Abstract
Here, we report on the isolation of bacterial isolates from Himalayan niches, which produced extracellular l-asparaginase with low/no glutaminase activity. From the 235 isolates, 85 asparaginase positive bacterial isolates were identified by qualitative screening using optimized chromogenic dyes assay. Optimized concentration of different dyes revealed maximum color visualization in phenol red (0.003%). The diversity analysis of asparaginase positive isolates revealed that Proteobacteria (83%) are the most dominant, followed by Actinobacteria (12%), Firmicutes (3%), and Bacteriodetes (2%). Eleven isolates, which represented seven Pseudomonas species, one species each of the genus Arthrobacter, Janthinobacterium, Lelliottia, and Rahnella, were selected for further studies based on highest zone ratio and novel aspects for l-asparaginase production. Of these, five isolates, namely, Pseudomonas sp. PCH133, Pseudomonas sp. PCH146, Pseudomonas sp. PCH182, Rahnella sp. PCH162, and Arthrobacter sp. PCH138, produced l-asparaginase without glutaminase activity after 55 h of growth with the former isolate showing the highest l-asparaginase activity (1.67 U/ml). Interestingly, this is the first report of l-asparaginase production by members of the genera Janthinobacterium, Rahnella, and Lelliottia.
Electronic supplementary material
The online version of this article (10.1007/s13205-019-1810-9) contains supplementary material, which is available to authorized users.
Keywords: l-asparaginase, Extracellular, Glutaminase free, Microbial diversity, Anti-leukemic
Introduction
l-Asparaginase (EC 3.5.1.1) hydrolyzes asparagine into aspartic acid and ammonia. The asparagine hydrolyzing property of the enzyme is used in the treatment of acute lymphocytic leukemia, and in food processing and bio-analytics (Batool et al. 2016; van den Berg 2011). It was first discovered in guinea pig sera which exhibited anti-neoplastic activity (Broome 1963; Kidd 1953). In 1966, the first clinical trial on L-asparaginase was reported (Dolowy et al. 1966). Even after 5 decades, it is still one of the key therapeutic enzymes and accounts for 40% of the total enzymes sale worldwide (El-Nagga et al. 2014). Escherichia coli (E. coli) asparaginase and Erwinia chrysanthemi (E. chrysanthemi) asparaginase are presently used as chemotherapeutic drugs in the treatment of leukemia. Elspar®, Oncaspar™, Erwinase, Spectrilla, and Calaspargase are dominating therapeutic formulations in the market. However, the strong immunogenic responses during and after the therapy in more than 50% of the cases (Killander et al. 1976; Vrooman et al. 2010), clinical resistance (Andrade et al. 2014), low substrate specificity, and neurotoxic effects attributed to glutaminase activity of the commercial l-asparaginases (Warrell et al. 1982) are limiting the therapeutic value. A pegylated form of native E. coli l-asparaginase (pegaspargase) has also been developed to counter the hypersensitive, immunogenic reactions, and resulting side effects. The modified commercial formulations have shown improved performances in the treatment compared to the E. coli wild type. Undoubtedly, bacterial l-asparaginases are currently considered as a reliable source for therapeutic applications and efforts were made in the past and in the current study to screen for better l-asparaginase, which exhibit less adverse side effects.
l-asparaginase is widely distributed in nature extending from plants, animals, and humans (Krishnapura et al. 2016). There are reports of l-asparaginases from many bacterial species such as Rhodosporidium toruloides (Ramakrishnan and Joseph 1996), Thermus thermophilus (Pritsa and Kyriakidis 2001), Pseudomonas aeruginosa (El-Bessoumy et al. 2004), Helicobacter pylori (Cappelletti et al. 2008), Pyrococcus furiosus (Bansal et al. 2010), Pectobacterium carotovora (Kumar et al. 2011), and Bacillus, licheniformis (Mahajan et al. 2014). However, a suitable l-asparaginase alternative for therapeutic applications with fewer side effects or allergic response is yet to be developed or discovered. Therefore, it is worthwhile to screen diverse and unexplored niches like high altitudes of the Himalayas for isolation of microorganisms capable of glutaminase-free l-asparaginase activity with unique properties.
The majority of the high altitudes niches in the Himalayas are still unexplored. The microbial life of high altitude niches (glacier surfaces, glacier waters, streams, and mineral soil) undergo exceptional physiological adaptations to cope with varying degree of stresses imposed by the environment (Ciccazzo et al. 2014). Microbes thriving in such niches are bestowed with remarkable properties of thermostability, survival in low-oxygen, salt tolerance, pH stability, low pressure, and UV radiations that can be of industrial applications (Kumar et al. 2018; Stres et al. 2013; Thakur et al. 2018). Hence, microbe inhabiting these niches acquires unique functions with novel activities. Therefore, we attempted to explore and screen rich microbial diversity of Indian trans-Himalaya for l-asparaginase, also, to find out the best chromogenic dyes for qualitative assay. The primary target of the present work is to find efficient and stable l-asparaginase for potential applications in therapeutics and food industry.
Materials and methods
Sample collection
The soil, rocks, and water samples were collected in sterile containers from a river, glacial lake, and glacial stream in the Pangi–Chamba region (PCH) of western Himalaya in Himachal Pradesh, India at the height of 2100–4500 m above sea level (Latitude 33°00ʹ20.2ʺN, Longitude 76°14ʹ23.2ʺE). The samples were transported to the lab in cool packs and stored at 4 °C until used.
Media, culturing and qualitative screening of l-asparaginase producers
1.0 g soil or 1.0 mL water samples were serially diluted and plated on M9 medium (g/L, Na2HPO4 2H2O 6.0 g, KH2PO4 3.0 g, NaCl 5.5 g, 2 mM MgSO4 7H2O, and 0.1 mM CaCl2 2H2O) agar plates supplemented with 0.5% (w/v) asparagine as a nitrogen source and 0.2% (w/v) glucose as a carbon source. The isolation of bacteria was also performed on M9 medium using enrichment technique. The colonies were further purified on M9 agar plates or slants and stored at 4 °C. The glycerol stocks of the bacterial isolates were prepared and stored at − 20 °C in a set of five for each isolate. Similarly, 111 previously isolated and characterized bacterial species obtained from high altitude niches in our lab were also screened for qualitative l-asparaginase activity (Kumar et al. 2018; Thakur et al. 2018). The qualitative screening of bacterial isolates was performed using phenol red/bromothymol blue/cresol red indicator dye plate assays. For initial screening, M9 agar plates (asparagine + glucose) supplemented with phenol red, bromothymol blue, and cresol red (0.009%, w/v) were prepared. The uninoculated plates were used as a control for the change of color. Furthermore, identified potential isolates were re-screened for asparaginase activity using different concentrations (0.003, 0.006, and 0.009%) of these dyes and were optimized for maximum visualization of color change. The bacterial isolates showing maximum color change were selected based on the zone of hydrolysis for subsequent analysis.
Zone of hydrolysis
The spot inoculation of bacterial cultures was done in the middle of M9 media agar plates. The plates were incubated at different temperatures (4, 10, 20, 28, and 37 °C) for different time intervals (12–72 h) and the change of color and zone formation was observed. The zone of hydrolysis was measured by the following formula: Zone of hydrolysis (ZH) = diameter of a zone formed (cm)/diameter of a bacterial colony (cm) (Ferbiyanto et al. 2015). The ZH was further divided into three sub-categories, i.e., strong (+++, ZH = 3–4), medium (++, ZH = 2–3), and low (+, ZH = 0.5–2.0).
Identification and taxonomy of the bacterial isolates
The asparaginase positive bacteria were selected for 16S rDNA sequencing and identification (Thakur et al. 2018) based on morphological and plate assay methods. The DNA sequencing was performed using ABI 3130XL Genetic Analyzer (Applied Biosystems). The sequences obtained after 16S rDNA sequencing were analyzed on EzTaxon server (http://www.eztaxon.org/). The partial 16S gene sequences were submitted to the GenBank database. Multiple sequence alignment of 16S rDNA sequences was performed by BioEdit program, and phylogenetic analysis was carried out using MEGA 6.06. Various diversity indices, i.e., Shannon diversity index (H′), Simpson’s index (D1), Evenness (E), and Dominance (D), were calculated using the PAST software for the total number of identified bacterial isolates and asparaginase positive isolates (Hammer et al. 2001).
Quantitative analysis of l-asparaginase
The asparaginase positive bacterial isolates were grown in M9 production medium at 28 °C and harvested at a different time starting from 24 to 72 h of cellular growth. Culture absorbance (460 nm) was taken at each 12–24 h intervals. The bacterial culture was harvested after centrifugation, and the supernatant was used as a source of extracellular asparaginase enzyme. The supernatant was assayed for asparaginase and glutaminase activity. Asparaginase/glutaminase activity was performed according to the earlier reports with minor modifications (Imada et al. 1973). Briefly, for enzyme activity assay, 450 µL of 0.05 M Tris–HCl buffer (pH 8.6), 50 µL of supernatant and 500 µL of 0.01 M asparagine/glutamine prepared in the same buffer were added in a reaction tube. In the control tube, 450 µL Tris–HCl buffer and 500 µL of 0.01 M asparagine were mixed. The reaction was incubated at 37 °C for 30 min. After incubation, 250 µL of 1.5 M trichloroacetic acid (TCA) was added to each tube to stop the reaction. In control, 50 µL of supernatant was added. The buffer was used as a blank. The reaction tubes were then centrifuged at 5000g for 5 min to remove the precipitates formed. The dilutions were made after centrifugation, 100 µL of Nessler’s reagent was added to each tube and were kept for 10 min at room temperature. The absorbance was read at 480 nm. The asparaginase and glutaminase activities were defined as micromoles of respective substrate converted into product per unit time.
Results and discussion
Isolation, identification, and taxonomy of the bacterial isolates
The present study was initiated to isolate low or glutaminase-free l-asparaginase-producing bacteria from high altitude niches of Pangi–Chamba Himalayan region. These niches are unique, unexplored, and known for its tough terrains/distinct geography that offers a stressed environment to the native life forms including microbes (Kumar et al. 2018; Thakur et al. 2018). The enrichment culture and serial dilution of soil/water samples were used for the isolation of asparaginase-producing bacteria. The enrichment of samples for bacterial isolation led to the predominance of mainly Proteobacteria even after repeated experiments. Therefore, direct plate screening without enrichment method was used for further isolation. A total of 235 pure bacterial isolates were screened including 111 isolates which were previously isolated in our lab (Kumar et al. 2018; Thakur et al. 2018) and 124 isolates obtained in the present study from soil and water samples of PCH region in the Indian Trans-Himalaya. These bacterial isolates were identified by 16S rDNA sequencing (Table S1). The bacterial isolates were mainly represented by four different phyla, namely, Proteobacteria (51%), Firmicutes (28%), Actinobacteria (13%), and Bacteroidetes (8%) representing 42 different genera and 123 different species (Fig. 1, Table S2). The diversity analysis of total bacteria revealed that Pseudomonas (29.78%), Bacillus (11.91%), Geobacillus (8.93%), Arthrobacter (4.68%), Stenotrophomonas (4.25%), Janthinobacterium (3.82%), Chryseobacterium (3.82%), and Delftia (3.40%) are the most dominant genera (Table S2). The total diversity of isolated bacteria at the genus level was assessed using Shannon index (H′) which signify the abundance of the species present and a value of 2.862, suggesting a fairly good diversity index (Morris et al. 2014). The values of Evenness (E) represent the extent to which individuals are split among species, and Simpson’s index (D1) denotes species diversity in a community (Kumar et al. 2018; Morris et al. 2014). The values of Evenness and Simpson’s index were statistically calculated to 0.406 and 0.882, respectively. The lower values were indicating that few genera dominate the community viz. Pseudomonas, Bacillus, Geobacillus, Arthrobacter, Janthinobacterium, Chryseobacterium, and Delftia (Tables 1 and S2).
Fig. 1.
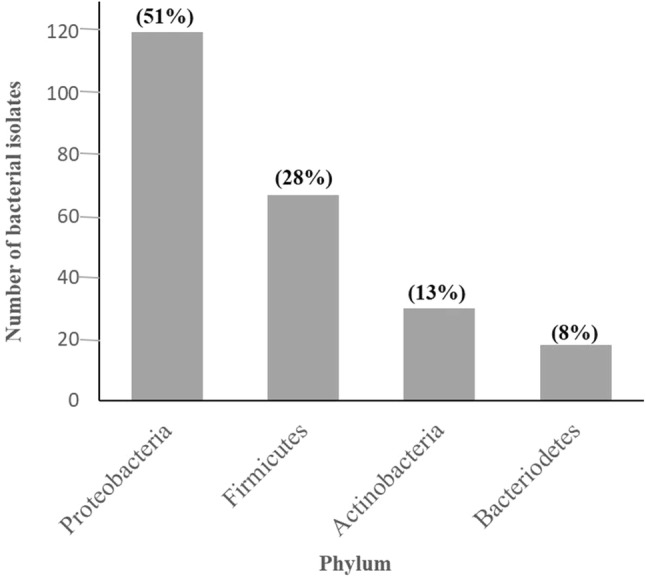
Diversity analysis of total bacterial isolates (235) and their percent distribution at phylum level (indicated in parentheses)
Table 1.
Diversity analysis among the total bacterial isolates (235) and within the l-asparaginase positive bacterial isolates (85)
| Attributes | Total bacteria | Asparaginase positive bacteria |
|---|---|---|
| Richness (S) | 43 | 15 |
| Individuals (N) | 235 | 85 |
| Dominance (D) | 0.116 | 0.377 |
| Simpson diversity index (D1) | 0.882 | 0.622 |
| Shannon index (H′) | 2.862 | 1.631 |
| Evenness (e^H/S) | 0.406 | 0.340 |
N number of individual bacteria identified, S number of genera based on 16S rDNA sequencing
H′ (Shannon index) = , where pi is the proportion of isolate to genus I, E (Evenness index) = H/lnS
D (Dominance) = , D1 (Simpson’s index) =
Qualitative screening of l-asparaginase producers
In the past, several indicator dyes were used to assess the presence of l-asparaginase activity in the microbes. The color of these dyes changes with the change in pH of media due to the ammonia production because of asparaginase activity (Gulati et al. 1997; Mahajan et al. 2013). The intensity of change in color of the dye is related to the production of the enzyme. In the present investigation, eighty-five (85) bacterial isolates were tested positive for asparaginase production on M9 media plates (Table S3) in the presence of 0.009% (w/v) indicator dye phenol red/bromothymol blue/cresol red (Gulati et al. 1997; Mahajan et al. 2013; Mihooliya et al. 2017). The purpose of using different dyes was to find out the best dye for the qualitative visualization of asparaginase activity. The change in the pH due to the production of ammonia in the presence of l-asparaginase activity results in the measurable color change on plates in the form of zones. Recently, Mihooliya et al. (2017) have screened different pH indicator dyes, i.e., phenol red, bromothymol blue, cresol red, methylene blue, bromocresol purple, bromocresol green, chlorophenol red, and litmus for the screening of l-asparaginase-producing bacteria. The zone of hydrolysis was recorded for each bacterial isolate (Table S3). Initially, the plates were incubated at different temperatures (4, 10, 20, 28, and 37 °C) to find out the best incubation temperature. It was observed that at 28 °C, maximum color development occurred within 48 h of incubation. Therefore, subsequent experiments were conducted at this temperature.
For the current study, initially, 0.009% (w/v) dye concentration was used, but after optimization of dye concentration, a maximum color and distinct zone formation was observed in 0.003% (w/v) dye concentration (Fig. S1a–c, Table 3). It was observed that maximum color intensity was recorded in 0.003% of phenol red (Fig. S1b, Table 3). The selection of best dye was based on the zone of hydrolysis and the time taken for color change. The zone of hydrolysis in 0.006% cresol red was comparable to the phenol red. However, the time required for optimum color visualization in phenol red was less than in cresol red. It was also observed that some of the bacteria did not produce any hydrolysis zone in cresol red or bromothymol blue, but was found in the presence of phenol red dye. The color intensity was poorly visible below 0.003% dye concentration. Earlier, researchers used 0.009% of dyes for the screening of l-asparaginase (Gulati et al. 1997; Mahajan et al. 2013; Mihooliya et al. 2017).
Table 3.
Qualitative screening of bacterial isolates using different concentrations of cresol red, phenol red, and bromothymol blue
| Bacterial isolate | Zone of hydrolysis (ZH) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cresol red (%) | Phenol red (%) | Bromothymol blue (%) | |||||||
| 0.003 | 0.006 | 0.009 | 0.003 | 0.006 | 0.009 | 0.003 | 0.006 | 0.009 | |
| Pseudomonas migulae PCH146 | + | ++ | +++ | ++ | ++ | +++ | ++ | ++ | +++ |
| Pseudomonas donghuensis PCH147 | + | ++ | +++ | ++ | ++ | +++ | ++ | ++ | +++ |
| Arthrobacter oryzae PCH138 | – | – | – | ++ | ++ | +++ | – | – | – |
| Janthinobacterium lividum PCH140 | + | + | + | ++ | ++ | +++ | ++ | ++ | +++ |
| Lelliottia nimipressuralis PCH72 | – | – | – | ++ | ++ | +++ | + | + | +++ |
| Pseudomonas frederiksbergensis PCH133 | + | ++ | +++ | + | ++ | +++ | ++ | ++ | +++ |
| Pseudomonas extremaustralis PCH157 | + | ++ | +++ | + | ++ | +++ | + | + | +++ |
| Rahnella aquatilis PCH162 | – | – | – | ++ | +++ | +++ | – | – | ++ |
| Pseudomonas antarctica PCH177 | + | + | ++ | ++ | +++ | +++ | ++ | ++ | ++ |
| Pseudomonas helmanticensis PCH182 | + | + | ++ | ++ | +++ | +++ | ++ | ++ | ++ |
| Pseudomonas orientalis PCH176 | + | + | ++ | ++ | +++ | +++ | ++ | ++ | ++ |
Symbols: +++, strong (ZH = 3–4); ++, medium (ZH = 2-3); +, low (ZH = 0.5–2)
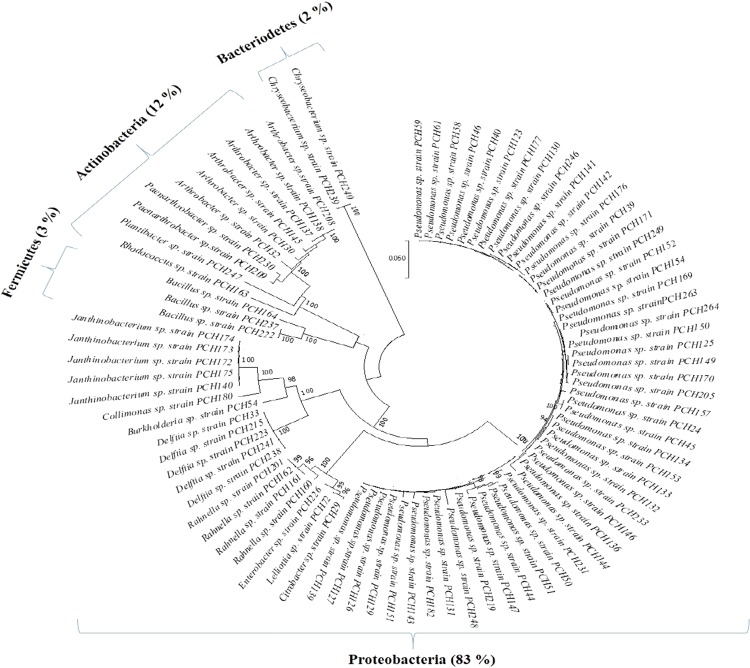
The diversity of asparaginase positive bacteria at the genus level was also assessed using Shannon index (H′) and a value of 1.631, suggesting an average diversity index. The values of Evenness (E) and Simpson’s index (D1) were statistically calculated to be 0.340 and 0.622, respectively, suggesting a very few genera dominate it (Table 1). The analysis of asparaginase positive bacteria reveals that Proteobacteria (83%) are the most dominant followed by Actinobacteria (12%), Firmicutes (3%), and Bacteriodetes (2%) (Fig. 2). Pseudomonas is the most dominant genus (60%) among the asparaginase positive bacteria (Table 2) and also the most dominant genera (29.78%) in the overall diversity (Table S2). The genera, namely, Pseudomonas, Arthrobacter, Enterobacter, Bacillus, Citrobacter, and Rhodococcus, are well documented for asparaginase-producing ability. However, the current study found that nine new genera viz. Paenarthrobacter, Plantibacter, Janthinobacterium, Burkholderia, Delftia, Lelliottia, Rahnella, Collimonas, and Chryseobacterium reported the first time for asparaginase-producing potential to the best of our knowledge (Table 2). Vimal and Kumar (2017) have reported screening of 21 fungal and 26 bacterial isolates for asparaginase production using phenol red as an indicator dye. Similarly, 165 isolates were found asparaginase positive upon screening of 240 actinomycetes in a separate study (Saxena et al. 2015). However, this is the first report of isolation of asparaginase-producing bacteria from high altitude PCH region of Western Himalaya. There is no report of diversity analysis of asparaginase-producing bacteria from Western Himalaya.
Fig. 2.
Phylogenetic tree based on 16S rDNA sequences of asparaginase positive bacterial isolates (85) using the neighbor-joining method and their percent distribution at phylum level (indicated in parentheses). Bootstrap values, indicated at the nodes, were obtained from 1000 bootstrap replicates. Bootstrap values > 94.99% were only included in the phylogenetic tree. The scale bar corresponds to 0.05 nucleotide changes per site
Table 2.
l-asparaginase positive isolates representing different genera
| Genus | Asparaginase positive isolates | Percent distribution (%) | Total number of species |
|---|---|---|---|
| Pseudomonas | 51 | 60.0 | 23 |
| Arthrobacter | 6 | 7.05 | 5 |
| Janthinobacterium | 5 | 5.88 | 2 |
| Delftia | 5 | 5.88 | 1 |
| Rahnella | 4 | 4.70 | 2 |
| Bacillus | 3 | 3.52 | 3 |
| Paenarthrobacter | 2 | 2.35 | 2 |
| Chryseobacterium | 2 | 2.35 | 2 |
| Plantibacter | 1 | 1.17 | 1 |
| Rhodococcus | 1 | 1.17 | 1 |
| Burkholderia | 1 | 1.17 | 1 |
| Collimonas | 1 | 1.17 | 1 |
| Citrobacter | 1 | 1.17 | 1 |
| Lelliottia | 1 | 1.17 | 1 |
| Enterobacter | 1 | 1.17 | 1 |
In the present study, 15 different genera and 47 species are reported as potential asparaginase producers (Table 2). Pseudomonas (Badoei-Dalfard 2016; Kuwabara et al. 2015), Arthrobacter (Munaganti et al. 2015), Enterobacter (Husain et al. 2016), Bacillus (Feng et al. 2017), and Citrobacter (Davidson et al. 1977) have been documented for production of asparaginase. However, Paenarthrobacter, Plantibacter, Janthinobacterium, Burkholderia, Delftia, Lelliottia, Rahnella, Collimonas, and Chryseobacterium have never been reported for asparaginase-producing ability. Besides, new species in the genus Pseudomonas have been identified which has the potential to produce l-asparaginase (Table S4). Asparaginase production by Pseudomonas pseudoalcaligenes (Badoei-Dalfard 2016), Pseudomonas fluorescens (Sindhu and Manonmani, 2018), Pseudomonas resinovorans strain IGS-131 (Mihooliya et al. 2017), Pseudomonas otitidis (Husain et al. 2016), and Pseudomonas aeruginosa (El-Bessoumy et al. 2004) have been reported in the past.
Quantitative analysis of l-asparaginase
The selection of bacterial isolates producing l-asparaginase for the quantitative assay was based on the zone of hydrolysis in qualitative screening and newly identified isolates having such potential. Based on the above criteria, 11 best bacterial isolates, namely, Pseudomonas sp. PCH182, Rahnella sp. PCH162, Pseudomonas sp. PCH133, Arthrobacter sp. PCH138, Pseudomonas sp. PCH157, Pseudomonas sp. PCH176, Pseudomonas sp. PCH177, Janthinobacterium sp. PCH140, Pseudomonas sp. PCH146, Pseudomonas sp. PCH147, and Lelliottia sp. PCH72 were selected and plated on M9 plates supplemented with different dyes to assess the zone of hydrolysis (Table 4). These isolates were further quantitatively assayed for asparaginase production in a liquid broth and samples were collected at different time intervals. After 35 h, Pseudomonas sp. PCH182, Arthrobacter sp. PCH138, and Pseudomonas sp. PCH146 exhibited asparaginase activity (0.17 IU/ml, 0.87 IU/ml, and 0.44 IU/ml, respectively) without showing any glutaminase activity. After 35 h, Pseudomonas sp. PCH157 showed maximum asparaginase activity (1.14 IU/ml) and highest glutaminase activity (1.73 IU/ml). Pseudomonas sp. PCH157, Pseudomonas sp. PCH176, and Pseudomonas sp. PCH177 have exhibited maximum asparaginase and glutaminase activity after 35 h of incubation (Table 4). In the current study, glutaminase activity in some of the isolates was observed at 24 h and 72 h, but not in 55 h timepoint of culture collection. The asparaginase and glutaminase activities have been assayed with the crude supernatants. The glutaminase activity in the supernatants is either due to glutaminase production along with asparaginase or due to the secondary activity of asparaginase towards glutamine. In addition, there are variable glutaminase or asparaginase enzymes in the bacterial systems which may be expressed during different growth phases. The enzyme production varies with the physiological growth phase of bacteria. Therefore, glutaminase activity in some of the isolates was observed at 24 h and 72 h, but not in 55 h. When the crude supernatant was assayed, there may be chances of glutaminase contamination. Hence, we selected the time course intentionally, where zero-glutaminase activity was observed. Therefore, the expression of asparaginase and glutaminase enzymes at different time intervals in various bacteria is expected, which is evident in Table 4. Streptomyces griseus NIOT-VKMA29 reported by Meena et al. (2015) possessed 5.36 IU/ml activity after the 6th day (144 h) and 0.181 IU/ml glutaminase activity. Enterobacter cloacae have 0.88 U/mg of protein asparaginase activity and zero-glutaminase activity (Husain et al. 2016). Asparaginase activity of 7.6 IU/ml was recorded in Enterobacter aerogenes MTCC111 (Erva et al. 2017).
Table 4.
Quantitative profiling of l-asparaginase positive bacteria
| S. No | Bacterial isolate | Asparaginase activity (IU/ml) | Glutaminase activity (IU/ml) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 24 h | 35 h | 55 h | 70 h | 24 h | 35 h | 55 h | 70 h | ||
| 1 | Pseudomonas helmanticensis PCH182 | 0 | 0.17 | 0.08 | 0 | 0 | 0 | 0 | 0 |
| 2 | Rahnella aquatilis PCH162 | 0 | 0.02 | 0.22 | 0 | 0 | 0.13 | 0 | 0.04 |
| 3 | Pseudomonas frederiksbergensis PCH133 | 0 | 0.51 | 1.67 | 1.5 | 0 | 1.25 | 0 | 0.66 |
| 4 | Arthrobacter oryzae PCH138 | 0.4 | 0.87 | 1.16 | 0 | 0.34 | 0 | 0 | 0.64 |
| 5 | Pseudomonas extremaustralis PCH157 | 0.98 | 1.14 | 0.16 | 0 | 0.7 | 1.73 | 0 | 0 |
| 6 | Pseudomonas orientalis PCH176 | 0.26 | 1.02 | 0 | 0 | 0.78 | 1.44 | 0.46 | 0 |
| 7 | Pseudomonas antarctica PCH177 | 0.24 | 1.05 | 0 | 0 | 1.02 | 1.15 | 0 | 0 |
| 8 | Janthinobacterium lividum PCH140 | 0 | 0.26 | 0.84 | 0.76 | 0 | 0.63 | 0 | 0.33 |
| 9 | Pseudomonas migulae PCH146 | 0.2 | 0.44 | 0.58 | 0 | 0.17 | 0 | 0 | 0.32 |
| 10 | Pseudomonas donghuensis PCH147 | 0.49 | 0.57 | 0.08 | 0 | 0.35 | 0.87 | 0 | 0 |
| 11 | Lelliottia nimipressuralis PCH72 | 0.13 | 0.52 | 0 | 0 | 0.39 | 0.73 | 0.23 | 0 |
Interestingly, the present study has identified some of the potential isolates which have shown zero-glutaminase activity. Pseudomonas sp. PCH133, Arthrobacter sp. PCH138, and Janthinobacterium sp. PCH140 have shown asparaginase activity of 1.67 IU/ml, 1.16 IU/ml, and 0.84 IU/ml, respectively, and zero-glutaminase activity after 55 h of incubation. Pseudomonas sp. PCH133 shows zero-glutaminase activity and highest asparaginase activity (1.67 IU/ml) after 55 h. Rahnella sp. PCH162, Janthinobacterium sp. PCH140, and Lelliottia sp. PCH72 is being reported for the first time for asparaginase production and also exhibited zero-glutaminase activity. Alrumman et al. (2019) reported L-asparaginase of Bacillus licheniformis with 36 U/mg enzyme activity and zero-glutaminase activity. l-asparaginase of Erwinia chrysanthemi and E. coli has 10% and 2% glutaminase activity (Nguyen et al. 2016). A comparative analysis of kinetic properties and representative glutaminase activity for some of the wild and recombinant l-asparaginases is given in Table S5. However, in the current study, Pseudomonas sp. PCH133 exhibited the highest asparaginase activity with zero-glutaminase activity at 55 h, which can be further selected as a potential candidate for the production of the enzyme. Furthermore, gene cloning, expression, and in vivo studies could be carried out to develop a possible candidate for chemotherapeutic and food applications.
Conclusion
The present study was designed to bioprospect novel sources of bacteria producing l-asparaginase with low/no glutaminase activity from Indian trans-Himalayas for applications in therapeutics and food industry, including analytical sector. The soil and water samples from high altitude regions were screened for isolation of bacteria with potentially l-asparaginase activity. The bacterial isolates belonging to different phyla were obtained after qualitative tests for asparaginase production using optimized chromogenic dyes. Phenol red was found to be the most appropriate dye for the qualitative screening of bacterial l-asparaginase. The bacterial isolates were identified, characterized, and evaluated by a color change in addition to the new source of asparaginase production. Eleven different bacteria were selected exhibiting high l-asparaginase activity with low/no glutaminase activity. Furthermore, they were assessed quantitatively for maximum l-asparaginase production in a time-dependent manner. The selected and potential isolates are now being tested for their possible applications in the treatment of acute lymphocytic leukemia, reduction of acrylamide in the foodstuffs, and detection of asparagine in the clinical and food samples.
Accession numbers: After the characterization of the isolates, the sequences were deposited in GenBank Database, maintained by the National Centre for Biotechnology Information (NCBI), with accession numbers KY628830–KY628834, KY628836–KY628839, KY628841–KY628891, KY628893–KY628940, MF774109–MF7741150, MF7741152–MF7741178, MH095996–MH095997, and MH095999–MH096056 (Tables S1, S3).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Authors duly acknowledge the technical assistance provided by Anil Kumar Chaudhary for 16S rDNA sequencing. This manuscript represents CSIR-IHBT Communication No. 4293.
Funding
DS duly acknowledge the financial support from CSIR, Govt. of India for CSIR-Fast Track Translation (FTT) project (33/BS/FTT/2016-PPD) and CSIR Network Project CeHAB (BSC0209).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Virender Kumar, Email: virender1188@gmail.com.
Subhash Kumar, Email: subhashkumar136@gmail.com.
Sanyukta Darnal, Email: sanyukta2306@gmail.com.
Vijeta Patial, Email: vijetapatial16@gmail.com.
Anju Singh, Email: anjusingh1993@gmail.com.
Vikas Thakur, Email: biotechvikas11@gmail.com.
Sanjay Kumar, Email: sanjaykumar@ihbt.res.in.
Dharam Singh, Phone: +91-1894-233339, Email: dharamsingh@ihbt.res.in.
References
- Alrumman SA, Mostafa YS, Kholood A, Al-izran Alfaifi MY, Taha TH, Elbehair SE. Production and anticancer activity of an l-asparaginase from Bacillus licheniformis isolated from the Red Sea, Saudi Arabia. Sci Rep. 2019;9:3756. doi: 10.1038/s41598-019-40512-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade FA, Borges SK, Silveira VS. Update on the use of l-asparaginase in infants and adolescent patients with acute lymphoblastic leukemia. Clin Med Insights Oncol. 2014;8:95–100. doi: 10.4137/CMO.S10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badoei-Dalfard A. l-asparaginase production in the Pseudomonas pseudoalcaligenes strain JHS-71 isolated from Jooshan Hot-spring. Mol Biol Res Commun. 2016;5:1–10. doi: 10.22099/MBRC.2016.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal S, Gnaneswari D, Mishra P, Kundu B. Structural stability and functional analysis of l-asparaginase from Pyrococcus furiosus. Biochem (Moscow) 2010;75:375–381. doi: 10.1134/S0006297910030144. [DOI] [PubMed] [Google Scholar]
- Batool T, Makky EA, Jalal M, Yusoff MM. A comprehensive review on l-asparaginase and its applications. Appl Biochem Biotechnol. 2016;178:900–923. doi: 10.1007/s12010-015-1917-3. [DOI] [PubMed] [Google Scholar]
- Broome J. Evidence that the l-asparaginase of guinea pig serum is responsible for its anti-lymphoma effects. J Exp Med. 1963;118:121–148. doi: 10.1007/s12010-015-1917-310.1084/jem.118.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappelletti D, Chiarelli LR, Pasquetto MV, et al. Helicobacter pyloril-asparaginase: a promising chemotherapeutic agent. Biochem Biophys Res Commun. 2008;377:1222–1226. doi: 10.1016/j.bbrc.2008.10.118. [DOI] [PubMed] [Google Scholar]
- Ciccazzo S, Esposito A, Rolli E, Zerbe S, Daffonchio D, Brusetti L. Safe-site effects on rhizosphere bacterial communities in a high-altitude alpine environment. Biomed Res Int. 2014;2014:1–9. doi: 10.1155/2014/480170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson L, Burkom M, Ahn S, Chang LC, Kitto B. l-asparaginases from Citrobacter freundii. Biochim Biophys Acta. 1977;480:282–294. doi: 10.1016/0005-2744(77)90341-2. [DOI] [PubMed] [Google Scholar]
- Dolowy WC, Henson D, Cornet J, Sellin H. Toxic and antineoplastic effects of l-asparaginase: study of mice with lymphoma and normal monkeys and report on a child with leukemia. Cancer. 1966;19:1813–1819. doi: 10.1002/1097-0142(196612)19:12<1813::AID-CNCR10>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Einsfeldt K, Baptista IC, Castanheira JC, et al. Recombinant l-asparaginase from Zymomonas mobilis: a potential new antileukemic agent produced in Escherichia coli. PLoS ONE. 2016;11(6):e0156692. doi: 10.1371/journal.pone.0156692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Bessoumy AA, Sarhan M, Mansour J. Production, isolation, and purification of l-asparaginase from Pseudomonas aeruginosa 50071 using solid-state fermentation. BMB Rep. 2004;37:387–393. doi: 10.5483/BMBRep.2004.37.4.387. [DOI] [PubMed] [Google Scholar]
- El-Nagga NE, El-Ewasy SM, El-Shweihy NM. Microbial l-asparaginase as a potential therapeutic agent for the treatment of acute lymphoblastic leukemia: the pros and cons. Int J Pharmacol. 2014;10:182–199. doi: 10.1007/s12010-015-1917-310.3923/ijp.2014.182.199. [DOI] [Google Scholar]
- Erva RR, Goswami AN, Suman P, Vedanabhatla R, Rajulapati SB. Optimization of l-asparaginase production from novel Enterobacter sp., by submerged fermentation using response surface methodology. Prep Biochem Biotechnol. 2017;16:219–228. doi: 10.1080/10826068.2016.1201683. [DOI] [PubMed] [Google Scholar]
- Feng Y, Liu S, Jiao Y, Gao H, Wang M, Du G, Chen J. Enhanced extracellular production of l-asparaginase from Bacillus subtilis 168 by B. subtilis WB600 through a combined strategy. Appl Microbiol Biotechnol. 2017;101:1509–1520. doi: 10.1007/s12010-015-1917-310.1007/s00253-016-7816-x. [DOI] [PubMed] [Google Scholar]
- Ferbiyanto A, Rusmana I, Raffiudin R. Characterization and identification of cellulolytic bacteria from gut of worker Macrotermes gilvus. HAYATI J Biosci. 2015;22:197–200. doi: 10.1016/j.hjb.2015.07.001. [DOI] [Google Scholar]
- Ghasemi A, Asad S, Kabiri M, Dabirmanesh B. Cloning and characterization of Halomonas elongatel-asparaginase, a promising chemotherapeutic agent. Appl Microbiol Biotechnol. 2017;101(19):7227–7238. doi: 10.1007/s00253-017-8456-5. [DOI] [PubMed] [Google Scholar]
- Gulati R, Saxena RK, Gupta R. A rapid plate assay for screening l-asparaginase producing micro-organisms. Lett Appl Microbiol. 1997;24:23–26. doi: 10.1046/j.1472-765X.1997.00331.x. [DOI] [PubMed] [Google Scholar]
- Hammer Ø, Harper DAT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4:1–9. [Google Scholar]
- Husain I, Sharma A, Kumar S, Malik F. Purification and characterization of glutaminase free asparaginase from Enterobacter cloacae: in vitro evaluation of cytotoxic potential against human myeloid leukemia HL-60 cells. PLoS One. 2016;11:1–27. doi: 10.1371/journal.pone.0148877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imada A, Igarasi S, Nakahama K, Isono M. Asparaginase and glutaminase activities of microorganisms. J Gen Microbiol. 1973;76:85–99. doi: 10.1099/00221287-76-1-85. [DOI] [PubMed] [Google Scholar]
- Kidd JG. Regression of transplanted lymphomas induced in vivo by means of normal guinea pig serum. Course of transplanted cancers of various kinds in mice and rats given guinea pig serum, horse serum, or rabbit serum. J Exp Med. 1953;98:565–582. doi: 10.1084/jem.98.6.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killander D, Dohlwitz A, Engstedt L, et al. Hypersensitive reactions and antibody formation during l-asparaginase treatment of children and adults with acute leukemia. Cancer. 1976;37:220–228. doi: 10.1002/1097-0142(197601)37:1<220::AID-CNCR23>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Krishnapura PR, Belur PD, Subramanya S. A critical review on properties and applications of microbial l-asparaginases. Crit Rev Microbiol. 2016;42:720–737. doi: 10.3109/1040841X.2015.1022505. [DOI] [PubMed] [Google Scholar]
- Kumar S, Venkata DV, Pakshirajan K. Purification and characterization of glutaminase-free l-Asparaginase from Pectobacterium carotovorum MTCC 1428. Bioresour Technol. 2011;102:2077–2082. doi: 10.1016/j.biortech.2010.07.114. [DOI] [PubMed] [Google Scholar]
- Kumar V, Thakur V, Ambika Kumar S, Singh D. Bioplastic reservoir of diverse bacterial communities revealed along altitude gradient of Pangi-Chamba trans-Himalayan region. FEMS Microbiol Lett. 2018 doi: 10.1093/femsle/fny144. [DOI] [PubMed] [Google Scholar]
- Kuwabara T, Asep A, Prihanto Wakayama M, Takagi K. Purification and characterization of Pseudomonas aeruginosa PAO1 asparaginase. Procedia Environ Sci. 2015;28:72–77. doi: 10.1016/j.proenv.2015.07.011. [DOI] [Google Scholar]
- Mahajan RV, Saran S, Saxena RK, Srivastava AK. A rapid, efficient and sensitive plate assay for detection and screening of l-asparaginase-producing microorganisms. FEMS Microbiol Lett. 2013;341:122–126. doi: 10.1111/1574-6968.12100. [DOI] [PubMed] [Google Scholar]
- Mahajan RV, Kumar V, Rajendran V, Saran S, Ghosh PC, Saxena RK. Purification and characterization of a novel and robust l-asparaginase having low-glutaminase activity from Bacillus licheniformis: in vitro evaluation of anti-cancerous properties. PLoS One. 2014 doi: 10.1371/journal.pone.0099037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meena B, Anburajan L, Sathish T, Vijaya Raghavan R, Dharani G, Vinithkumar NV, Kirubagaran R. l-Asparaginase from Streptomyces griseus NIOT-VKMA29: optimization of process variables using factorial designs and molecular characterization of l-asparaginase gene. Sci Rep. 2015;5:12404. doi: 10.1038/srep12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihooliya NK, Nandal J, Swami L, Verma H, Chopra L, Sahoo DK. A new pH indicator dye-based method for rapid and efficient screening of l-asparaginase producing microorganisms. Enz Microb Technol. 2017;107:72–81. doi: 10.1016/j.enzmictec.2017.08.004. [DOI] [PubMed] [Google Scholar]
- Morris EK, Caruso T, Buscot F. Choosing and using diversity indices: insights for ecological applications from the German biodiversity exploratories. Ecol Evol. 2014;4:3514–3524. doi: 10.1002/ece3.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munaganti RK, Muvva V, Indupalli MD. Studies on optimization of l-Asparaginase production by Arthrobacter kerguelensis VL-RK_09 isolated from Mango orchards. Int J Pharm Pharm Sci. 2015;7:112–115. [Google Scholar]
- Nguyen HA, Su Y, Lavie A. Design and characterization of Erwinia chrysanthemil-asparaginase variants with diminished l-glutaminase activity. J Biol Chem. 2016;291:17664–17676. doi: 10.1074/jbc.M116.728485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritsa AA, Kyriakidis DA. l-Asparaginase of Thermus thermophilus: purification properties and identification of essential amino acids for its catalytic activity. Mol Cell Biochem. 2001;216:93–101. doi: 10.1023/A:1011066129771. [DOI] [PubMed] [Google Scholar]
- Radha R, Arumugam N, Sathyanarayana Gummadi N. Glutaminase free l-asparaginase from Vibrio cholerae: heterologous expression, purification and biochemical characterization. Int J Biol Macromol. 2018;111:129–138. doi: 10.1016/j.ijbiomac.2017.12.165. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan MS, Joseph R. Characterization of an extracellular asparaginase of Rhodosporidium toruloides CBS14 exhibiting unique physicochemical properties. Can J Microbiol. 1996;42:316–325. doi: 10.1139/m96-047. [DOI] [Google Scholar]
- Saxena A, Upadhyay R, Kango N. Isolation and identification of actinomycetes for production of novel extracellular glutaminase free l-asparaginase. Ind J Exp Biol. 2015;53:786–793. [PubMed] [Google Scholar]
- Sindhu R, Manonmani HK. Expression and characterization of recombinant l-asparaginase from Pseudomonas fluorescens. Protein Expr Purif. 2018;143:83–91. doi: 10.1016/j.pep.2017.09.009. [DOI] [PubMed] [Google Scholar]
- Stres B, Sul WJ, Murovec B, Tiedje JM. Recently deglaciated high-altitude soils of the Himalaya: diverse environments, heterogeneous bacterial communities and long-range dust inputs from the upper troposphere. PLoS One. 2013;8:e76440. doi: 10.1371/journal.pone.0076440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur V, Kumar V, Kumar S, Singh D. Diverse culturable bacterial communities with cellulolytic potential revealed from pristine habitat in Indian trans-Himalaya. Can J Microbiol. 2018;64:798–808. doi: 10.1139/cjm-2017-0754. [DOI] [PubMed] [Google Scholar]
- van den Berg H. Asparaginase revisited. Leuk Lymphoma. 2011;52:168–178. doi: 10.3109/10428194.2010.537796. [DOI] [PubMed] [Google Scholar]
- Vimal A, Kumar A. In vitro screening and in silico validation revealed key microbes for higher production of significant therapeutic enzyme l-asparaginase. Enz Microb Technol. 2017;98:9–17. doi: 10.1016/j.enzmictec.2016.12.001. [DOI] [PubMed] [Google Scholar]
- Vrooman LM, Supko JG, Neuberg DS, et al. Erwinia asparaginase after allergy to E. coli asparaginase in children with acute lymphoblastic leukemia. Pediatr Blood Cancer. 2010;54:199–205. doi: 10.1002/pbc.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrell RP, Arlin ZA, Gee TS. Clinical evaluation of succinylated Acinetobacter glutaminase-asparaginase in adult leukemia. Cancer Treat Rep. 1982;66:1479–1485. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.