Abstract
Maternally-transmitted endosymbiotic bacteria are ubiquitous in insects. Among other influential phenotypes, many heritable symbionts of arthropods are notorious for manipulating host reproduction through one of four reproductive syndromes, which are generally exerted during early developmental stages of the host: male feminization; parthenogenesis induction; male killing; and cytoplasmic incompatibility (CI). Major advances have been achieved in understanding mechanisms and identifying symbiont factors involved in reproductive manipulation, particularly male killing and cytoplasmic incompatibility. Nonetheless, whether cytoplasmically-transmitted bacteria influence the maternally-loaded components of the egg or early embryo has not been examined. In the present study, we investigated whether heritable endosymbionts that cause different reproductive phenotypes in Drosophila melanogaster influence the mRNA transcriptome of early embryos. We used mRNA-seq to evaluate differential expression in Drosophila embryos lacking endosymbionts (control) to those harbouring the male-killing Spiroplasma poulsonii strain MSRO-Br, the CI-inducing Wolbachia strain wMel, or Spiroplasma poulsonii strain Hyd1; a strain that lacks a reproductive phenotype and is naturally associated with Drosophila hydei. We found no consistent evidence of influence of symbiont on mRNA composition of early embryos, suggesting that the reproductive manipulation mechanism does not involve alteration of maternally-loaded transcripts. In addition, we capitalized on several available mRNA-seq datasets derived from Spiroplasma-infected Drosophila melanogaster embryos, to search for signals of depurination of rRNA, consistent with the activity of Ribosome Inactivating Proteins (RIPs) encoded by Spiroplasma poulsonii. We found small but statistically significant signals of depurination of Drosophila rRNA in the Spiroplasma treatments (both strains), but not in the symbiont-free control or Wolbachia treatment, consistent with the action of RIPs. The depurination signal was slightly stronger in the treatment with the male-killing strain. This result supports a recent report that RIP-induced damage contributes to male embryo death.
Subject terms: Gene expression, Bacteria, Entomology
Introduction
Heritable associations between arthropods and endosymbiotic bacteria are widespread and influential to their hosts1 and communities2. With few exceptions3, inheritance is achieved via the mother, generating an asymmetry in the interests of the symbiont regarding the host’s sex, whereby males are generally dead-ends for the symbiont. Consistent with this inequality, numerous heritable bacteria manipulate host reproduction in favour of symbiont-bearing females. Four such reproductive phenotypes have been described4. Feminization occurs when genetic males develop and function as females. Parthenogenesis-induction occurs in haplo-diploid systems, where unfertilized eggs, which would otherwise develop as males, develop instead into reproductively functional females. In male-killing (or son-killing), the symbiont eliminates infected males to the presumed advantage of surviving infected female siblings5. In cytoplasmic incompatibility (CI)6, matings between symbiont-infected males and uninfected females result in death of offspring at the embryonic stage. The CI mechanism involves symbiont-mediated damage to the male sperm that is rescued in the presence of a compatible symbiont strain in the egg4.
Most maternally inherited symbionts are transmitted to a new host through the egg cytoplasm7. As such, they may manipulate the female host’s reproductive system or usurp her cellular machinery in order to invade developing oocytes8–10. Infection-induced changes during oogenesis may thus have effects on the composition of an egg (e.g. Wolbachia reduces the maternal transmission of the gypsy endogenous retrovirus11). Drosophila melanogaster eggs contain maternal RNAs that are exclusively expressed during early development (prior to embryonic stage 5 or ~2 h after egg deposition; AED). Zygotic transcription is silent during this period and therefore maternal mRNAs play a crucial role in early embryonic development12. The egg, in a sense, is a point of convergence between the existing host, symbiont, and new host, and consequently could undergo symbiont-induced changes that could lay the foundation for the occupation of the symbiont within the new host. Furthermore, it is possible that infection by a reproductive parasite could cause changes in maternally-derived components that are necessary to induce a reproductive phenotype. As hosts of two independent lineages of maternally transmitted bacteria, Wolbachia and Spiroplasma13, members of the genus Drosophila, have emerged as a model system for heritable symbiosis e.g.14–23.
The wall-less bacterial genus Spiroplasma (class Mollicutes) is generally associated with arthropods and plants, and can reside intra- and extra-cellularly24. The nature of Spiroplasma-host associations ranges from pathogenic to mutualistic, although the fitness consequences of the majority of Spiroplasma strains remain unknown. Most strains of Spiroplasma known to date appear to transmit horizontally from the environment or via a vector (e.g. several insect-vectored plant pathogens). A few strains of Spiroplasma, however, are maternally-transmitted by their arthropod hosts. Among these, ~20 species in the genus Drosophila are reported to harbour Spiroplasma13,25,26. A subset of maternally-transmitted Spiroplasma of Drosophila and other arthropods are male killers. All of the Drosophila-associated Spiroplasma male-killing strains that have been genetically characterized to date fall within the poulsonii clade; one of the four Spiroplasma clades that independently invaded the genus Drosophila26. The poulsonii clade also contains non-male-killing strains such as “Hyd1” and “sNeo”; naturally-occurring defensive mutualists of Drosophila hydei and Drosophila neotestacea, respectively25,27, as well as several strains that lost the ability to kill males in the lab15,28,29.
The mechanism by which Spiroplasma exerts death of Drosophila male embryos is not fully understood, but several aspects have been elucidated15,30–35. First, a functional dosage compensation complex (DCC; also known as the male-specific lethal complex) is required, as mutants of components of this complex that are infected with male-killing Spiroplasma, fail to express male-killing32. Secondly, the DCC, which normally acetylates X chromatin in males, mis-localizes to other regions of the nucleus immediately prior to the killing stage35. This phenomenon is accompanied by inappropriate histone acetylation, and genome-wide transcription misregulation35. Thirdly, abnormal massive apoptosis and neural disorganization occurs30,31,34. The male X chromosome exhibits signs of DNA damage (chromatin bridges and segregation defects), and bridge breakage triggers sex-specific abnormal apoptosis via p53-dependent pathways30. Recently Harumoto and Lemaitre15 identified spaid, a Spiroplasma-encoded gene that appears to be responsible for male killing. Overexpression of spaid in D. melanogaster embryos causes death of males, but not females, and induces massive apoptosis and neural defects, reminiscent of the Spiroplasma-induced male-killing phenomenon in pattern, but in a somewhat delayed fashion (i.e., developmental arrest at embryonic stage 12–1331,33 in Spiroplasma-infected wild-type D. melanogaster vs. at the second larval instar with expression of the spaid transgene in Spiroplasma-free D. melanogaster15). Spaid contains an OTU (ovarian tumor) deubiquitinase domain and ankyrin repeats (ANK). Harumoto and Lemaitre15 propose that the OTU domain promotes nuclear localization of Spaid (in both female and male embryos), while the ankyrin repeats interact with DCC complex itself or with its associated histone modifications. How this leads to DNA damage and segregation defects of the male X chromosome, as well as to other phenotypes associated with male-killing described above, and which are the host cellular targets of Spaid remain unknown.
In addition to Spaid, a different type of toxin has been reported in Spiroplasma. Ribosome inactivating proteins (RIPs) are plant- (e.g. ricin and saporin) and bacteria-encoded (e.g. Shiga toxin) enzymes that cleave an adenine base (hereafter “depurinate”) from a specific position within a motif of the 28S rRNA that is universally conserved across eukaryotes. This motif is known as the sarcin-ricin loop (SRL). Depurination of this site renders the ribosome incapable of protein synthesis36. Several confirmed and predicted RIPs are encoded in the genomes of Spiroplasma strains that associate with Drosophila, including the male-killing strain native to D. melanogaster (MSRO) and the closely-related non-male-killing strains sNeo17,37,38 and Hyd139. These poulsonii-clade strains are also known for their defensive abilities against certain parasitic wasps and nematodes of Drosophila21,22,27,40,41. Evidence of SRL depurination consistent with the action of RIP has been detected in nematodes and wasps exposed to Drosophila harbouring Spiroplasma17,37, and has led to the hypothesis that RIPs play a major role in the Spiroplasma-mediated defence against wasps and nematodes. Depurination of Drosophila ribosomes in Spiroplasma-infected treatments had been detected in larvae, but was mostly restricted to “cell-free hemolymph”, suggesting only ribosomes outside the cell were targets37. Furthermore, despite detecting significant levels of depurinated Drosophila ribosomes in Spiroplasma-infected flies, this phenomenon is generally not accompanied by a significant decrease in intact (i.e., non-depurinated) ribosomes17,37. This, along with the observation that the detected levels of depurination were not associated with larva-to-adult fly mortality, was interpreted as RIP activity having a negligible direct effect on fly fitness17,37. A more recent study, however, revealed that Spiroplasma-mediated depurination of Drosophila ribosomes varies widely by life stage. It is strongest in embryos and old adults38, but also not accompanied by a detectable decrease in intact ribosomes. In addition, significant ribosome depurination (but not significant depletion of intact ribosomes) occurs under heterologous expression of two Spiroplasma RIP genes in D. melanogaster, confirming their RIP activity. Their expression was also associated with embryo mortality (male mortality was higher), and with a reduction in fly lifespan and in adult hemocyte number38.
In this study we examined whether Spiroplasma influences the composition of maternally loaded transcripts of host eggs (i.e., prior to fertilization), particularly in a manner that may facilitate male-killing. To do so we compared the effects of three heritable endosymbionts (a male-killing Spiroplasma, a non-male-killing Spiroplasma, and a CI-inducing Wolbachia) on the composition of mRNA transcripts of early embryos of Drosophila melanogaster. Furthermore, we used data from this study and published RNA-seq data to analyse whether there is a difference in signals of rRNA depurination of host embryo ribosomes in the presence of male-killing and non-male-killing symbionts.
Materials and Methods
Generation of symbiont treatments
We used three infection treatments, one Wolbachia and two Spiroplasma strains, and a symbiont-free control (Fig. 1). Laboratory stocks of D. melanogaster (Canton S strain; CS) that naturally harbour the Wolbachia strain wMel were used to generate the Wolbachia treatment (W + S−). Positive infection for wMel was confirmed based on PCR with Wolbachia-specific primers targeting the wsp gene42. The same stock was reared in tetracycline food (final concentration 0.02 g/ml) for two generations, followed by three generations of antibiotic-free food to generate a Wolbachia-free (W−) stock. The W− flies served as the symbiont-free control. The Spiroplasma infection treatments (W−S+) were generated by artificially infecting Wolbachia-free (W−) flies with the strain MSRO-BR (Red 42) native to D. melanogaster43 or Hyd1 (TEN-104-106) native to D. hydei13. Fifteen Wolbachia-free (W−) females (15 lines) were infected per Spiroplasma strain. These artificially infected lines were maintained for 3–5 generations before being used for the experiment. Spiroplasma-infected (W−S+) lines were selected every generation to ensure positive infection status, based on PCR with Spiroplasma-specific 16S ribosomal DNA primers43. MSRO treatment lines were backcrossed to Wolbachia-free (W−) CS males every generation, as male-killing by this strain is nearly perfect. A minimum of four infected lines was combined, per replicate, at the start of the experiment, to create a total of three biological replicates for each Spiroplasma treatment. The biological replicates for the Wolbachia treatment and control were maintained as three different populations for four generations prior to the start of the experiment.
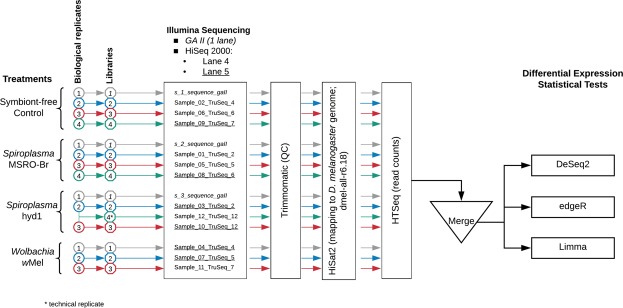
Figure 1.
Experimental design and workflow for data analysis. The biological replicates, corresponding libraries, and sequence file labels are distinguished by colours and font type. Italics = samples run on Illumina GAII. Non-italics = samples run on Illumina HiSeq 2000; non-underlined samples were pooled into one sequencing lane (4), whereas underlined samples were pooled into another lane (5). Library labelled “Sample_12_TruSeq_12” is the result of combining total RNA from Hyd1 biological replicates 2 and 3, and thus considered a technical, rather than biological, replicate. This library was excluded from the differential expression analyses.
Embryo collection
Approximately 40–50 three-day-old virgin females, from each replicate of each treatment, were allowed to mate in cages with Wolbachia-free (W−) CS males during the collection period, and allowed to lay eggs on cornmeal food plates. The initial batch of eggs was discarded to improve the chances of collecting fertilized eggs from the same stage44. Thereafter, egg laying was monitored and cornmeal plates were changed approximately every 45 min, so as to collect embryos that were on average ~60–75 min old. Eggs were collected from each replicate with a small brush 6–8 times over a 2-day period for each treatment and the control. The eggs were placed in sterile RNase-free 1.7 ml microtubes, and immediately put on dry ice during the collection period, after which they were transferred to −80 °C for storage.
RNA extraction, library preparation, and sequencing
Three to four biological replicates per condition (MSRO, Hyd1, wMel, and control) were used for the extractions (see Fig. 1). RNA was extracted per collection tube of eggs (mentioned above) with Trizol® Plus RNA Purification System (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. RNA from tubes that belonged to the same biological replicate within a treatment was pooled. All RNA samples were DNase-treated with Ambion® DNA- free (Invitrogen) to remove any DNA contamination. Total RNA was quantified with a NanoDrop® ND-1000 UV spectrophotometer (NanoDrop Technologies, Wilmington, DE), and sample quality and integrity were further tested with the Agilent 2100 BioAnalyzer (Agilent Inc., Santa Clara, CA).
Total RNA was submitted to the Texas AgriLife Genomics and Bioinfomatics Services facility for library preparation, sequencing, and demultiplexing. Three biological replicates were subjected to multiplex NuGen library preparation and pooled into a single sequencing lane of the Illumina (San Diego, CA) Genome Analyzer II platform (single-end; 76 bp read length; italicized samples in Fig. 1). The remaining biological replicates were subjected to the Illumina mRNA TruSeq kit library preparation protocol (12 libraries). Half of the libraries were pooled into one lane of the Illumina HiSeq 2000 platform and the other half were pooled into another lane (single-end; 100 bp read length; see Fig. 1).
Analyses of differential expression (DE)
Raw reads have been deposited in the NCBI SRA Database under Accession Numbers SRR7279355-SRR7279369 (BioProject PRJNA474708; BioSample SAMN09370647). Command lines, as well as input and output files needed to replicate our analyses, from the different pipelines used are available in Supporting Data Files S1–S4. Raw RNA sequence files were first processed with Trimmomatic (v 0.35)45 to remove adapters and low quality reads (see Fig. 1). The processed runs were then mapped to the Flybase Drosophila genome (version dmel-all-r6.18) with HiSat2 (v.2.0.5)46 to obtain bam files. The files containing the mapped read information were then analysed with HTseq (0.6.1)47 to obtain read counts for both genes and exon regions specified in a corresponding gff file for the dmel-all-r6.18 genome sequence. The resulting count files were then manually formatted into a count matrix suitable for differential expression analysis (Supporting Data File S2).
Differential expression analyses were performed with three different pipelines edgeR v.3.20.548 and Limma + Voom v. limma_3.34.549 and DeSeq2 v.1.18.150. Command line and relevant input files for these analyses are provided in Supporting Data Files S1–S3. All programs conducted their respective tests on genes/exons with 10 or more mapped reads per replicate within at least one of two the treatments being compared. For the EdgeR pipeline both the glm and QFglm models were used for DE. For Deseq2, the wald statistical test was used for DE analysis. Before analysis in both Limma-Voom and EdgeR pipeline, two genes with extremely high counts, 16S rRNA (FBgn0013686) and ef-1α (FBgn0284245), were removed. Genes with an adjusted p-value (or false discovery rate; FDR) of 0.05 or less were considered to be potentially differentially expressed (DE).
Power analyses for differential expression
To identify possible limitations of our experimental design in the detection of DE genes, we performed a statistical power analysis with the method of Hart et al.51. The parameters (and rationale) used to run the simulations are provided in Table 1 and Fig. 2.
Table 1.
Parameters used to run the simulations of the power analyses. All possible combinations of these parameters (60 total) were run.
| Coverage (X) | Coefficient of variation (CV)c | Number of replicatesd | Effect size (log-fold change)e | Alfa (threshold p-value) |
|---|---|---|---|---|
| 20a | 0.2 | 3 | 1.5 | 0.05 |
| 10b | 0.3 | 4 | 1.75 | |
| 0.4 | 2 | |||
| 3 | ||||
| 4 |
abased on the Lander/Waterman estimate of coverage for our data; i.e., mean coverage of 23X per replicate (range 19–28 among our replicates).
bbased on our minimum number of reads per replicate (in one treatment) required for a DE test (see text)
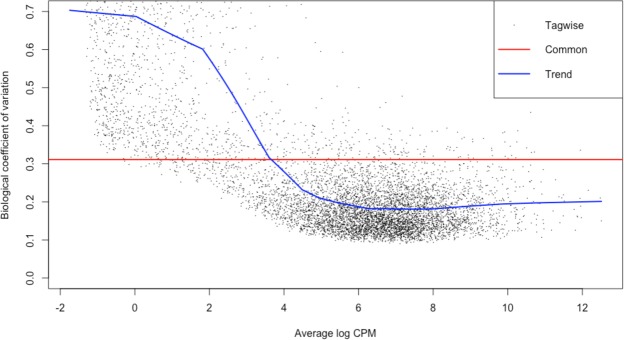
cThese values encompass the range of most of the values of Biological Coefficient of Variation (BCV; estimated by edgeR) obtained (i.e., a “Common” value of ~0.31 and a stabilized “Trend” value of ~0.2; see Fig. 2).
dreflects the range of replicates per treatment used in this study
epower was high and stable beyond an effect size of 4 (see Fig. 5). Thus, higher effect sizes are not reported.
Figure 2.
Biological coefficient of variation (BCV) for our data plotted against average log CPM (copies per million), as estimated by edgeR.
Analyses of depurination signal in the sarcin-ricin loop (SRL) of the 28 S rRNA of Drosophila
RIP toxins remove a specific adenine present in the sarcin-ricin loop (SRL) of the 28 S rRNA leaving an abasic site (i.e., the backbone remains intact)52. When a reverse transcriptase encounters an abasic site, it preferentially adds an adenine in the nascent complementary (c)DNA strand53. This property, which results in an incorrect base at the RIP-depurinated site in the cDNA and all subsequent PCR amplification steps, can be used to detect evidence of RIP activity in any procedure that relies on reverse transcription (e.g. RNA-seq or reverse-transcription qPCR). To examine whether a signal of depurination consistent with RIP activity was detectable in Spiroplasma-infected flies, we used the RNA-seq data generated in the present study, as well as RNA-seq data (also derived from poly-A-tail-enriched RNA libraries) from two published studies that also compared patterns of gene expression from Spiroplasma-infected and Spiroplasma-free D. melanogaster embryos: NCBI Acc. No. PRJDB446930; and PRJNA31837335. The Spiroplasma strain examined by35 and30 is substrain UG (for Uganda) of MSRO. Bowtie2 v.2.1.054 and Samtools 0.1.1955 were used to map and extract, respectively, the reads from the three studies to the 28S rRNA gene of D. melanogaster (Acc. No. NR_133562.1 positions 2591–3970). We verified that the reads that mapped to NR_133562.1:2591–3970 (hereafter “28S reads”) did not belong to other parts of the Drosophila genome or to the genomes of Spiroplasma or Wolbachia, as follows. We used HISAT2 v.2.0.2-beta46 (for Drosophila) and Bowtie2 v. 3.4.254 (for the prokaryotic genomes) to attempt mapping the “28S reads” to the following reference genomes: (1) the D. melanogaster genome (BDGP6); (2) MSRO-UG (NZ_PETG00000000.1); (3) a draft genome of Spiroplasma Hyd1 (unpublished data); and (4) Wolbachia wMel (NC_002978). As expected, the “28S reads” only mapped to (1), and specifically to regions whose annotation terms included “28S rRNA”. To visualize and count the shift from A to T (or other bases) in the source RNA pool, the “28S reads” were mapped again to the NR_133562.1:2591–3970 in Geneious v.11.1.2 (Biomatters Inc., Newark, NJ; “low sensitivity mode”; maximum gap size = 3; iterate up to 25 times). The number of reads containing each of the four bases or a gap at the target site was counted by selecting the position at all the reads to be counted, and recording the counts reported by Geneious under the “Nucleotide Statistics” option (gapped reads were excluded from subsequent analyses). The proportion of reads with an A at the target site (i.e., putatively intact rRNA) was calculated and compared among treatments and replicates. We used a mixed model in JMP Pro 13 (SAS Institute Inc., Cary, NC) to examine the effect of symbiont treatment (fixed) on the proportion of adenines (arcsine square root transformed) at the target position (excluding gaps from total number of reads). Source Study was treated as a random effect.
Results
The present study aimed at determining whether heritable symbionts alter the composition of maternally-loaded mRNAs in D. melanogaster. To achieve this, we used mRNA-seq to evaluate differential expression (DE) in Drosophila embryos lacking endosymbionts (control) to those harbouring one of the following heritable endosymbionts: the male-killing Spiroplasma poulsonii strain MSRO-Br; the CI-inducing Wolbachia strain wMel; and the non-male-killing Spiroplasma poulsonii strain Hyd1 (native to D. hydei). A power analysis was performed to determine the limitations of our experimental design. A secondary goal of this study capitalized on several available mRNA-seq datasets derived from Spiroplasma-infected Drosophila melanogaster embryos, to search for signals of damage (i.e., depurination) to Drosophila rRNA, consistent with the activity of Ribosome Inactivating Proteins (RIPs), which are encoded in the genomes of several Spiroplasma strains. This assay examined the proportion of adenines at the RIP target position (indicative of intact rRNA) in Spiroplasma-infected vs. Spiroplasma-free treatments.
Differential expression
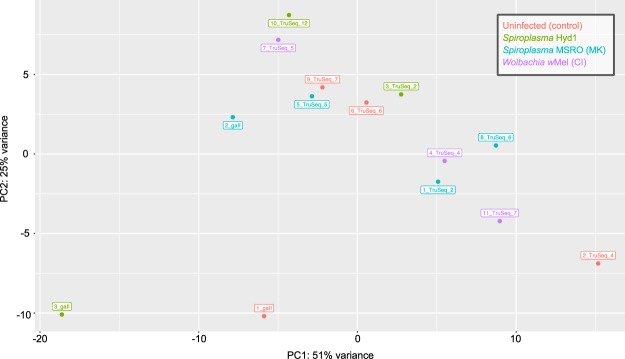
The number of reads that was obtained, passed QC, and mapped to the Drosophila genome is shown in Table 2. The PCA plot did not reveal any particular grouping of samples according to treatment (Fig. 3). PC1 explained 51% of the variance and revealed a slight “batch” effect, where the “GAII” replicates (i.e., 1_gall, 2_gall, and 3_gall) fell to the left of the other replicates. Nonetheless, removal of these replicates did not lead to a better grouping of replicates within each treatment (not shown). Thus, we considered that exclusion of the “GAII” replicates was not justified. Inclusion or exclusion of the Hyd1 technical replicate (e.g. n = 4 vs. 3 replicates, respectively) did not affect the differential expression analyses (only n = 3 is shown).
Table 2.
Number of reads obtained, retained after quality control, and mapped to the reference genome.
| Treatment | Replicate | Total reads | Reads after quality control | Reads mapped to reference genome | ||
|---|---|---|---|---|---|---|
| Number | Number | % | Number | % | ||
| Control | 1_gaII | 38,623,190 | 38,595,804 | 99.93 | 36,658,295 | 94.98 |
| Control | 2_TruSeq_4 | 32,414,845 | 32,413,140 | 99.99 | 31,382,402 | 96.82 |
| Control | 6_TruSeq_6 | 28,893,141 | 28,891,426 | 99.99 | 27,923,563 | 96.65 |
| Control | 9_TruSeq_7 | 31,672,204 | 31,672,204 | 100.00 | 30,671,362 | 96.84 |
| Hyd1 | 3_gaII | 39,050,068 | 39,031,083 | 99.95 | 36,931,211 | 94.62 |
| Hyd1 | 3_TruSeq_2 | 28,074,997 | 28,074,997 | 100.00 | 27,151,330 | 96.71 |
| Hyd1 | 10_TruSeq_12 | 31,084,062 | 31,084,062 | 100.00 | 30,070,722 | 96.74 |
| MSRO | 2_gaII | 36,838,900 | 36,824,546 | 99.96 | 34,640,850 | 94.07 |
| MSRO | 1_TruSeq_2 | 31,841,684 | 31,832,465 | 99.97 | 30,867,941 | 96.97 |
| MSRO | 5_TruSeq_5 | 30,537,077 | 30,535,822 | 100.00 | 29,442,640 | 96.42 |
| MSRO | 8_TruSeq_6 | 30,124,318 | 30,124,318 | 100.00 | 29,172,390 | 96.84 |
| wMel | 4_TruSeq_4 | 32,691,282 | 32,691,282 | 100.00 | 31,449,013 | 96.20 |
| wMel | 7_TruSeq_5 | 27,032,887 | 27,032,887 | 100.00 | 26,073,220 | 96.45 |
| wMel | 11_TruSeq_7 | 28,550,836 | 28,548,722 | 99.99 | 27,595,195 | 96.66 |
Figure 3.
DESeq-generated principle component analysis (PCA) with VSD transformation on the 14 biological replicates (see Fig. 1). Each treatment is labelled by a different colour. MK = male killing; CI = cytoplasmic incompatibility. The technical replicate “Sample_12_TruSeq_12” (not shown) was intermediate between “Sample_3_TruSeq_2” and “Sample_10_TruSeq_12”.
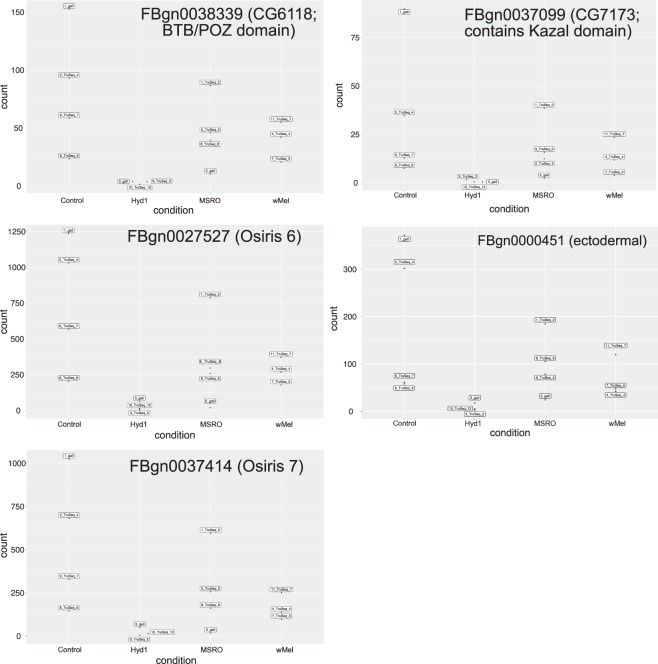
All differential expression (DE) tests were performed by comparing each symbiont treatment to the symbiont-free control. None of the eight pipelines used detected differentially expressed (DE) genes in the MSRO (male-killing Spiroplasma) and wMel (CI-inducing Wolbachia) treatments (Table 3). For the Hyd1 (non-male-killing Spiroplasma) treatment, half of the pipelines did not detect DE genes, whereas the other half detected a few DE genes (Table 3). The Deseq2 pipeline and the edgeR pipeline (glm model only) detected 5–12 differentially expressed (DE) genes in the Hyd1 treatment based on ‘by gene’ and ‘by exon’ analyses (Table 3). Only five DE genes (ect, Osi6, Osi7, FBgn0038339, and FBgn0037099) were found in common among these pipelines. For all of these, the Hyd1 treatment exhibited lower expression, and lower variation among replicates, than the other treatments (Table 3 and Fig. 4).
Table 3.
The number, identity (Flybase gene number), log fold-change (logFC), adjusted p-value or FDR, of genes identified as differentially expressed (DE) between each treatment and the symbiont-free control, by each of the four methods utilized. Results are shown for analyses of both genes and exons. Only the Hyd1 treatment exhibited significant DE genes.
| Deseq 2 | EdgeR | EdgeR | LimmaVoom | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| by Gene | by Exon | glm (TMM) by gene | glm (TMM) by exon | qlglm (TMM) by gene | qlglm (TMM) by exon | TMM + voom by gene | TMM + voom by exon | |||||||||
| Treatment (number of replicates) | ||||||||||||||||
| MSRO (4) | None | None | None | None | None | None | None | None | ||||||||
| wMel (3) | None | None | None | None | None | None | None | None | ||||||||
| Hyd1 (3) | 7 | 12 | 5 | 5 | None | None | None | None | ||||||||
| FlyBaseGeneNo. | Log FC | adjusted p | FlyBaseGeneNo. | Log FC | adjusted p | FlyBaseGeneNo. | Log FC | FDR | FlyBaseGeneNo. | logFC | FDR | |||||
| FBgn0038339 | −4.97 | 0.000087 | FBgn0038339 | −4.96 | 0.000086 | FBgn0038339 | −4.90 | 0.002497 | FBgn0038339 | −4.89 | 0.002948 | |||||
| FBgn0000451 | −4.57 | 0.000151 | FBgn0000451 | −4.53 | 0.000253 | FBgn0000451 | −4.54 | 0.030333 | FBgn0000451 | −4.50 | 0.030877 | |||||
| FBgn0027527 | −4.81 | 0.006198 | FBgn0027527 | −4.81 | 0.005639 | FBgn0027527 | −4.77 | 0.030344 | FBgn0027527 | −4.78 | 0.030877 | |||||
| FBgn0037099 | −6.66 | 0.027599 | FBgn0037099 | −6.66 | 0.021794 | FBgn0037099 | −6.29 | 0.030344 | FBgn0037099 | −6.30 | 0.030877 | |||||
| FBgn0037414 | −4.56 | 0.008411 | FBgn0037414 | −4.55 | 0.006991 | FBgn0037414 | −4.53 | 0.035559 | FBgn0037414 | −4.52 | 0.041176 | |||||
| FBgn0001254 | −3.51 | 0.006703 | FBgn0001254 | −3.51 | 0.005761 | FBgn0001254* | −3.50 | 0.076620 | ||||||||
| FBgn0001256 | −3.98 | 0.009660 | FBgn0001256 | −3.98 | 0.007762 | FBgn0001256* | −3.96 | 0.084605 | ||||||||
| FBgn0029807 | −4.23 | 0.026208 | ||||||||||||||
| FBgn0000568 | −0.99 | 0.049803 | ||||||||||||||
| FBgn0262366 | 2.90 | 0.049803 | ||||||||||||||
| FBgn0035430 | −3.80 | 0.049803 | ||||||||||||||
| FBgn0033275 | −3.77 | 0.005761 | ||||||||||||||
*Not significant at alfa = 0.05.
Figure 4.
Normalized counts, as estimated by Deseq2, for the five genes detected as differentially expressed (DE) by a subset of the pipelines (see text and Table 3). The wMel treatment (i.e., Wolbachia-infected) represents the treatment comparable to FlyBase-reported expression values. Nonetheless, the RPKM values reported by FlyBase (and presented in Table 4) are not directly comparable to our normalized counts, for the equivalent treatment.
Power analyses for differential expression
A plot of the power analyses is shown in Fig. 5 and the rationale for the parameters assumed is provided in Table 1. Assuming a coverage of 20X and a coefficient of variation of 0.3, we should expect to detect: ~100% of genes differentially expressed by log-fold changes ≥3; at least 60% of genes with log-fold changes ≥2, and at least ~40% of genes with log-fold changes ≥1.75. Therefore, these results suggest that our design and data have sufficient power to detect log-fold changes ≥3, but more limited power below that log-fold change value.
Figure 5.
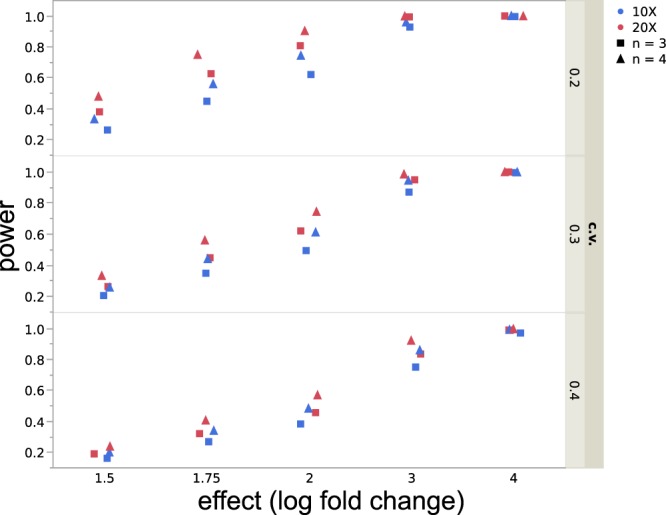
Power Analyses. Estimation of power for alfa = 0.05, at five different effect values ( = log-fold change: 1.5–4); number of replicates (n = 3 and 4), coverage values (X = 10 and 20), and coefficient of variation (c.v. = 0.2, 0.3, 0.4).
Signals of depurination
The number of reads mapped to the target site, as well as the proportion of adenines (representative of non-depurinated rRNA) by treatment replicate, are shown Fig. 6 and Table S1. Symbiont treatment had a significant effect on the proportion of adenines (F4,26 = 29.8; p < 0.0001), whereas source study did not (Wald p = 0.4895). Both MSRO (UG and BR) treatments had a significantly (p ≤ 0.001) smaller proportion of As (mean = 96.78%) than the control (mean = 99.79%) and wMel (mean = 100%) treatments. Hyd1 exhibited intermediate proportions of As (mean = 98.53%; Fig. 6). In other words, we detected a mean depurination of 3.22% for MSRO and 1.47% for Hyd1, and effectively no depurination for the control or wMel treatment. Pooling of the two substrains of MSRO (i.e., MSRO-UG and MSRO-BR) into one treatment produced similar results (not shown). The Harumoto et al.30 dataset, which included two embryo stages and differentiated between male and female embryos, did not exhibit substantially different depurination levels between stages or sex (e.g. mean %A was 98.27 for females and 98.49 for males; see Table S1).
Figure 6.
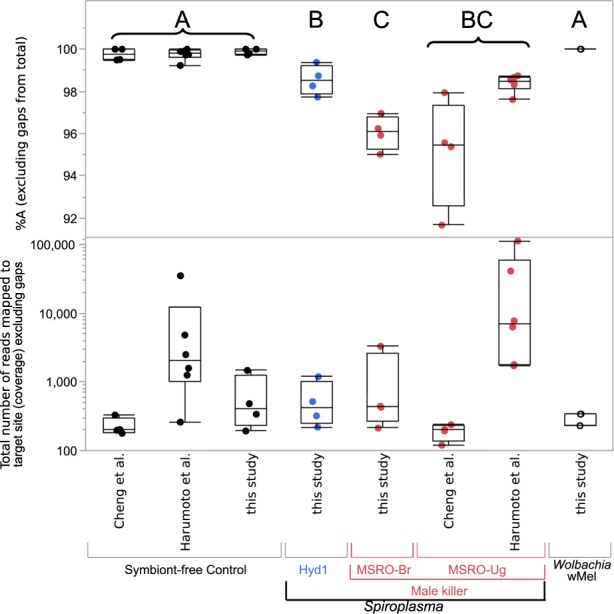
Presence of male-killing Spiroplasma (MSRO) results in a significant signal of depurination. Top Panel. Percent of adenines (i.e., the expected base in the absence of depurination) at the target site of Ribosome Inactivating Proteins (RIPs). Different letters indicate significant Posthoc tests (at P-value < 0.04). Bottom Panel. Coverage (number of reads) that mapped to the target depurination sites. Jitter points (replicates) and box plots per treatment (symbiont and source study) are shown.
Outcome of mapping mRNAseq reads to symbiont genomes
To determine the degree to which poly-A-tailed enriched RNAseq data yield reads assignable to the bacterial symbionts, we used Bowtie2 (v.2.3.4)54 with default settings to map the reads from each symbiont-infected replicate to the closest symbiont reference genome available. The number of reads per replicate mapped to the symbiont genome ranged from 5 (for a Hyd1 replicate) to 8619 (for a wMel replicate; Table S2), representing a very small fraction of the total reads. The percentage of these putative symbiont-derived reads that mapped to the symbiont ribosomal RNA genes was quite variable (range 3.94–100%; mean 52%). The above numbers of symbiont-derived genes are inadequate for analyses of differential expression (DE) of the symbiont. Collectively these findings suggest that libraries generated via a poly-A-tail enrichment process tend to yield a very low proportion of reads of bacterial origin; a result that is expected given that polyadenylation is not part of the process of bacterial mRNA maturation.
Discussion
Effects of symbionts on early embryo mRNA composition
The present study aimed to test whether infection by a heritable bacterium strain that kills males during the embryonic stage influences the composition of mRNA transcripts present in the early embryo (ca. 60–75 min post-oviposition), and thus, before the onset of zygotic transcription. Based on several analysis pipelines and assumptions, we did not detect any differential expression in genes among embryos harbouring the male-killing Spiroplasma strain (MSRO-Br), the CI-inducing Wolbachia strain, and the symbiont-free control. These results suggest that these symbionts do not influence the composition of initial maternally-loaded transcripts or their degradation up to the examined stage, unless they do so to a degree below what was detectable by our experimental design (e.g. below a log-fold change of 3; see Power Analyses). Furthermore, it is possible, that such cytoplasmically transmitted symbionts could influence the composition of maternally-loaded proteins, or regulate the protein complement of the early embryo in a transcriptionally-independent manner (e.g. through post-transcriptional or post-translational controls). To our knowledge, these features have not been compared in Spiroplasma-infected vs. uninfected embryos.
Our results showing no effect of the male-killing Spiroplasma on maternally-loaded mRNA composition are consistent with what is known about the male-killing mechanism. A Spiroplasma-encoded protein (Spaid) appears to interact with the DCC, which assembles onto the X chromosome of wild-type males but not females, and is accompanied by DNA damage and abnormal apoptosis15. This model does not require “priming” of the egg during oogenesis by the symbiont. Nonetheless, the delayed expression of male death in males flies over-expressing wild type Spaid (2nd larval instar), compared to those harbouring male-killing Spiroplasma, requires further investigation and may indicate that other Spiroplasma-encoded factors are relevant to the male-killing phenotype (e.g. RIP)38.
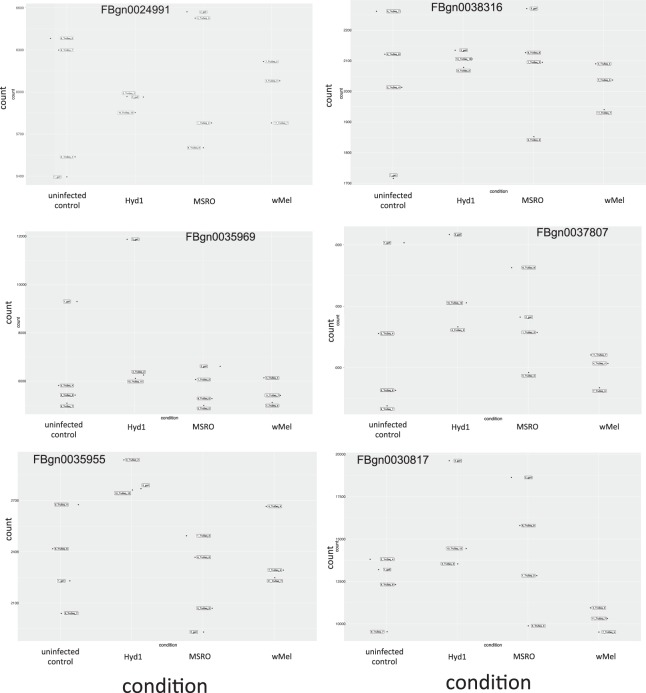
The differential expression analyses of the comparisons involving the Hyd1 treatment (i.e., the non-male-killing Spiroplasma strain native to D. hydei), yielded equivocal results, with four pipelines revealing no differentially expressed genes, and the other four pipelines revealing an intersection of five DE genes (ect, Osi6, Osi7, FBgn0038339, and FBgn0037099). All of these five genes had a lower expression level in the Hyd1 treatment compared to the other three treatments (Fig. 4). The time course of expression levels reported in FlyBase for these genes56, which is based on Wolbachia-infected flies57, indicates that early embryos have lower levels than later embryo stages (see Table 4). Based on this pattern, a plausible hypothesis is that the Hyd1 treatment exhibits a developmental delay. To address this, we compared the expression levels among treatments of six genes that should exhibit high levels at the 0–2 h embryo and substantially lower levels at the 2–4 h embryo (see Table 4). Our rationale was that if the Hyd1 treatment were developmentally delayed, we might detect a trend of higher expression levels at these genes compared to the other treatments. Nonetheless, Hyd1 exhibited no such a pattern; except for one of these genes (i.e., FBgn0035955; not significant; Fig. 7). Therefore, collectively, the patterns of expression of Hyd1-infected embryos are inconsistent with a developmental delay.
Table 4.
RPKM counts reported in FlyBase for the five DE genes found in our study and for six additional genes, “Developmental Markers” whose expression is higher in the 0–2 h embryo than in later stages. Gene name or Annotation Symbol in parenthesis.
| FlyBase report for wMel infected flies (RPKM)a | |||||
|---|---|---|---|---|---|
| 0–2 h embryo | 2–4 h embryo | 4–6 h embryo | 6–8 h embryo | ||
| DE Genes in this study | FBgn0038339 (CG6118) | 1 | 1 | 1 | 3 |
| FBgn0000451 (ect) | 1 | 1 | 1 | 5 | |
| FBgn0027527 (Osi6) | 10 | 7 | 8 | 31 | |
| FBgn0037414 (Osi7) | 5 | 3 | 4 | 16 | |
| FBgn0037099 (CG7173) | 0 | 0 | 1 | 2 | |
| “Developmental markers” (high 0–2 h; much lower after) | FBgn0024991 (CG2694) | 56 | 9 | 4 | 7 |
| FBgn0035969 (CG4476) | 85 | 2 | 1 | 1 | |
| FBgn0035955 (CG5194) | 56 | 2 | 1 | 1 | |
| FBgn0038316 (CG6276) | 55 | 9 | 2 | 2 | |
| FBgn0037807 (CG6293) | 94 | 7 | 1 | 1 | |
| FBgn0030817 (CG4991) | 156 | 4 | 1 | 1 | |
aNote: RPKM values reported in FlyBase are not expected to be directly comparable to the normalized count values reported in the present study.
Figure 7.
Normalized counts for each treatment and replicate, as estimated by Deseq2, for the six genes regarded as “Developmental markers” because they have high expression in the 0-2 h stage and much lower expression at subsequent stages. FlyBase-reported expression levels (in RPKM) are given in Table 4.
Available information on the function of the five genes at which the Hyd1 treatment had significantly lower expression provides little insight into the possible causes or consequences. Based on FlyBase, ectodermal (ect; FBgn0000451) is expected to have very low expression (FlyBase RPKM = 1) at the stage we examined. It is expressed later (Stages 13–16 ≈ embryo 14–16 h) in several tissue types: foregut, epidermis, trachea, and hindgut, with possible roles in cuticle development and tubular formation (e.g. tracheal tubes)58,59. Osiris 6 (Osi6; FBgn0027527) and Osiris 7 (Osi7; FBgn0037414) are also expected to have very low expression (FlyBase RPKM = 10 and 5, respectively) at the stage examined; with the highest expression at later embryonic stages (i.e., 14–16 h). Both genes are physically close to each other and belong to the Osiris gene cluster (OSI; FBgg0000612). The OSI family of genes shares a highly conserved protein domain is present, and retains high synteny, in all insects examined to date, and appears to have evolved via gene duplications at the base of the Insecta60,61. In D. melanogaster, these genes are within the dosage-sensitive Triploid-lethal (Tpl) locus; individuals with one or three copies of Tpl die as late embryos or early first instar larvae60,62. Although Osiris genes are generally considered of unknown function, based on expression patterns or knock-down experiments, they have been associated with cuticle formation, metamorphosis, digestion, resistance/tolerance to toxins and viruses, wing formation, and phenotypic plasticity (reviewed in Smith, et al.63). FBgn0038339 (CG6118) contains a BTB/POZ domain, and appears to be involved in regulation of transcription by RNA polymerase II. FBgn0037099 (CG7173) contains a Kazal domain (i.e., a type of serine proteinase inhibitor). Both of these genes are expected to have very low expression (FlyBase RPKM = 1 and 0, respectively) at the stage we examined, with peak expression levels occurring in the 14–16 h embryo.
Assuming that the significant results for the five genes are repeatable, they imply more perturbation of gene expression by Hyd1 than by the other symbiont strains; a finding that could reflect that D. melanogaster is outside the fundamental niche of Hyd1, as also suggested by its lower vertical transmission efficiency in this host (80%64), and by the difficulty of maintaining this artificial association in the lab (personal observation and Kageyama, et al.65). Reports of additional phenotypic effects of Hyd1 in D. melanogaster have been anecdotal or contradictory. Concerning adult longevity, Kageyama, et al.65 reported substantially shorter adult life spans of both males and females, whereas Silva, et al.66 detected little to no adult mortality. The study of Silva, et al.66 is expected to more closely represent the conditions and specific D. melanogaster (i.e., Canton S) and Hyd1 (i.e., sub-strain TEN104–106) genetic backgrounds of the present transcriptomics study. Concerning fly fecundity, both Kageyama, et al.65 and Hutchence, et al.64 report anecdotal evidence of lower female fecundity in Hyd1-infected D. melanogaster than their uninfected counterparts; Silva, et al.66 did not examine this trait. Consistent with the detrimental effect of Hyd1 on fecundity of D. melanogaster, the microarray-based study of Hutchence, et al.64 (focused on adults), revealed downregulation of a cluster of genes involved in egg production and fertilization. Nonetheless, in general, Hutchence, et al.64 found that D. melanogaster adults infected with Hyd1 exhibited less perturbation of gene expression than those infected by MSRO-Br and Spiroplasma poulsonii strain NSRO64 (a close relative of MSRO that also kills males and naturally occurs in Drosophila nebulosa). The apparently different patterns of gene expression perturbation by Hyd1 between our study and that of Hutchence, et al.64 could be due to the different fly stages examined (embryos vs. adults), different experimental tools (RNA-seq vs. microarrays), or other factors (e.g. all of Hutchence et al.’s treatments were also infected with Wolbachia).
The lack of an effect of wMel, a CI-inducing strain, on D. melanogaster early embryo mRNA composition, is not unexpected given what is known about the CI mechanism. Essentially, wMel encodes two contiguous protein coding genes (cifA and cifB)18. Each of these proteins is capable of causing CI when expressed in the male germline, whereas only cifA (when expressed in embryos) is able to rescue CI19. Our results suggest that wMel does not “prime” the egg for rescue by manipulating maternally-loaded transcripts or their degradation.
Depurination patterns
Our study capitalized on the presence of (non-target) rRNA reads in RNAseq datasets prepared by poly-A-tail enrichment, to estimate levels of depurination at the RIP target position of the sarcin-ricin loop (SLR) in the 28 SrRNA of eukaryotes. We acknowledge that the levels of depurination estimated from mRNAseq data may be downwardly biased because rRNA depurinated by RIPs is highly prone to hydrolysis of the sugar-phosphate backbone at the lesion site67. Furthermore, the freezing to which these samples were subjected between collection and library preparation might have decreased the detectability of depurination53. In addition, it is not known whether the polyA-RNA enrichment protocol used prior to library preparation, which is aimed at depleting rRNA in the sample, could bias representation of depurinated vs. intact rRNA. Only one study has compared inferences of ribosome depurination from mRNAseq vs. qPCR assays in the same system. Based on mRNAseq, Hamilton et al.17 detected ~3.8% depurination (i.e., ~96.2% adenine) in the nematode Howardula infesting adult Drosophila neotestacea harbouring the non-male-killing Spiroplasma sNeo strain. Comparatively, using the qPCR approach, the abundance of depurinated template representing RIP-induced depurination increased ~20-fold, whereas the levels of intact nematode rRNA were reduced ~six-fold in the presence of Spiroplasma17. Notwithstanding the potential biases, the consistent finding of no depurination in Spiroplasma-free treatments vs. depurination in Spiroplasma-present treatments across three independent studies, lends credibility to the mRNAseq-based approach we employed for inference of depurination.
The significantly lower proportion of adenines at the RIP target site for the Spiroplasma treatments vs. the Wolbachia treatment and the control is consistent with the action of a Spiroplasma-encoded RIP, and supports recent findings that: (1) MSRO-UG infection causes ribosome depurination in D. melanogaster embryos; (2) that heterologous expression of a Spiroplasma RIP1 and RIP2 genes (separately) also causes ribosome depurination in D. melanogaster embryos (measured by a qPCR approach); and (3) that the degree of ribosome depurination (“RIP activity” sensu Garcia-Arraez, et al.38) in the different Spiroplasma treatments or RIP transgene constructs is positively correlated with embryo mortality38. Importantly, despite detecting significantly higher levels of depurinated template with the qPCR approach, none of the qPCR assays revealed significantly lower levels of intact template38. This suggests that detectable depletion of intact template in the qPCR approach is not a pre-requisite for detecting a phenotypic effect of depurination.
Our results suggest that MSRO substrain Brazil (MSRO-BR) causes comparable levels of depurination to the Uganda substrain, which is consistent with its strong male-killing effect43,66, and with the identical content of RIP-encoding genes (unpublished data). Interestingly, the male-killer MSRO generally exhibited a higher signal of depurination than the non-male-killer Hyd1 (MSRO range >1–8%; Hyd1 range < 1– < 3%; Fig. 6). This difference could be the result of differences in titers of MSRO vs. Hyd1 at the embryonic stage. Densities of these strains in D. melanogaster Canton-S background at the adult stage are lower for Hyd1 than for MSRO-Br, but only immediately after adult eclosion66; densities at the embryonic stage have not been compared. Alternatively, it is possible that the RIP genes encoded in the MSRO genome are more actively expressed or secreted, or are more efficient than the putative RIP genes detected in the genome of Hyd139. Levels of fly ribosome depurination in the presence of sNeo, the non male-killing strain native to D. neotestacea, have not been assayed in embryos, but a qPCR assay of D. neotestacea ovaries indicated a small, albeit non significant, signal of depurination17. Similarly, an mRNA-seq experiment of adult D. neotestacea revealed 0.4% depurination (i.e., %A = 99.6%) in the presence of sNeo17. Therefore, the three members of the poulsonii clade examined to date (MSRO, Hyd1, and sNeo) encode RIPs capable of depurination of Drosophila ribosomes, but the male-killing strain exhibits the highest levels depurination, a phenomenon that appears to contribute to the male-killing mechanism38. The general patterns, however, indicate that Spiroplasma RIPs are particularly efficient at depurinating the ribosomes of natural enemies of Drosophila (i.e., parasitic wasps and nematodes), leading to the hypothesis that RIP-induced depurination plays an important role in defence mechanism17,37. The higher depurination levels of fly ribosomes detected in embryos (and ovaries) compared to other fly stages could be due to greater exposure of ribosomes to RIP prior to cellularization, after which Spiroplasma becomes effectively extra-cellular38. The relatively high levels of depurination in old adults appears to be the result of higher Spiroplasma densities at that stage38. Additional roles have been attributed to RIPs, which could contribute to the male-killing phenotype. For example, several RIPs are reported to cause DNA damage68,69. Thus, one or more Spiroplasma-encoded RIP might directly contribute to the DNA damage reported during the process of male-killing30.
Conclusions
This study employed a transcriptomics approach to examine whether cytoplasmically-transmitted bacteria, including reproductive manipulators that strongly impact survival of the embryonic stage of Drosophila, influence composition maternally-loaded mRNAs. The results revealed that mRNA composition does not differ significantly among the embryos harbouring the reproductive parasites Spiroplasma MSRO and Wolbachia wMel and those lacking endosymbionts. Only the symbiont Spiroplasma Hyd1, which does not manipulate reproduction, appeared to alter expression levels of a handful of genes (5–12), but not all analytical approaches supported this finding. Our power analyses indicated that our experimental design should be able to detect most of the genes exhibiting ≥ 3-log-fold change in expression among treatments. Collectively, our results suggest that these cytoplasmically-transmitted bacteria do not alter the composition of mRNAs of the early embryo, and are thus unlikely to use this mechanism to exert their reproductive phenotypes. Capitalizing on several transcriptomics datasets, this study also detected signals of Spiroplasma-induced damage to ribosomes in early Drosophila embryos, with greater damage caused by the male-killer (MSRO) than the non-male-killer (Hyd1), consistent with recent results implicating this mechanism in male killing.
Supplementary information
Acknowledgements
Library preparation and Illumina sequencing were performed by the Texas AgriLife Genomics and Bioinformatics facility. Portions of this research were conducted with high performance research computing resources provided by Texas A&M University (https://hprc.tamu.edu) and UNAM-CCG (litza server). This work was supported by National Institutes of Health grant R03 AI078348 to MM. Lacie L. Güenther and Jialei Xie provided technical assistance.
Author Contributions
Conceived and designed study: N.O.S., M.M., R.A., J.W.E., Conducted the experiments: N.O.S., M.M., Analysed the data: M.M., N.O.S., P.R., V.M.H.A., R.A., Interpreted results: M.M., N.O.S., P.R., V.M.H.A., R.A., J.W.E., Drafted the manuscript: M.M., N.O.S., P.R., V.M.H.A., Reviewed and edited the manuscript: M.M., N.O.S., P.R., V.M.H.A., R.A., J.W.E.
Data Availability
The datasets generated during and/or analysed during the current study are available as Supporting Data or at NCBI’s Sequence Read Archive (SRA) database under Accession Numbers SRR7279355–SRR7279369 (BioProject PRJNA474708; BioSample SAMN09370647).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-45371-0.
References
- 1.Duron O, Hurst GD. Arthropods and inherited bacteria: from counting the symbionts to understanding how symbionts count. BMC Biol. 2013;11:45. doi: 10.1186/1741-7007-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kraft LJ, Kopco J, Harmon JP, Oliver KM. Aphid symbionts and endogenous resistance traits mediate competition between rival parasitoids. PLoS One. 2017;12:e0180729. doi: 10.1371/journal.pone.0180729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watanabe K, Yukuhiro F, Matsuura Y, Fukatsu T, Noda H. Intrasperm vertical symbiont transmission. Proc Natl Acad Sci USA. 2014;111:7433–7437. doi: 10.1073/pnas.1402476111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 2008;6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- 5.Hurst G, Jiggins F. Male-killing bacteria in insects: mechanisms, incidence, and implications. Emerg. Infect. Dis. 2000;6:329–336. doi: 10.3201/eid0604.000402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yen JH, Barr AR. New hypothesis of the cause of cytoplasmic incompatibility in Culex pipiens L. Nature. 1971;232:657–658. doi: 10.1038/232657a0. [DOI] [PubMed] [Google Scholar]
- 7.Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 8.Ferree PM, et al. Wolbachia utilizes host microtubules and Dynein for anterior localization in the Drosophila oocyte. PLoS Pathog. 2005;1:e14. doi: 10.1371/journal.ppat.0010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fast EM, et al. Wolbachia enhance Drosophila stem cell proliferation and target the germline stem cell niche. Science. 2011;334:990–992. doi: 10.1126/science.1209609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herren JK, Paredes JC, Schupfer F, Lemaitre B. Vertical transmission of a Drosophila endosymbiont via cooption of the yolk transport and internalization machinery. MBio. 2013;4:e00532–00512. doi: 10.1128/mBio.00532-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Touret F, Guiguen F, Terzian C. Wolbachia influences the maternal transmission of the gypsy endogenous retrovirus in Drosophila melanogaster. MBio. 2014;5:e01529–01514. doi: 10.1128/mBio.01529-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136:3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- 13.Mateos M, et al. Heritable endosymbionts of Drosophila. Genetics. 2006;174:363–376. doi: 10.1534/genetics.106.058818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anbutsu H, Fukatsu T. Spiroplasma as a model insect endosymbiont. Environ Microbiol Rep. 2011;3:144–153. doi: 10.1111/j.1758-2229.2010.00240.x. [DOI] [PubMed] [Google Scholar]
- 15.Harumoto T, Lemaitre B. Male-killing toxin in a bacterial symbiont of Drosophila. Nature. 2018 doi: 10.1038/s41586-018-0086-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masson, F., Calderon Copete, S., Schüpfer, F., Garcia-Arraez, G. & Lemaitre, B. In vitro culture of the insect endosymbiont Spiroplasma poulsonii highlights bacterial genes involved in host-symbiont interaction. mBio9, 10.1128/mBio.00024-18 (2018). [DOI] [PMC free article] [PubMed]
- 17.Hamilton PT, Peng F, Boulanger MJ, Perlman SJ. A ribosome-inactivating protein in a Drosophila defensive symbiont. Proc. Natl. Acad. Sci. USA. 2016;113:350–355. doi: 10.1073/pnas.1518648113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LePage DP, et al. Prophage WO genes recapitulate and enhance Wolbachia-induced cytoplasmic incompatibility. Nature. 2017;543:243–247. doi: 10.1038/nature21391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shropshire JD, On J, Layton EM, Zhou H, Bordenstein SR. One prophage WO gene rescues cytoplasmic incompatibility in Drosophila melanogaster. Proc Natl Acad Sci USA. 2018;115:4987–4991. doi: 10.1073/pnas.1800650115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paredes JC, Herren JK, Schupfer F, Lemaitre B. The role of lipid competition for endosymbiont-mediated protection against parasitoid wasps in Drosophila. MBio. 2016;7:e01006–01016. doi: 10.1128/mBio.01006-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mateos M, et al. Independent origins of resistance or susceptibility of parasitic wasps to a defensive symbiont. Ecol. Evol. 2016;6:2679–2687. doi: 10.1002/ece3.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaenike J, Unckless R, Cockburn SN, Boelio LM, Perlman SJ. Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science. 2010;329:212–215. doi: 10.1126/science.1188235. [DOI] [PubMed] [Google Scholar]
- 23.Turelli M, Hoffmann AA. Rapid spread of an inherited incompatibility factor in California Drosophila. Nature. 1991;353:440–442. doi: 10.1038/353440a0. [DOI] [PubMed] [Google Scholar]
- 24.Gasparich GE. Spiroplasmas and phytoplasmas: Microbes associated with plant hosts. Biologicals. 2010;38:193–203. doi: 10.1016/J.Biologicals.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 25.Jaenike J, Stahlhut JK, Boelio LM, Unckless RL. Association between Wolbachia and Spiroplasma within Drosophila neotestacea: an emerging symbiotic mutualism? Mol. Ecol. 2010;19:414–425. doi: 10.1111/j.1365-294X.2009.04448.x. [DOI] [PubMed] [Google Scholar]
- 26.Haselkorn TS, Markow TA, Moran NA. Multiple introductions of the Spiroplasma bacterial endosymbiont into. Drosophila. Mol. Ecol. 2009;18:1294–1305. doi: 10.1111/j.1365-294X.2009.04085.x. [DOI] [PubMed] [Google Scholar]
- 27.Xie J, Vilchez I, Mateos M. Spiroplasma bacteria enhance survival of Drosophila hydei attacked by the parasitic wasp Leptopilina heterotoma. PLoS ONE. 2010;5:e12149. doi: 10.1371/journal.pone.0012149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen A, Williamson D, Oishi K. SpV3 viruses of Drosophila spiroplasmas. Isr. J. Med. Sci. 1987;23:429–433. [PubMed] [Google Scholar]
- 29.Anbutsu H, Fukatsu T. Population dynamics of male-killing and non-male-killing Spiroplasmas in Drosophila melanogaster. Appl. Environ. Microbiol. 2003;69:1428–1434. doi: 10.1128/AEM.69.3.1428-1434.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harumoto T, Anbutsu H, Lemaitre B, Fukatsu T. Male-killing symbiont damages host’s dosage-compensated sex chromosome to induce embryonic apoptosis. Nat. Commun. 2016;7:12781. doi: 10.1038/ncomms12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harumoto T, Anbutsu H, Fukatsu T. Male-killing Spiroplasma induces sex-specific cell death via host apoptotic pathway. PLoS Pathog. 2014;10:e1003956. doi: 10.1371/journal.ppat.1003956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veneti Z, Bentley JK, Koana T, Braig HR, Hurst GD. A functional dosage compensation complex required for male killing in Drosophila. Science. 2005;307:1461–1463. doi: 10.1126/science.1107182. [DOI] [PubMed] [Google Scholar]
- 33.Bentley JK, Veneti Z, Heraty J, Hurst GDD. The pathology of embryo death caused by the male-killing Spiroplasma bacterium in Drosophila nebulosa. BMC Biol. 2007;5:9. doi: 10.1186/1741-7007-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin J, Chong T, Ferree PM. Male killing Spiroplasma preferentially disrupts neural development in the Drosophila melanogaster embryo. PLoS ONE. 2013;8:e79368. doi: 10.1371/journal.pone.0079368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng B, Kuppanda N, Aldrich JC, Akbari OS, Ferree PM. Male-killing Spiroplasma alters behavior of the dosage compensation complex during Drosophila melanogaster embryogenesis. Curr. Biol. 2016;26:1339–1345. doi: 10.1016/j.cub.2016.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Endo Y. Mechanism of action of ricin and related toxins on the inactivation of eukaryotic ribosomes. Cancer Treat. Res. 1988;37:75–89. doi: 10.1007/978-1-4613-1083-9_5. [DOI] [PubMed] [Google Scholar]
- 37.Ballinger MJ, Perlman SJ. Generality of toxins in defensive symbiosis: Ribosome-inactivating proteins and defense against parasitic wasps in. Drosophila. PLoS Pathog. 2017;13:e1006431. doi: 10.1371/journal.ppat.1006431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia-Arraez MG, Masson F, Escobar JCP, Lemaitre B. Functional analysis of RIP toxins from the Drosophila endosymbiont Spiroplasma poulsonii. BMC Microbiol. 2019;19:46. doi: 10.1186/s12866-019-1410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinez-Montoya, H. Spiroplasma and its interaction with Drosophila: genome sequencing and analysis of potential fitness effects of naturally infected populations Ph.D. thesis, Texas A&M University (2017).
- 40.Xie J, Butler S, Sanchez G, Mateos M. Male killing Spiroplasma protects Drosophila melanogaster against two parasitoid wasps. Heredity. 2014;112:399–408. doi: 10.1038/hdy.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haselkorn TS, Jaenike J. Macroevolutionary persistence of heritable endosymbionts: acquisition, retention and expression of adaptive phenotypes in. Spiroplasma. Mol. Ecol. 2015;24:3752–3765. doi: 10.1111/mec.13261. [DOI] [PubMed] [Google Scholar]
- 42.Jeyaprakash A, Hoy MA. Long PCR improves Wolbachia DNA amplification: wsp sequences found in 76% of sixty-three arthropod species. Insect Mol. Biol. 2000;9:393–405. doi: 10.1046/j.1365-2583.2000.00203.x. [DOI] [PubMed] [Google Scholar]
- 43.Montenegro H, Solferini VN, Klaczko LB, Hurst GDD. Male-killing Spiroplasma naturally infecting Drosophila melanogaster. Insect Mol. Biol. 2005;14:281–288. doi: 10.1111/j.1365-2583.2005.00558.x. [DOI] [PubMed] [Google Scholar]
- 44.Dobson SL. Transfection of Wolbachia pipientis into Drosophila embryos. Current Protocols in Microbiology. 2007;5:3A.4.1–3A.4.11. doi: 10.1002/9780471729259.mc03a04s05. [DOI] [PubMed] [Google Scholar]
- 45.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Law CW, Alhamdoosh M, Su S, Smyth GK, Ritchie ME. RNA-seq analysis is easy as 1-2-3 with limma. Glimma and edgeR. F1000Research. 2016;5:1408. doi: 10.12688/f1000research.9005.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq 2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hart SN, Therneau TM, Zhang Y, Poland GA, Kocher JP. Calculating sample size estimates for RNA sequencing data. J. Comput. Biol. 2013;20:970–978. doi: 10.1089/cmb.2012.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walsh MJ, Dodd JE, Hautbergue GM. Ribosome-inactivating proteins: potent poisons and molecular tools. Virulence. 2013;4:774–784. doi: 10.4161/viru.26399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Melchior WB, Jr., Tolleson WH. A functional quantitative polymerase chain reaction assay for ricin, Shiga toxin, and related ribosome-inactivating proteins. Anal. Biochem. 2010;396:204–211. doi: 10.1016/j.ab.2009.09.024. [DOI] [PubMed] [Google Scholar]
- 54.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Graveley BR, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gutzwiller F, et al. Dynamics of Wolbachia pipientis gene expression across the Drosophila melanogaster life cycle. G3 (Bethesda) 2015;5:2843–2856. doi: 10.1534/g3.115.021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raha D, Nguyen QD, Garen A. Molecular and developmental analyses of the protein encoded by the Drosophila gene ectodermal. Dev. Genet. 1990;11:310–317. doi: 10.1002/dvg.1020110410. [DOI] [PubMed] [Google Scholar]
- 59.Rosa JB, Metzstein MM, Ghabrial AS. An Ichor-dependent apical extracellular matrix regulates seamless tube shape and integrity. PLoS Genet. 2018;14:e1007146. doi: 10.1371/journal.pgen.1007146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shah N, Dorer DR, Moriyama EN, Christensen AC. Evolution of a large, conserved, and syntenic gene family in insects. G3 (Bethesda) 2012;2:313–319. doi: 10.1534/g3.111.001412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Misof B, et al. Phylogenomics resolves the timing and pattern of insect evolution. Science. 2014;346:763–767. doi: 10.1126/science.1257570. [DOI] [PubMed] [Google Scholar]
- 62.Dorer DR, Rudnick JA, Moriyama EN, Christensen AC. A family of genes clustered at the Triplo-lethal locus of Drosophila melanogaster has an unusual evolutionary history and significant synteny with Anopheles gambiae. Genetics. 2003;165:613–621. doi: 10.1093/genetics/165.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith CR, Morandin C, Noureddine M, Pant S. Conserved roles of Osiris genes in insect development, polymorphism and protection. J. Evol. Biol. 2018;31:516–529. doi: 10.1111/jeb.13238. [DOI] [PubMed] [Google Scholar]
- 64.Hutchence KJ, Fischer B, Paterson S, Hurst GDD. How do insects react to novel inherited symbionts? A microarray analysis of Drosophila melanogaster response to the presence of natural and introduced. Spiroplasma. Mol. Ecol. 2011;20:950–958. doi: 10.1111/j.1365-294X.2010.04974.x. [DOI] [PubMed] [Google Scholar]
- 65.Kageyama D, et al. Prevalence of a non-male-killing Spiroplasma in natural populations of Drosophila hydei. Appl. Environ. Microbiol. 2006;72:6667–6673. doi: 10.1128/AEM.00803-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Silva NO, Guenther LL, Xie J, Mateos M. Infection densities of three Spiroplasma strains in the host Drosophila melanogaster. Symbiosis. 2012;57:83–93. doi: 10.1007/S13199-012-0181-3. [DOI] [Google Scholar]
- 67.Endo, Y. & Tsurugi, K. Mechanism of action of ricin and related toxic lectins on eukaryotic ribosomes. Nucleic Acids Symp. Ser., 187–190 (1986). [PubMed]
- 68.Bolognesi A, et al. Endocytosis and intracellular localisation of type 1 ribosome-inactivating protein saporin-s6. J. Biol. Regul. Homeost. Agents. 2012;26:97–109. [PubMed] [Google Scholar]
- 69.Bertholdo-Vargas LR, et al. Type 1 ribosome-inactivating proteins—Entomotoxic, oxidative and genotoxic action on Anticarsia gemmatalis (Hübner) and Spodoptera frugiperda (JE Smith) (Lepidoptera: Noctuidae) J. Insect Physiol. 2009;55:51–58. doi: 10.1016/j.jinsphys.2008.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available as Supporting Data or at NCBI’s Sequence Read Archive (SRA) database under Accession Numbers SRR7279355–SRR7279369 (BioProject PRJNA474708; BioSample SAMN09370647).