomal RNA gene analysis was performed on stored samples.
Results
Thirteen bronchoalveolar samples from infants with CF and nine from control infants, collected at median ages of 1.8-months (25th-75th percentile 1.5 to 3.1-months) and 5-months (25th-75th percentile 2.9 to 8.2-months) respectively, provided 16S rRNA gene data. Bacterial biomass was positively associated with inflammation. Alpha diversity was reduced in infants with CF and between-group compositional differences were apparent. These differences were driven by increased Staphylococcus and decreased Fusobacterium and were most apparent in symptomatic infants with CF.
Conclusion
In CF lung disease, differences in lower airway microbial community composition and structure are established by age 6-months.
Keywords: Cystic fibrosis, microbiota, 16S ribosomal RNA, bronchoalveolar lavage fluid, inflammation, infant
Summary
Differences in the composition and structure of the lower airway microbiota of infants with and without cystic fibrosis are established in the first six months of life, and are most apparent in symptomatic infants.
INTRODUCTION
Over the past two decades, the widespread application of molecular analysis techniques, including 16S ribosomal RNA (16S rRNA) gene sequencing, has changed our understanding of the human microbiome. In health, the lungs are not sterile, and the host-microbiota relationship is likely to have an important role in the development and modulation of the immune response, and therefore, in the early pathogenesis of disease [1, 2].
Unfortunately, the “healthy” or “typical” lower airway microbiota in infancy and early childhood remains unknown [3]. Lower airway samples from infants and young children can only be reliably obtained via bronchoscopy, requiring general anaesthesia. Consequently, the lower airway microbiota in this important population has not been described.
Cystic fibrosis (CF) lung disease begins in infancy [4]. Culture-based studies suggest that lower airway infection is a key trigger for inflammation, however elevated inflammatory markers have been reported in the setting of negative culture results [5–7]. There are significant differences between the nasopharyngeal microbiota of infants with CF and healthy controls at the time of diagnosis by newborn screening and before antibiotic exposure, most notably an increased prevalence of Staphylococcus species [8, 9]. However, these observations cannot be inferred to represent the lower airway microbiota [10–12]. Similarly, microbiota composition changes and diversity decreases with age and advancing disease [13], and findings from sputum samples of older children and adults with CF cannot be extrapolated to infants.
We have previously explored the development of the lower airway microbiota over time in a birth cohort of infants and young children with CF [14]. In order to explore the role of the lower airway microbiota in the earliest pathogenesis of CF lung disease, we now present a subset of this study population, infants who had lower airway samples obtained at younger than age 6-months. We compare the composition and structure of their lower airway microbiota to that of a control group of infants without lung disease, who had lower airway samples obtained during the same time period. We hypothesised that there would be differences in microbiota composition in infants with CF, and that these differences would be most pronounced in infants with respiratory symptoms.
METHODS
Participants
Infants newly diagnosed with CF, either following newborn screening or presentation with meconium ileus and sweat chloride concentrations >60mmol/L were recruited into a single-centre study in Melbourne, Australia, from 1992–2001. Otherwise healthy infants, undergoing bronchoscopy for investigation of congenital stridor during the same time period, who were free of other symptoms and not taking antibiotics in the previous 14-days, were recruited as controls. Bronchoscopy and bronchoalveolar lavage (BAL) were performed at study entry. Samples collected from infants with CF aged <6-months and from control infants aged <12-months were included in this analysis. Infants with CF were not routinely prescribed prophylactic antibiotics.
BAL samples were obtained from the right middle lobe and lingula, and except for a small subset of infants with CF [15], pooled at the time of collection, as previously reported [5, 14, 16] and described in the Online Supplement. Inflammatory markers, interleukin-8 (IL-8) and neutrophil elastase (NE), were measured and quantitative bacterial cultures were performed contemporaneously. Whole BAL samples were stored at −70°C and 16S rRNA gene sequencing analysis was performed on available samples. The Royal Children’s Hospital Melbourne Human Research Ethics Committee approved this study. Written informed consent was obtained from parents/guardians.
16S ribosomal RNA gene sequencing
16S rRNA gene sequences were generated on the Roche 454 platform, using V1–3 variable regions, following Human Microbiome Project protocols [17], as reported previously [14] and detailed in the Online Supplement. Reads passing through quality control were classified using Ribosomal Database Project naïve Bayesian classifier [18], V2.2 with training set 6. Sequences were classified to the lowest taxonomic level that could be assigned with confidence values >0.5.
Statistical analysis
Statistical analysis was performed using ‘R’ [19], packages ‘vegan’ [20] and ‘metagenomeSeq’ [21]. BAL samples from infants with CF were designated as “unwell” or “stable” based on the presence or absence of respiratory symptoms and/or antibiotic use at the time of collection.
NE values below the assay detection limit of 5 mcg/mL were assigned a value of 2.5 mcg/mL. IL-8 and NE values were logarithmically transformed for analysis. Chi square tests, t-tests and Mann-Whitney tests were used to compare categorical, and parametric and non-parametric continuous variables respectively.
BAL samples that returned a minimum of 1000 reads were included for analysis of microbiota diversity and composition (Figure E1). Sequences not classified to genus level were removed prior to analysis. Alpha-diversity was assessed with Shannon Diversity Index (SDI), richness and Pielou’s evenness index.
The data were rarefied to 1000 reads per sample for analysis of beta-diversity, using the Bray-Curtis dissimilarity index and principal coordinate analysis [20]. Permutational multivariate analysis of variance, multivariate homogeneity of groups dispersion and Tukey’s Honest Significant Difference methods were used to compare beta-diversity between subject groups [20]. Constrained ordination using redundancy analysis with analysis of variance testing was utilised to explore the impact of disease status on microbiota composition [20].
Data were normalised using ‘MetagenomeSeq’ to account for sparse high-throughput data from low volume and potentially low concentration samples [21]. Relative abundance of genera in individual samples, odds ratios for presence of individual genera and logarithmic fold changes in their prevalence between infants with CF and controls were calculated. P-values were adjusted using the Benjamini-Hochberg correction.
RESULTS
Participants
Twenty-one BAL samples from 21 infants with CF and ten samples from ten control infants, collected at median ages of 1.8-months (25th-75th percentile 1.5 to 2.5-months) and 5.6-months (2.9–8.6-months) respectively underwent 16S rRNA gene sequencing, including 13 samples from infants with CF (62%) and nine samples from controls (90%) that yielded greater than 1000 reads (Table 1; Figure E2 and Table E1). Control infants were diagnosed with laryngeal or subglottic pathology based on bronchoscopic findings [laryngomalacia, n=9 (90%); stridor not otherwise specified, n=1 (10%)]. Of the infants with CF whose BAL samples yielded above 1000 reads, eight (62%) were stable and five (38%) were unwell (with respiratory symptoms and/or antibiotic use) at the time of BAL. There were no differences in age or cystic fibrosis transmembrane regulator genotype between these groups, although more of the unwell infants with CF had prior antibiotic exposure (4/5 [80%] compared with 3/8 [38%] stable infants, p=0.36). There were no significant differences in the clinical characteristics of infants with CF whose BAL samples yielded above or below 1000 reads.
TABLE 1:
Subjects’ clinical and bronchoalveolar lavage characteristics.
| Infants with CF* whose BAL samples yielded > 1000 16S rRNA gene reads (n=13) | Infants with CF whose BAL samples yielded < 1000 16S rRNA gene reads (n = 8) | p† | Control infants whose BAL samples yielded > 1000 16S rRNA gene reads (n=9) | p‡ | |
|---|---|---|---|---|---|
| Clinical characteristics | |||||
| Male, n (%) | 7 (54%) | 4 (50%) | 1 | 7 (78%) | 0.57 |
| Age (months), median (25th-75th ile) | 1.8 (1.5–3.1) | 2.0 (1.7–2.4) | 0.54 | 5.0 (2.9 – 8.2) | 0.007 |
| P.Phe508del homozygous, n (%) |
11 (85%) | 5 (63%) | 0.53 | - | - |
| Diagnosis by newborn screening, n (%) |
11 (85%) | 6 (75%) | 1 | - | - |
| Prior antibiotic exposure, n (%) | 7 (54%) | 4 (50%) | 1 | - | - |
| Respiratory symptoms at BAL, n (%) | 5 (38%) | 1 (13%) | 0.43 | ||
| Antibiotics at BAL, n (%) | 4 (31%) | 0 | 0.24 | ||
| Bronchoalveolar lavage features | |||||
| Bacterial colony forming units/mL, median (25th – 75th ile) | 1.94 × 104 (6 × 103 – 6.3 × 105) | 645 (0.75–1635) | 0.008 | 5.83 × 104 (1.11 × 104— 2.02 × 105) | 0.9 |
| Interleukin-8 (pg/mL), median (25th – 75th ile) | 79 (56–195.6) | 30 (25–36) § | 0.03 | 41 (22.1–71) § | 0.21 |
| Neutrophil elastase (mcg/mL), median (25th – 75th ile) | 6.2 (2.5–16.1) | 4.2(2.5–8.6) ** | 0.37 | 2.5 (2.5–5.1) § | 0.33 |
CF: Cystic fibrosis
Comparison between infants with CF whose bronchoalveolar lavage samples yielded greater than or fewer than 1000 16S rRNA gene sequence reads respectively. Chi-square test for categorical variables; t-test for parametric continuous variables; Whitney test for non-parametric continuous variables
Comparison between infants with CF and control infants whose bronchoalveolar lavage samples yielded greater than 1000 16S rRNA gene sequence reads. Chi-squared test for comparison of categorical variables; Mann Whitney test for comparison of continuous variables
Missing data from three participants
Missing data from one participant
Missing data from two participants
Quantitative culture and airway inflammation
BAL samples yielding fewer than 1000 reads had reduced bacterial growth on quantitative culture compared with the samples yielding greater than 1000 reads (median 790 colony forming units (CFU)/mL (25th-75th percentile 1–2340 CFU/mL) versus 36765 CFU/mL (6301–223300 CFU/mL) respectively, p=0.004); Figure E3).
BAL samples yielding fewer than 1000 16S rRNA gene reads, suggestive of lower bacterial biomass, had reduced IL-8 compared to samples yielding greater than 1000 reads (median pg/mL (25th-75th percentile 26.3–35 pg/mL) versus 66.4 pg/mL (41.1–195.3 pg/mL respectively, t-test (log10 IL-8): p=0.03), although there was no difference in NE concentration between the groups (median 5.9 mcg/mL (2.5–7.8 mcg/mL) versus 2.5 mcg/mL (2.5–12.1 mcg/mL) respectively, t-test (log10 NE): p=0.6; Figure E4).
Alpha-diversity
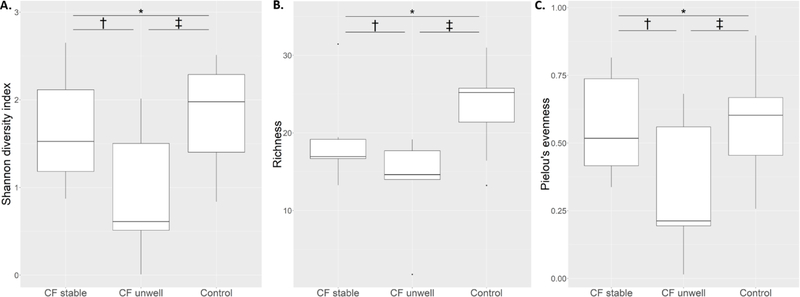
Microbial richness was reduced in the lower airway samples of infants with CF compared with controls regardless of clinical status at the time of BAL (95% confidence interval (CI) of difference between infants with CF and controls: 1.13 – 11.83, p=0.02). There were no differences in either Shannon diversity index or Pielou’s evenness between the groups overall (Shannon index: 95% CI of difference −0.21 – 1.0, p=0.19; Pielou’s evenness: 95% CI of difference −0.1 – 0.29, p=0.31). Although there were reductions in both indices in unwell infants with CF, these were not statistically significant (Figure 1). Alpha-diversity indices were not correlated with lower airway inflammatory markers (Figure E5).
Figure 1: Alpha-diversity indices in bronchoalveolar lavage samples of stable infants with cystic fibrosis, unwell infants with cystic fibrosis and control infants.
1A: Shannon diversity index. *95% confidence interval (CI) of difference −0.53 – 0.76, p = 0.7; †95% CI of difference −0.3 – 1.72, p = 0.14 ; ‡95% CI of difference −0.16 – 1.82, p = 0.09. 1B: Microbial richness. *95% CI of difference −1.26 – 10.17, p = 0.12; †95% CI of difference −3.18 – 13.9, p = 0.18; ‡95% CI of difference 1.32 – 18.31, p = 0.03; 1C: Pielou’s evenness: *95% CI of difference - 0.19 – 0.21, p = 0.91; †95% CI of difference −0.12 – 0.57, p = 0.16; ‡95% CI of difference −0.1 – 0.56, p = 0.14.
Beta-diversity
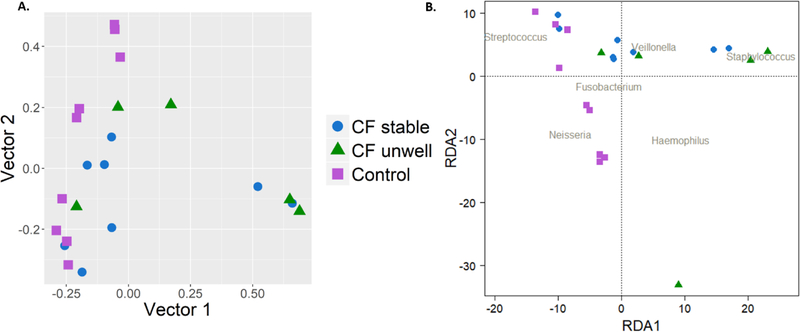
The Bray-Curtis dissimilarity index and principal coordinate analysis were used to quantify compositional differences in the microbiota of individual BAL samples (Figure 2A). Permutational multivariate analysis of variance using distance matrices demonstrated significant differences between the subject groups (p=0.01).
Figure 2:
Beta diversity of individual bronchoalveolar lavage samples. 2A: Principal coordinate analysis of bronchoalveolar lavage samples from stable and unwell infants with cystic fibrosis and control infants. 2B: Redundancy analysis plot of bronchoalveolar lavage samples, constrained by disease status.
Constrained ordination was then performed using redundancy analysis to assess the compositional variation in lower airway microbiota attributable to disease status (CF stable, CF unwell and control; ANOVA, p=0.04). Results are shown in Figure 2B.
Average dispersion from the centroid was increased in BAL samples from unwell infants with CF (0.56) compared with controls (0.39; 95% CI of difference between means: −0.34 – 0.01, p=0.07), indicating greater inter-individual differences amongst the former group. There were no differences in the beta-diversity of lower airway samples of control infants compared to stable infants with CF (0.47; 95% CI of difference between means: −0.23 – 0.08, p=0.43), or between unwell and stable infants with CF (95% CI of difference between means: −0.09 – 0.27, p=0.43; Figure E6).
Microbiota composition
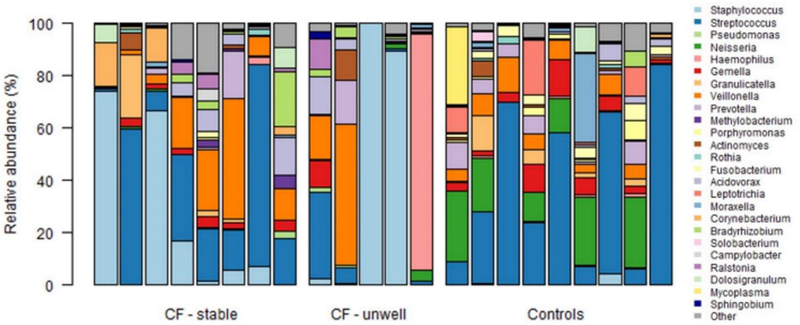
Normalisation methods to account for sparse high-throughput data were employed to enable comparison of both the relative abundance of genera within individual BAL samples and the differential abundance of individual genera in BAL samples of infants with CF and controls (Figure 3).
Figure 3: Composition of the lower airway microbiota of individual stable infants with cystic fibrosis, unwell infants with cystic fibrosis and control infants.
Each column represents a single bronchoalveolar lavage sample. The relative abundance of the individual genera, calculated from normalized data, is displayed in the stacked column graphs (range 0–100%), with each colour representing an individual genus. Genera with a mean relative abundance > 0.5% in any of the three subject groups are shown (see legend).
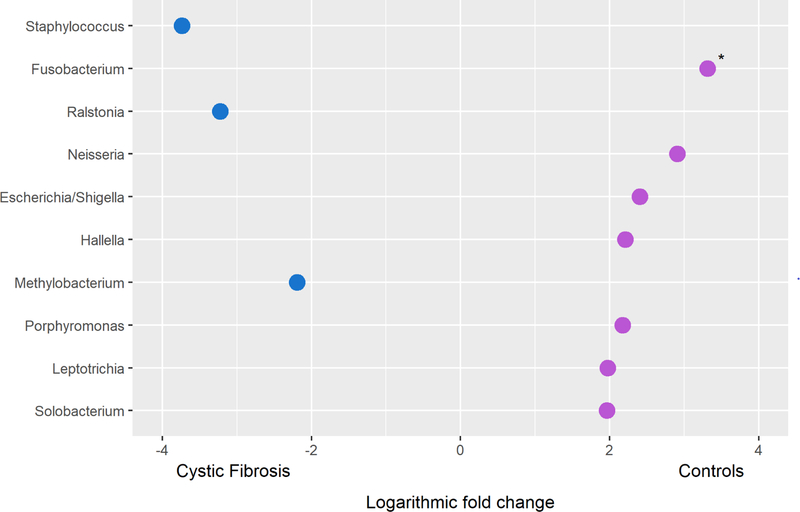
In a linear fit model assessing the differential abundance of individual genera in BAL samples from infants with CF and control infants, Staphylococcus, Ralstonia and Methylobacterium were increased in the former with logarithmic (log2) fold increases of 3.7, 3.2 and 2.2 respectively. Fusobacterium, Neisseria and Escherichia/Shigella were the most increased genera in the lower airway samples of control infants with logarithmic (log2) fold increases of 3.3, 2.9 and 2.4 respectively; Figure 4; Table E2).
Figure 4: The differential abundance of individual genera in the lower airways of infants with cystic fibrosis compared to control infants.
The logarithmic (log2) fold change of the top 10 ranked genera in a linear model fit assessing between-group differences of bronchoalveolar lavage samples is presented. Genera with increased relative abundance in infants with cystic fibrosis and controls are indicated in blue (●) and purple (●) respectively. * p < 0.05.
Differences in median relative abundance of individual genera in the three subject groups are further highlighted in Supplementary Figure E7. Differences in the composition of the lower airway microbiota in infants with CF were most pronounced in those who were unwell at the time of BAL, with the greatest increase in Staphylococcus and decrease in Streptococcus in this group.
Antibiotic exposure
In infants with CF, previous or current exposure to treatment antibiotics had no impact on bacterial load in quantitative culture, 16S rRNA gene sequence reads, alpha-diversity or beta-diversity indices when compared to BAL samples collected from antibiotic-naïve infants (Table E3; Figure E8).
DISCUSSION
This study revealed substantial differences between the early lower airway microbiota of infants with and without CF that were most marked in symptomatic infants. Both structural and compositional differences were apparent in the first months of life: richness was reduced in infants with CF; Staphylococcus was increased and Fusobacterium was decreased. In the presence of active respiratory disease, the relative abundance of recognised CF pathogens increased at the expense of a diverse range of typical bacteria. We also found a relationship between lower airway bacterial biomass and inflammation, such that those BAL samples which failed to amplify to 1000 16S rRNA gene sequence reads had less bacterial growth in quantitative culture and lower IL-8 concentrations compared with the BAL samples that yielded greater than 1000 sequence reads.
This study provides the first description of the lower airway microbiota of infants without significant lung pathology. By comparing such “healthy” lower airway microbiota to that of very young infants with CF, diagnosed predominantly by newborn screening, not prescribed anti-staphylococcal prophylaxis and managed in a single CF centre with minimal treatment variation, this study offers unique insights into the microbiological origins of CF lung disease. As has been reported in the nasopharynx [8, 9], the most substantial difference between the airway microbiota of infants with CF and controls was the increased prevalence of Staphylococcus in the former. It has long been established that S. aureus is a major pathogen in early CF lung disease. Its detection using classical culture methodology is associated with increased neutrophilic inflammation, earlier onset of structural lung disease and poorer lung function outcomes [4–6, 16, 22–25]. Using molecular analysis techniques its prevalence has now been confirmed in both the upper and lower airways in the first months of life, soon after diagnosis by newborn screening, and prior to antibiotic exposure [8, 9, 14]. This study highlighted both the relative absence of Staphylococcus from the lower airways of healthy infants and its dominance in symptomatic infants with CF, thus emphasising its central and early role in the pathogenesis of CF lung disease.
This study provides further evidence that CF lung disease begins early, with profound differences in the lower airway microbiota driven by disease state and established by age 6-months. We previously demonstrated that in infants and young children with CF not prescribed antibiotic prophylaxis, both reduced alpha-diversity in the lower airway microbiota and dominance of recognised CF pathogens were associated with increased lower airway inflammatory markers [14], which are risk factors for the earlier development of structural lung disease. In that longitudinal study, including the CF BAL samples presented in this analysis, microbial community composition was dynamic, with considerable changes in its composition over the first years of life.
Age-related changes in the lower airway microbiota of preschool children with CF have been further described in two recent studies. The first, involving 136 children (median age 11-years (range 0.2–20-years) undergoing clinically indicated bronchoscopy at 13 North American CF centres, reported that in participants younger than 2-years, non-traditional taxa, particularly Streptococcus, comprised approximately half of the lower airway microbiota, while in those older than 6-years, the microbiota was frequently dominated by traditional pathogenic taxa [13]. Similarly, in a contemporary Australian cohort of 46 preschool children (median age 1.95-years (25th-75th percentile 1.13–4.06-years) routinely prescribed antibiotic prophylaxis until age 2-years, a distinct progression of the lower airway microbiota was described; from relative sterility in infancy, to dominance of bacterial sequences common to the oropharynx by age 2-years and dominance of traditional CF pathogens in participants older than 4-years [26]. Our data in infants not prescribed routine antibiotic prophylaxis identified a different, likely pathogenic microbiota even in the first six months of life in infants with CF.
The influence of prophylactic antibiotics on the early lower airway microbiota is highlighted in our recent comparison of BAL samples from 17 Australian infants prescribed antibiotic prophylaxis (age 3.5±0.9-months) and 15 North American infants (age 6±1.6-months) [27]. In that study, alpha-diversity was lower in the youngest infants and in those prescribed anti-staphylococcal prophylaxis, and decreased alpha-diversity was associated with decreased airway inflammation. Neither alpha-diversity nor microbiota composition was influenced by intermittent use of treatment antibiotics. Unlike the current study however, these studies lacked non-CF controls.
The present study systematically described the lower airway microbiota of non-CF infants without significant lower airway pathology. Not unexpectedly, the composition of the lower airway microbiota differed substantially from previous descriptions of the healthy upper airway microbiota [28–30]. The most prevalent genera in the lower airways of infants without CF were Streptococcus and Neisseria, with relative absence of the skin flora typically encountered in the nasopharynx, specifically Staphylococcus, Dolosigranulum and Corynebacterium. Moraxella and Haemophilus, which are also commonly reported in upper airway samples, had a median relative abundance of 0.2% and 0.09% respectively in control BAL samples. While these BAL samples were obtained from infants with stridor, without evidence of concomitant infection, they emphasise the significant distinctions between the upper and lower airway microbiological communities in health.
This study has a number of important limitations. Firstly, it involved the retrospective analysis of BAL samples collected approximately 20-years earlier and is limited by small sample size, including of antibiotic-naïve CF infants, and a 3.2-month difference in median age between the CF and control populations. If the lung microbiota follows a similar ontogeny to that of the human gut, with increase in bacterial diversity and changes in response to weaning and diet, then the older age of the subjects without CF may have favoured a more diverse lung microbiota. However, maturation of the human lung microbiota during infancy has not been studied.
Samples were subject to strict storage conditions and analysis criteria to minimise the impact of time on the quality of 16S rRNA gene data. We have previously demonstrated that these data correlated both with culture results obtained contemporaneously on the same BAL samples [14] and with descriptions of the lower airway microbiota in older participants, suggesting that they accurately reflect the lower airway microbiota. We have been unable to differentiate 16S rRNA gene reads corresponding to the Staphylococcus genus to a species level, however our earlier analysis demonstrated correlation between the detection of Staphylococcus by molecular analysis and the finding of S. aureus in the corresponding BAL culture and the presence of pulmonary inflammation [14]. While quantitative PCR was not performed, normalisation techniques were employed to minimise bias in analysis of differential abundance of individual genera in these small volume, low bacterial load samples [21].
16S rRNA gene analysis of eight BAL samples from infants with CF returned fewer than 1000 reads and could not be included for analysis of microbiota composition. While potential differences in dilution were not controlled for, the sequences obtained from these BAL samples more closely represent the negative laboratory controls than the BAL samples of control infants, suggesting low bacterial biomass as a likely explanation for the low reads. Consistent with this, these BAL samples had lower total bacteria in quantitative culture and reduced IL-8, raising the possibility that the resident lower airway microbiota of “healthier” infants with CF had not yet become established, and emphasising that infection is indeed an important trigger for inflammation in early CF lung disease.
Lastly, this study included BAL samples from a control population of infants with stridor, primarily laryngomalacia, who were older than the study group of infants with CF. Obtaining lavage samples from healthy infants not otherwise requiring anaesthesia is ethically unjustifiable. We hypothesise that the lower airway microbiota of infants with laryngeal or subglottic pathology would potentially be more similar to that of infants with CF than would be that of completely healthy infants and thus, the differences in lower airway microbiota between our infant cohorts highlight the significant alterations in the lower airway microbiota present early in CF lung disease.
This study provides further evidence that CF lung disease begins very early in life, with substantial differences in the lower airway microbiota established soon after birth, and most pronounced in symptomatic infants. The lower airway microbiota is dynamic and there is increasing evidence that age and/or disease progression and antibiotic prophylaxis influence its composition. It will be important to determine how the earliest changes in the lower airway microbiota described in this study are associated, or even implicated, with the development of lung damage in CF. It is also possible that altering the lower airway microbiota in CF infants to have a similar composition and structure to that of the healthy infant’s lower respiratory tract could be a potential therapeutic strategy to prevent progression of early lung disease. Further longitudinal studies, including long-term outcome measures in both infants with CF and age-matched controls are required to better understand the clinical significance of these changes.
Supplementary Material
HIGHLIGHTS.
The lower airway microbiota is altered substantially in infants with cystic fibrosis
Differences appear within the first six months of life
Differences are most apparent in symptomatic infants with cystic fibrosis
Airway inflammation is associated with bacterial biomass
ACKNOWLEDGEMENTS
The authors wish to thank Shu Mei Teo, Centre for Systems Genomics, the University of Melbourne, for her advice regarding statistical analysis, and Suzanna Vidmar, Murdoch Children’s Research Institute, Melbourne for her assistance with data collection and cleaning.
FUNDING SUPPORT
16S rRNA gene sequencing was funded by grants from the Murdoch Childrens Research Institute “65km for CF” and the Royal Children’s Hospital Cystic Fibrosis Research Trust (RCH CFRT). KBF was supported by the Thoracic Society of Australia and New Zealand Vertex Cystic Fibrosis Paediatric Clinical Fellowship, the RCH CFRT, the Australian Cystic Fibrosis Research Trust Postgraduate Studentship, an Australian National Health and Medical Research Council (NHMRC) postgraduate scholarship, a Royal Australasian College of Physicians Paediatrics and Child Health Division NHMRC Award for Excellence (Top-up), the Clifford Family PhD Scholarship and an Australian Government Research Training Program Scholarship. TWF, SDD, SCR, GAS, and KMW were supported by the National Institutes of Health (NIH) grant, HL116211 and NHMRC award, GNT1043768. These bodies had no role in the study design, data analysis, interpretation or reporting of results, and were not involved in the decision to submit this manuscript for publication.
ABBREVIATIONS
- 16S rRNA
16S ribosomal RNA
- BAL
Bronchoalveolar lavage
- CF
Cystic fibrosis
- IL-8
Interleukin 8
- NE
Neutrophil elastase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST STATEMENT
The authors have no conflicts of interest.
REFERENCES
- 1.Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science 2016: 352(6285): 539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shima K, Coopmeiners J, Graspeuntner S, Dalhoff K, Rupp J. Impact of micro-environmental changes on respiratory tract infections caused by intracellular bacteria. FEBS Lett 2016: 590(21): 3887–3904. [DOI] [PubMed] [Google Scholar]
- 3.Frayman KB, Armstrong DS, Grimwood K, Ranganathan SC. The airway microbiota in early cystic fibrosis lung disease. Pediatr Pulmonol 2017: 52(11): 1384–1404. [DOI] [PubMed] [Google Scholar]
- 4.Sly PD, Gangell CL, Chen L, Ware RS, Ranganathan SC, Mott LS, Murray CP, Stick SM, investigators AC. Risk factors for bronchiectasis in children with cystic fibrosis. N Eng J Med 2013: 368(21): 1963–1970. [DOI] [PubMed] [Google Scholar]
- 5.Armstrong DS, Hook SM, Jamsen KM, Nixon GM, Carzino R, Carlin JB, Robertson CF, Grimwood K. Lower airway inflammation in infants with cystic fibrosis detected by newborn screening. Pediatr Pulmonol 2005: 40(6): 500–510. [DOI] [PubMed] [Google Scholar]
- 6.Armstrong DS, Grimwood K, Carlin JB, Carzino R, Gutierrez JP, Hull J, Olinsky A, Phelan EM, Robertson CF, Phelan PD. Lower airway inflammation in infants and young children with cystic fibrosis. Am J Respir Crit Care Med 1997: 156(4 Pt 1): 1197–1204. [DOI] [PubMed] [Google Scholar]
- 7.Khan TZ, Wagener JS, Bost T, Martinez J, Accurso FJ, Riches DW. Early pulmonary inflammation in infants with cystic fibrosis. Am J Respir Crit Care Med 1995: 151(4): 1075–1082. [DOI] [PubMed] [Google Scholar]
- 8.Prevaes SMPJ, de Winter-de Groot KM, Janssens HM, de Steenhuijsen Piters WAA, Tramper-Stranders GA, Wyllie AL, Hasrat R, Tiddens HA, van Westreenen M, van der Ent CK, Sanders EAM, Bogaert D. Development of the nasopharyngeal microbiota in infants with cystic fibrosis. Am J Respir Crit Care Med 2016: 193(5): 504–515. [DOI] [PubMed] [Google Scholar]
- 9.Mika M, Korten I, Qi W, Regamey N, Frey U, Casaulta C, Latzin P, Hilty M, group oboSs. The nasal microbiota in infants with cystic fibrosis in the first year of life: a prospective cohort study. Lancet Respir Med 2016: 4(8): 627–635. [DOI] [PubMed] [Google Scholar]
- 10.Kloepfer KM, Deschamp AR, Ross SE, Peterson-Carmichael SL, Hemmerich CM, Rusch DB, Davis SD. In children, the microbiota of the nasopharynx and brochoalveolar lavage fluid are both similar and different. Pediatr Pulmonol 2018: 53(4): 475–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marsh RL, Kaestli M, Chang AB, Binks MJ, Pope CE, Hoffman LR, Smith-Vaughan HC. The microbiota in bronchoalveolar lavage from young children with chronic lung disease includes taxa present in both the oropharynx and nasopharynx. Microbiome 2016: 4(1): 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prevaes SM, de Steenhuijsen Piters WA, de Winter-de Groot KM, Janssens HM, Tramper-Stranders GA, Chu ML, Tiddens HA, van Westreenen M, van der Ent CK, Sanders EA, Bogaert D. Concordance between upper and lower airway microbiota in infants with cystic fibrosis. Eur Respir J 2017: 49(3): 1602235. [DOI] [PubMed] [Google Scholar]
- 13.Zemanick ET, Wagner BD, Robertson CE, Ahrens RC, Chmiel JF, Clancy JP, Gibson RL, Harris WT, Kurland G, Laguna TA, McColley SA, McCoy K, Retsch-Bogart G, Sobush KT, Zeitlin PL, Stevens MJ, Accurso FJ, Sagel SD, Harris JK. Airway microbiota across age and disease spectrum in cystic fibrosis. Eur Respir J 2017: 50(5): 1700832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frayman KB, Armstrong DS, Carzino R, Ferkol TW, Grimwood K, Storch GA, Teo SM, Wylie K, Ranganathan S. The lower airway microbiota in early cystic fibrosis lung disease: a longitudinal analysis. Thorax 2017: 72(12): 1104–1112. [DOI] [PubMed] [Google Scholar]
- 15.Gutierrez JP, Grimwood K, Armstrong DS, Carlin JB, Carzino R, Olinsky A, Robertson CF, Phelan PD. Interlobar differences in bronchoalveolar lavage fluid from children with cystic fibrosis [DOI] [PubMed]
- 16.Armstrong DS, Grimwood K, Carlin JB, Carzino R, Olinsky A, Phelan PD. Bronchoalveolar lavage or oropharyngeal cultures to identify lower respiratory pathogens in infants with cystic fibrosis. Pediatr Pulmonol 1996: 21(5): 267–275. [DOI] [PubMed] [Google Scholar]
- 17.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012: 486: 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007: 73(16): 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.R Core Team. R: A language and environment for statistical computing R Foundation for Statistical Computing, Vienna, Austria, 2017. [Google Scholar]
- 20.Okansen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Henry M, Stevens H, Szoecs E, Wagner H. Vegan: Community Ecology Package R package version; 2.4–6 ed, 2018. [Google Scholar]
- 21.Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nature Methods 2013: 10(12): 1200–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gangell C, Gard S, Douglas T, Park J, de Klerk N, Keil T, Brennan S, Ranganathan SC, Robins-Browne RM, Sly PD, CF A. Inflammatory responses to individual microorganisms in the lungs of children with cystic fibrosis. Clin Infect Dis 2011: 53(5): 425–432. [DOI] [PubMed] [Google Scholar]
- 23.Pillarisetti N, Williamson E, Linnane B, Skoric B, Robertson CF, Robinson P, Massie J, Hall GL, Sly P, Stick S, Ranganathan S, CF A. Infection, inflammation, and lung function decline in infants with cystic fibrosis. Am J Respir Crit Care Med 2011: 184(1): 75–81. [DOI] [PubMed] [Google Scholar]
- 24.Ramsey KA, Ranganathan S, Park J, Skoric B, Adams AM, Simpson SJ, Robins-Browne RM, Franklin PJ, de Klerk N, Sly P, Stick SM, Hall GL, CF A. Early respiratory infection is associated with reduced spirometry in children with cystic fibrosis. Am J Respir Crit Care Med 2014: 190(10): 1111–1116. [DOI] [PubMed] [Google Scholar]
- 25.Rosenfeld M, Farrell PM, Kloster M, Swanson JO, Vu T, Brumback L, Acton JD, Castile RG, Colin AA, Conrad CK, Hart MA, Kerby GS, Hiatt PW, Mogayzel PJ, Johnson RC, Davis SD. Association of lung function, chest radiographs and clinical features in infants with cystic fibrosis. Eur Respir J 2013: 42(6): 1545–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muhlebach MS, Zorn BT, Esther CR, Hatch JE, Murray CP, Turkovic L, Ranganathan SC, Boucher RC, Stick SM, Wolfgang MC. Initial acquisition and succession of the cystic fibrosis lung microbiome is associated with disease progression in infants and preschool children. PLoS Pathog 2018: 14(1): e1006798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pittman JE, Wylie KM, Akers K, Storch GA, Hatch J, Quante J, Frayman KB, Clarke N, Davis M, Stick SM, Hall GL, Montgomery G, Ranganathan S, Davis SD, Ferkol TW, CF. A. Association of antibiotics, airway microbiome and inflammation in infants with cystic fibrosis. Ann Am Thorac Soc 2017: 14(10): 1548–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biesbroek G, Tsivtsivadze E, Sanders EA, Montijn R, Veenhoven RH, Keijser BJ, Bogaert D. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med 2014: 190(11): 1283–1292. [DOI] [PubMed] [Google Scholar]
- 29.Mika M, Mack J, Korten I, Qi W, Aebi S, Frey U, Latzin P, Hilty M. Dynamics of the nasal microbiota in infancy: A prospective cohort study. J Allergy Clin Immunol 2015: 135(4): 905–912. [DOI] [PubMed] [Google Scholar]
- 30.Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, Holt BJ, Hales BJ, Walker ML, Hollams E, Bochkov YA, Grindle K, Johnston SL, Gern JE, Sly PD, Holt PG, Holt KE, Inouye M. The Infant Nasopharyngeal Microbiome Impacts Severity of Lower Respiratory Infection and Risk of Asthma Development. Cell Host Microbe 2015: 17(5): 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.