Abstract
Purpose:
Brain malformations caused by 17p13.3 deletions include lissencephaly with deletions of the larger Miller-Dieker syndrome region or smaller deletions of only PAFAH1B1, white matter changes, and a distinct syndrome due to deletions including YWHAE and CRK but sparing PAFAH1B1. We sought to understand the significance of 17p13.3 deletions between the YWHAE/CRK and PAFAH1B1 loci.
Methods:
We analyzed the clinical features of six individuals from five families with 17p13.3 deletions between and not including YWHAE/CRK and PAFAH1B1 identified among individuals undergoing clinical chromosomal microarray testing or research genome sequencing.
Results:
Five individuals from four families have multi-focal white matter lesions while a sixth had a normal MRI. A combination of our individuals and a review of those in the literature with white matter changes and deletions in this chromosomal region narrows the overlapping region for this brain phenotype to ~345 kb, including 11 RefSeq genes, with RTN4RL1 haploinsufficiency as the best candidate for causing this phenotype.
Conclusion:
While previous literature has hypothesized dysmorphic features and white matter changes related to YWHAE, our cohort contributes evidence to the presence of additional genetic changes within 17p13.3 required for proper brain development.
Keywords: 17p13.3 microdeletion, leukoencephalopathy, chromosomal microarray, white matter
Introduction:
There is a spectrum of microdeletion syndromes associated with 17p13.3 deletions. Miller-Dieker syndrome (OMIM#247200), associated with severe lissencephaly, dysmorphic features and multiple congenital anomalies, occurs when a deletion includes YWHAE and PAFAH1B1 (LIS1). Isolated lissencephaly sequence (OMIM#607432), with primarily only lissencephaly occurs with smaller deletions involving PAFAH1B1 but not YWHAE.1 Individuals with smaller deletions including YWHAE and CRK but sparing LIS1 have growth restriction, cognitive impairment, dysmorphic features, and various brain abnormalities.2–4 White matter abnormalities have been reported in some individuals with 17p13.3 microdeletions, and previous literature has hypothesized that this may be related to YWHAE haploinsufficiency.2–5 We report a cohort of children with primarily white matter changes on brain MRI but no significant cognitive impairment. All have small interstitial 17p13.3 microdeletions proximal to CRK, which has been hypothesized to be associated with short stature,2,3 and distal to PAFAH1B1, associated with lissencephaly.1
Materials & Methods:
Subject ascertainment
This study was approved by the Baylor College of Medicine Institutional Review Board for Human Subjects Research. Individual 1 enrolled in the Undiagnosed Diseases Network (UDN) with a known chromosomal deletion involving 17p13.3, since the pathogenicity of the deletion and the etiology of her disease was not well established. Review of Baylor Genetics cytogenetic database for individuals with similar deletions limited to the region between YWHAE/CRK and PAFAH1B1 identified three other individuals with white matter changes and one individual with a normal brain MRI. Two of these individuals were brothers (Subjects 2–3) and also enrolled in the UDN. Subject 5 was referred after an investigator (AV) learned learned of the study cohort. A literature search with 17p13 search terms allowed identification of other reports of similar white matter changes in individuals with deletions involving this region. No individuals in the DECIPHER database with deletions in the same region showed similar white matter changes, although MRI information was only available for a single individual (2726). Informed consent was obtained to publish identifiable clinical data presented in this paper.
Chromosomal microarray testing
V8, V9, and V10 chromosomal microarrays (CMAs) were designed by Baylor Medical Genetics Laboratories and manufactured by Agilent (Santa Clara, CA, USA). Subjects 2, 4, and 6 were studied by the V8 array, which included approximately 180,000 oligonucleotides and covered ~1,714 genes with an average of 4.2 probes per exon, excluding low-copy repeats and other repetitive sequences.6 Subjects 3 and 1 were studied by the V9 and V10 arrays respectively, which targeted over 4,900 genes at the exon level plus 60,000 probes used for SNP analysis for the detection of uniparental disomy and absence of heterozygosity.7
Exome sequencing
Subjects 1 and 2 had exome sequencing according to previously described methods.8
Genome sequencing
Subject 5 had genome sequencing through the Illumina Clinical Services Laboratory TruGenome Undiganosed Disease test. Sequencing libraries were prepared using Illumina’s TruSeq DNA PCR-Free kit and sequenced on a HiSeqX. Variant calling was performed using the Illumina ISAS pipeline, with single nucleotide variants (SNVs) and indels called using the Strelka germline caller9 and copy number variants (CNVs) identified with Canvas.10 All variants were classified according to the American College of Medical Genetics standards and guidelines for interpretation of genetic sequence variants.
Results
Subject 1 was born full-term and required phototherapy for hyperbilirubinemia. She met all of her early milestones and attends regular classes but has difficulty with short-term memory.
At age 7 years she had recurrent vomiting after a viral illness. Additional symptoms included headaches, dizziness, inability to concentrate, and tingling in the legs. Brain and spine MRI showed significant white matter hyperintensities in the brain (Figure 1A), a Chiari I malformation, and a tethered cord. The white matter lesions remained unchanged on subsequent MRIs. Additional anomalies include a patent ductus arteriosus, a low IgA screen for celiac disease, and joint laxity (Beighton score 4/9). On examination she is normocephalic and nondysmorphic. Genetic workup included normal metabolic testing for arylsulfatase A, galactocerebrosidase, and thymidine levels. CSF analysis showed normal protein, glucose, and lactate. Infectious workup for tuberculosis, enterovirus and HSV was negative. CMA showed a 0.7 Mb de novo 17p13.3 deletion (Figure 1E, Table 1). Exome sequencing did not show any pathogenic variants or candidate genes associated with white matter changes, nor rare variants in the non-deleted 17p13.3 alleles.
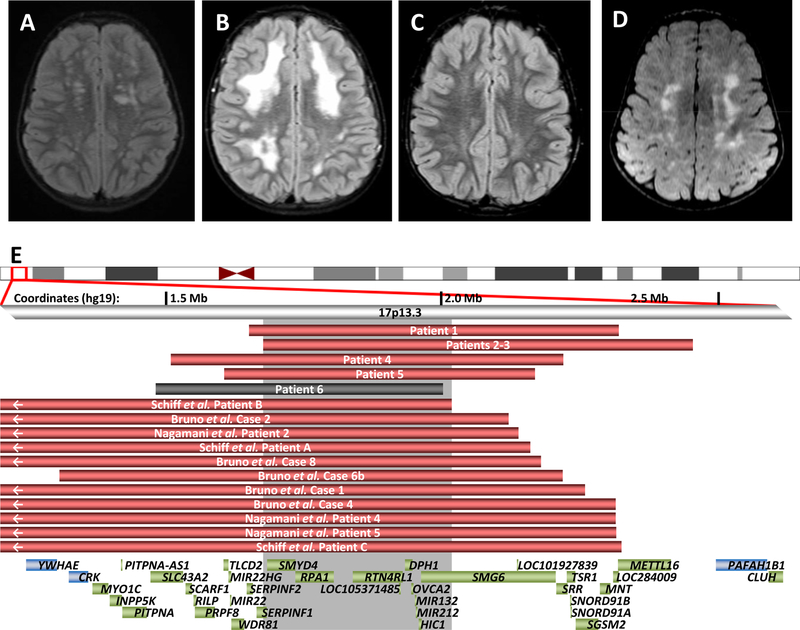
Figure 1. Leukoencephalopathies in individuals with 17p13.3 deletions.
Axial T2 weighted FLAIR (fluid attenuated inversion recovery) images demonstrate bilateral T2 hyperintensities in Subject 1 (A), Subject 2 (B), Subject 3 (C) and Subject 5 (D). (E) Deletions associated with abnormal MRIs in our cohort and review of the literature are red; the individual with a normal MRI is shown in gray. Gray shaded area represents the smallest region of overlap among cases with characteristic leukoencephalopathy. Genes in the region are shown as green boxes, with the critical genes of note flanking the region shown in blue.
Table 1.
Genotypes and phenotypes in individuals with 17p13.3 deletions between YWHAE/CRK and PAFAH1B1.
| Patient | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| Deletion (hg19) | 1646282–2322673 | 1674154–2456949 | 1509667–2219036 | 1607541–2167224 | 1480995–2002424 | |
| Inheritance | De novo | Paternal | Maternal | De novo | De novo | |
| Sex, Age | Female, 8 years | Male, 8 years | Male, 5 years | Male, 3 years | Male, 3 years | Female, 7 years |
| Weight/Height/ OFC | 17th/31st/10th | 95th/89th/97th | 87th/78th/99th (+2.45SDs) | NS | <1st/<1st/43th | <5th/<5th/14th |
| MRI findings | Chiari I, focal white matter lesions | Multifocal white matter lesions | Multifocal white matter lesions | Periventricular leukomalacia | T2 bilateral hyperintensities | Normal MRI |
| Development | Normal | Fine motor delays, regression?, normal IQ | Normal | NS | Normal | Normal |
| Behavior | Normal | ADHD, tics/ convulsions, anxiety, oppositional | ADHD, impulsive, disinhibited | NS | Normal | Normal |
| Additional neurologic findings | Mild dysarthria, tethered cord, poor concentration & short-term memory | Hypotonia | Hypotonia | NS | None | Learning difficulties, febrile seizures |
| Joint laxity | Mild | Present; pes planus | Present; pes planus | Present | Absent | Absent |
| Dysmorphisms | Mild synophrys, protruding ears | Pointed chin | None | NS | None | Short nose, pointed chin |
| Additional features | Patent ductus arteriosus, chronic constipation | Normal echo; scoliosis | Normal echo | NS | GH deficiency | Normal echo, clinodactyly |
ADHD, attention deficit hyperactivity disorder; GH, growth hormone, NS, not specified
Subject 2 was born at 33 weeks secondary to preterm labor. He was in the neonatal intensive care unit for 24 days and had a grade I intraventricular hemorrhage. He had early developmental delays but a normal IQ and ADHD at age 6. He underwent a brain MRI for worsening behaviors without regression that showed extensive bilateral multifocal T2 hyperintensities (Figure 1B). Multiple subsequent brain MRIs were stable. Physical exam showed macrocephaly with head circumference >95%, frontal bossing and hypermobile joints (Beighton score 6/9). Echocardiogram and eye exams were normal. Genetic workup included normal urine mucopolysaccharide screening, serum very long chain fatty acids and arylsulfatase activity. CMA showed a 0.8 Mb 17p13.3 deletion (Figure 1E, Table 1) and a small deletion on 15q11.2 including only non-coding exons of SNRPN. The 17p13.3 deletion was inherited from his father and shared with a younger brother (Individual 3). Their father may also be similarly affected. He had problems with rolling his ankles when he was younger, as well as behavior issues. He has a cranial metal plate after a motor vehicle accident and therefore is unable to obtain a brain MRI, but a CT scan at the time of the accident showed old white matter changes; additional details are not available. In Individual 2, exome sequencing did not show any pathogenic variants or candidate genes associated with white matter changes, nor rare variants in the non-deleted 17p13.3 alleles.
Subject 3 was born full-term with normal delivery and newborn course. He met all of his early milestones. Brain MRI at 3 years, performed due to macrocephaly and his brother’s MRI findings, showed patchy punctate bilateral multifocal T2 hyperintensities (Figure 1C) and 2 small arachnoid cysts. Multiple subsequent brain MRIs were stable. His exam is significant for head circumference >98% with frontal bossing and hypermobile joints (Beighton score 6/9). Echocardiogram showed mild mitral and tricuspid regurgitation. He was diagnosed with ADHD at age 5.
Subject 4 is a 3-year-old male with limited additional information secondary to loss of follow-up. MRI reported periventricular leukomalacia. CMA showed a 0.7 Mb maternally inherited 17p13.3 deletion (Figure 1E, Table 1).
Subject 5 was born full-term with no neonatal complications. Early motor and language development were normal. He underwent brain MRI due to low IGF-1 and growth hormone deficiency. He has a normal pituitary but incidentally found bilateral T2 hyperintensities in the brain (Figure 1D). An extensive evaluation for leukodystrophy was unremarkable apart from a de novo 0.6 Mb 17p13.3 deletion (Figure 1D, Table 1) detected through a research genome. At age three years, the individual has normal development and neurological exam.
Subject 6 is a 7-year-old female referred to genetics for short stature. She has relative microcephaly with dysmorphic features including deep-set eyes, upslanting palpebral fissures, mild midface hypoplasia, and a pointed chin. Normal laboratory workup included thyroid studies, urinalysis and IGF-1. Brain MRI performed at 9 years did not show white matter abnormalities. CMA showed a 0.5 Mb de novo 17p13.3 deletion (Figure 1E, Table 1).
Discussion
Microdeletion syndromes involving 17p13.3 have a range of phenotypes, with the most severe being Miller-Dieker syndrome, with primary lissencephaly, growth failure, intellectual disability and dysmorphic features.1 Genotype-phenotype correlations have elucidated the roles of genes: PAFAH1B1 involved in brain development1 and CRK in short stature.2,3 In reviewing the literature, similar white matter changes have also been reported in eleven other individuals with microdeletions in this region (Figure 1E, Table S1).2–4 We report a small cohort of children with white matter lesions and normal cognition with small 17p13.3 microdeletions between and not including YWHAE/CRK and PAFAH1B1. Previous literature has speculated a possible role for YWHAE in the brain anomalies, including the white matter changes.2–5 Our cohort suggests a different gene is likely responsible for the white matter findings, although it is likely that multiple 17p13.3 genes contribute to brain development. Due to the normal MRI in individual 6 whose deletion includes all genes in the smallest region of overlap (SRO) for this phenotype (Figure 1E), reduced penetrance is likely for these white matter changes.
White matter changes in the brain are often concerning for possible neurodegenerative leukodystrophy. Our cohort demonstrates that white matter changes can be associated with static, less severe conditions. The lesions in individuals 1–3 appear to be static based on multiple brain MRIs in each individual and the likely presence of similar findings in the father of individuals 2 and 3. The white matter changes in our cohort and those previously reported who have MRI images to review are similar, located mostly in the subcortical regions suggestive of enlarged perivascular spaces. There is no evidence of more severe brain injury such as gliosis that may have been associated with prematurity or an infectious process.
There are eleven RefSeq genes within our newly defined SRO (Figure 1D) for these white matter changes, and none has been previously associated with such brain findings. It is unclear whether individual genetic changes are responsible for white matter abnormalities in these individuals or if these are a feature of CNV in general. Similar white matter changes have been described in individuals with other CNVs, although for the most part no causative genes have been confirmed within CNVs.11 Exceptions for example include haploinsufficiency of MBP (myelin basic protein) in 18q23 deletions which have been linked to white matter changes on MRI.12
Most of these genes found in our SRO are not associated with Mendelian disorders, although RTN4RL1, SMG6, MIR132 and MIR212 are expressed in the brain. SMG6 and RTN4RL1 have the highest pLIs (probabilities of loss-of-function intolerance,13 1.00 and 0.79, respectively) of the SRO genes. RTN4RL1 regulates axonal and dendritic growth,14 and it may serve as a receptor for Nogo-66, a myelin-associated inhibitor.15 Of note, the Nogo-66 receptor gene RTN4R is located in 22q11.21, recurrent deletions of which can cause white matter abnormalities, a phenotype which has been hypothesized to be further modulated by polymorphisms in the non-deleted RTN4R allele.16 SMG6 is involved in mRNA decay.17 As microRNAs MIR132 and MIR212 are noncoding, pLI scores are not applicable, although attenuation of Mir132 in mice reduces neurite outgrowth.18
It has been hypothesized that HIC1 haploinsufficiency (pLI score not available) contributes to the facial features and heart and gastrointestinal anomalies in Miller-Dieker syndrome,19 although these features are not prominent in our cohort. Given HIC1’s role in precartilaginous tissues and muscle development,19 this could be a candidate for the hypermobility in some of our individuals. Additionally, as HIC1 acts as a tumor suppressor,20 future monitoring of individuals with HIC1 deletions for risk of neoplasia is possibly warranted. The remainder of the genes in the SRO have low pLI scores, and two (SERPINF1 and DPH1) are associated with recessive disease (OMIM#613982 and #616901), so it is less likely that these genes are haploinsufficient. However, as these white matter changes may have minimal outward phenotypic impacts, we cannot rule out the possibility that one of these other genes with more tolerance to loss-of-function contributes to the MRI findings.
When white matter changes are found, long-term follow-up is sometimes recommended to evaluate whether the leukoencephalopathy is static. Given the likely presence of such changes in an adult (father of individuals 2–3) and normal development in our cohort overtime, reassurance may be given to families with similar 17p13.3 deletions regarding the likely evolution of their white matter changes over time, and repeated MRIs should not be mandatory.
Our cohort contributes evidence to the presence of multiple genes within 17p13.3 required for normal brain development. Identification of additional individuals with deletions and pathogenic variants within this region may help to identify specific genes responsible. Additional studies, including diffusion tensor imaging and downstream techniques such as RNAseq to evaluate for abnormal expression of genes within and adjacent to the deletion and in relevant developmental pathways, may assist with elucidating genetic etiologies of abnormal myelination.
ACKNOWLEDGEMENTS
We thank the individuals and their families for their participation in this research. Research reported in this manuscript was supported by the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director under Award Number U01 HG007709–01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was also supported by the BCM Intellectual and Developmental Disabilities Research Center (HD024064) from the Eunice Kennedy Shriver National Institute of Child Health and NIGMS T32 GM007526 (BL, LE, MJ, LB, JB). LCB is also supported by a Career Award for Medical Scientists from the Burroughs Wellcome Fund and NIH K08DK106453.
The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing conducted at Baylor Genetics, including chromosomal microarray testing. John W. Belmont, Ryan Taft, Denise Perry and Krista Bluske are employees and shareholders of Illumina Inc. Honey Nagakura is an employee of LabCorp.
Footnotes
Conflict of interest notification
The remaining authors have no conflicts to declare.
REFERENCES
- 1.Dobyns WB, Das S. LIS1-Associated Lissencephaly/Subcortical Band Heterotopia. In: Adam MP, Ardinger HH, Pagon RA, et al. , eds. GeneReviews((R)) Seattle (WA)1993. [Google Scholar]
- 2.Nagamani SC, Zhang F, Shchelochkov OA, et al. Microdeletions including YWHAE in the Miller-Dieker syndrome region on chromosome 17p13.3 result in facial dysmorphisms, growth restriction, and cognitive impairment. J Med Genet December 2009;46(12):825–833. [DOI] [PubMed] [Google Scholar]
- 3.Bruno DL, Anderlid BM, Lindstrand A, et al. Further molecular and clinical delineation of co-locating 17p13.3 microdeletions and microduplications that show distinctive phenotypes. J Med Genet May 2010;47(5):299–311. [DOI] [PubMed] [Google Scholar]
- 4.Schiff M, Delahaye A, Andrieux J, et al. Further delineation of the 17p13.3 microdeletion involving YWHAE but distal to PAFAH1B1: four additional individuals. Eur J Med Genet Sep-Oct 2010;53(5):303–308. [DOI] [PubMed] [Google Scholar]
- 5.Noor A, Bogatan S, Watkins N, Meschino WS, Stavropoulos DJ. Disruption of YWHAE gene at 17p13.3 causes learning disabilities and brain abnormalities. Clin Genet February 2018;93(2):365–367. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhry A, Noor A, Degagne B, et al. Phenotypic spectrum associated with PTCHD1 deletions and truncating mutations includes intellectual disability and autism spectrum disorder. Clin Genet September 2015;88(3):224–233. [DOI] [PubMed] [Google Scholar]
- 7.Wiszniewska J, Bi W, Shaw C, et al. Combined array CGH plus SNP genome analyses in a single assay for optimized clinical testing. Eur J Hum Genet January 2014;22(1):79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y, Muzny DM, Xia F, et al. Molecular findings among individuals referred for clinical whole-exome sequencing. JAMA : the journal of the American Medical Association November 12 2014;312(18):1870–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics July 15 2012;28(14):1811–1817. [DOI] [PubMed] [Google Scholar]
- 10.Ivakhno S, Roller E, Colombo C, Tedder P, Cox AJ. Canvas SPW: calling de novo copy number variants in pedigrees. Bioinformatics September 27 2017. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Cazorla A, Sans A, Baquero M, et al. White matter alterations associated with chromosomal disorders. Dev Med Child Neurol March 2004;46(3):148–153. [DOI] [PubMed] [Google Scholar]
- 12.Loevner LA, Shapiro RM, Grossman RI, Overhauser J, Kamholz J. White matter changes associated with deletions of the long arm of chromosome 18 (18q- syndrome): a dysmyelinating disorder? AJNR. American journal of neuroradiology Nov-Dec 1996;17(10):1843–1848. [PMC free article] [PubMed] [Google Scholar]
- 13.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature August 18 2016;536(7616):285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pignot V, Hein AE, Barske C, et al. Characterization of two novel proteins, NgRH1 and NgRH2, structurally and biochemically homologous to the Nogo-66 receptor. Journal of neurochemistry May 2003;85(3):717–728. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L, Kuang X, Zhang J. Nogo receptor 3, a paralog of Nogo-66 receptor 1 (NgR1), may function as a NgR1 co-receptor for Nogo-66. Journal of genetics and genomics = Yi chuan xue bao November 20 2011;38(11):515–523. [DOI] [PubMed] [Google Scholar]
- 16.Perlstein MD, Chohan MR, Coman IL, et al. White matter abnormalities in 22q11.2 deletion syndrome: preliminary associations with the Nogo-66 receptor gene and symptoms of psychosis. Schizophrenia research January 2014;152(1):117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ottens F, Boehm V, Sibley CR, Ule J, Gehring NH. Transcript-specific characteristics determine the contribution of endo- and exonucleolytic decay pathways during the degradation of nonsense-mediated decay substrates. RNA August 2017;23(8):1224–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vo N, Klein ME, Varlamova O, et al. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci U S A November 8 2005;102(45):16426–16431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grimm C, Sporle R, Schmid TE, et al. Isolation and embryonic expression of the novel mouse gene Hic1, the homologue of HIC1, a candidate gene for the Miller-Dieker syndrome. Hum Mol Genet April 1999;8(4):697–710. [DOI] [PubMed] [Google Scholar]
- 20.Fleuriel C, Touka M, Boulay G, Guerardel C, Rood BR, Leprince D. HIC1 (Hypermethylated in Cancer 1) epigenetic silencing in tumors. Int J Biochem Cell Biol January 2009;41(1):26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]