Abstract
Petunia × hybrida cv ‘Mitchell Diploid’ floral volatile benzenoid/phenylpropanoid (FVBP) biosynthesis ultimately produces floral volatiles derived sequentially from phenylalanine, cinnamic acid, and p-coumaric acid. In an attempt to better understand biochemical steps after p-coumaric acid production, we cloned and characterized three petunia transcripts with high similarity to p-coumarate 3-hydroxylase (C3H), hydroxycinnamoyl-CoA:shikimate/quinate hydroxycinnamoyl transferase (HCT), and caffeoyl shikimate esterase (CSE). Transcript accumulation of PhC3H and PhHCT was highest in flower limb tissue during open flower stages. PhCSE transcript accumulation was also highest in flower limb tissue, but it was detected earlier at initial flower opening with a bell-shaped distribution pattern. Down regulation of endogenous PhC3H transcript resulted in altered transcript accumulation of many other FVBP network transcripts, a reduction in floral volatiles, and the emission of a novel floral volatile. Down regulation of PhHCT transcript did not have as large of an effect on floral volatiles as was observed for PhC3H down regulation, but eugenol and isoeugenol emissions were significantly reduced on the downstream floral volatiles. Together these results indicate that PhC3H is involved in FVBP biosynthesis and the reduction of PhC3H transcript influences FVBP metabolism at the network level. Additional research is required to illustrate PhHCT and PhCSE functions of petunia.
Subject terms: Plant physiology, Metabolomics
Introduction
Plants have ability to produce specialized metabolites classified as phenylpropanoids. Thousands of distinct chemical structures are present in this class of organic compounds, ultimately all derived from the aromatic amino acid phenylalanine1,2. Many of these compounds and their respective conjugates are crucial for various function in plants: e.g. lignins, suberins, tannins (biopolymers - structural support, integrity, and pathogen resistance); flavonoids, isoflavonoids (pigments, sunscreens, and biotic interactions), and benzenoids/phenylpropenes (pollinator attractants, florivore repellents, and antimicrobials). Levels of these phenylpropanoid compounds can fluctuate drastically in response to abiotic and biotic stimuli1.
The initial enzymatic sequence of the phenylpropanoid pathway consists of phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), and 4-coumaric acid CoA ligase (4CL)3. PAL catalyzes the non-oxidative deamination of L-phenylalanine to cinnamic acid and ammonia4–6. Distinct PAL gene variants demonstrate differential transcript accumulation in tissue specific patterns, throughout development, and after specific stimuli. This suggests discrete and individual functions for each of the corresponding PAL isozymes7–10. PAL proteins are, for the most part, soluble and localized to the cytosol11–15. Functional PAL enzymes are usually identified as a homotetrameric protein aggregate1,16,17.
C4H is a cytochrome P450-dependent monooxygenase and consumes molecular oxygen and utilizes NADPH to catalyze the hydroxylation of cinnamic acid to produce p-coumaric acid18. Transcript accumulation of C4H is identified in most plant tissues, at some level, and can be regulated by numerous abiotic and biotic stimuli5,19–22. C4H protein is localized to the endoplasmic reticulum (ER) where it is anchored to the cytoplasmic surface of the membrane by an N-terminal leader/targeting peptide11,12,23, and functional C4H enzyme is usually identified in a homo/heterodimeric form, although some evidence exists for heterotrimeric interactions with other ER bound proteins11,24.
4CL catalyzes the reactions of cinnamic acid derivatives into the corresponding CoA thioesters including p-coumaroyl-CoA, caffeoyl-CoA, feruloyl-CoA, 5-hydroxyferuloyl-CoA, and sinapoyl-CoA9,25. Multiple 4CL genes detected in several species were regulated differentially according to the tissues, developmental stage, and environmental stress, which lead to specific biological processes such as monolignol biosynthesis and flavonoid production9,26. The substrate preference for each 4CL isoform has not been elucidated yet25, and 4CL protein is localized to the cytosol and re-localized through interactions with other proteins including C3H, HCT near to the ER15,25.
Petunia × hybrida cv ‘Mitchell Diploid (MD)’ is a well-characterized model plant to study flower volatiles, and many genes/transcripts/proteins involved in floral volatile benzenoid/phenylpropanoid (FVBP) biosynthesis have been characterized using this model (Fig. 1)27–30. Several steps downstream of 4CL have been identified and the functions of caffeoyl-CoA O-methyltransferase (CCoAOMT1)31, cinnamoyl-CoA reductase (CCR)29,32, cinnamyl alcohol dehydrogenase (CAD)32, coniferyl alcohol acyltransferase (CFAT)33, eugenol synthase1 (EGS1)34, and isoeugenol synthase1 (IGS1)34 have been reported for the biosynthesis of the phenylpropenes, eugenol and isoeugenol. Although several genes were characterized in other plant species, the biochemical steps from p-coumaric acid to caffeoyl-CoA have not yet been clearly investigated in Petunia.
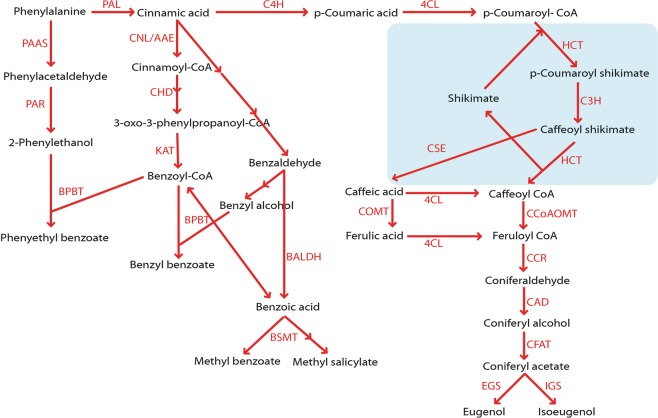
Figure 1.
Proposed FBVP biosynthesis in Petunia ‘MD’. BALDH, benzaldehyde dehydrogenase; BPBT, benzoyl-CoA:benzylalcohol/2-phenylethanol benzoyltransferase; BSMT, benzoic acid/salicylic acid carboxyl methyltransferase; 4CL, 4-coumaroyl-CoA ligase; C3H, p-coumarate 3-hydroxylase; C4H, cinnamate 4-hydroxylase; CAD, cinnamyl alcohol dehydrogenase; CCoAOMT, caffeoyl-CoA 3-O-methyltransferase; CCR, cinnamoyl-CoA reductase; CFAT, coniferyl alcohol acetyltransferase; CHD, cinnamoyl-CoA hydratase-dehydrogenase; CNL/AAE, cinnamate:CoA ligase/acyl-activating enzyme; COMT, caffeic/5-hydroxyferulic acid O-methyltransferase; CSE, caffeoyl shikimate esterase; EGS, eugenol synthase; HCT, hydroxycinnamoyl transferase; IGS, isoeugenol synthase; KAT, 3-ketoacyl-CoA thiolase; PAAS, phenylacetaldehyde synthase; PAL, phenylalanine ammonialyase; PAR, phenylalanine reductase.
The p-coumarate 3-hydroxylase (C3H) converts p-coumaroyl shikimate/quinate to caffeoyl shikimate/quinate35 and hydroxycinnamoyl transferase (HCT) participates in a reaction from p-coumaroyl CoA to caffeoyl CoA by exchanging CoA with shikimate or quinate as intermediates36,37. Recently, caffeoyl shikimate esterase (CSE) was also shown to be involved in phenylpropanoid biosynthesis. CSE converts caffeoyl shikimate into caffeic acid and then is combined with 4CL making caffeoyl-CoA, which can provide a bypass of the second reaction of HCT38,39.
Protein-protein associations are well known in these phenylpropanoid biosynthesis genes. The positive linear relations between PALs and C4Hs in Arabidopsis40, tobacco11,41, petunia20, and hybrid poplar14 have been established. Poplar C4H and C3H were reported to form heterodimeric and heterotrimeric membrane protein complexes24 and Arabidopsis C3H suppression is accompanied by reduced expression of PAL1, C4H, and HCT42. Bassard et al.12 showed co-localization and co-streaming of Arabidopsis C4H (CYP73A5) and C3H (CYP98A3) in the ER. The connecting HCT was also partially associated with the ER and C3H was important in driving the protein associations in Arabidopsis. Therefore, we hypothesized that the effects of PhC3H down regulation on the transcript accumulation of phenylpropanoid biosynthetic genes and that modification of PhC3H and/or PhHCT would alter the profile of petunia floral volatiles. To elucidate the role of the genes, we made knock-out constructs and transformed each construct into petunia. This research shows the reduction of PhC3H or PhHCT transcript resulted in change of the transcript level of FVBP genes and flower volatiles. Although additional research is required to illustrate PhHCT and PhCSE functions, we report the possibility of C3H as an important regulator of petunia MD FVBP biosynthesis.
Results
Identification of C3H, HCT, and CSE in Petunia
The cloned cDNA for PhC3H was 1790 base pairs (bp) in total length and included a 1533 bp open reading frame (ORF), which was predicted to encode for a protein composed of 511 residues with a predicted molecular weight of 58.3 kDa. The cloned cDNA for PhHCT was 1618 bp, contained a predicted 1305 bp ORF, which would result in 435 residues with a predicted molecular weight of 48.4 kDa. The cloned cDNA for PhCSE was 1149 bp in total length and included a predicted 933 bp ORF, which was predicted to encode for a protein composed of 311 residues with a predicted molecular weight of 35.0 kDa. The sequences for PhC3H, PhHCT, and PhCSE were submitted to NCBI under accession number KY679148, KY679147, and MF421742, respectively.
The predicted amino acid sequence of PhC3H shared 94% homology to cytochrome P450 98A2-like protein from Capsicum annuum (NP_001311496), 85% identity to C3H from Populus tomentosa (AFZ78540), and 81% homology to CYP98A3 from Arabidopsis thaliana (OAP09214) (Fig. 2). PhC3H included all conserved domains of oxygen binding and activation (A/G-G-X-E/D-T-T/S), ERR triad (E-X-X-R…..R), and heme binding (F-X-X-G-X-R-X-C-X-G) for cytochrome P450 families43. PhC3H was predicted to localize to the endoplasmic reticulum (TargetP 1.1 Server) and had an N-terminal membrane anchoring peptide (TMHMM Server v.2.0). The predicted amino acid sequence of PhHCT shared 94% homology to HCT of Nicotiana tabacum (NP_001312552), 94% identity to shikimate O-HCT of Capsicum annuum (NP_001311756), and 78% homology to HCT of Arabidopsis thaliana (NP_199704) (see Supplementary Fig. S1). PhHCT contained a HHXXXDG and DFGWG motifs for BAHD superfamily and closed to hydroxycinnamoyl-CoA:shikimate/quinate hydroxycinnamoyl transferase (PLN02663) in a condensation superfamily37,44,45. The predicted subcellular localization for PhHCT was the cytoplasm (Plant-mPLoc). The predicted amino acid sequence of PhCSE shared 84% homology to predicted CSE of Nicotiana tabacum (XP_016470669), 82% identity to predicted CSE of Solanum lycopersicum (XP_004235722), and 75% homology to LysoPL2 of Arabidopsis thaliana (OAP14317) (see Supplementary Fig. S2). PhCSE belongs to α/β-hydrolase superfamily and contains nucleophile (GX-Nuc-XG)-acid-histidine catalytic triad, which is conserved motif among members of α/β-hydrolase family (see Supplementary Fig. S2)46. The predicted subcellular localization for PhCSE was in the cytoplasm (WoLF PSORT).
Figure 2.
Predicted amino acid sequence alignment for C3H from various plants. The predicted amino acids of PhC3H contain all conserved domains of oxygen binding and activation (A/G-G-X-E/D-T-T/S), ERR triad (E-X-X-R…..R), and heme binding (F-X-X-G-X-R-X-C-X-G) for cytochrome P450 families. AkC3H, Acacia koa; AtCYP98A3, Arabidopsis thaliana; BeC3H, Bambusa emeiensis; CaC3H, Capsicum annuum; ClC3H, Cunninghamia lanceolata; FtC3H, Fagopyrum tataricum; NtC3H, Narcissus tazetta; PgC3H, Platycodon grandifloras; PhC3H, Petunia × hybrida; PtC3H, Populus tomentosa; ShC3H, Sinopodophyllum hexandrum; SiC3H, Sesamum indicum; TcC3H, Theobroma cacao.
Transcript accumulation of PhC3H, PhHCT, and PhCSE in MD
Since C3H, HCT, and CSE are involved in phenylpropanoid biosynthesis in other plant species, we hypothesized that relative transcript accumulation of PhC3H, PhHCT, and PhCSE would be highest in open flower and petal limb tissue, consistent with previously characterized FVBP genes25,47–50. Transcript accumulation was assayed using the ∆ΔCt qRT-PCR method with total RNA extracted from a spatial series of MD tissues: root, stem, stigma, anther, leaf, petal tube, petal limb, and sepal; along with total RNA from a staged floral developmental series of tissues including 11 consecutive stages MD flowers48.
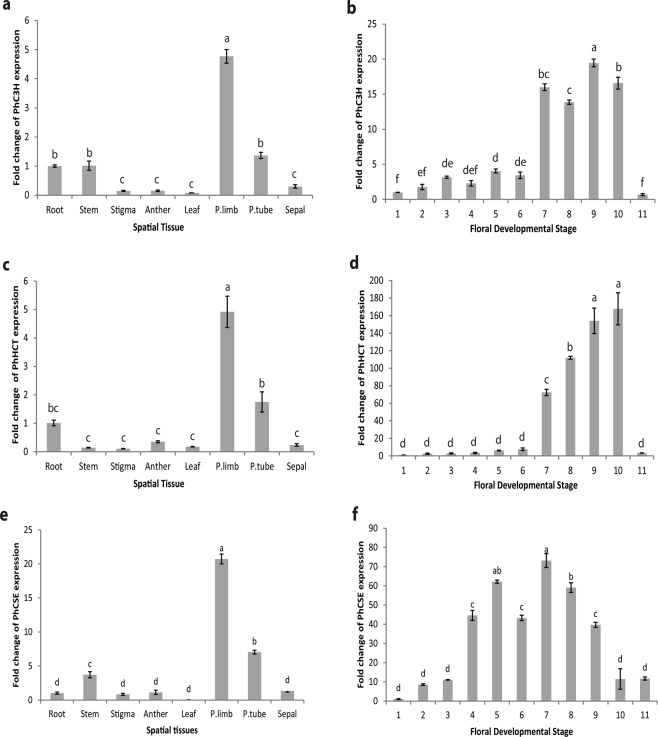
Spatial transcript accumulation was calculated based on that of root tissue where most transcripts of petunia volatile genes were lowest in previous studies31,33,48,51. The transcript accumulation of PhC3H, PhHCT, and PhCSE was highest in petal limb tissue while lowest in reproductive organs, leaf and sepal tissues (Fig. 3a,c,e). An approximate five-fold increase of PhC3H and PhHCT transcript was detected in petal limb tissue compared to root tissue; whereas, an approximate twenty-fold increase of PhCSE transcript was detected for the same tissue comparison. The transcript accumulation level of PhC3H and PhHCT in root tissue relatively elevated compared to stigma, anther, leaf, and sepal tissue. These results do not consistent with the profiles of known FVBP genes of petunia31,33,48,51.
Figure 3.
Transcript accumulation analyses of PhC3H, PhHCT, and PhCSE for spatial (a,c,e) and developmental stages (b,d,f). (q)RT-PCR was performed with ubiquitin as an internal reference. Error bar represents standard error and means separation was analyzed by Tukey test (one-way ANOVA, p < 0.05).
Floral developmental analyses demonstrated high levels of transcript accumulation only in open flowers from stage 7 to stage 10 for both PhC3H and PhHCT (Fig. 3b,d), while PhCSE transcript accumulation occurred earlier during flower development and showed a bell-shape with a peak at the flower opening (stage 7) (Fig. 3f). Focusing on flower developmental stage 9 (fully open corolla and sexually receptive flower), PhC3H transcript was accumulated approximately 20-fold more compared to stage 1 (initial flower bud). Similar trends were observed in the accumulation of PhHCT and PhCSE with an approximate 100-fold and 40-fold increase respectively. However, PhCSE accumulation peaked earlier, at flower stage 7 rather than stage 9, with approximately a 70-fold increase compared to stage 1.
Transgenic ir-PhC3H and ir-PhHCT lines
Transgenic petunia lines were generated with reduced endogenous transcript for PhC3H and PhHCT using standard transformation and inverted repeat (ir) RNAi techniques driven by a constitutive promoter, pFMV33,50–54. Changes in overall plant growth, reproductive, or other morphological phenotypes were not observed. Transcript levels of endogenous PhC3H were decreased between 75.6 and 98.2% in flowers of representative, independent ir-PhC3H plants (ir-PhC3H-7, ir-PhC3H-20, and ir-PhC3H-38) compared to flowers from MD control plants (Fig. 4a). Endogenous PhHCT transcript levels were decreased in flower tissue of representative, independent ir-PhHCT plants (ir-PhHCT-14, ir-PhHCT-24, and ir-PhHCT-34) by approximately 48.7 to 82.3% compared to MD (Fig. 4b).
Figure 4.
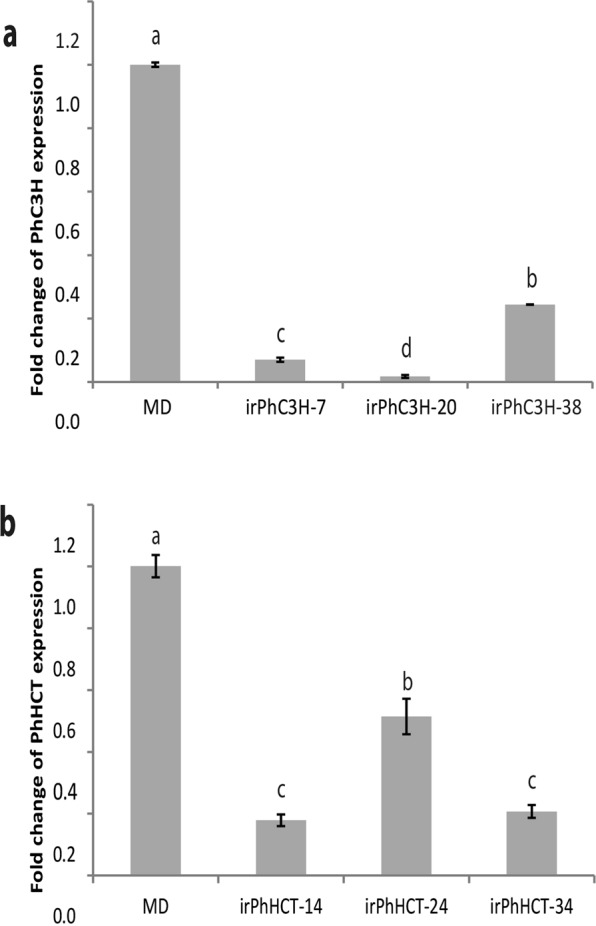
Transcript accumulation analyses in ir-PhC3H plants (a) and ir-PhHCT plants (b). (q)RT-PCR was performed with PhFBP1 as an internal reference. Error bars represent standard error and means separation was analyzed by Tukey test (one-way ANOVA, p < 0.05).
Volatile analyses of ir-PhC3H and ir-PhHCT flowers
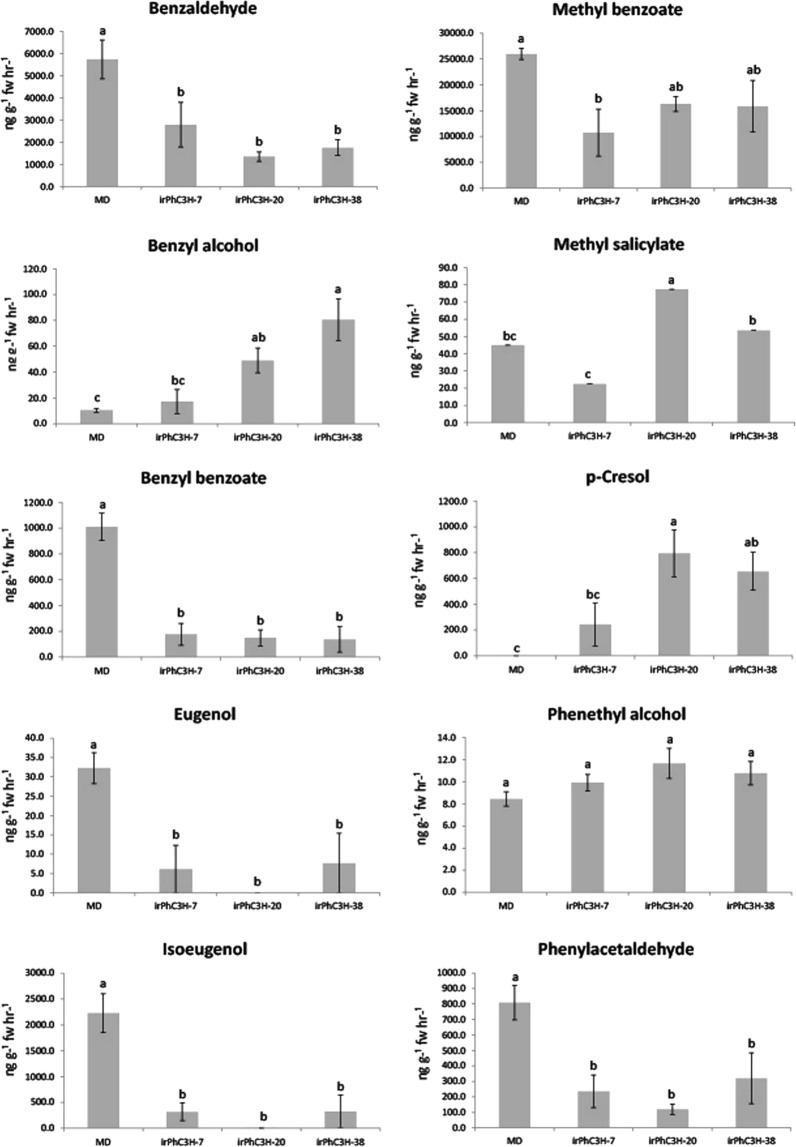
Standard volatile collection and GC-MS analysis methods were used to compare floral volatile profiles of ir-PhC3H and ir-PhHCT lines to control MD plants. In general, ir-PhC3H lines showed reduced emission for five major petunia floral volatiles: eugenol, isoeugenol, benzyl benzoate, benzaldehyde, and phenylacetaldehyde. Compared to MD, ir-PhHCT lines were reduced in phenylpropene and benzyl benzoate emission and showed a trend toward increased emission of benzaldehyde and phenylacetaldehyde (Fig. 5, see Supplementary Fig. S3). The most abundant constituent of the MD floral volatile profile, methyl benzoate, was also reduced in ir-PhC3H-7 flowers, but to a lesser extent. Phenethyl alcohol was the only floral volatile not significantly different in all three ir-PhC3H lines compared to MD. The benzenoid p-cresol, which is a novel volatile molecule for the MD genetic background, was detected from flowers of ir-PhC3H lines (Fig. 5).
Figure 5.
Effects of down-regulation of PhC3H on the floral volatiles. The open flowers (developmental stage 8) were used to collect volatiles at 18.0 h for 1 hr and the calculated emission was compared to MD plants. Error bars represnt standard error and means separation was analyzed by Duncan’s multiple range test (one-way ANOVA, p < 0.05).
Floral volatile emission from ir-PhHCT lines were significantly reduced for the phenylpropene volatiles, eugenol and isoeugenol, and the two conjugated volatiles benzyl benzoate and phenethyl benzoate (see Supplementary Fig. S3). The ir-PhHCT lines exhibited elevated emission of benzaldehyde, benzyl alcohol, and phenylacetaldehyde compared to MD. Methyl benzoate was also increased in ir-PhHCT-14 flowers, but to a lesser extent. P-cresol was not detected from any ir-PhHCT flowers (data not shown).
Transcript accumulation of FVBP related genes in ir-PhC3H and ir-PhHCT flowers
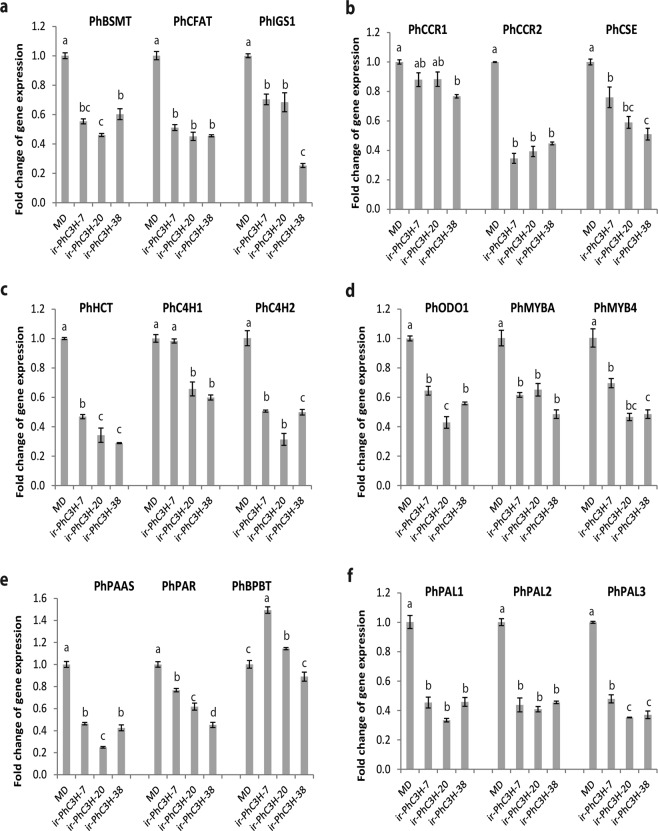
C3H is reported as a key protein concerning regulation of metabolically related protein aggregates12. As PhC3H transcript levels were low in petunia flowers, most of the volatiles were reduced (Fig. 5). Transcript accumulation of the metabolically related genes, PhC4Hs, PhPALs, and PhHCT showed decreased levels in ir-PhC3H flowers compared to MD. Many other FVBP genes including PhBSMT, PhCCR2, PhCFAT, PhCSE, PhIGS1, PhMYBA, PhMYB4, PhODO1, PhPAAS, and PhPAR demonstrated reduced transcript accumulation in ir-PhC3H flowers (Fig. 6). However, down regulation of PhHCT did not affect transcript accumulation of PhCSE as much as that of PhC3H (see Supplementary Fig. S4).
Figure 6.
Transcript accumulation analyses of phenylpropanoid biosynthesis genes in ir-PhC3H plants. (q)RT-PCR was done with PhFBP1 as an internal reference. Error bars represnent standard error and means separation was analyzed by Tukey test (one-way ANOVA, p < 0.05).
Discussion
The transcript accumulation profile, enzyme function, protein localization, and protein-protein interactions of coumarate 3-hydroxylase (C3H) have been investigated in model plant systems like Arabidopsis12,35,42,43,55,56 and poplar24,57. C3H is an integral protein feature of the cellular machinery leading to produce monolignols which serve as precursors to lignin and lignin production in plants58. Coniferyl alcohol, a common monolignol, can serve as a precursor to volatile phenylpropene biosynthesis in petunia floral tissue where the requirement for lignin is relatively low59. Around 7:00 PM, a petunia flower can emit over 10 ug*gFW−1*h−1 of a volatile phenylpropene, isoeugenol48,50, and the rate of phenylpropene and benzenoid emission is least influenced by a limiting pool of phenylalanine, the initial substrate for the phenylpropanoid pathway, indicating a strong regulation of carbon flux toward monolignol production52,60. When arogenate dehydratase, which converts arogenate to phenylalanine, was down regulated in petunia, the emission of many petunia volatiles was reduced60.
Throughout this work, three transcripts were cloned and sequenced from Petunia × hybrida cv ‘MD’ flower tissue. Single and unique transcripts with homology to C3H, HCT, and CSE were identified using publicly available genomic and transcriptomic databases, although the possibilities for the multiple copies of each gene family member could not be excluded (Fig. 2, see Supplementary Figs S1 and S2). All three petunia transcripts accumulated to their relatively highest levels in flower limb tissue compared to other parts of the plant (Fig. 3), which is in line with FVBP network transcripts48. PhC3H and PhHCT exhibited typical accumulation patterns of genes involved in FVBP biosynthesis during floral development20,25,29,31,33,48,49,51–53,60. The transcript levels were relatively low in young or developing floral buds, highest in open flowers, and then showed a dramatic reduction at senescence (Fig. 3)48. Compared to PhC3H, PhHCT and other known genes in the FVBP biosynthesis pathway, PhCSE transcript accumulated earlier during development and displayed an almost normal distribution rather than a developmentally-delayed distribution (Fig. 3). Interestingly, PhCSE followed a developmental bell-shaped accumulation profile similar to that of the petunia R2R3-MYB transcription factor, PhEOBII, which appears to have a positive regulatory effect on flower opening and FVBP biosynthesis61,62.
Petunia RNAi (ir) lines for PhC3H and PhHCT were generated to test the effects of reduced transcript levels of each gene on FVBP pathway. Multiple lines of ir-PhC3H showed a reduction of endogenous PhC3H transcript by 75.6–98.2% in floral tissue, and ir-PhHCT lines were reduced in endogenous transcript by 48.7–82.3% (Fig. 4). It is unclear whether a reduction of PhHCT transcript beyond 82.3% is lethal, but no obvious growth phenotypes were observed any of the ir-PhHCT or ir-PhC3H lines. Floral volatile analysis of the ir-PhC3H lines demonstrated a clear but unexpected volatile phenotype compared to the control petunia volatile emission phenotype (Fig. 5). The reduction of endogenous PhC3H resulted in very low levels of emitted phenylpropenes, isoeugenol and eugenol, as expected. Additionally, most of the emitted benzenoid volatiles were also reduced, along with phenylacetaldehyde which is generated directly from the initial substrate of the phenylpropanoid pathway, phenylalanine. A novel floral volatile molecule, p-cresol was detected from ir-PhC3H lines at considerable concentrations (~200–800 ng*gFW-1*h−1). It was an unexpected product because p-cresol has not been reported in FVBP biosynthesis of MD before but detected in other floral volatile profiles such as Petunia × hybrida (V26) having purple flowers, Satyrium pumilum (African orchid), and Jasminum polyanthum (Pink Jasmin)63–65. P-cresol is known to be converted from 4-hydroxyphenylacetic acid which is derived from tyrosine65,66.
The volatile phenotype of the ir-PhC3H lines suggested that a disruption of C3H activity cause a downregulation of the majority of the phenylpropanoid pathway in petal tissue of petunia flowers. Based on the transcript accumulation assay, 16 of the 18 FVBP related genes demonstrated clear reductions in the ir-PhC3H lines compared to controls (Figs 1, 6). Known positive and negative regulating transcription factors20,61,66, core phenylpropanoid pathway enzymes20,25, enzymes that produce specific FVBP compounds33,67, and even enzymes responsible for biochemical steps after C3H29,31 were all significantly reduced in transcript accumulation. For example, the small family of phenylalanine ammonia-lyase (PAL1, 2, 3) transcripts was reduced 52.3–66.8%, which would severely limit phenylpropanoid metabolism in petal tissue, especially at the elevated rates normally found in petunia flower petal tissue.
It is unclear at this point what mechanisms are involved in the downregulation of the general phenylpropanoid pathway. In Arabidopsis and poplar, C3H appears to be a major driver of protein-protein interaction at the ER, where C4H and C3H form homodimer and heterodimer protein complexes with elevated enzymatic activity12,24,37,43. The P450 protein complexes can associate with soluble phenylpropanoid pathway enzymes like PALs, 4CLs, and HCT to form supramolecular structures. These large, ER tethered protein aggregates are thought to concentrate required enzymes to accommodate for a high metabolic demand of the phenylpropanoid pathway in specific conditions or tissues12. We demonstrated that the down regulation of PhC3H in petunia resulted in the change of flower volatiles and expression of related FVBP genes, but further studies are required to elucidate the role of PhC3H in the stability of a large phenylpropanoid related protein complex in petunia flower limb tissue.
Materials and Methods
Plant materials and Cloning
Petunia × hybrida cv ‘MD’ was used as a control and genetic background for all experiments. Plants were grown in glass greenhouses as previously described by Dexter et al.33.
Multiple data sources including the National Center for Biotechnology Information (NCBI - http://www.ncbi.nlm.nih.gov), the Sol Genomics Network (SGN - https://solgenomics.net), and the 454 petunia database (http://biosrv.cab.unina.it/454petuniadb/protocol.php) were employed to search for petunia nucleotide sequences with similarity to Arabidopsis thaliana p-coumarate 3-hydroxylase (AtC3H, AT2G40890), Nicotiana tabacum hydroxycinnamoyl transferase (NtHCT, AJ507825), and Arabidopsis thaliana caffeoyl shikimate esterase (AtCSE, AT1G52760). The target petunia sequences were collected and assembled into contigs using a software package (Vector NTI Advance™ 11.3) and a SMARTer™ RACE cDNA Amplification Kit (Clontech Laboratories, Inc., Mountain View, CA) according to the manufacturer’s protocol. This approach resulted in three in silico candidate sequences for PhC3H, PhHCT, and PhCSE. The full sequences were amplified using PfuTurbo DNA Polymerase (Agilent Technologies, Santa Clara, CA) (primers on Table S1) and cloned into a pGEM-T easy vector (Promega, Madison, WI) using similar methods as Colquhoun et al.20. Nucleotide sequencing with multiple clones from multiple amplifications was performed at an on campus Sanger sequencing core (Interdisciplinary Center for Biotechnology Research, University of Florida, FL) using Big Dye V1–2. The resulted high-quality sequence was then used as a query to search the petunia genome database at SGN (https://solgenomics.net), which supported that each sequence most likely originating from a single locus.
Transgenic PhC3H RNAi (ir-PhC3H) and PhHCT RNAi (ir-PhHCT) plants
A 302 bp sequence of PhC3H and a 335 bp sequence of PhHCT were amplified for RNAi vector construction (primers on Table S1). The RNAi gene driven by a flower specific constitutive promoter, pFMV in pHK vector was introduced into MD leaf discs using Agrobacterium-mediated transformation methods68. Detailed methods for this procedure have been described by Dexter et al.33 and Underwood et al.50. All T0 plant tissues were collected for floral volatile analyses and transcript accumulation analyses, and then the flowers were self-pollinated.
Analyses of transcript accumulation
To observe transcript accumulation based on spatial and flower development, petunia MD tissues were collected following the method of Colquhoun et al.48. The spatial series consisted of root, stem, stigma, anther, leaf, petal tube, petal limb, and sepal. The developmental series included 11 stages of flowers, bud < 0.5 cm (stage 1); bud 0.5 to 1.5 cm (stage 2); bud 1.5 to 3.0 cm (stage 3); bud 3.0 to 5.0 cm (stage 4); bud fully elongated, 5.0 to 6.5 cm (stage 5); flower opening 0 to 2 cm limb diameter (stage 6); flower fully open days 0 (stage 7), 1 (stage 8), 2 (stage 9), and 3 (stage 10); senescing flower (flower open day 7 for MD) (stage 11). All tissues were collected in liquid N2 at 16.00 h and stored at -80 °C with two biological replications.
Total RNA was extracted as previously described67 using TriZOLTM (ThermoFisher Scientific, Waltham, MA), treated with TURBOTM DNA-freeTM (Ambion Inc., Austin, TX), and then purified using the RNeasy® Mini protocol (Qiagen Co., Valencia, CA). 50 ng µL−1 of RNA was prepared after measuring the concentration using a NanoDropTM 2000c spectrophotometer (ThermoFisher Scientific, Waltham, MA). Transcript accumulation was analyzed with semi-quantitative (sq)RT-PCR using a One-step RT-PCR kit (Qiagen Co., Valencia, CA) and with ∆ΔCt quantitative (q)RT-PCR using Power SYBR® Green RNA-to-CTTM 1-Step kit and StepOnePlusTM real-time PCR system (ThermoFisher Scientific, Waltham, MA). Based on the nucleotide arrangement of full sequences, primers were designed using Primer3 (http://biotools.umassmed.edu/bioapps/primer3_www.cgi) (primers on Table S1). To analyze transcript accumulation of other FVBP genes in ir-PhC3H plants, ∆ΔCt quantitative (q)RT-PCR was performed with petunia benzoyl-CoA:benzylalcohol/2-phenylethanol benzoyltransferase (PhBPBT; AY611496), benzoic acid/salicylic acid carboxyl methyltransferase (PhBSMT, AY233465), cinnamoyl-CoA reductases (PhCCR1, KF040494 and PhCCR2), PhC4H1 (HM447144), PhC4H2 (HM447145), PhCFAT (DQ767969), PhCSE (AT1G52760), PhHCT (KY679147), PhIGS1 (DQ372813), PhMYBA (EU374207), PhMYB4 (HM447143), ODORANT1 (PhODO1, AY705977), phenylacetaldehyde synthase (PhPAAS, DQ243784), PhPALs (PhPAL1, AY705976; PhPAL2, CO805160; PhPAL3), and phenylalanine reductase (PhPAR) genes. ∆ΔCt (q)RT-PCR of PhCSE was also performed in ir-PhHCT plants to test an effect of reduced PhHCT transcript on other related FVBP biosynthesis genes. The PhFBP1 (M91190) or PhUbiq (SGN-U207515) was used as an internal standard to compare expression of each gene. All ∆ΔCt (q)RT-PCR data was analyzed using 2−∆ΔCt method69, while nonparametric statistical analyses were conducted using Kruskal-Wallis test with the JMP Pro v.12 statistical software package (SAS Institute Inc., Cary, NC). P-values were computed at significance level (alpha = 0.05).
Volatile collection
Petunia flowers were harvested at 16.00 h and volatiles were collected for 1 hour in glass tubes using a push-pull dynamic headspace collection system as previously described33,51,70. Volatiles collected from at least three biological replicate flowers on glass columns containing approximately 50 mg HaySep Q 80–100 porous polymer adsorbent (Hayes Separations Inc., Bandera, TX) were eluted with methylene chloride. Quantification of volatiles from the elution matrix was performed on an Agilent 7890A Series gas chromatograph (GC) equipped with an Agilent 5977A single quadrupole mass spectrum detector (MSD). Parameters of the GC were used as follows: Helium carrier gas fixed at 11.5 psi, split injector at 20:1 split, inlet temperature 220 °C, injection volume 2 uL, and the syringe wash solvents were acetone and hexane. Sample analytes were separated using an equipped DB-5 column (Agilent Technologies, Santa Clara, CA, USA). Oven temperatures were programmed as follows: the initial oven temperature of 40 °C was held for 0.5 minutes then ramped 5 °C*minute−1 to 250 °C and held for 4 minutes. The MSD was equipped with an extractor ion source and tuned for sensitivity and mass accuracy just prior to sample analysis. Parameters for the MSD were maintained as follows: MSD transfer line temperature 280 °C, MS source temperature 230 °C, MS quad temperature 150 °C, solvent delay of 4.40 minutes, mass scan range 40–205 m/z with a threshold of 150. Data was acquired using Agilent MassHunter Workstation Acquisition (version, Agilent Technologies, Santa Clara, CA) and processed using Agilent’s MassHunter Quantitative Analysis program. Compound identity was verified by extracting and comparing the mass spectral data of each compound peak to the 2011 NIST mass spectral library and comparing retention time and mass spectral profiles to authentic standards run under identical machine parameters. Volatile mass emission rates (ng*gFW−1*hr−1) were calculated based on each compound individual peak area relative to the peak area of an elution standard, nonyl acetate, within each sample and standardized for each sample corresponding biological mass. Dilutions for volatile standards were run on the GC-MS in duplicate to obtain a response factor for each compound that was used in the calculation of volatile emission mass. Mean separation and comparison of ir-PhC3H and ir-PhHCT floral volatiles to MD controls were performed using Duncan’s multiple range test (one-way ANOVA, P < 0.05) with the JMP Pro v.12 statistical software package (SAS Institute Inc., Cary, NC).
Supplementary information
Acknowledgements
This work was supported by grants from the American Floral Endowment, the USDA-ARS Floriculture and Nursery Research Initiative, the Fred C. Gloeckner Foundation, and the National Horticulture Foundation.
Author Contributions
T.C. and J.K. designed the experiments and J.K., T.J., R.S., M.A., and K.C. performed the experiments. J.K. and T.C. wrote, and T.J., K.C., and D.C. edited the manuscript. All authors reviewed the manuscript.
Data Availability
All submitted manuscripts including figures and tables are available on Scientific report.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-45183-2.
References
- 1.Tohge T, Watanabe M, Hoefgen R, Fernie AR. Shikimate and phenylalanine biosynthesis in the green lineage. Front. Plant Sci. 2013;4:62. doi: 10.3389/fpls.2013.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogt T. Phenylpropanoid biosynthesis. Mol. Plant. 2010;3:2–20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- 3.Fraser, C. M. & Chapple, C. The phenylpropanoid pathway in Arabidopsis. The Arabidopsis Book. e0152 (2011). [DOI] [PMC free article] [PubMed]
- 4.Koukol J, Conn EE. The metabolism of aromatic compounds in higher plants IV. Purification and properties of the phenylalanine deaminase of Hordeum vulgare. J. Biol. Chem. 1961;236:2692–2698. [PubMed] [Google Scholar]
- 5.Docimo T, Consonni R, Coraggio I, Mattana M. Early phenylpropanoid biosynthetic steps in Cannabis sativa: link between genes and metabolites. Int. J. Mol. Sci. 2013;14:13626–13644. doi: 10.3390/ijms140713626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang J, et al. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010;153:1526–1538. doi: 10.1104/pp.110.157370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar A, Ellis BE. The phenylalanine ammonia-lyase gene family in raspberry. Structure, expression, and evolution. Plant Physiol. 2001;127:230–239. doi: 10.1104/pp.127.1.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osakabe Y, Osakabe K, Chiang VL. Characterization of the tissue-specific expression of phenylalanine ammonia-lyase gene promoter from loblolly pine (Pinus taeda) in Nicotiana tabacum. Plant Cell Rep. 2009;28:1309–1317. doi: 10.1007/s00299-009-0707-1. [DOI] [PubMed] [Google Scholar]
- 9.Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 2003;133:1051–1071. doi: 10.1104/pp.103.026484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X, Gou M, Liu CJ. Arabidopsis Kelch repeat F-box proteins regulate phenylpropanoid biosynthesis via controlling the turnover of phenylalanine ammonia-lyase. Plant Cell. 2013;25:4994–5010. doi: 10.1105/tpc.113.119644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Achnine L, Blancaflor EB, Rasmussen S, Dixon RA. Colocalization of L-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase for metabolic channeling in phenylpropanoid biosynthesis. Plant Cell. 2004;16:3098–3109. doi: 10.1105/tpc.104.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bassard JE, et al. Protein–protein and protein–membrane associations in the lignin pathway. Plant Cell. 2012;24:4465–4482. doi: 10.1105/tpc.112.102566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cochrane FC, Davin LB, Lewis NG. The Arabidopsis phenylalanine ammonia lyase gene family: kinetic characterization of the four PAL isoforms. Phytochem. 2004;65:1557–1564. doi: 10.1016/j.phytochem.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 14.Sato T, Takabe K, Fujita M. Immunolocalization of phenylalanine ammonia-lyase and cinnamate-4-hydroxylase in differentiating xylem of poplar. Comptes Rendus Biol. 2004;327:827–836. doi: 10.1016/j.crvi.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Wanner LA, Li G, Ware D, Somssich IE, Davis KR. The phenylalanine ammonia-lyase gene family in Arabidopsis thaliana. Plant Mol. Biol. 1995;27:327–338. doi: 10.1007/BF00020187. [DOI] [PubMed] [Google Scholar]
- 16.Hyun MW, Yun YH, Kim JY, Kim SH. Fungal and plant phenylalanine ammonia-lyase. Mycobiol. 2011;39:257–265. doi: 10.5941/MYCO.2011.39.4.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rasmussen S, Dixon RA. Transgene-mediated and elicitor-induced perturbation of metabolic channeling at the entry point into the phenylpropanoid pathway. Plant Cell. 1999;11:1537–1551. doi: 10.1105/tpc.11.8.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russell DW. The metabolism of aromatic compounds in higher plants X. Properties of the cinnamic acid 4-hydroxylase of pea seedlings and some aspects of its metabolic and developmental control. J. Biol. Chem. 1971;246:3870–3878. [PubMed] [Google Scholar]
- 19.Bell-Lelong DA, Cusumano JC, Meyer K, Chapple C. Cinnamate-4-hydroxylase expression in Arabidopsis (regulation in response to development and the environment) Plant Physiol. 1997;113:729–738. doi: 10.1104/pp.113.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colquhoun TA, et al. PhMYB4 fine-tunes the floral volatile signature of Petunia × hybrida through PhC4H. J. Exp. Bot. 2011;62:1133–1143. doi: 10.1093/jxb/erq342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hahlbrock K, Scheel D. Physiology and molecular biology of phenylpropanoid metabolism. Annu. Rev. Plant Biol. 1989;40:347–369. doi: 10.1146/annurev.pp.40.060189.002023. [DOI] [Google Scholar]
- 22.Jin H, et al. Transcriptional repression by AtMYB4 controls production of UV‐protecting sunscreens in Arabidopsis. EMBO J. 2000;19:6150–6161. doi: 10.1093/emboj/19.22.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bayburt TH, Sligar SG. Single-molecule height measurements on microsomal cytochrome P450 in nanometer-scale phospholipid bilayer disks. Proc. Nat. Aca. Sci. 2002;99:6725–6730. doi: 10.1073/pnas.062565599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen HC, et al. Membrane protein complexes catalyze both 4-and 3-hydroxylation of cinnamic acid derivatives in monolignol biosynthesis. Proc. Nat. Aca. Sci. 2011;108:21253–21258. doi: 10.1073/pnas.1116416109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klempien A, et al. Contribution of CoA ligases to benzenoid biosynthesis in petunia flowers. Plant Cell. 2012;24:2015–2030. doi: 10.1105/tpc.112.097519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun H, et al. Analysis of five rice 4-coumarate: coenzyme A ligase enzyme activity and stress response for potential roles in lignin and flavonoid biosynthesis in rice. Biochem. Biophys. Res. Commun. 2013;430:1151–1156. doi: 10.1016/j.bbrc.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 27.Colquhoun TA, Clark DG. Unraveling the regulation of floral fragrance biosynthesis. Plant Signal Behav. 2011;6:378–381. doi: 10.4161/psb.6.3.14339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dudareva N, Klempien A, Muhlemann JK, Kaplan I. Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 2013;198:16–32. doi: 10.1111/nph.12145. [DOI] [PubMed] [Google Scholar]
- 29.Muhlemann JK, Woodworth BD, Morgan JA, Dudareva N. The monolignol pathway contributes to the biosynthesis of volatile phenylpropenes in flowers. New Phytol. 2014;204:661–670. doi: 10.1111/nph.12913. [DOI] [PubMed] [Google Scholar]
- 30.Schuurink RC, Haring MA, Clark DG. Regulation of volatile benzenoid biosynthesis in petunia flowers. Trends Plant Sci. 2006;11:20–25. doi: 10.1016/j.tplants.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 31.Shaipulah NF, et al. CCoAOMT downregulation activates anthocyanin biosynthesis in petunia. Plant Physiol. 2016;170:717–731. doi: 10.1104/pp.15.01646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan H, et al. Structural studies of cinnamoyl-CoA reductase and cinnamyl-alcohol dehydrogenase, key enzymes of monolignol biosynthesis. Plant Cell. 2014;26:3709–3727. doi: 10.1105/tpc.114.127399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dexter R, et al. Characterization of a petunia acetyltransferase involved in the biosynthesis of the floral volatile isoeugenol. Plant J. 2007;49:265–275. doi: 10.1111/j.1365-313X.2006.02954.x. [DOI] [PubMed] [Google Scholar]
- 34.Koeduka T, et al. Eugenol and isoeugenol, characteristic aromatic constituents of spices, are biosynthesized via reduction of a coniferyl alcohol ester. Proc. Nat. Aca. Sci. 2006;103:10128–10133. doi: 10.1073/pnas.0603732103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schoch G, et al. CYP98A3 from Arabidopsis thaliana is a 3′-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J. Biol. Chem. 2001;276:36566–36574. doi: 10.1074/jbc.M104047200. [DOI] [PubMed] [Google Scholar]
- 36.Hoffmann L, Maury S, Martz F, Geoffroy P, Legrand M. Purification, cloning, and properties of an acyltransferase controlling shikimate and quinate ester intermediates in phenylpropanoid metabolism. J. Biol. Chem. 2003;278:95–103. doi: 10.1074/jbc.M209362200. [DOI] [PubMed] [Google Scholar]
- 37.Li X, Bonawitz ND, Weng JK, Chapple C. The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids. Plant Cell. 2010;22:1620–1632. doi: 10.1105/tpc.110.074161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vanholme R, et al. Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science. 2013;341:1103–1106. doi: 10.1126/science.1241602. [DOI] [PubMed] [Google Scholar]
- 39.Vargas L, et al. Improving total saccharification yield of Arabidopsis plants by vessel-specific complementation of caffeoyl shikimate esterase (cse) mutants. Biotech. Biofuels. 2016;9:139. doi: 10.1186/s13068-016-0551-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000;12:2383–2393. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blount JW, et al. Altering expression of cinnamic acid 4-hydroxylase in transgenic plants provides evidence for a feedback loop at the entry point into the phenylpropanoid pathway. Plant Physiol. 2000;122:107–116. doi: 10.1104/pp.122.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abdulrazzak N, et al. A coumaroyl-ester-3-hydroxylase insertion mutant reveals the existence of nonredundant meta-hydroxylation pathways and essential roles for phenolic precursors in cell expansion and plant growth. Plant Physiol. 2006;140:30–48. doi: 10.1104/pp.105.069690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bak Søren, Beisson Fred, Bishop Gerard, Hamberger Björn, Höfer René, Paquette Suzanne, Werck-Reichhart Danièle. Cytochromes P450. The Arabidopsis Book. 2011;9:e0144. doi: 10.1199/tab.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D’Auria JC. Acyltransferases in plants: a good time to be BAHD. Curr. Opin. Plant Biol. 2006;9:331–340. doi: 10.1016/j.pbi.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 45.Stachelhaus T, Mootz HD, Bergendahl V, Marahiel MA. Peptide bond formation in nonribosomal peptide biosynthesis catalytic role of the condensation domain. J. Biol. Chem. 1998;28:22773–22781. doi: 10.1074/jbc.273.35.22773. [DOI] [PubMed] [Google Scholar]
- 46.Mei GY, Yan XX, Turak A, Luo ZQ, Zhang LQ. AidH, an alpha/beta-hydrolase fold family member from an Ochrobactrum sp. strain, is a novel N-acylhomoserine lactonase. App. Environ. Microbiol. 2010;76:4933–4942. doi: 10.1128/AEM.00477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adebesin F, et al. Emission of volatile organic compounds from petunia flowers is facilitated by an ABC transporter. Science. 2017;356:1386–1388. doi: 10.1126/science.aan0826. [DOI] [PubMed] [Google Scholar]
- 48.Colquhoun TA, et al. Petunia floral volatile benzenoid/phenylpropanoid genes are regulated in a similar manner. Phytochem. 2010;71:158–167. doi: 10.1016/j.phytochem.2009.09.036. [DOI] [PubMed] [Google Scholar]
- 49.Qualley AV, Widhalm JR, Adebesin F, Kish CM, Dudareva N. Completion of the core β-oxidative pathway of benzoic acid biosynthesis in plants. Proc. Nat. Aca. Sci. 2012;109:16383–16388. doi: 10.1073/pnas.1211001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Underwood BA, et al. Ethylene-regulated floral volatile synthesis in petunia corollas. Plant Physiol. 2005;138:255–266. doi: 10.1104/pp.104.051144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langer MK, et al. PhDAPH1 is required for floral volatile benzenoid/Phenylpropanoid biosynthesis in Petunia x hybrid cv ‘Mitchell Diploid’. Phytochem. 2014;103:22–31. doi: 10.1016/j.phytochem.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 52.Colquhoun TA, et al. A petunia chorismate mutase specialized for the production of floral volatiles. Plant J. 2010;61:145–155. doi: 10.1111/j.1365-313X.2009.04042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Colquhoun TA, et al. A peroxisomally localized acyl-activating enzyme is required for volatile benzenoid formation in a Petunia × hybrida cv. ‘Mitchell Diploid’ flower. J. Exp. Bot. 2012;63:4821–4833. doi: 10.1093/jxb/ers153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tieman DM, Loucas HM, Kim JY, Clark DG, Klee HJ. Tomato phenylacetaldehyde reductases catalyze the last step in the synthesis of the aroma volatile 2-phenylethanol. Phytochem. 2007;68:2660–2669. doi: 10.1016/j.phytochem.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 55.Franke R, et al. Changes in secondary metabolism and deposition of an unusual lignin in the ref8 mutant of Arabidopsis. Plant J. 2002;30:47–59. doi: 10.1046/j.1365-313X.2002.01267.x. [DOI] [PubMed] [Google Scholar]
- 56.Kim JI, Ciesielski PN, Donohoe BS, Chapple C, Li X. Chemically induced conditional rescue of the reduced epidermal fluorescence8 mutant of Arabidopsis reveals rapid restoration of growth and selective turnover of secondary metabolite pools. Plant Physiol. 2014;164:584–595. doi: 10.1104/pp.113.229393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Coleman HD, Park JY, Nair R, Chapple C, Mansfield SD. RNAi-mediated suppression of p-coumaroyl-CoA 3′-hydroxylase in hybrid polar impacts lignin deposition and soluble secondary metabolism. Proc. Nat. Aca. Sci. 2008;105:4501–4506. doi: 10.1073/pnas.0706537105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y, Chantreau M, Sibout R, Hawkins S. Plant cell wall lignification and monolignol metabolism. Front. Plant Sci. 2013;4:220. doi: 10.3389/fpls.2013.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muhlemann JK, Klempien A, Dudareva N. Floral volatiles: from biosynthesis to function. Plant Cell Environ. 2014;37:1936–1949. doi: 10.1111/pce.12314. [DOI] [PubMed] [Google Scholar]
- 60.Maeda H, et al. RNAi suppression of Arogenate Dehydratase1 reveals that phenylalanine is synthesized predominantly via the arogenate pathway in petunia petals. Plant Cell. 2010;22:832–849. doi: 10.1105/tpc.109.073247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Colquhoun TA, et al. EOBII controls flower opening by functioning as a general transcriptomic switch. Plant Physiol. 2011;156:974–984. doi: 10.1104/pp.111.176248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van Moerkercke A, Haring MA, Schuurink RC. The transcription factor EMISSION OF BENZENOIDS II activates the MYB ODORANT1 promoter at a MYB binding site specific for fragrant petunias. Plant J. 2011;67:917–928. doi: 10.1111/j.1365-313X.2011.04644.x. [DOI] [PubMed] [Google Scholar]
- 63.Christensen LP, Jakobsen HB, Kristiansen K, Møller J. Volatiles emitted from flowers of γ-radiated and nonradiated Jasminum polyanthum Franch. in situ. J. Agric. Food Chem. 1997;45:2199–2203. doi: 10.1021/jf960961q. [DOI] [Google Scholar]
- 64.Van der Niet T, Hansen DM, Johnson SD. Carrion mimicry in a South African orchid: flowers attract a narrow subset of the fly assemblage on animal carcasses. Annals. Bot. 2011;107:981–992. doi: 10.1093/aob/mcr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oliva M, et al. Enhanced formation of aromatic amino acids increases fragrance without affecting flower longevity or pigmentation in Petunia × hybrida. Plant Biotech. J. 2015;13:125–136. doi: 10.1111/pbi.12253. [DOI] [PubMed] [Google Scholar]
- 66.Clayton TA. Metabolic differences underlying two distinct rat urinary phenotypes, a suggested role for gut microbial metabolism of phenylalanine and a possible connection to autism. FEBS letters. 2012;586:956–961. doi: 10.1016/j.febslet.2012.01.049. [DOI] [PubMed] [Google Scholar]
- 67.Verdonk JC, et al. Regulation of floral scent production in petunia revealed by targeted metabolomics. Phytochem. 2003;62:997–1008. doi: 10.1016/S0031-9422(02)00707-0. [DOI] [PubMed] [Google Scholar]
- 68.Jorgensen RA, Cluster PD, English J, Que Q, Napoli CA. Chalcone synthase cosuppression phenotypes in petunia flowers: comparison of sense vs. antisense constructs and single-copy vs. complex T-DNA sequences. Plant Mol. Biol. 1996;31:957–973. doi: 10.1007/BF00040715. [DOI] [PubMed] [Google Scholar]
- 69.Livak KJ, Schmittgen TD. Analysis of relative gene expression data. Method. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 70.Johnson TS, et al. Lilium floral fragrance: A biochemical and genetic resource for aroma and flavor. Phytochem. 2016;122:103–112. doi: 10.1016/j.phytochem.2015.11.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All submitted manuscripts including figures and tables are available on Scientific report.