Abstract
The stringent response to nutrient deprivation is a stress response found throughout the bacterial domain of life. Although first described in proteobacteria for matching ribosome synthesis to the cell’s translation status and for preventing formation of defective ribosomal particles, the response is actually much broader, regulating many hundreds of genes—some positively, some negatively. Utilization of the signaling molecules ppGpp and pppGpp for this purpose is ubiquitous in bacterial evolution, although the mechanisms employed vary. In proteobacteria, the signaling molecules typically bind to two sites on RNA polymerase, one at the interface of the β′ and ω subunits and one at the interface of β′ secondary channel and the transcription factor DksA. The β′ secondary channel is targeted by other transcription regulators as well. Although studies on the transcriptional outputs of the stringent response date back at least 50 years, the mechanisms responsible are only now coming into focus.
Keywords: stringent response, promoter, ppGpp, DksA, TraR, stress response
1. HISTORICAL PERSPECTIVE
Early investigators of bacterial metabolism proposed that the major activities of the cell are balanced and coregulated so that certain important cell constituents increase synchronously. In the early 1950s, it was reported that a reduction in amino acid availability resulted in a severe decline in “pentose nucleic acid” (rRNA and tRNA) synthesis (104), coupling what would later be recognized as ribosome synthesis to protein synthesis. We now know that nutrient deprivation in bacterial cells results in a stress response, called the stringent response, that involves hundreds of genes. The term initially referred only to a response to amino acid limitation, but it is now recognized that the stimuli that induce the stringent response are much broader and the mechanisms involved differ in different branches of the bacterial domain. The term now generally refers to changes resulting from increases in production of the signaling molecules guanosine-5′, 3′-tetraphosphate (ppGpp) and/or guanosine-5′, 3′-pentaphosphate (pppGpp). Although the ratios of the two species vary with growth conditions, in Escherichia coli the tetraphosphate is usually the more abundant species (81), whereas in Bacillus subtilis the pentaphosphate is often more abundant (120). For brevity, we use ppGpp to refer to both the tetraphosphate and the pentaphosphate.
ppGpp production has dramatic effects on rRNA and tRNA synthesis, but even early on it was clear that the stringent response regulates expression of many other gene products as well, some of which decrease and some of which increase (reviewed in 51, 93). There is also an extensive literature on ppGpp binding to many other cellular targets besides RNA polymerase (RNAP) (22, 29, 60, 76, 127). Other recent reviews have focused on the signaling pathway, i.e., on ppGpp synthesis itself and its regulation, on the distribution of ppGpp synthesizing enzymes in evolution, on the role of ppGpp in specific bacterial subgroups, and on the role of ppGpp in bacterial persistence (5, 36, 41, 48, 52, 105).
The goal here is not to repeat these reviews. Rather, after some brief comments about induction of ppGpp synthesis and its regulation, we focus on the interaction of ppGpp with RNAP in proteobacteria and on identification of the direct effects of this interaction on transcription. Even on this limited aspect of the stringent response, the literature is voluminous—too large to cover here in its entirety. Thus we try to provide an update on what is known about the mechanism by which ppGpp directly regulates gene expression while touching only briefly on other important aspects of ppGpp function.
2. ppGpp SYNTHESIS AND ITS REGULATION
relA encodes the major synthase responsible for synthesizing ppGpp in E. coli (16, 21). relA mutants, cells in which the strict dependence of RNA synthesis on translation is relaxed, are unable to adjust efficiently to changing nutritional conditions (35). Later it was demonstrated that SpoT is the major ppGpp degradase, and it also has a weak synthase activity (123). Separate RelA and SpoT proteins are generally found in beta- and gammaproteobacteria, whereas in most other bacteria the synthase and degradase activities are combined in a single RSH (RelA SpoT Homolog) protein (5, 52, 54, 80).
Some phyla, e.g., Firmicutes, also contain SAS proteins (small alarmone synthetases) that can synthesize ppGpp. There are also examples of SAH proteins (small alarmone hydrolases). Both the RSH and SAS enzymes are subject to allosteric regulation by ppGpp itself. The regulation of RSH and SAS enzyme activity in different bacterial species has been the subject of several excellent reviews (36, 52, 105). Here we wish to make only a few comments.
Recent structures of RelA bound to the ribosome (3, 19, 78) indicate that the C-terminal autoregulatory domain of RelA interacts directly with the C terminus of uncharged tRNAs in the ribosomal A-site, suggesting that this sequestration activates the ppGpp synthase activity. Conversely, when the tRNA is charged with an amino acid at its C terminus, the covalently bound amino acid prevents the interaction of RelA with the tRNA, turning off ppGpp synthesis. RelA interacts with the Sarcin-Ricin loop of 23S rRNA, consistent with the model that RelA senses uncharged tRNAs and is activated to produce ppGpp when bound to the ribosome (49, 120a) and not consistent with an alternative proposal that RelA is released from the ribosome in response to the uncharged tRNA and then produces ppGpp (32).
The regulation of SpoT, the bifunctional ppGpp synthase/degradase responsible for the residual ppGpp levels in relA mutant cells, is much less clear. Additional work is needed to determine the interactions and molecular mechanisms underlying the shift in the equilibrium between the hydrolase and synthase activities. Some appealing proposals have been made in which SpoT interactions with other cellular constituents are responsible for turning on/off SpoT hydrolase/synthase activity, thereby adjusting ppGpp levels to stresses unrelated to translation.
For example, it has been proposed that SpoT might couple ppGpp synthesis to cellular fatty acid metabolism by interacting with acyl carrier protein, ACP, and triggering a conformational switch in SpoT that leads to ppGpp accumulation (10, 11). Likewise, it has been proposed that carbohydrate starvation induces ppGpp synthesis through SpoT (22). However, ppGpp induction by alpha methyl glucoside (which prevents glucose uptake) is dependent on relA (85), suggesting that glucose starvation might in fact be sensed through uncharged tRNA occupancy of the ribosomal A-site. Finally, SpoT interacts with the 50S ribosomal subunit-associated factor ObgE, suggesting that SpoT might be ribosome associated after all (57, 122).
3. REGULATION OF RIBOSOME SYNTHESIS BY ppGpp AND NTP CONCENTRATIONS IN E. COLI
The role of ppGpp in the regulation of ribosome synthesis was debated for many years. In retrospect, the use of different kinds of assays by different investigators for measuring effects on rRNA transcription in vivo and the inability to observe large regulatory effects of ppGpp in vitro resulted in differences in interpretation. We can divide the in vivo studies into two classes: (a) measurements of gene expression within minutes of a nutritional shift and (b) steady-state assays in which the induction of ppGpp in response to a given nutritional condition (or mutagenic event) is separated from the measurement of its effect on transcription by multiple cell generations.
ppGpp is induced by a variety of stresses, both nutritional and environmental. The closer in time the measurement of the response is to the generation of the signal, the less chance there is for a secondary event (e.g., a mutation or a change in metabolism) to obscure a direct response of the transcriptional machinery and to complicate examination of the effects of the initial stimulus.
Studies in which rRNA synthesis was measured soon after changes in nutritional status generally led to similar conclusions: Increases or decreases in ppGpp concentration led to decreases or increases, respectively, in rRNA promoter activity not only in the proteobacterium E. coli (85), but also in B. subtilis (64), species separated evolutionarily by more than a billion years. Conversely, steady-state assays (typically measuring rRNA, tRNA, or rRNA promoter-lacZ fusions in strains containing or lacking ppGpp) have led to results that differ among investigators. For example, it was reported that strains lacking ppGpp display almost wild type–growth rate–dependent control of rRNA synthesis (7, 40), whereas another report using different growth conditions concluded that regulation was defective (94). The variables responsible for the opposite conclusions in these studies were not defined, but their identification might prove useful for determining secondary metabolic routes that cells utilize to compensate for loss of the ppGpp signaling pathway. Because we now know that the direct effects of ppGpp are even more extensive than originally predicted, understanding the cause and effect relationships responsible for the differences in results will require systematic analysis of the signaling pathways and the kinetics of the responses. These secondary events are no less important to cell physiology than the direct mechanisms, but they are likely to be complicated and difficult to unravel.
It is worth noting that not all upshifts and downshifts in rRNA transcription result from changes in ppGpp levels. For example, ppGpp levels are so low when cells are in prolonged stationary phase that a small further decrease in ppGpp concentration from addition of new medium would probably be insufficient to cause an increase in rRNA promoter activity. However, there is a large increase in the concentrations of NTPs during outgrowth. Promoters that form unstable complexes with RNAP, such as ribosomal RNA and some ribosomal protein promoters (20a, 84, 85), depend on high concentrations of the initiating NTP to drive transcription initiation forward by mass action. Thus, there is a rapid increase in rRNA transcription during outgrowth from stationary phase that is not dependent on a decrease in ppGpp synthesis but rather results from a large increase in NTP concentration (20a, 84, 85).
Likewise, ppGpp concentrations spike as cells approach and enter stationary phase, but within a few hours, ppGpp concentrations return to levels similar to those found in exponentially growing cultures. We suggest that the low concentrations of ppGpp present in prolonged stationary phase are insufficient to inhibit rRNA promoters. Rather, rRNA transcription is inhibited by the low concentrations of the initiating nucleotides present under these conditions, ATP and GTP for the rrn P1 promoters and CTP for the rrn P2 promoters (84, 85). Once NTP concentrations increase during outgrowth and saturate rRNA promoter complexes, changes in the level of ppGpp, resulting from the status of charged tRNA, rather than NTP, concentrations, become the dominant means of rRNA promoter regulation.
Finally, the activities of E. coli rRNA promoters are also dependent on the transcription factor Fis, which binds to sites upstream of the P1 promoters in each of the seven rRNA operons, stimulating transcription primarily by helping recruit RNAP to the promoter (53a). The promoter for the fis gene itself and thus Fis protein levels are regulated by the concentrations of NTPs and by ppGpp (79a). Finally, fis expression is increased by a translational enhancer (85a) whose efficiency could in theory contribute to its regulation and ultimately to nutritional control of rRNA expression.
4. DksA MODIFIES RNAP, GREATLY INCREASING THE RESPONSE OF RNAP TO ppGpp
Evidence for ppGpp acting directly on RNAP to regulate transcription dates back more than 40 years (6, 7, 58, 62, 63, 75). However, attempts to identify the direct target(s) of ppGpp on transcription and its mechanism of action were confounded by the small effects of ppGpp on transcription in vitro. Identification of the 151–amino acid DksA as a transcription factor required for responses to ppGpp was a major step forward for reproduction of the effects of the stringent response in a purified system.
DksA was originally identified as a multicopy suppressor of a dnaK mutant, presumably because it somehow alleviated a protein-folding defect (59), and it was later proposed to function at the level of translation (20). However, a chaperone activity has never been found associated with DksA (23, 24), and at its native concentration, DksA protein was found to bind instead to RNAP (90, 92). Inclusion of both DksA and ppGpp together decreased transcription from rRNA promoters by as much as 20 fold in vitro relative to the level of transcription observed in the absence of both ppGpp and DksA or to the ~3-fold decrease observed with ppGpp but not DksA (90). Furthermore, inclusion of both DksA and ppGpp together in transcription reactions in vitro resulted in stimulation of the subset of amino acid biosynthesis and transport promoters previously identified as stimulated by ppGpp in vivo (91). Cells lacking the dksA gene are defective in responses to ppGpp, and like strains unable to make ppGpp (∆relA∆spoT mutants), they are unable to grow in a defined medium lacking amino acids (20, 83, 103). This defect in transcription from the amino acid biosynthesis/transport promoters could account for the amino acid requirement of ∆relA∆spoT and ∆dksA mutants, although the effect might be less direct.
Several parts of DksA are important for its function. DksA, like the transcription elongation factors GreA and GreB, contains a globular domain and a long coiled-coil with aspartate residues at the coiled-coil tip (30, 92) (Figure 1). Otherwise, DksA has little or no sequence homology to the Gre factors: It has a zinc-binding fold in its globular domain not found in the Gre factors, and it has a C-terminal helix (CTH) not present in the Gre factors (92). Finally, in contrast to Gre factors, which function on transcription elongation (17, 34), DksA functions primarily on initiation (90). However, DksA does share limited homology with TraR and its homologs, a widely distributed family of transcription factors about half its length that can mimic effects of DksA and ppGpp together (15, 45; see Section 10).
Figure 1.

Structural models of transcription factors DksA (151 residues) and TraR (73 residues) adapted from PDB1TJL (92) and PDB5W1S (82), respectively. Each protein contains a zinc ion bound in the globular domain and a C-terminal helix (CTH). The N terminus of each protein is indicated. Residues shown in red interact with RNA polymerase (RNAP) near the active site and are required for function but not for binding to RNAP (Asp74, Ala76 in the coiled-coil tip of DksA; Asp6, Ala8 at the N-terminal end of TraR) (45, 70). Residues shown in blue in DksA (Leu95, Lys98, Arg129, Lys139) form part of ppGpp Binding Site 2 but are not required for DksA binding to RNAP (99). Residues in blue in TraR (Ile20, Glu66) are required for function but not for binding to RNAP (45).
Extensive genetic and biochemical analyses demonstrated that DksA docks on the secondary channel rim helices of the β′ subunit, and the coiled-coil tip of DksA approaches the RNAP active site (38, 70, 72, 74, 90, 92, 102). The RNAP trigger loop is required for DksA function (74, 86, 103), as is a DksA CTH interaction with β SI1, a species-specific subdomain near the N terminus of the β subunit (89; see Section 6, below).
From bioinformatic comparisons, DksA homologs appear to be present in most proteobacterial species (37, 39, 89, 92, 99, 121), with a few notable exceptions (e.g., 28). DksA does not appear to be present in Firmicutes or Thermophiles, but it should be emphasized that such analyses are not definitive. For example, DksA was not identified as a homolog of the Gre factors until its coiled-coil domain structure was solved and the structural similarity became apparent (92). Many DksA homologs have been identified based on the presence of a zinc-binding motif. It is now apparent that there are DksA homologs in other species [e.g., Pseudomonas aeruginosa (13, 37) and Rhodobacter sphaeroides (73)] with similar globular domain folds that do not contain a zinc finger and that directly regulate transcription in vitro. There are also proteins identified as DksA homologs because they have zinc finger domains but do not function like DksA (64, 73, 121).
5. IDENTIFICATION OF THE ppGpp BINDING SITES ON RNAP
Many suppressors of ∆relA, ∆relA∆spoT, or ∆dksA phenotypes have been identified (e.g., 9, 83, 103, 113). The suppressor mutations mimic the effects of ppGpp and/or DksA, and all map in rpoB, rpoC, or rpoD (103). One study reported a ppGpp-resistant RNAP, but the mutation was never mapped (110). Most of the suppressors map to parts of RNAP lining the main DNA channel and result in destabilization of RNAP interactions with rRNA promoters and/or elongation complexes (6, 103).
Attempts to locate a binding site for ppGpp on RNAP using biochemical approaches go back at least 30 years (26, 88, 107, 112). A ppGpp binding site was reported in a Thermus thermophilus RNAP cocrystal (4). However, it was later found that T. thermophilus RNAP was unaffected by ppGpp in vivo or in vitro (61, 118). Substitutions for the residues in E. coli RNAP that corresponded to those interacting with ppGpp in the T. thermophilus RNAP cocrystal had no effects on responses of E. coli RNAP to ppGpp in vitro (118).
Therefore, a detailed cross-linking study was undertaken to identify the ppGpp binding site on E. coli RNAP (100). A cross-link to 32P-6-thio-ppGpp mapped to a section of RNAP near the junction of the β′ and ω subunits, an extensive set of single substitutions was created in the vicinity of this cross-link, and RNAPs containing these substitutions were purified and analyzed for loss of cross-linking to ppGpp and loss of the effect of ppGpp on transcription. This study revealed a potential ppGpp binding pocket at the interface formed by two parts of the β′ subunit and the N-terminal region of ω (100). ppGpp binding to this site was then defined with greater precision by high-resolution X-ray structures in which ppGpp was diffused into crystals of E. coli RNAP (81, 128).
Residues contributing to the ppGpp binding pocket (β′ R362, R417; K619, D622, and the N-terminal residues of ω) are generally much less conserved in bacterial species outside the proteobacteria, consistent with the absence of effects of ppGpp on T. thermophilus and B. subtilis RNAP in vitro (64, 118). The results also explained the requirement for ω for regulation of E. coli RNAP by ppGpp in vitro (119).
However, some properties of the RNAPs containing substitutions in the ppGpp binding pocket were puzzling. Even the combination of multiple ppGpp binding site substitutions had relatively mild effects on the recovery of cells from downshifts to defined medium from rich medium or to amino acid limitation (100), and previous studies on cells with deletions of the rpoZ gene (coding for ω) did not detect defects in the stringent response in vivo (42). Furthermore, although RNAPs without the ppGpp binding site were unaffected by ppGpp in vitro, ppGpp inhibited rRNA transcription with wild-type RNAP only to about 25–30% of its activity in the absence of ppGpp (7, 119), a much weaker effect than the inhibition of rRNA promoter activity observed in vivo after amino acid limitation (85, 90).
Taken together, the inability to fully recapitulate the effect of ppGpp on rRNA transcription in vitro that was observed in vivo, the failure of RNAP to show positive regulation by ppGpp in vitro, and the weak phenotypes of strains lacking the ppGpp binding site suggested that another cellular component might be needed for full responses of RNAP to ppGpp. DksA was an obvious candidate for the missing component, since as noted above, DksA greatly increases the inhibitory effect of ppGpp on rRNA promoters in vitro, and ppGpp and DksA together stimulate amino acid biosynthesis promoters in vitro (90, 91). Furthermore, DksA rescued the ppGpp-unresponsiveness of RNAP lacking ω in vivo (119).
Effects of ppGpp on an rRNA promoter (rrnB P1) in vitro were compared using either wild-type RNAP or an RNAP lacking the ppGpp binding site (as a result of deletion of the ω subunit), with or without DksA (Figure 2). Although RNAP lacking ω was not inhibited by ppGpp in the absence of DksA (100), inclusion of both DksA and ppGpp together resulted in an approximately fivefold decrease in transcription by RNAP lacking ω relative to that observed with RNAP containing neither DksA nor ω. Taken together, these results suggested a model in which ppGpp inhibits transcription by approximately three- to fourfold by binding to the site at the β′–ω interface (hereafter called Site 1) and another approximately fivefold by binding to a second site (hereafter called Site 2) that requires DksA, accounting for the full inhibition of rrnB P1 observed in vivo (99) (Figure 2). 32P-6-thio-ppGpp did not cross-link to the RNAP lacking Site 1 in the absence of DksA, but it cross-linked to DksA itself when RNAP was present in the reaction, supporting the model that there might be a second binding site for ppGpp and that it might be at an interface of DksA and RNAP. In contrast to the requirement of both sites for full inhibition of rRNA transcription, activation of transcription required only the binding site associated with DksA (Site 2).
Figure 2.

Transcription in vitro was performed with ppGpp and/or DksA and with Escherichia coli RNA polymerase (RNAP) containing or lacking the ω subunit, as indicated. The resulting RNAPs thereby contain the ppGpp binding sites indicated below the bars. Reactions were performed on supercoiled plasmid DNA templates containing (a) the rrnB P1 promoter or (b) the iraP promoter. The ppGpp concentration was 200 µM, and the DksA concentration was 2 µM. Transcription with RNAP lacking both ppGpp binding sites is set at 1.0 (data are from Reference 99).
To identify the residues responsible for forming ppGpp Binding Site 2, a higher-resolution model of the DksA-RNAP interface was created based on genetic information (70, 74, 103), cross-linking data (74, 89), and a structure of T. thermophilus RNAP in complex with Gfh1, a Gre factor homolog (109). Analyses of mutant RNAPs and mutant DksA proteins for effects of ppGpp on transcription resulted in identification of a potential ppGpp binding pocket, and a direct binding assay (97) confirmed that RNAP variants or DksA variants with substitutions in the site failed to bind ppGpp (99).
A surface representation showing the positions of the two ppGpp binding sites on E. coli RNAP is shown in Figure 3. The two sites are more than 60 Å apart on the surface of RNAP, and each is more than 30 Å from the active site. Since a dissociable factor contributes to ppGpp binding in each case (ω for Site 1 and DksA for Site 2), the actual affinities of ppGpp for the two sites in vivo depend on the concentrations of ω and DksA. Studies to determine these concentrations are under way. Neither site corresponds to a site proposed on the basis of the T. thermophilus RNAP cocrystal (4) or to a different site on E. coli RNAP proposed solely on the basis of cross-linking to 8-azido ppGpp (108).
Figure 3.

Surface representation of Escherichia coli RNAP showing locations of ppGpp Binding Site 1 (ω-β′ interface) and Binding Site 2 (DksA-β′ interface). Adapted in Pymol from PDB4JKR and PDB1TJL. Site 1 position as in PDB4JKR; Site 2 position modeled based on genetic/biochemical data as described Reference 99. RNAP subunits, RNAP β SI1, secondary channel, and active site are indicated. Abbreviations: NTD, N-terminal domain; RNAP, RNA polymerase.
6. MECHANISM OF ACTION OF ppGpp/DksA ON TRANSCRIPTION
6.1. Effects on Transcription Initiation
Studies dating at least as far back as the 1970s implicated ppGpp in direct and specific inhibition of promoters for rRNAs in vitro (58, 75). Effects of ppGpp on templates that differed only in the sequence of the promoter and not in the transcribed region, as well as the use of assays that included only steps prior to NTP incorporation, suggested that initiation was the targeted kinetic step (6, 7). Because these experiments were performed in the absence of DksA, the observed regulation was the result of ppGpp binding to Site 1. Negative and positive regulation by ppGpp have now been explored in the presence of DksA (99). Sites 1 and 2 are both required for full inhibition of negatively regulated promoters, whereas Site 2 is sufficient for positive regulation in vitro of several amino acid biosynthesis promoters as well as for the promoter for IraP, a small anti-adaptor protein that regulates the stability of the stationary phase sigma factor σS (17a, 99) (Figure 2).
The mechanism by which ppGpp and DksA affect transcription is an active area of study. The rates of formation and decay of RNAP-promoter complexes, as well as escape of RNAP from the promoter, differ significantly between all promoters—not just the ones regulated by ppGpp/DksA (101). We proposed that the specific kinetics of the transcription initiation reaction at different promoters determines whether or not a promoter is regulated by ppGpp/DksA (6, 7, 51, 90, 91). In this model, the assumption is that ppGpp and DksA bind to all promoter complexes in the same manner, but the DNA sequences of the promoter determine whether the ppGpp/DksA responsive step(s) play a role (are rate determining) for transcription output.
ppGpp by itself at Site 1 and DksA and ppGpp together at Site 2 affect step(s) after initial binding of RNAP to the promoter (7, 90, 91). Studies on the kinetic properties of the negatively regulated rrnB P1 promoter indicated that it associates rapidly with RNAP but forms an unstable open complex that is in rapid equilibrium with earlier conformational intermediates (7, 45, 50, 51, 94a, 103). DksA and/or ppGpp shifts the occupancy of rrnB P1 by RNAP to an intermediate in which promoter DNA is not correctly positioned in the active site and cannot initiate transcription (7, 103). ppGpp and DksA therefore function at step(s) after initial closed complex formation but before formation of the first phosphodiester bond. In contrast, activated promoters have slow rates of isomerization that are increased by DksA/ppGpp. However, their open complexes, once formed, are relatively stable and insensitive to the inhibitory effects of DksA/ppGpp (7, 91, 103).
ppGpp Site 1, which affects inhibition but not activation, is located at the interface connecting two rigid-body “modules” in RNAP, the core and shelf modules (109). Its location suggests that ppGpp binding might block hinge-like motions between the modules (100, 128). Further studies will be needed to understand why this reduces transcription initiation.
Although DksA alone can inhibit transcription to some extent, ppGpp (when bound to Site 2) greatly enhances DksA’s inhibitory effect. In theory, ppGpp could increase DksA occupancy of the promoter complex by improving the DksA-RNAP interaction. Kd determinations for DksA binding to RNAP indicate that the presence of ppGpp results in a small (2.5-fold) increase in affinity (82). However, there is still a requirement for ppGpp for positive control even at very high concentrations of DksA or with the tight-binding DksA variant N88I (14), suggesting that an increase in DksA binding to RNAP is not sufficient to account for the effect of ppGpp at Site 2.
Residues at the coiled-coil tip of DksA, D74, and A76 are critical for effects of DksA on transcription initiation (70, 74, 90, 103). In addition, residues at the junction of the DksA globular domain and CTH (L95, K98, R129, and K139) are required for ppGpp binding and function at Site 2 but not for DksA binding to RNAP (99) (Figure 1). Substitutions for residues N680 and K681 on the rim helices of β′ are also specifically defective for the response to ppGpp, suggesting that these residues in β′ along with L95, K98, R129, and K139 in DksA make up Site 2 (99). A recent cocrystal structure of RNAP, DksA, and ppGpp confirmed the location of Site 2 (82).
The DksA CTH cross-links to a sequence insertion in proteobacterial RNAPs near the N terminus of the β subunit, β SI1, also known as βi4 (67). β SI1 is critical for regulation of transcription by DksA (89). We therefore speculate that ppGpp affects the position of the CTH with respect to β SI1 and that this interaction plays an important role in the effect of ppGpp at Site 2. Likewise, the RNAP trigger loop is necessary for the effect of DksA/ppGpp (103).
Positive regulation requires DksA and ppGpp binding to Site 2 in vitro (Figure 2b), and these factors together affect isomerization to the open complex (91). However, two additional mechanisms could also contribute to positive regulation by ppGpp/DksA at specific promoters in vivo. Because rRNA transcription accounts for as much as 70% of all transcription in vivo (18), positive control could derive indirectly in vivo from the decrease in rRNA transcription caused by ppGpp/DksA. Supporting this hypothesis, several reports have implicated the contribution of increased availability of RNAP to positive control during starvation conditions (6, 43, 69, 79). It was also proposed that positive regulation might sometimes derive from effects of ppGpp/DksA on reducing the stability of promoter complexes, which might facilitate escape of RNAP from promoters to which it bound very tightly (47).
6.2. Effects of ppGpp/DksA on Elongation
We emphasize that identification of the effects of Sites 1 and 2 on transcription initiation in vitro did not rule out potential additional effects of ppGpp on transcription elongation. In fact, there is an extensive literature on effects of ppGpp as an inhibitor of elongation in vitro and in vivo (62, 63, 116, 117). Like the early studies on initiation, early studies on elongation in vitro did not include DksA, suggesting that ppGpp binding to Site 1 might account for at least some effects during elongation. More recently, effects of DksA on elongation have also been reported (38, 98), although it has also been argued that effects of DksA on elongation might be excluded by the trigger loop insertion in β′ (39). In any case, slowing of elongation might help couple transcription and translation and/or improve transcriptional fidelity (124, 126, 128).
In summary, a network of interactions between DksA, ppGpp, and multiple parts of RNAP mediate the effects of ppGpp and DksA on transcription. Although the interactions between ppGpp and DksA with RNAP are becoming clearer, the mechanism of regulation is far from understood. Presumably, a more detailed picture will become possible when the conformational changes that RNAP undergoes during transcription are themselves better understood.
7. ppGpp/DksA AND GLOBAL TRANSCRIPTION REGULATION
Global studies of the effects of ppGpp on transcription were originally performed using expression microarrays after treating cells with serine hydroxamate, an inhibitor of aminoacylation of serine tRNA (111a), to induce the stringent response (31). Expression microarrays were also used to compare wild-type, isoleucine-depleted wild-type, and ∆relA∆spoT cells (115). Each of these studies identified 700–800 transcripts regulated positively or negatively by ppGpp. Another study compared transcripts differentially expressed in wild-type and ∆dksA or ∆relA∆spoT strains (1). Although the different reports identified some transcripts in common, there was considerable variation. Furthermore, because amino acid limitation has profound ppGpp-independent effects on cellular metabolism, and genetic changes can have indirect consequences, it was difficult to distinguish effects in these studies caused by direct ppGpp binding to RNAP from those resulting in metabolic changes that in turn caused changes in transcription.
In an attempt to clarify direct from indirect effects of ppGpp, a recent study was performed using direct RNA sequencing (RNA-seq) on RNA produced after induction of ppGpp without concurrent amino acid limitation (103a). This study resulted in the identification of more than 700 transcripts that changed within 5 min of ppGpp induction, ~400 inhibited and ~300 stimulated. The number swelled to more than 1,200 transcripts by 10 min after ppGpp induction. A large majority of the transcripts that changed in the first 5 min were not identified in the earlier study (31). In order to further separate direct from indirect effects of ppGpp, the promoters for more than 100 of the targets identified in vivo were tested by in vitro transcription with the major E. coli RNAP holoenzyme (Eσ70), ppGpp, and DksA. The large majority of promoters tested in this way showed correlating effects in vitro: Transcripts that were inhibited by ppGpp induction in vivo were inhibited in vitro, and transcripts that were stimulated by ppGpp induction in vivo were stimulated in vitro (103a).
RNA-seq was also performed on the strain containing the RNAP that lacked the ppGpp binding sites. None of the >700 transcripts regulated by ppGpp after five minutes of RelA induction responded normally in the mutant strain. These results indicate that these transcriptional responses to ppGpp in E. coli all result from ppGpp binding to RNAP and not from ppGpp binding to transcription factors or other gene products that influence transcription (103a).
In addition to their effects on promoters recognized by Eσ70, ppGpp and DksA directly or indirectly regulate some promoters transcribed by other holoenzymes, e.g., EσS, EσE, and EσF (27, 43, 44, 71, 95). There are also promoters for small RNAs that are regulated by ppGpp/DksA (43). Finally, there are reports that DksA and ppGpp can have separate or even opposing effects (1, 2, 44, 79), although in most cases the mechanisms responsible have yet to be elucidated.
8. DNA SIGNATURES FOR REGULATION OF TRANSCRIPTION INITIATION BY ppGpp/DksA
Identification of the DNA signatures for regulation of transcription initiation by ppGpp/DksA has remained a challenge. Promoters regulated by transcription factors can usually be identified by their proximity to a transcription factor–DNA binding site sequence. In contrast, promoters regulated by RNAP-binding transcription factors do not have an obvious DNA signature. The promoter characteristics that make a particular promoter susceptible to regulation by ppGpp/DksA are the kinetic properties of the promoter complex, and these are usually determined by interactions between multiple parts of the promoter with RNAP (51).
Early studies on promoter requirements for stringent control identified a G+C-rich discriminator region as a key determinant (114). It was later shown that a guanine residue on the nontemplate strand just downstream from the −10 hexamer fits into a pocket in σ region 1.2, and it is the absence of this guanine residue in rRNA promoters that contributes to the short half-life needed for regulation of the promoter complex by ppGpp/DksA (8, 33, 50, 125). In addition, mutations in this or other positions in the discriminator that make it A+T rich also eliminate regulation by facilitating opening of the transcription bubble. Thus, promoters with A+T-rich discriminator sequences or a guanine that interacts with σ region 1.2 are generally resistant to negative control by ppGpp.
Analysis of rrnB P1 promoter mutations has shown that sequences in other parts of the promoter can also eliminate regulation by ppGpp. In general, mutations that increase the lifetime of the promoter complex (e.g., creating a better extended −10 sequence, a better −35 hexamer sequence, or a 17-bp −10/−35 spacer sequence) reduce regulation of rrnB P1 by ppGpp/DksA (8, 50).
The sequence requirements for positive regulation are even more complex. Positively regulated promoters often contain an A+T-rich discriminator region sequence, but this sequence is insufficient for positive regulation in other promoter contexts (47, 96, 103a). Positively regulated promoters make relatively long-lived complexes with RNAP (6), but what causes them to be activated by ppGpp/DksA, as opposed to simply not inhibited, remains to be determined. It is clear, however, that positive regulation derives primarily from effects of ppGpp/DksA on steps after closed complex formation (91). It is possible that ppGpp/DksA might stimulate different steps for different positively regulated promoters. Although it was reported that ppGpp/DksA can stimulate promoter escape (47), that study did not distinguish between effects on isomerization steps versus promoter escape.
The genome-wide analysis of E. coli promoters responding to ppGpp provided a much larger dataset of confirmed directly regulated promoter sequences than available previously (103a). The results of those studies are consistent with the idea that there is no single consensus promoter sequence, either negative or positive, that correlates with regulation by ppGpp/DksA, although it did hint at some specific sequence motifs not identified previously. In any case, these studies, as well as previous genome-wide reports (1, 31, 115), demonstrated just how broad the direct effects of ppGpp are. It appears that the numbers of transcripts that are regulated by direct binding of ppGpp to RNAP in proteobacteria might approach the number affected by all DNA-binding transcription factors combined.
9. OTHER PROTEINS THAT BIND ppGpp IN E. coli AND OTHER BACTERIAL SPECIES
Studies as far back as the early 1970s identified effects of ppGpp on cellular processes as diverse as replication, translation, and central metabolism (reviewed in 22, 41a, 56, 60, 76, 77). Briefly, among the proteins proposed as direct targets of ppGpp binding are DNA primase (DnaG), translation initiation factor 2 (IF2), elongation factor G (EF-G), ribosome release factor 3 (RF3), the GTPase ObgE, and the ribosome assembly factor BipA. Other metabolic enzymes reported to bind ppGpp include Gpt, Hpt, Apt, GuaB, PurA, and LdcI. Interactions of ppGpp with many of these enzymes were recently detected using direct binding assays (127).
Many of the identified targets of ppGpp are GTP-binding enzymes in which the GTP and ppGpp binding sites overlap. Thus, the in vivo relevance of ppGpp binding to each of these proteins will depend on measurements of their relative affinities for GTP and ppGpp as well as their concentrations in cells.
In B. subtilis, the importance of ppGpp binding to enzymes responsible for nucleotide metabolism is well established. B. subtilis RNAP lacks critical residues in β′ and ω that form Site 1 in proteobacterial RNAPs, and B. subtilis does not appear to contain either a DksA homolog or the residues in the β′ rim helices that contribute to Site 2 (64, 99, 100). However, as with E. coli, B. subtilis rRNA promoters form short-lived open complexes. Furthermore, rRNA promoters start with GTP and require high concentrations of GTP for transcription initiation (64). Thus, by binding to and inhibiting enzymes required for GTP synthesis, like GMK and HPRT (65, 77), ppGpp regulates rRNA transcription indirectly (Figure 4). Whether there are additional mechanisms for regulating rRNA transcription in B. subtilis awaits further investigation, but in any case, ppGpp appears to play a critical role in regulating GTP levels in many Firmicutes species (12, 41, 76).
Figure 4.
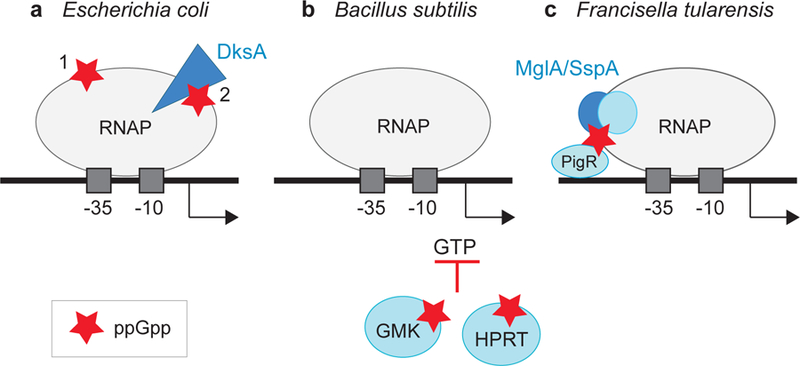
Three paradigms for regulation of transcription initiation by ppGpp: (a) In Escherichia coli, ppGpp (red star) binds directly to two sites on RNA polymerase (RNAP, gray oval). DksA is represented by a blue triangle (99). (b) In Bacillus subtilis, ppGpp binds to enzymes responsible for GTP biosynthesis (e.g., GMK, HPRT, blue circles), indirectly inhibiting transcription from rRNA operons by reducing the concentration of the initiating nucleotide GTP (64, 77). (c) In Francisella tularensis, ppGpp binds to the RNAP-associated heterodimer MglA/SspA (dark blue circles), leading to its interaction with the transcriptional activator PigR (blue oval) (25, 28).
Although both binding sites for ppGpp on E. coli RNAP appear to be conserved in the proteobacteria, exceptions have been reported. In the gammaproteobacterium Francisella tularensis, the causative agent of tularemia, a dksA-like gene has not been identified, suggesting that F. tularensis RNAP lacks Site 2. Rather, ppGpp binds to a heterodimeric complex of two RNAP-associated transcription factors, MglA and SspA, and facilitates their interaction with a third transcription factor, PigR, that is proposed to bind DNA and stimulate transcription from promoters for virulence factors during infection (25, 28) (Figure 4).
It was reported that the rRNA synthesis rate in Synechococcus decreases in the dark, but this decrease required factors other than ppGpp (55). However, ppGpp did regulate the expression of a number of translation-related genes, including ribosomal hibernation-promoting factor (Hpf), which causes ribosomes to dimerize in the dark, decreasing translation. Thus, ppGpp may regulate ribosome synthesis or ribosome activity in different ways in cyanobacteria compared to Proteobacteria or Firmicutes.
Although genes for ppGpp synthases are not present in most eukaryotes, RelA homologs have been found in Arabidopsis chloroplasts, and ppGpp regulates chloroplast function to influence growth and development (106). A SpoT ortholog that plays a role in stress responses was also reported in Drosophila (107). Furthermore, although yeast do not naturally make ppGpp, it was reported that Saccharomyces cerevisiae engineered to make ppGpp stopped growing (53), and in another study, ppGpp production in S. cerevisiae resulted in enhanced tolerance to various stresses (87). Although studies of ppGpp in eukaryotes are in their infancy, these results suggest the potential for significant gene regulation by ppGpp.
10. DksA HOMOLOGS THAT TARGET THE RIM HELICES OF RNAP AND MIMIC THE EFFECTS OF DksA/ppGpp
The 73–amino acid E. coli F element–encoded protein TraR is a distant homolog of DksA, although it is only half its size (15) (Figure 1). TraR acts as a global regulator, inhibiting some promoters and activating others by binding to the secondary channel of RNAP using interactions similar, but not identical, to those of DksA (45). TraR inhibits rRNA transcription as strongly as DksA and ppGpp combined and much more strongly than DksA alone. Unlike DksA, it activates transcription even in the absence of ppGpp (15, 45).
Although TraR lacks the residues in DksA that interact with ppGpp, it utilizes the same residues in the β′ rim helices that contribute to Site 2 in the DksA-ppGpp-RNAP complex. By binding directly to the region of the RNAP secondary channel that otherwise binds ppGpp, its N-terminal region (residues D3, D6, A8), like the coiled-coil tip of DksA, appears to engage the active site region of RNAP and affect transcription allosterically (45). Binding of TraR to the secondary channel of RNAP and interaction of TraR with the residues in RNAP that form Site 2 were confirmed by a recent TraR-RNAP X-ray structure (82).
The role of TraR in F element biology is unclear, but two nonexclusive models have been proposed. In one model, the conjugation process disrupts the bacterial outer membrane, activating an extracytoplasmic stress response in which TraR turns on σE-dependent promoters independent of the standard DegS/RseA signal transduction cascade (46). In a second model, TraR inhibits and activates promoters typically regulated by ppGpp/DksA during a stringent response but under conditions in which ppGpp is not induced. In this way, TraR could help switch the cell’s transcription machinery to conjugation functions by inhibiting rRNA transcription and activating amino acid synthesis (15, 45).
TraR-like proteins appear to be ubiquitous in bacteria, even in phyla quite distant from Proteobacteriaceae (Figure 5). Genes encoding TraR-like proteins are often associated with extrachromosomal elements such as conjugal plasmids or bacteriophages. Although the functions of these TraR-like proteins will require further investigation, their widespread distribution attests to their importance as well as to the model that the rim helices are a major target of bacterial transcription factors.
Figure 5.

Distribution of TraR homologs in bacteria and bacteriophages. TraR-like proteins were identified by NCBI Blast (https://www.ncbi.nlm.nih.gov/blast), and a phylogenetic tree depicting their distribution was constructed using phyloT (http://phylot.biobyte.de/). Only representative species are shown. Red font is used for phyla or families in which there are proteins approximately the same length as TraR with key conserved residues. Blue font is used for phyla in which TraR-like proteins were not identified. A list of species with TraR homologs is in Reference 45.
The TraR class of transcription factors should be distinguished from proteins with a short coiled-coil domain that do not have a DksA-like activity, e.g., E. coli Rnk (66), R. sphaeroides Rsp066 (73), and Sinorhizobium meliloti SMc00049 (121). These and proteins like T. thermophilus Gfh1 (68) could potentially compete with Gre factors or DksA for the secondary channel. However, heterologous overexpression of a factor can result in competition that is nonphysiologically significant (102). Information about the residency time of the factor on RNAP at its native concentration will be needed to evaluate models in which competition is invoked. For example, single-molecule measurements showed recently that the residence time of GreB in the secondary channel of elongation complexes is very short, suggesting that GreB does not compete with GreA or DksA and explaining why DksA and GreB can both bind to the same site on RNAP without interfering with each other’s activities during elongation (111).
11. OPEN QUESTIONS
This review has focused almost exclusively on transcriptional responses to production of ppGpp in E. coli, specifically, how it binds to RNAP and how promoters are affected by its binding. ppGpp is remarkably conserved, although the mechanisms by which it is utilized for the stringent response are quite diverse. It is now clear that there are at least three general paradigms for regulation of transcription by ppGpp (Figure 4): (a) the E. coli model, in which ppGpp binds to two sites on RNAP and allosterically regulates transcription, (b) the B. subtilis model, in which ppGpp regulates enzymes responsible for nucleotide synthesis, indirectly inhibiting transcription initiation, and (c) the F. tularensis model, in which ppGpp binds to transcription factors that themselves interact with RNAP. With the vast diversity of the bacterial domain, it seems very unlikely that these are the only scenarios. Furthermore, there appears to be a universe of small proteins like TraR that can bind to the secondary channel rim helices in RNAP and regulate transcription by mimicking the effects of ppGpp/DksA.
With respect to E.coli RNAP, single-particle cryo-EM structures of ppGpp/DksA/RNAP and the TraR/RNAP promoter complexes will become available shortly. It is likely that this structural information will help elucidate the conformational changes in the transcription apparatus caused by the regulatory factors. With that information in hand, we should be equipped to tackle the next level of complexity: How are different promoter sequences able to respond in different ways to ppGpp? In addition, are there times when only one ppGpp binding site or the other is filled, and if so, are there consequences on global gene expression patterns? For even the most heavily studied ppGpp model system, there are many unanswered questions.
ACKNOWLEDGMENTS
Work in our lab is funded by the National Institutes of Health (GM37048). We apologize to colleagues whose important contributions were not included as a result of space limitations.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Aberg A, Fernández-Vázquez J, Cabrer-Panes JD, Sánchez A, Balsalobre C. 2009. Similar and divergent effects of ppGpp and DksA deficiencies on transcription in Escherichia coli. J. Bacteriol 191:3226–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aberg A, Shingler V, Balsalobre C. 2008. Regulation of the fimB promoter: a case of differential regulation by ppGpp and DksA in vivo. Mol. Microbiol 67:1223–41 [DOI] [PubMed] [Google Scholar]
- 3.Arenz S, Abdelshahid M, Sohmen D, Payoe R, Starosta AL, et al. 2016. The stringent factor RelA adopts an open conformation on the ribosome to stimulate ppGpp synthesis. Nucleic Acids Res 44:6471–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Artsimovitch I, Patlan V, Sekine S, Vassylyeva MN, Hosaka T, et al. 2004. Structural basis for transcription regulation by alarmone ppGpp. Cell 117:299–310 [DOI] [PubMed] [Google Scholar]
- 5.Atkinson GC, Tenson T, Hauryliuk V. 2011. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLOS ONE 6:e23479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barker MM, Gaal T, Gourse RL. 2001. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J. Mol. Biol 305:689–702 [DOI] [PubMed] [Google Scholar]
- 7.Barker MM, Gaal T, Josaitis CA, Gourse RL. 2001. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol 305:673–88 [DOI] [PubMed] [Google Scholar]
- 8.Barker MM, Gourse RL. 2001. Regulation of rRNA transcription correlates with nucleoside triphosphate sensing. J. Bacteriol 183:6315–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartlett MS, Gaal T, Ross W, Gourse RL. 1998. RNA polymerase mutants that destabilize RNA polymerase-promoter complexes alter NTP-sensing by rrn P1 promoters. J. Mol. Biol 279:331–45 [DOI] [PubMed] [Google Scholar]
- 10.Battesti A, Bouveret E. 2006. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol. Microbiol 62:1048–63 [DOI] [PubMed] [Google Scholar]
- 11.Battesti A, Bouveret E. 2009. Bacteria possessing two RelA/SpoT-like proteins have evolved a specific stringent response involving the acyl carrier protein-SpoT interaction. J. Bacteriol 191:616–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bittner AN, Kriel A, Wang JD. 2014. Lowering GTP level increases survival of amino acid starvation but slows growth rate for Bacillus subtilis cells lacking (p)ppGpp. J Bacteriol 196:2067–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blaby-Haas CE, Furman R, Rodionov DA, Artsimovitch I, de Crécy-Lagard V. 2011. Role of a Zn-independent DksA in Zn homeostasis and stringent response. Mol. Microbiol 79:700–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blankschien MD, Lee JH, Grace ED, Lennon CW, Halliday JA, et al. 2009. Super DksAs: substitutions in DksA enhancing its effects on transcription initiation. EMBO J 28:1720–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blankschien MD, Potrykus K, Grace E, Choudhary A, Vinella D, et al. 2009. TraR, a homolog of a RNAP secondary channel interactor, modulates transcription. PLOS Genet 5(1):e1000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borek EK, Rockenbach J, Ryan A. 1956. Studies on a mutant of Escherichia coli with unbalanced ribonucleic acid synthesis. J. Bacteriol 71:318–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borukhov S, Sagitov V, Goldfarb A. 1993. Transcript cleavage factors from E. coli. Cell 72:459–66 [DOI] [PubMed] [Google Scholar]
- 17a.Bougdour A, Wickner S, Gottesman S. 2006. Modulating RssB activity: IraP, a novel regulator of σS stability in Escherichia coli. Genes Dev 20:884–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bremer H, Dennis P. 2008. Modulation of chemical composition and other parameters of the cell at different exponential growth rates. EcoSal Plus 10.1128/ecosal.5.2.3 [DOI] [PubMed]
- 19.Brown A, Fernandez IS, Gordiyenko Y, Ramakrishnan V. 2016. Ribosome dependent activation of stringent control. Nature 534:277–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown L, Gentry D, Elliott T, Cashel M. 2002. DksA affects ppGpp induction of RpoS at a translational level. J. Bacteriol 184:4455–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Burgos HL, O’Connor K, Sanchez-Vazquez P, Gourse RL. 2017. Roles of transcriptional and translational control mechanisms in regulation of ribosomal protein synthesis in Escherichia coli. J. Bacteriol 199: e00407–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cashel M, Gallant J. 1969. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature 221:838–41 [DOI] [PubMed] [Google Scholar]
- 22.Cashel M, Gentry D, Hernandez VJ, Vinella D. 1996. The stringent response. In Escherichia coli and Salmonella: Cellular and Molecular Biology, Vol. 1, ed. Neidhardt FC, pp. 1458–96. Washington, DC: ASM Press [Google Scholar]
- 23.Chandrangsu P, Lemke JJ, Gourse RL. 2011. The dksA promoter is negatively feedback regulated by DksA and ppGpp. Mol. Microbiol 80:1337–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandrangsu P, Wang L, Choi S-H, Gourse RL. 2012. Suppression of a dnaKJ deletion by multicopy dksA results from non-feedback regulated transcripts that originate upstream of the major dksA promoter. J. Bacteriol 194:1437–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charity JC, Blalock LT, Costante-Hamm MM, Kasper DL, Dove SL. 2009. Small molecule control of virulence gene expression in Francisella tularensis. PLOS Pathog 5(10):e1000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chatterji D, Fujita N, Ishihama A. 1998. The mediator for stringent control, ppGpp, binds to the beta-subunit of Escherichia coli RNA polymerase. Genes Cells 3:279–87 [DOI] [PubMed] [Google Scholar]
- 27.Costanzo A, Nicoloff H, Barchinger S, Banta A, Gourse RL, Ades SE. 2008. Mechanism of regulation of the extracytoplasmic stress factor σE in Escherichia coli by DksA and the alarmone ppGpp. Mol. Microbiol 67:619–32 [DOI] [PubMed] [Google Scholar]
- 28.Cuthbert BJ, Ross W, Rohlfing AE, Dove SL, Gourse RL, et al. 2017. Dissection of the molecular circuitry controlling virulence in Francisella tularensis. Genes Dev 31:1549–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dalebroux ZD, Swanson MS. 2012. ppGpp: magic beyond RNA polymerase. Nat. Rev. Microbiol 10:203–12 [DOI] [PubMed] [Google Scholar]
- 30.Darst SA, Stebbins CE, Borukhov S, Orlova M, Feng G, et al. 1994. Crystallization of GreA, a transcript cleavage factor from Escherichia coli. J. Mol. Biol 242(4):582–85 [DOI] [PubMed] [Google Scholar]
- 31.Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ. 2008. Transcription profiling of the stringent response in Escherichia coli. J. Bacteriol 190:1084–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.English BP, Hauryliuk V, Sanamrad A, Tankov S, Dekker NH, Elf J. 2011. Single-molecule investigations of the stringent response machinery in living bacterial cells. PNAS 108:E365–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feklistov A, Barinova N, Sevostyanova A, Heyduk E, Bass I, et al. 2006. A basal promoter element recognized by free RNA polymerase sigma subunit determines promoter recognition by RNA polymerase holoenzyme. Mol. Cell 23:97–107 [DOI] [PubMed] [Google Scholar]
- 34.Feng GH, Lee DN, Wang D, Chan CL, Landick R. 1994. GreA-induced transcript cleavage in transcription complexes containing Escherichia coli RNA polymerase is controlled by multiple factors, including nascent transcript location and structure. J. Biol. Chem 269:22282–94 [PubMed] [Google Scholar]
- 35.Fiil N, Friesen JD. 1968. Isolation of “relaxed” mutants of Escherichia coli. J. Bacteriol 95:729–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fung DK, Anderson BW, Tse JL, Wang JD. 2018. Nucleotide second messengers: (p)ppGpp and cyclic dinucleotides. In Bacillus: Cellular and Molecular Biology, ed. Graumann P. 3rd ed. In press
- 37.Furman R, Biswas T, Danhart EM, Foster MP, Tsodikov OV, Artsimovitch I. 2013. DksA2, a zinc-independent structural analog of the transcription factor DksA. FEBS Lett 587:614–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Furman R, Sevostyanova A, Artsimovitch I. 2012. Transcription initiation factor DksA has diverse effects on RNA chain elongation. Nucleic Acids Res 40(8):3392–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Furman R, Tsodikov OV, Wolf YI, Artsimovitch I. 2013. An insertion in the catalytic trigger loop gates the secondary channel of RNA polymerase. J. Mol. Biol 425:82–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaal T, Gourse RL. 1990. Guanosine 3′-diphosphate 5′-diphosphate is not required for growth rate-dependent control of rRNA synthesis in Escherichia coli. PNAS 87:5533–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaca AO, Kajfasz JK, Miller JH, Liu K, Wang JD, et al. 2013. Basal levels of (p)ppGpp in Enterococcus faecalis: the magic beyond the stringent response. mBio 4(5):e00646–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a.Gallant JA. 1979. Stringent control in E. coli. Annu. Rev. Genet 13:393–415 [DOI] [PubMed] [Google Scholar]
- 42.Gentry D, Xiao H, Burgess R, Cashel M. 1991. The ω subunit of Escherichia coli K-12 RNA polymerase is not required for stringent RNA control in vivo. J. Bacteriol 173:3901–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Girard ME, Gopalkrishnan S, Grace ED, Halliday JA, Gourse RL, Herman C. 2018. DksA and ppGpp regulate the σS stress response by activating promoters for the small RNA DsrA and the anti-adapter protein IraP. J. Bacteriol 200:e00463–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gopalkrishnan S, Nicoloff H, Ades SE. 2014. Co-ordinated regulation of the extracytoplasmic stress factor, sigmaE, with other Escherichia coli sigma factors by (p)ppGpp and DksA may be achieved by specific regulation of individual holoenzymes. Mol. Microbiol 93:479–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gopalkrishnan S, Ross W, Chen AY, Gourse RL. 2017. TraR directly regulates transcription initiation by mimicking the combined effects of the global regulators DksA and ppGpp. PNAS 114:E5539–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grace ED, Gopalkrishnan S, Girard ME, Blankschien MS, Ross W, et al. 2015. Activation of the σE-dependent stress pathway by conjugative TraR may anticipate conjugational stress. J. Bacteriol 197:924–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gummesson B, Lovmar M, Nyström T. 2013. A proximal promoter element required for positive transcriptional control by guanosine tetraphosphate and DksA protein during the stringent response. J. Biol. Chem 288:21055–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harms A, Fino C, Sørensen MA, Semsey S, Gerdes K. 2017. Prophages and growth dynamics confound experimental results with antibiotic-tolerant persister cells. mBio 8:e01964–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haseltine WA, Block R. 1973. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. PNAS 70(5):1564–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haugen SP, Berkmen MB, Ross W, Gaal T, Ward C, Gourse RL. 2006. rRNA promoter regulation by nonoptimal binding of sigma region 1.2: an additional recognition element for RNA polymerase. Cell 125:1069–82 [DOI] [PubMed] [Google Scholar]
- 51.Haugen SP, Ross W, Gourse RL. 2008. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat. Rev. Microbiol 6:507–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. 2015. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol 13:298–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hesketh A, Vergnano M, Wan C, Oliver SG. 2017. Bacterial signaling nucleotides inhibit yeast cell growth by impacting mitochondrial and other specifically eukaryotic functions. mBio 8:e01047–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53a.Hirvonen CA, Ross W, Wozniak CE, Marasco E, Anthony JR, et al. 2001. Contributions of UP elements and the transcription factor FIS to expression from the seven rrn P1 promoters in Escherichia coli. J. Bacteriol 183:6305–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hogg T, Mechold U, Malke H, Cashel M, Hilgenfeld R. 2004. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response. Cell 117(1):57–68 [DOI] [PubMed] [Google Scholar]
- 55.Hood RD, Higgins SA, Flamholz A, Nichols RJ, Savage DF. 2016. The stringent response regulates adaptation to darkness in the cyanobacterium Synechococcus elongatus. PNAS 113:E4867–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Irving SE, Corrigan RM. 2018. Triggering the stringent response: signals responsible for activating (p)ppGpp synthesis in bacteria. Microbiology 164:268–76 [DOI] [PubMed] [Google Scholar]
- 57.Jiang M, Datta K, Walker A, Strahler J, Bagamasbad P, et al. 2006. The Escherichia coli GTPase CgtAE is involved in late steps of large ribosome assembly. J. Bacteriol 188:6757–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kajitani M, Ishihama A. 1984. Promoter selectivity of Escherichia coli RNA polymerase: differential stringent control of the multiple promoters from ribosomal RNA and protein operons. J. Biol. Chem 259:1951–57 [PubMed] [Google Scholar]
- 59.Kang PJ, Craig EA. 1990. Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a dnaK deletion mutation. J. Bacteriol 172:2055–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanjee U, Ogata K, Houry WA. 2012. Direct binding targets of the stringent response alarmone (p)ppGpp. Mol. Microbiol 85:1029–43 [DOI] [PubMed] [Google Scholar]
- 61.Kasai K, Nishizawa T, Takahashi K, Hosaka T, Aoki H, Ochi K. 2006. Physiological analysis of the stringent response elicited in an extreme thermophilic bacterium, Thermus thermophilus. J. Bacteriol 188:7111–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kingston RE, Chamberlin MJ. 1981. Pausing and attenuation of in vitro transcription in the rrnB operon of E. coli. Cell 27(3 Part 2):523–31 [DOI] [PubMed] [Google Scholar]
- 63.Kingston RE, Nierman WC, Chamberlin MJ. 1981. A direct effect of guanosine tetraphosphate on pausing of Escherichia coli RNA polymerase during RNA chain elongation. J. Biol. Chem 256:2787–97 [PubMed] [Google Scholar]
- 64.Krasny L, Gourse RL. 2004. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J 23:4473–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kriel A, Bittner AN, Kim SH, Liu K, Tehranchi AK, et al. 2012. Direct regulation of GTP homeostasis by (p)ppGpp: a critical component of viability and stress resistance. Mol. Cell 48:231–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lamour V, Hogan BP, Erie DA, Darst SA. 2006. Crystal structure of Thermus aquaticus Gfh1, a Gre-factor paralog that inhibits rather than stimulates transcript cleavage. J. Mol. Biol 356:179–88 [DOI] [PubMed] [Google Scholar]
- 67.Lane WJ, Darst SA. 2010. Molecular evolution of multisubunit RNA polymerases: sequence analysis. J. Mol. Biol 395:671–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Laptenko O, Kim SS, Lee J, Starodubtseva M, Cava F, et al. 2006. pH-dependent conformational switch activates the inhibitor of transcription elongation. EMBO J 25:2131–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Laurie AD, Bernardo LM, Sze CC, Skarfstad E, Szalewska-Palasz A, et al. 2003. The role of the alarmone (p)ppGpp in sigma N competition for core RNA polymerase. J. Biol. Chem 278:1494–503 [DOI] [PubMed] [Google Scholar]
- 70.Lee J-H, Lennon CW, Ross W, Gourse RL. 2012. Role of the coiled-coil tip of Escherichia coli DksA in promoter control. J. Mol. Biol 416:503–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lemke JJ, Durfee T, Gourse RL. 2009. DksA and ppGpp directly regulate transcription of the Escherichia coli flagellar cascade. Mol. Microbiol 74:1368–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lennon CW, Gaal T, Ross W, Gourse RL. 2009. Escherichia coli DksA binds to free RNA polymerase with higher affinity than to RNA polymerase in an open complex. J. Bacteriol 191:5854–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lennon CW, Lemmer KC, Irons JL, Sellman MI, Donohue TJ, et al. 2014. A protein mechanistically similar to Escherichia coli DksA regulates photosynthetic growth of Rhodobacter sphaeroides. mBio 5:e01105–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lennon CW, Ross W, Martin-Tumasz S, Toulokhonov I, Vrentas CE, et al. 2012. Direct interactions between the coiled coil tip of DksA and the trigger loop of RNA polymerase mediate transcriptional regulation. Genes Dev 26:2634–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lindahl L, Post L, Nomura M. 1976. DNA-dependent in vitro synthesis of rRNA, ribosomal proteins, protein elongation factors, and RNA polymerase subunit alpha: inhibition by ppGpp. Cell 9:439–48 [DOI] [PubMed] [Google Scholar]
- 76.Liu K, Bittner AN, Wang JD. 2015. Diversity in (p)ppGpp metabolism and effectors. Curr. Opin. Microbiol 24:72–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu K, Myers AR, Pisithkul T, Claas KR, Satyshur KA, et al. 2015. Molecular mechanism and evolution of guanylate kinase regulation by (p)ppGpp. Mol. Cell 57:735–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Loveland AB, Bah E, Madireddy R, Zhang Y, Brilot AF, et al. 2016. Ribosome·RelA structures reveal the mechanism of stringent response activation. eLife 5:e17029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Magnusson LU, Gummesson B, Joksimović P, Farewell A, Nyström T. 2007. Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. J. Bacteriol 189:5193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79a.Mallik P, Paul BJ, Rutherford ST, Gourse RL, Osuna R. 2006. DksA is required for growth phase-dependent regulation, growth rate-dependent control, and stringent control of fis expression in Escherichia coli. J. Bacteriol 188:5775–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mechold U, Murphy H, Brown L, Cashel M. 2002. Intramolecular regulation of the opposing (p)ppGpp catalytic activities of Rel(Seq), the Rel/Spo enzyme from Streptococcus equisimilis. J. Bacteriol 184:2878–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mechold U, Potrykus K, Murphy H, Murakami KS, Cashel M. 2013. Differential regulation by ppGpp versus pppGpp in Escherichia coli. Nucleic Acids Res 41:6175–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Molodtsov V, Sineva E, Zhang L, Huang X, Cashel M, et al. 2018. Allosteric effector ppGpp potentiates the inhibition of transcript initiation by DksA. Mol. Cell 69:828–39.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Murphy H, Cashel M. 2003. Isolation of RNA polymerase suppressors of a (p)ppGpp deficiency. Methods Enzymol 371:596–601 [DOI] [PubMed] [Google Scholar]
- 84.Murray HD, Gourse RL. 2004. Unique roles of the rrn P2 rRNA promoters in Escherichia coli. Mol. Microbiol 52:1375–87 [DOI] [PubMed] [Google Scholar]
- 85.Murray HD, Schneider DA, Gourse RL. 2003. Control of rRNA expression by small molecules is dynamic and nonredundant. Mol. Cell 12:125–34 [DOI] [PubMed] [Google Scholar]
- 85a.Nafissi M, Chau J, Xu J, Johnson RC. 2012. Robust translation of the nucleoid protein Fis requires a remote upstream AU element and is enhanced by RNA secondary structure. J. Bacteriol 194:2458–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nayak D, Voss M, Windgassen T, Mooney RA, Landick R. 2013. Cys-pair reporters detect a constrained trigger loop in a paused RNA polymerase. Mol. Cell 50:882–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ochi K, Nishizawa T, Inaoka T, Yamada A, Hashimoto K, et al. 2012. Heterologous expression of a plant RelA-SpoT homologue results in increased stress tolerance in Saccharomyces cerevisiae by accumulation of the bacterial alarmone ppGpp. Microbiology 158:2213–24 [DOI] [PubMed] [Google Scholar]
- 88.Owens JR, Woody AY, Haley BE. 1987. Characterization of the guanosine-3′-diphosphate-5′-diphosphate binding site on E. coli RNA polymerase using a photoprobe, 8-azidoguanosine-3′−5′-bisphosphate. Biochem. Biophys. Res. Commun 142:964–71 [DOI] [PubMed] [Google Scholar]
- 89.Parshin A, Shiver AL, Lee J, Ozerova M, Schneidman-Duhovny D, et al. 2015. DksA regulates RNA polymerase in Escherichia coli through a network of interactions in the secondary channel that includes Sequence Insertion 1. PNAS 112:e6862–71. Correction. 2016. PNAS 113:E103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, et al. 2004. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118:311–22 [DOI] [PubMed] [Google Scholar]
- 91.Paul BJ, Berkmen MB, Gourse RL. 2005. DksA potentiates direct activation of amino acid promoters by ppGpp. PNAS 102:7823–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, et al. 2004. Regulation through the secondary channel–structural framework for ppGpp-DksA synergism during transcription. Cell 118:297–309 [DOI] [PubMed] [Google Scholar]
- 93.Potrykus K, Cashel M. 2008. (p)ppGpp: still magical? Annu. Rev. Microbiol 62:35–51 [DOI] [PubMed] [Google Scholar]
- 94.Potrykus K, Murphy H, Philippe N, Cashel M. 2011. ppGpp is the major source of growth rate control in E. coli. Environ. Microbiol 13:563–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94a.Rao L, Ross W, Appleman JA, Gaal T, Leirmo S, et al. 1994. Factor independent activation of rrnB P1: an “extended” promoter with an upstream element that dramatically increases promoter strength. J. Mol. Biol 235:1421–35 [DOI] [PubMed] [Google Scholar]
- 95.Repoila F, Majdalani N, Gottesman S. 2003. Small non-coding RNAs, co-ordinators of adaptation processes in Escherichia coli: the RpoS paradigm. Mol. Microbiol 48:855–61 [DOI] [PubMed] [Google Scholar]
- 96.Riggs DL, Mueller RD, Kwan HS, Artz SW. 1986. Promoter domain mediates guanosine tetraphosphate activation of the histidine operon. PNAS 83:9333–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Roelofs KG, Wang J, Sintim HO, Lee VT. 2011. Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. PNAS 108:15528–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Roghanian M, Zenkin N, Yuzenkova Y. 2015. Bacterial global regulators DksA/ppGpp increase fidelity of transcription. Nucleic Acids Res 43:1529–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ross W, Sanchez-Vazquez P, Chen AY, Lee JH, Burgos HL, Gourse RL. 2016. ppGpp binding to a site at the RNAP-DksA interface accounts for its dramatic effects on transcription initiation during the stringent response. Mol. Cell 62:811–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ross W, Vrentas CE, Sanchez-Vazquez P, Gaal T, Gourse RL. 2013. The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol. Cell 50:420–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ruff EF, Record MT Jr., Artsimovitch I. 2015. Initial events in bacterial transcription initiation. Biomolecules 5:1035–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rutherford ST, Lemke JJ, Vrentas CE, Gaal T, Ross W, Gourse RL. 2007. Effects of DksA, GreA, and GreB on transcription initiation: insights into the mechanisms of factors that bind in the secondary channel of RNA polymerase. J. Mol. Biol 366:1243–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rutherford ST, Villers CL, Lee J-H, Ross W, Gourse RL. 2009. Allosteric control of Escherichia coli rRNA promoter complexes by DksA. Genes Dev 23:236–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103a.Sanchez-Vazquez P. 2018. Genome-wide effects of ppGpp binding to RNA polymerase on E. coli gene expression. PhD Thesis, Univ. Wisconsin–Madison [Google Scholar]
- 104.Sands MK, Roberts RB. 1952. The effects of a tryptophan-histidine deficiency in a mutant of Escherichia coli. J. Bacteriol 63:505–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Steinchen W, Bange G. 2016. The magic dance of the alarmones (p)ppGpp. Mol. Microbiol 101:531–44 [DOI] [PubMed] [Google Scholar]
- 106.Sugliani M, Abdelkefi H, Ke H, Bouveret E, Robaglia C, et al. 2016. An ancient bacterial signaling pathway regulates chloroplast function to influence growth and development in Arabidopsis. Plant Cell 28:661–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sun D, Lee G, Lee JH, Kim HY, Rhee HW, et al. 2010. A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses. Nat. Struct. Mol. Biol 17:1188–94 [DOI] [PubMed] [Google Scholar]
- 108.Syal K, Chatterji D. 2015. Differential binding of ppGpp and pppGpp to E. coli RNA polymerase: photo-labeling and mass spectral studies. Genes Cells 20:1006–16 [DOI] [PubMed] [Google Scholar]
- 109.Tagami S, Sekine S, Kumarevel T, Hino N, Murayama Y, et al. 2010. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature 468:978–82 [DOI] [PubMed] [Google Scholar]
- 110.Tedin K, Bremer H. 1992. Toxic effects of high levels of ppGpp in Escherichia coli are relieved by rpoB mutations. J. Biol. Chem 267:2337–44 [PubMed] [Google Scholar]
- 111.Tetone LE, Friedman LJ, Osborne ML, Ravi H, Kyzer S, et al. 2017. Dynamics of GreB-RNA polymerase interaction allow a proofreading accessory protein to patrol for transcription complexes needing rescue. PNAS 114:E1081–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111a.Tosa T, Pizer LI. 1971. Biochemical bases for the antimetabolite action of l-serine hydroxamate. J. Bacteriol 106:972–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Toulokhonov II, Shulgina I, Hernandez VJ. 2001. Binding of the transcription effector ppGpp to Escherichia coli RNA polymerase is allosteric, modular, and occurs near the N terminus of the beta′-subunit. J. Biol. Chem 276:1220–25 [DOI] [PubMed] [Google Scholar]
- 113.Trautinger BW, Lloyd RG. 2002. Modulation of DNA repair by mutations flanking the DNA channel through RNA polymerase. EMBO J 21:6944–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Travers AA. 1980. Promoter sequence for stringent control of bacterial ribonucleic acid synthesis. J. Bacteriol 141:973–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Traxler MF, Summers SM 1, Nguyen HT, Zacharia VM, Hightower GA, et al. 2008. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol 68:1128–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vogel U, Jensen KF. 1994. Effects of guanosine 3′,5′-bisdiphosphate (ppGpp) on rate of transcription elongation in isoleucine-starved Escherichia coli. J. Biol. Chem 269:16236–41 [PubMed] [Google Scholar]
- 117.Vogel U, Jensen KF. 1997. NusA is required for ribosomal antitermination and for modulation of the transcription elongation rate of both antiterminated RNA and mRNA. J. Biol. Chem 272:12265–71 [DOI] [PubMed] [Google Scholar]
- 118.Vrentas CE, Gaal T, Berkmen MB, Rutherford ST, Haugen SP, et al. 2008. Still looking for the magic spot: The crystallographically-defined binding site for ppGpp on RNA polymerase is unlikely to be responsible for rRNA transcription regulation. J. Mol. Biol 377:551–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vrentas CE, Gaal T, Ross W, Ebright RH, Gourse RL. 2005. Response of RNA polymerase to ppGpp: requirement for omega subunit and relief of this requirement by DksA. Genes Dev 19:2378–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang JD, Sanders GM, Grossman AD. 2007. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell 128:865–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120a.Winther KS, Roghanian M, Gerdes K. 2018. Activation of the stringent response by loading of RelA-tRNA complexes at the ribosomal A-site. Mol. Cell 70:95–105 [DOI] [PubMed] [Google Scholar]
- 121.Wippel K, Long SR. 2016. Contributions of Sinorhizobium meliloti transcriptional regulator DksA to bacterial growth and efficient symbiosis with Medicago sativa. J. Bacteriol 198:1374–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wout P, Pu K, Sullivan SM, Reese V, Zhou S, et al. 2004. The Escherichia coli GTPase CgtAE cofractionates with the 50S ribosomal subunit and interacts with SpoT, a ppGpp synthetase/hydrolase. J. Bacteriol 186:5249–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem 266:5980–90 [PubMed] [Google Scholar]
- 124.Zenkin N, Yuzenkova Y. 2015. New insights into the functions of transcription factors that bind the RNA polymerase secondary channel. Biomolecules 5:1195–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang Y, Feng Y, Chatterjee S, Tuske S, Ho MX, et al. 2012. Structural basis of transcription initiation. Science 338:1076–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang Y, Mooney RA, Grass JA, Sivaramakrishnan P, Herman C, et al. 2014. DksA guards elongating RNA polymerase against ribosome-stalling-induced arrest. Mol. Cell 53:766–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang Y, Zbornikova E, Rejman D, Gerdes K. 2018. Novel (p)ppGpp binding and metabolizing proteins of Escherichia coli. mBio 9:e02188–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zuo Y, Wang Y, Steitz TA. 2013. The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex. Mol. Cell 50:430–36 [DOI] [PMC free article] [PubMed] [Google Scholar]


