Abstract
G protein-coupled receptors (GPCRs) in the G protein-coupled active state have higher affinity for agonists compared to when they are in the inactive state, but the molecular basis for this is unclear. We have determined four active-state structures of the β1-adrenoceptor (β1AR) bound to conformation-specific nanobodies in the presence of agonists of varying efficacy. Comparison with inactive-state structures of β1AR bound to the identical ligands showed a 24-42% reduction in the volume of the orthosteric binding site. Potential hydrogen bonds were also shorter, and there was up to a 30% increase in the number of atomic contacts between the receptor and ligand. This explains the increase in agonist affinity of GPCRs in the active state for a wide range of structurally distinct agonists.
GPCRs exist in an ensemble of conformations that can be selectively stabilized by the binding of a ligand and through interactions with signaling molecules such as G proteins (1, 2). Pharmacology has characterized at least two distinct states of GPCRs, an active state with high affinity for agonists when coupled to G proteins and an inactive state with low affinity for agonists in the absence of G proteins (1), although a plethora of sub-states can also exist between these two extremes (3–7). The reason why the active state has a high affinity for agonists is unclear, because receptor structures in the inactive and active states have been determined bound to different ligands, such as for the β2-adrenoceptor (β2AR) (8–11). Here we present structures of β1AR in the active state and compare them to inactive state structures (12) bound to the identical ligand to define the structural differences in the orthosteric binding site.
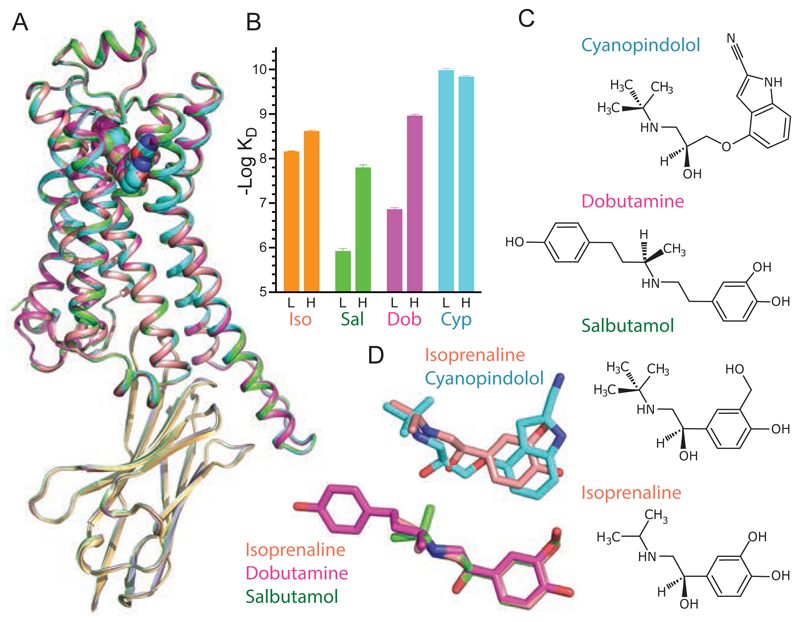
Four crystal structures with overall resolutions between 3.0 Å - 3.2 Å (Table S1) were determined of β1AR bound to either Nb80 or Nb6B9, and the overall structures were virtually identical (Fig. 1; 0.2-0.3 Å RMSD for Cα atoms). Nb80 and Nb6B9 are nanobodies originally developed to stabilize the active state of β2AR (8, 10) and bind to β1AR due to the high sequence conservation of the receptors. Structures were determined bound to a full agonist (isoprenaline), partial agonists (salbutamol, dobutamine) and a weak partial agonist (cyanopindolol). Isoprenaline, salbutamol and dobutamine showed an increase in affinity when β1AR was coupled to the engineered G protein mini-Gs, whereas cyanopindolol bound with similar high affinity in both the presence and absence of mini-Gs (Fig. 1). The overall structure of the β1AR-nanobody complexes bound to either agonist or partial agonists is virtually identical to that of the agonist-bound Nb6B9-β2AR complex (0.5 Å RMSD of 1601 atoms) and the overall conformational changes are virtually identical. These changes result in the partial occlusion of the orthosteric binding pocket (Fig. 2), which is consistent with observations on nanobody-bound β2AR (8, 10).
Fig. 1. Structure of the active state of agonist-bound β1AR-nanobody complex.
(A) Superposition of four structures of β1AR-nanobody complexes bound to ligands shown in (C). (B) Affinities of β1AR in the low affinity state, L, and high affinity state coupled to mini-Gs, H, for the ligands co-crystallised with the receptor. Data are in Tables S2 and S3, and Fig. S6. Results are the mean of 2-4 experiments performed in duplicate with error bars representing the SEM. (C) Structures of the ligands co-crystallised in the β1AR complexes. (D) Disposition of the ligands after superposition of the receptors, using the same colour coding as in (B). See Fig. S9 for ligand densities.
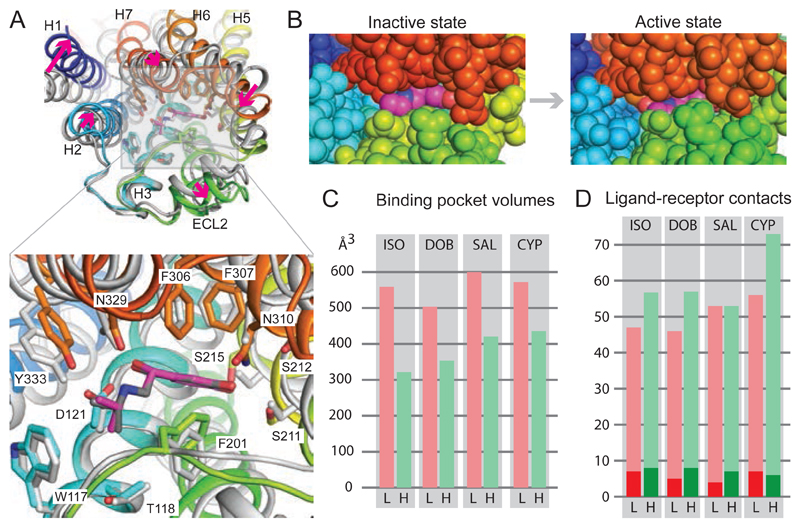
Fig. 2. Conformational changes in isoprenaline-bound β1AR.
(A), Superposition of isoprenaline-bound β1AR in the inactive state (grey, PDB ID 2Y03) with isoprenaline-bound β1AR in the active state (rainbow colouration). Arrows (magenta) indicate the transitions from the inactive to active state. Alignment was performed based on the isoprenaline molecules using PyMol (magenta, isoprenaline bound to active state β1AR). (B) View of the orthosteric binding site from the extracellular surface with atoms shown as space filling models: isoprenaline (magenta, carbon atoms); β1AR: H1, dark blue; H2, light blue; H5, yellow; ECL2, green; ECL3 and parts of H6 and H7, red. (C) Volumes of the orthosteric binding site in the low-affinity inactive state (L, pink bars) compared to the high-affinity active state (H, green bars). (D) Number of atomic contacts (Database S1) between the respective ligands and β1AR in the low-affinity inactive state (L, pink bars) compared to the high-affinity active state (H, green bars). The dark shades represent the number of polar interactions. Ligand abbreviations are shown in Fig. 1.
Detailed comparisons were made between the inactive state structures of β1AR with the respective active state structures bound to same ligand (Figs. 2 and 3). In all cases, there was a decrease in the volume (13) of the orthosteric binding site that varied depending on the ligand (Fig 2, Fig. S1). The largest decrease was observed for the full agonist isoprenaline (42%) and the smallest decrease was observed for the weak partial agonist cyanopindolol (24%). The decrease in the volume of the orthosteric binding site when isoprenaline was bound was due primarily to the inward movement of the extracellular ends of H6 and H7, an inward movement and an increase in the H5 bulge at Ser2155.46 and the reorientation of residues Phe201ECL2 and Phe3257.35. The magnitude of these changes was greatest for the full agonist isoprenaline and smallest for the weak partial agonist cyanopindolol. The pincer-like movement of Phe201ECL2 and Phe3257.35 towards the ligand has the largest effect on reducing the volume of the orthosteric binding pocket, with the maximal shift observed in the isoprenaline structure of 3.1 Å for Phe201ECL2 and 2.5 Å for Phe3257.35 (measured at the CZ atom of the side chain). The movement of Phe201ECL2 appeared to correlate with the structure of the ligand bound, because in all cases it formed van der Waals contacts with the ligand. In contrast, Phe3257.35 was not within van der Waals contact with any of the four ligands and moved as a consequence of the inward tilt of H7.
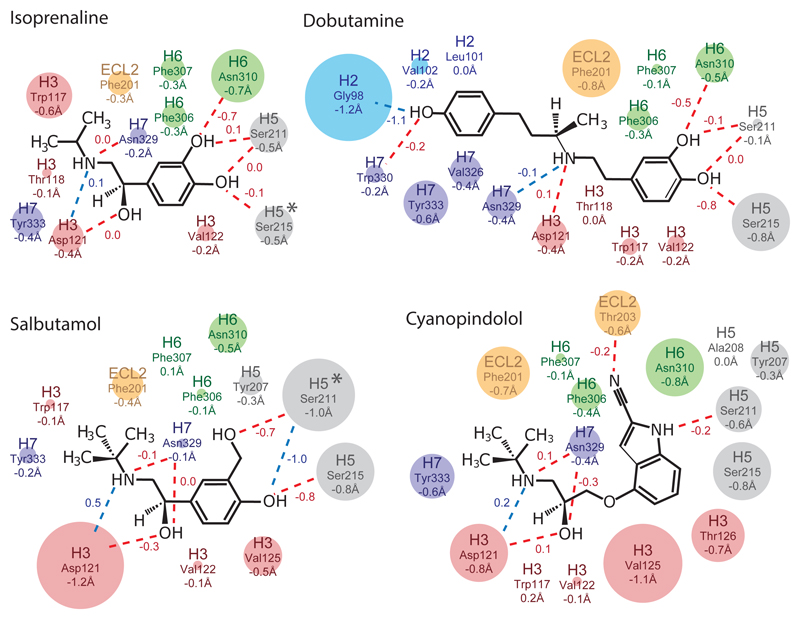
Fig. 3. Changes in β1AR-ligand contact distances.
The maximal changes in contact distances between ligands and atoms in β1AR from the inactive to active states are depicted. Amino acid side chains making contact to the ligands are indicated and coloured according to where they are in β1AR (blue, H2; red, H3; orange, ECL2; grey, H5; green, H6; purple, H7) with the diameter of the circle representing the magnitude of the distance change (shown as numbers below the amino acid residue). Numbers next to the lines indicate the change in length of polar contacts (blue dashed lines) and hydrogen bonds (red dashed lines; determined using HBPLUS). Negative numbers imply a decrease in distance between the ligand and receptor in the transition from the inactive state to the active state. An asterisk indicates a significant rotamer change between the inactive and active states. For the details of additional contacts made by each side chain, see Fig. S2.
The reduction in the volume of the orthosteric binding pocket correlated with an overall reduction in the average distance between atoms in the ligand and receptor by 0.1-0.3 Å. Amino acid residues in H3, H5, H6, H7 and ECL2 (and H2 for dobutamine) were all involved in contributing to ligand-receptor contacts, but there was no clear pattern to which regions of the receptor changed most significantly (Fig. 3, Fig. S2). Side chains were up to 1.2 Å closer to the ligand in the active state compared to the inactive state. In a number of instances, changes resulted in the strengthening of hydrogen bonds. For example, Asn3106.55 was predicted to make a weak hydrogen bond to the para-hydroxyl group of isoprenaline (3.5 Å between donor and acceptor) in the inactive state, which changed to 2.8 Å in the active state. In the active-state structures containing dobutamine and salbutamol, the distance to Ser2155.46 was 0.8 Å shorter for both ligands allowing hydrogen bond formation; the hydrogen bond to Ser2115.42 also shortened by 0.7 Å to the para hydroxyl in salbutamol, but remained unchanged to the meta hydroxyl in dobutamine. Most of the observed differences are due to the contraction of the binding pocket, whereas the significant shortening of the hydrogen bond between Ser2115.42 and salbutamol is due to a rotamer change. Although all the ligand binding pockets contracted upon receptor activation, the changes in ligand-receptor contacts were not conserved, despite the similarity in chemotypes amongst the four ligands studied. There was a weak correlation between the decrease in volume of the orthosteric binding site and ligand efficacy (Fig. S3), particularly if the most similar chemtoypic ligands were compared (cyanopindolol, salbutamol, isoprenaline). However, there was no correlation between efficacy and the magnitude of ligand affinity increase on receptor activation or the increase in the number of ligand-receptor atomic contacts (Fig. S3). It was particularly notable that cyanopindolol bound to β1AR with similar affinity in both the presence and absence of a coupled G protein (Fig. 1) despite the contraction of the binding pocket and increase in receptor ligand contacts upon activation. This may be a consequence of constraints on the possible conformation change imposed by the rigidity of cyanopindol that prevents the full contraction of the ligand binding pocket by preventing the movement of H7 and the bulge in H5 observed in the other structures (Fig. S1).
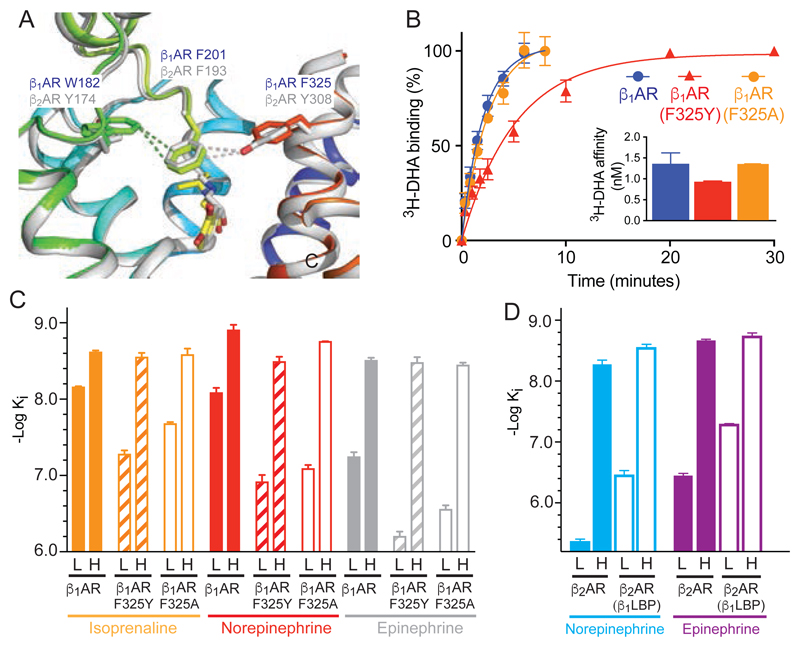
The role of the partial occlusion of the orthosteric binding site upon activation of β1AR was tested by mutagenesis inspired from the active state structure of β2AR (8, 10). In β2AR, it was proposed that the occlusion of the binding site was a significant factor in increasing agonist affinity upon G protein coupling (14). In particular, Tyr3087.35 was within van der Waals distance of Phe193ECL2 on the opposite side of the entrance to the orthosteric binding pocket and had a major effect on decreasing the rates of association and dissociation of ligands in the active state compared to the inactive state (14). The β2AR residues Phe193ECL2/Tyr3087.35 are equivalent to Phe201ECL2/Phe3257.35 in β1AR, which are not in van der Waals contact (Fig. 4). Thus the mutation F325Y7.35 in β1AR was predicted to occlude the entrance to the orthosteric binding pocket and decrease the rate of ligand association, and conversely, F325A7.35 was predicted to make the entrance wider and increase the rate of ligand association. When the initial rate of 3H-dihydroalprenolol (3H-DHA) association was measured (Fig. 4), β1AR(F325A) had the same rate as β1AR, but β1AR(F325Y) had a considerably slower rate of association. However, the affinities (Fig. 4) of epinephrine and isoprenaline for the high affinity state of β1AR and β1AR(F325Y) were identical and there was only a minor difference with norepinephrine. Comparisons of affinities (Fig. 4) for the inactive states of β1AR and β1AR(F325Y) showed a large decrease in affinities for norepinephrine (14.8-fold), epinephrine (11.2-fold) and isoprenaline (7.6-fold), which implied that the greater agonist affinity shift observed in β1AR(F325Y) compared to β1AR was due to destabilisation of the inactive state and not stabilisation of the active state. This suggested that partial occlusion of the ligand binding pocket in β1AR(F325Y) during formation of the active state played little role in the increase of agonist affinity on G protein coupling.
Fig. 4. Comparisons between β1AR and β2AR.
(A) Alignment of the active state structures of β1AR (rainbow colouration) and β2AR (grey, PDB ID 4LDO). Ligands are shown as sticks; isoprenaline, yellow; adrenaline, grey. (B) Rate of association of the radioligand 3H-DHA on to β1AR (blue circles), β1AR(F325Y) (red triangles) and β1AR(F325A) (orange circles). The inset shows the affinities of 3H-DHA (same colour code). (C, D) Affinities of β1AR, β2AR and their respective mutants in the low affinity state, L, and high affinity state coupled to mini-Gs, H. All data are in Tables S2 and S3 and representative graphs of affinity shifts are in Figure S6. Results are the mean of 2-6 experiments performed in duplicate with error bars representing the SEM.
The destabilising effect of the F325Y mutation in β1AR on the agonist-bound inactive state suggested that converting the extracellular surface of β2AR to make it similar to β1AR would increase the affinity of the inactive state and leave the affinity of the G protein-coupled activated state approximately unchanged. The β2AR mutant constructed, β2AR(β1LBP), did indeed show these characteristics (Fig S4; see Methods for the rationale of the four mutations used: Y174WECL2, H296N6.58, K305D7.32 and Y308F7.35). In addition, the accessibility of the β1AR orthosteric binding pocket in the G protein-coupled state to 125I-cyanopindolol was greater than that observed for β2AR (Fig. S5). The four mutations in β2AR(β1LBP) converted the behaviour of β2AR to that of β1AR. Adding the converse residues from β2AR into β1AR, to make the mutant β1AR(β2LBP), converted the accessibility of the orthosteric binding site in β1AR to that of β2AR (Fig. S5).
The increase in affinity of agonist binding to the active state of β1AR arises from the increase in the number and/or strength of ligand-receptor contacts (a thermodynamic effect). This is consequently associated with a decrease in the rate of ligand dissociation. Where a native ligand is a large peptide that interacts with extracellular domains, this thermodynamic effect may be the major contributor to the decrease in the rate of ligand dissociation. However, where the ligand is small, such as adrenaline or acetylcholine, the rate of ligand dissociation may also be decreased due to a purely steric blockage of the entrance to the ligand binding pocket (a kinetic effect) (14). Given the high conservation of both the structure and function of GPCRs (15, 16), these considerations are likely to apply to other GPCRs that bind diffusible ligands.
Supplementary Material
One Sentence Summary.
High affinity agonist binding to G protein-coupled GPCRs results from an increase in the number and strength of protein-ligand interactions.
Acknowledgments
We thank the beamline staff at the European Synchrotron Radiation Facility (beamlines ID23-2, ID30-A3, ID29, ID30B and MASSIF-1) and at Diamond Light Source (beamline I24).
Funding: This work was supported by core funding from the Medical Research Council [MRC U105197215 and U105184325] and a grant from the ERC (EMPSI 339995)
Footnotes
Author contributions: T.W. performed receptor and nanobody expression, purification, crystallization, cryo-cooling of the crystals, data collection, data processing and structure refinement. T.W. also performed the pharmacological analyses. P.C.E. purified mini-Gs and A.D.S assisted with structure solution. A.G.W.L. was involved in data processing and structure solution, refinement and analysis. Manuscript preparation was performed by T.W., A.G.W.L. and C.G.T. The overall project management was by C.G.T.
Competing interests: C.G.T. is a shareholder, consultant and member of the Scientific Advisory Board of Heptares Therapeutics, who also partly funded this work
Data and materials availability: The co-ordinates and structure factors for all the structures determined have been deposited at the PDB with the following accession codes (ligand co-crystallised in parentheses): 6H7J (isoprenaline), 6H7L (dobutamine), 6H7M (salbutamol), 6H7O, cyanopindolol.
References and Notes
- 1.Gether U. Uncovering molecular mechanisms involved in activation of G protein-coupled receptors. Endocr Rev. 2000;21:90–113. doi: 10.1210/edrv.21.1.0390. [DOI] [PubMed] [Google Scholar]
- 2.Rosenbaum DM, Rasmussen SG, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vauquelin G, Van Liefde I. G protein-coupled receptors: a count of 1001 conformations. Fundam Clin Pharmacol. 2005;19:45–56. doi: 10.1111/j.1472-8206.2005.00319.x. [DOI] [PubMed] [Google Scholar]
- 4.Gregorio GG, et al. Single-molecule analysis of ligand efficacy in beta2AR-G-protein activation. Nature. 2017;547:68–73. doi: 10.1038/nature22354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao XJ, et al. The effect of ligand efficacy on the formation and stability of a GPCR-G protein complex. Proc Natl Acad Sci U S A. 2009;106:9501–9506. doi: 10.1073/pnas.0811437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manglik A, et al. Structural Insights into the Dynamic Process of beta2-Adrenergic Receptor Signaling. Cell. 2015;161:1101–1111. doi: 10.1016/j.cell.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye L, Van Eps N, Zimmer M, Ernst OP, Prosser RS. Activation of the A2A adenosine G-protein-coupled receptor by conformational selection. Nature. 2016;533:265–268. doi: 10.1038/nature17668. [DOI] [PubMed] [Google Scholar]
- 8.Rasmussen SG, et al. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rasmussen SG, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ring AM, et al. Adrenaline-activated structure of beta2-adrenoceptor stabilized by an engineered nanobody. Nature. 2013;502:575–579. doi: 10.1038/nature12572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenbaum DM, et al. Structure and function of an irreversible agonist-beta(2) adrenoceptor complex. Nature. 2011;469:236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warne T, et al. The structural basis for agonist and partial agonist action on a beta(1)-adrenergic receptor. Nature. 2011;469:241–244. doi: 10.1038/nature09746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durrant JD, Votapka L, Sorensen J, Amaro RE. POVME 2.0: An Enhanced Tool for Determining Pocket Shape and Volume Characteristics. J Chem Theory Comput. 2014;10:5047–5056. doi: 10.1021/ct500381c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeVree BT, et al. Allosteric coupling from G protein to the agonist-binding pocket in GPCRs. Nature. 2016;535:182–186. doi: 10.1038/nature18324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venkatakrishnan AJ, et al. Diverse activation pathways in class A GPCRs converge near the G-protein-coupling region. Nature. 2016;536:484–487. doi: 10.1038/nature19107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venkatakrishnan AJ, et al. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 17.Carpenter B, Tate CG. Engineering a minimal G Protein to facilitate crystallisation of G protein coupled receptors in their active conformation. Protein Eng Design Sel. 2016;29:583–593. doi: 10.1093/protein/gzw049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warne T, Chirnside J, Schertler GF. Expression and purification of truncated, non-glycosylated turkey beta-adrenergic receptors for crystallization. Biochim Biophys Acta. 2003;1610:140. doi: 10.1016/s0005-2736(02)00716-2. [DOI] [PubMed] [Google Scholar]
- 19.Warne T, Serrano-Vega MJ, Tate CG, Schertler GF. Development and crystallization of a minimal thermostabilised G protein-coupled receptor. Protein Expr Purif. 2009;65:204–213. doi: 10.1016/j.pep.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 20.Urbani A, Warne T. A colorimetric determination for glycosidic and bile salt-based detergents: applications in membrane protein research. Anal Biochem. 2005;336:117–124. doi: 10.1016/j.ab.2004.09.040. [DOI] [PubMed] [Google Scholar]
- 21.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCoy AJ, et al. Phaser crystallographic software. Journal of applied crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Carpenter B, Tate CG. Expression and Purification of Mini G Proteins from Escherichia coli. Bio Protoc. 2017;7 doi: 10.21769/BioProtoc.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baker JG, Proudman RG, Hill SJ. Identification of key residues in transmembrane 4 responsible for the secondary, low-affinity conformation of the human beta1-adrenoceptor. Mol Pharmacol. 2014;85:811–829. doi: 10.1124/mol.114.091587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baker JG, Proudman RG, Tate CG. The pharmacological effects of the thermostabilising (m23) mutations and intra and extracellular (beta36) deletions essential for crystallisation of the turkey beta-adrenoceptor. Naunyn Schmiedebergs Arch Pharmacol. 2011;384:71–91. doi: 10.1007/s00210-011-0648-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.