Abstract
Nucleic acid aptamers are often referred to as chemical antibodies. Because they possess several advantages, like their smaller size, temperature stability, ease of chemical modification, lack of immunogenicity and toxicity, and lower cost of production, aptamers are promising tools for clinical applications. Aptamers against cell surface protein biomarkers are of particular interest for cancer diagnosis and targeted therapy. In this study, we identified and characterized RNA aptamers targeting cells expressing integrin α5β1. This αβ heterodimeric cell surface receptor is implicated in tumor angiogenesis and solid tumor aggressiveness. In glioblastoma, integrin α5β1 expression is associated with an aggressive phenotype and a decrease in patient survival. We used a complex and original hybrid SELEX (selective evolution of ligands by exponential enrichment) strategy combining protein-SELEX cycles on the recombinant α5β1 protein, surrounded by cell-SELEX cycles using two different cell lines. We identified aptamer H02, able to differentiate, in cyto- and histofluorescence assays, glioblastoma cell lines, and tissues from patient-derived tumor xenografts according to their α5 expression levels. Aptamer H02 is therefore an interesting tool for glioblastoma tumor characterization.
Keywords: integrin α5β1, aptamer, SELEX, glioblastoma, cell-surface biomarker, cytofluorescence, aptahistofluorescence, therapeutic payloads delivery
Graphical Abstract
Introduction
Glioblastoma (GBM), the highest-grade glioma tumor (grade IV), is the most aggressive and the most common malignant form of astrocytoma. Standard therapy consists of surgical resection to an extent that is safely feasible, followed by radiotherapy and concomitant chemotherapy with temozolomide (Stupp protocol).1 Despite these therapies, patients with GBM rarely live longer than 2 years.2 Histological features that characterize GBM are the presence of necrosis and abnormal growth of blood vessels around the tumor. Defining molecular profiles aims to develop molecularly guided approaches for the treatment of patients. The 2016 World Health Organization (WHO) classification scheme3 integrated phenotypic and genotypic parameters for CNS tumor classification. GBMs are divided into isocitrate dehydrogenase (IDH) 1 wild-type (about 90% of cases; corresponds to the most frequent primary or de novo GBM) and IDH-mutant GBM (about 10% of cases; corresponds to secondary GBM). Some of the GBM biomarkers that have been and are being discovered4 are cell surface protein biomarkers.5, 6, 7, 8 Expression of cell surface protein is often remodeled in cancers. Genetic and epigenetic features altered in cancer8 include modification of copy number (under- or overexpression), truncations, mutations, and post-translational modifications. These modified proteins are major clinical targets for diagnosis and therapies, considering their accessibility for pharmacological compounds.
Tumor-specific tools such as aptamers can be used as recognition ligands to discriminate a tumor cell from another cell, as agonists or antagonists, or as carriers to deliver therapeutic payloads to targeted tumor cells.9, 10, 11, 12, 13 Aptamers are single-stranded DNA or RNA molecules that constitute an alternative class of molecules emerging as cancer-specific therapeutic, diagnostic, and theranostic tools.9, 10, 14, 15, 16, 17, 18 They are selected through an in vitro selection process, published for the first time in 1990 by three independent research groups,19, 20, 21 known as SELEX (selective evolution of ligands by exponential enrichment).20 Aptamers19 from the Latin aptāre (to fit) and from the ancient Greek meros (part) are often referred to as chemical antibodies13 because they bind to their specific targets with high affinity and specificity. Aptamers possess numerous advantages over antibodies, like smaller size, temperature stability, self-refolding, fewer side effects for immunotherapy, lack of immunogenicity and toxicity, more efficient penetration into biological compartments, chemical synthesis with high batch fidelity, and the option of site-specific and flexible introduction of linkers, reporters, functional groups, small interfering RNA (siRNA), nanoparticles, drugs, and so forth.10, 11, 14 Aptamers toward a wide variety of targets have been identified, the most common ones remaining proteins. We recently reviewed aptamers to more than 30 different tumor cell surface protein biomarkers,22 a few of them being heterodimeric receptors, such as tyrosine kinase receptors and cell adhesion molecules. However, selection of aptamers to cell surface proteins remains a complex process.
Among cell surface biomarkers, integrins are heterodimeric cell surface receptors for cell migration, differentiation, and survival,23 composed of α and β subunits; their deregulation leads to cancer progression and therapy resistance.24 In mammals, 24 distinct integrins are formed by the combination of 18 α and 8 β subunits. Specific heterodimers preferentially bind to distinct extracellular matrix proteins. Integrin α5β1, the fibronectin receptor, belongs to the arginine, glycine, and aspartate (RGD) -binding integrin family. Overexpressed on tumor neovessels and on solid tumor cells, integrin α5β1 is implicated in tumor angiogenesis and solid tumor aggressiveness. We and others have shown that α5β1 integrin is a pertinent therapeutic target for GBM25 through its active role in tumor proliferation, migration, invasion, and resistance to chemotherapy.26, 27, 28, 29, 30 At the mRNA level, high α5β1 integrin expression is associated with more aggressive tumors in patients with glioma.26 At the protein level, to date, only a few in situ analyses of GBM tumor section have been described.31, 32 Further investigation of α5β1 expression in GBM tumor cells as a potential prognostic factor and/or biomarker for diagnosis requires rapid and accurate tools.
In our study, aptamers to integrin α5β1-expressing cells were selected by an original and complex hybrid SELEX process. This SELEX combines three rounds of protein-SELEX surrounded by 15 rounds of cell-SELEX on two different cell lines genetically modified to overexpress integrin α5, the human GBM U87MG cell line, and the Chinese hamster ovaries CHO-B2 cell line. Counterselection steps were performed on isogenic cell lines underexpressing α5 for U87MG or ones that do not express α5 for CHO-B2. We identified and characterized an aptamer named H02. Directly coupled to the cyanine 5 fluorophore, aptamer H02 was able to discriminate between 10 GBM cell lines expressing high and low levels of integrin α5. Aptamer H02 is internalized at 37°C. As a proof of concept, we also demonstrated that aptamer H02 is very efficient in apta-fluorescence assays to distinguish GBM tumor tissues from patient-derived tumor xenografts expressing high levels of α5 from GBM tumor tissues expressing low levels of α5.
Results
Selection Strategy Combining Cell-SELEX on Two Cell Lines Expressing Integrin α5β1 and Protein-SELEX on the Recombinant α5β1 Integrin
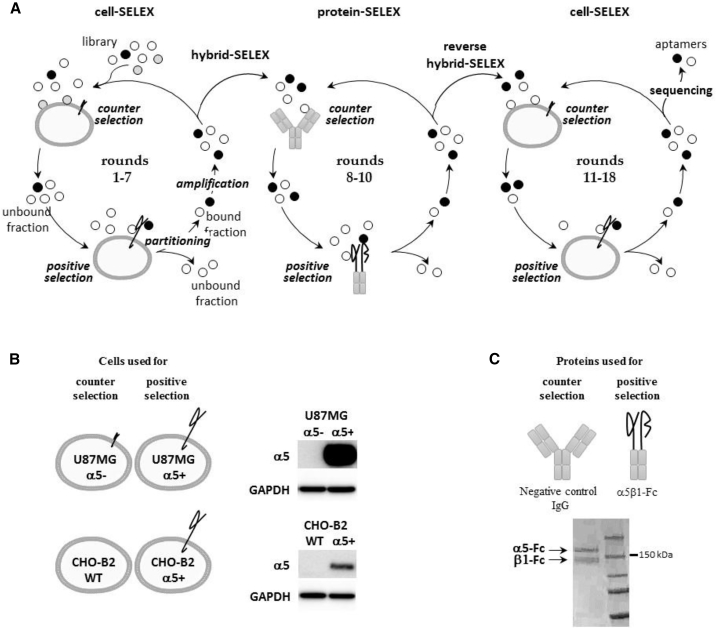
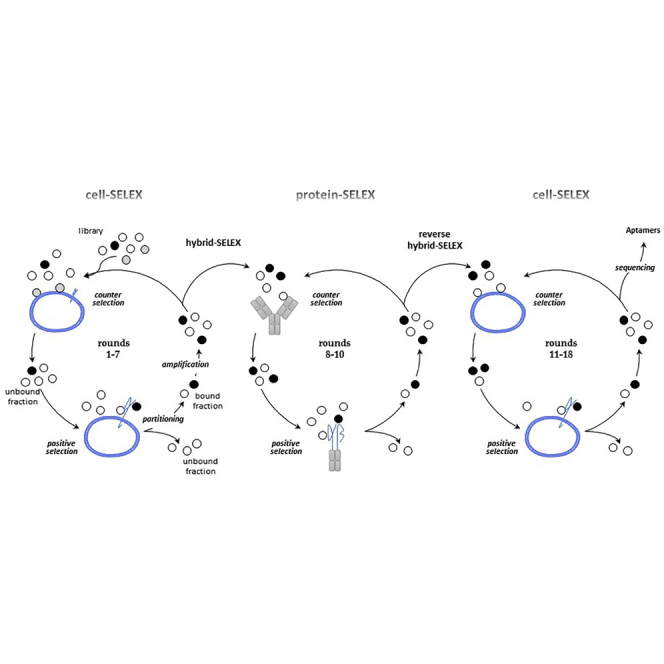
To guide the selection toward α5β1, we used an original hybrid SELEX strategy that alternates protein and cell-SELEX. Our strategy is divided into three steps, symbolized by three circles in Figure 1A, using two different cell lines (Figure 1B) and a recombinant α5β1 protein (Figure 1C). The first seven rounds of cell-SELEX were performed on human GBM U87MG cells overexpressing the α5 subunit (U87MG α5+).26 An RNA library comprising 1014 different molecules was incubated with U87MG α5+ cells plated in a cell culture dish. Isogenic U87MG cells modified to underexpress the α5 subunit (U87MG α5−)26 were used for counterselection steps. Although the α5 subunit has never been detect by western blot (Figure 1B) in these cells, to avoid any loss of potential α5 binders, counterselection steps were not introduced before the fourth round of selection and were performed only during rounds 4–6. The next three rounds of selection were performed by a protein-based SELEX process (rounds 8–10) on the protein A-purified form of integrin α5β1. The recombinant protein α5(E951) β1(D708)-Fc (α5β1-Fc) is composed of human α5 and β1 ectodomains fused to Fcγ1 knobs mutated into their CH3 domains to increase the likelihood of heterodimerization between α5 and β1 chains.33 Negative and counterselection steps preceding positive selection steps were performed on protein A-Sepharose beads (rounds 8–10) and on an antibody presenting the same γ1 isotype as the Fc fused to α5β1 ectodomains (rounds 9 and 10). Finally, cell-SELEX rounds were implemented on two different cell lines: on U87MG cells (round 11, as for rounds 1–7) and CHO-B2 cells (rounds 12–18). CHO-B2 cells, which do not naturally express α5,34 were used for counterselection. For positive selection, CHO-B2 cells were manipulated to generate positive α5 cells by overexpressing human ITGA5. During the course of the SELEX process, the stringency was progressively increased by decreasing and increasing the incubation time for positive and counterselection, respectively by introducing competitor yeast tRNAs and increasing the number of washes (Table S1). Enrichment of RNA pools during selection was followed by qRT-PCR. The RNA pool at round 18 showed significant amplification compared with pools of preceding rounds (Figure S1) and was cloned. Of 82 sequenced molecules, five aptamers, named H02, H03, G10, B03, and G11, were selected for further characterization.
Figure 1.
SELEX Strategy
(A) Scheme of the cell- and protein-based SELEX strategy used for aptamer selection. Briefly, one round of SELEX first involves a selection step. The nucleic acid library is incubated with a target (positive selection), which can be preceded by counterselection to remove non-specific nucleic acid molecules. During the partitioning step, bound and unbound fractions are separated. The bound fraction is amplified to obtain an enriched pool for the next round of selection. First, cell-SELEX processes were performed (rounds 1–7), followed by protein-SELEX (rounds 8–10) and then by cell-SELEX (rounds 11–18). The combination of cell- and protein-based SELEX is called hybrid SELEX and reverse hybrid SELEX. At the end of selection, nucleic acid molecules were cloned and sequenced. Individual sequences are aptamers. (B) Description of cells used for counterselection and positive selection (rounds 1–7 and 11–18). On the right, western blots show the level of expression of α5 in the different cell lines used for the SELEX strategy. (C) Description of proteins used for counterselection and positive selection (rounds 8–10). Counterselection was also performed on protein-A Sepharose beads alone in rounds 8–10. Shown below is a denaturing SDS polyacrylamide gel loaded with the protein A-purified recombinant α5β1-Fc protein and Coomassie blue stained.
Identification of Aptamers Binding to α5-Expressing Cells
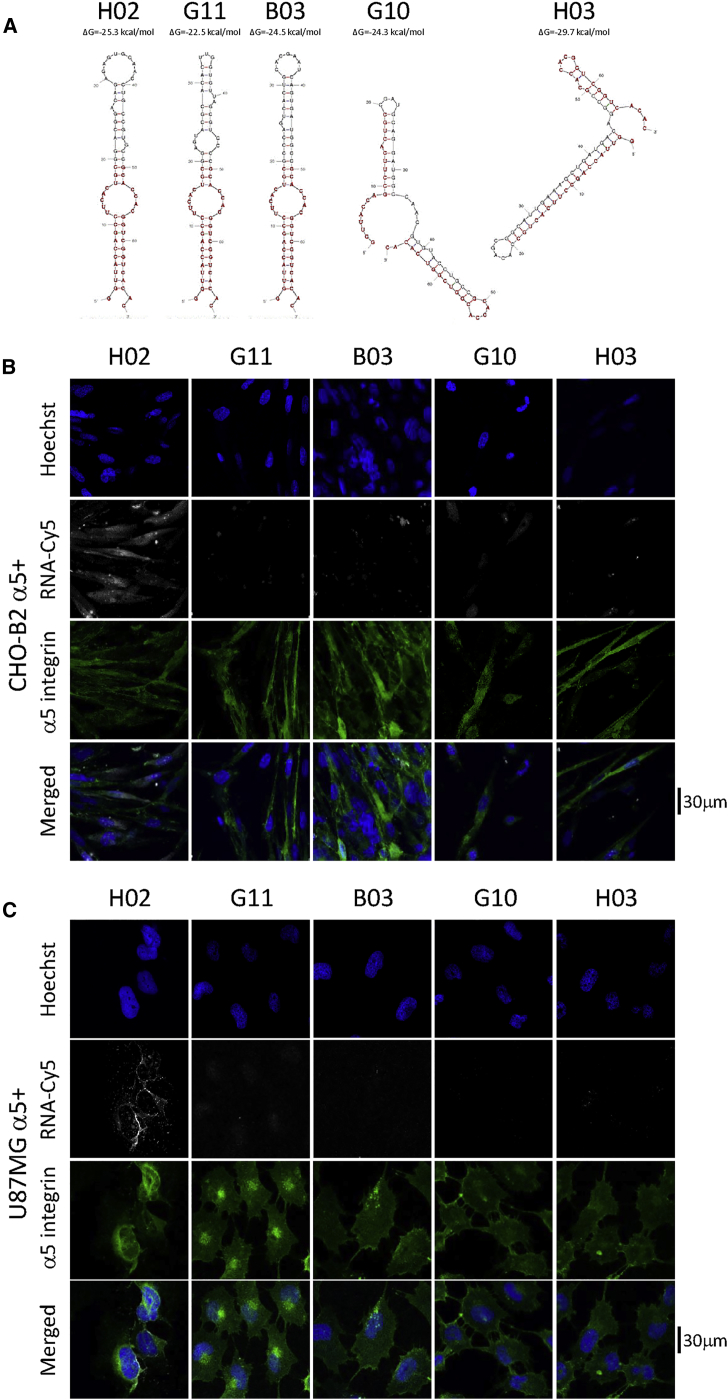
Aptamer H02 was the most frequently represented over all sequences (6 times over 82 sequences, 7.3%). Aptamers H03, G10, and G11 represented 3.7% of all sequenced molecules and aptamer B03 1.2%. The predicted secondary structures of the five aptamers (H02, G11, B03, G10, and H03) are shown in Figure 2A. Fixed regions (shown in dark red in Figure 2A) were designed to display partial complementarity and pre-organize aptamers in hairpin structures.35 The secondary structure predictions of aptamers H02, G11, and B03 are highly similar and very different from those of G10 and H03.
Figure 2.
Aptamers Predicted Secondary Structures and Binding to α5-Expressing Cells
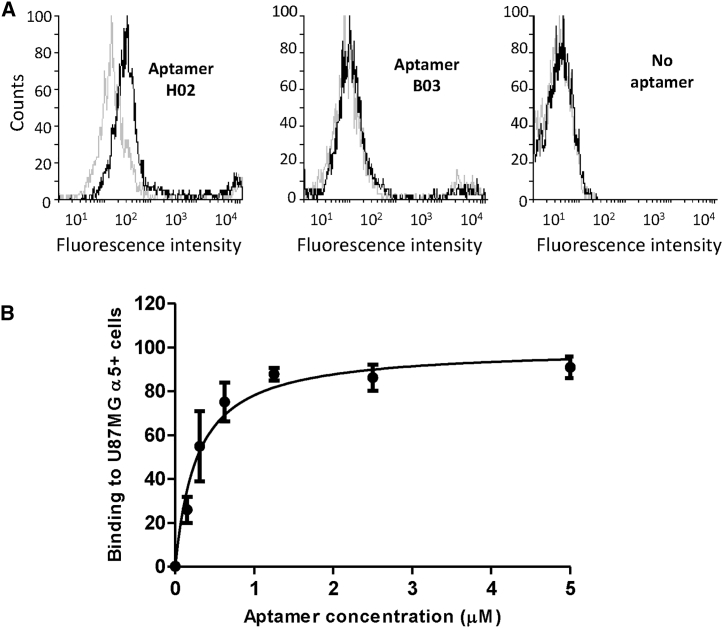
(A) Predicted secondary structure of aptamers H02, G11, B03, G10, and H03. Structures were predicted using the mfold web server.67 Nucleotides 1−19 and 50−68 correspond to fixed flanks of the candidate sequences. They are shown in dark red. ΔG values are noted above the structures. (B and C) Monitoring of the binding of five Cy5-labeled aptamers (H02, G11, B03, G10, and H03) at 5 μM on CHO-B2 and U87MG cell lines at 4°C using confocal microscopy. Nuclei, counterstained with Hoechst, are shown in royal blue. Aptamers, coupled to Cy5, are shown in white. (B) Binding on CHO-B2 α5+. α5 integrin is visualized, using the GFP-fused protein, in green. (C) Binding on U87MG α5+. α5 integrin was labeled with the IIA1 antibody and is shown in green. Images were captured at the same setting to allow direct comparison of staining patterns.
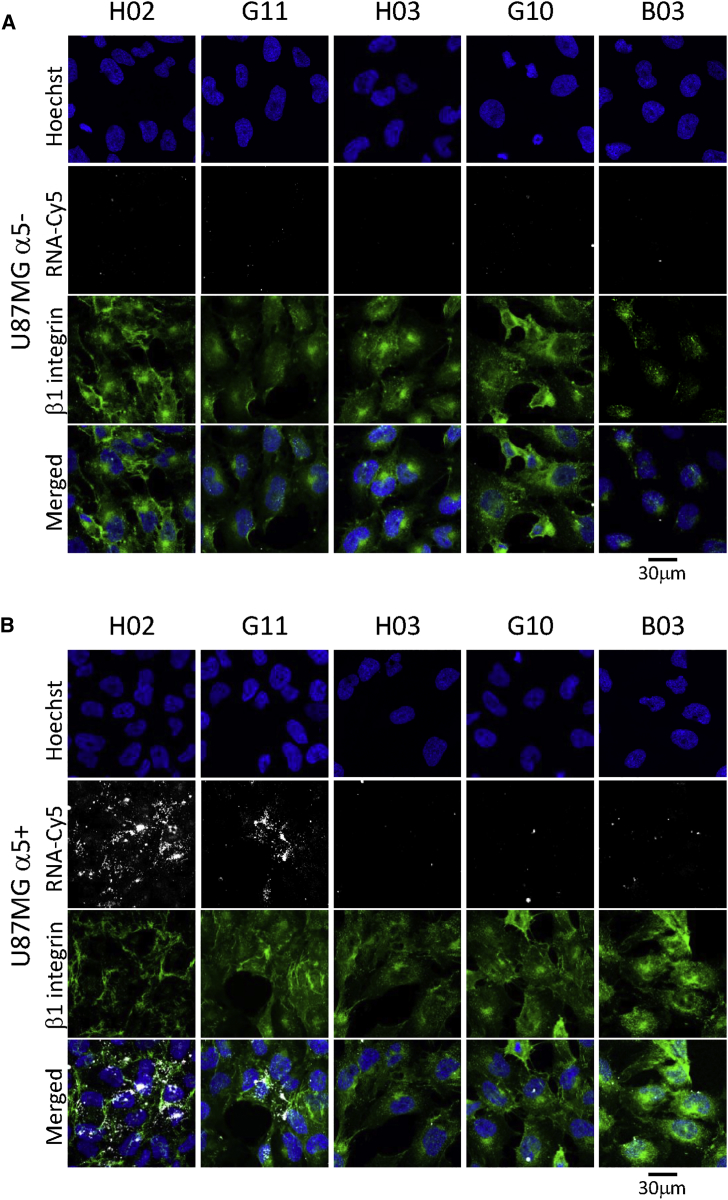
Identification of α5β1-binding aptamers was performed using confocal fluorescence microscopy by incubating cyanine-5 (Cy5)-labeled aptamers at 4°C with the cells used for the cell-SELEX process. None of the five aptamers binds to U87MG α5− and CHO-B2 cells used for the counterselection steps (Figures S2A and S2B). Only aptamer H02 binds to the U87MG α5+ and CHO-B2 α5+ cells used for positive selection steps (Figures 2B and 2C). We next checked whether, under the same methodological conditions, these aptamers were able to bind to U87MG cells at 37°C, the temperature used for the cell-SELEX process. H02, G11, B03, G10, and B03 did not bind to U87MG α5− cells (Figure 3A). On U87MG α5+ cells, we observed strong binding not only of aptamer H02 but also, to a lesser extent, of aptamer G11 (Figure 3B). Because of its binding to U87MG α5+ cells at 4°C and at 37°C, subsequent analyses were performed using aptamer H02.
Figure 3.
Binding of Aptamers to U87MG Cells
(A and B) Monitoring the binding of five Cy5-labeled aptamers (H02, G11, B03, G10, and H03; 5 μM) at 37°C, using confocal microscopy, on U87MG α5− (A) and U87MG α5+ cells (B). Nuclei, counterstained with Hoechst, are shown in royal blue. Aptamers, coupled to Cy5, are shown in white. The anti-β1 TS2/16 antibody (Ab) labeling is shown in green. Merged images are shown. Images were captured at the same setting to allow direct comparison of staining patterns.
Validation of Integrin α5β1 as the Target of Aptamer H02
The SELEX process was performed to guide the selection toward integrin α5β1. However, cell-SELEX-based strategies have already led to the selection of aptamers against “undesirable targets,” meaning other proteins than the expected pre-identified targets.36, 37
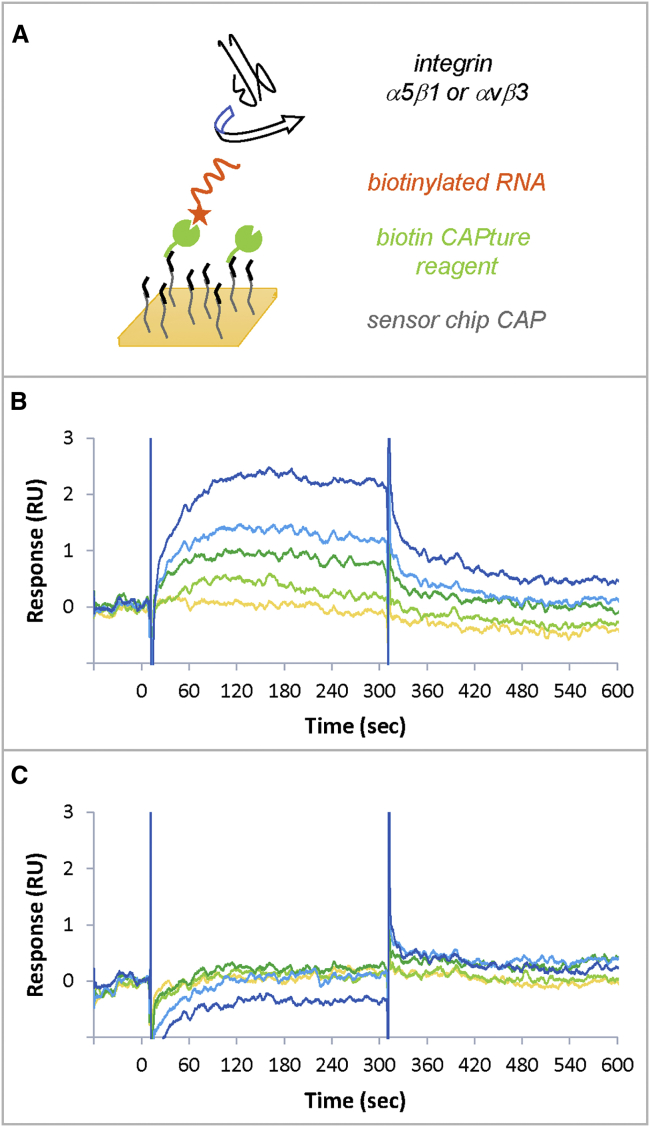
To validate integrin α5β1 as the target of our SELEX process, surface plasmon resonance (SPR) experiments were performed on a Biacore T200 instrument. Figure 4A shows the experimental scheme. Aptamer H02, with 2′-fluoro pyrimidines to increase its stability in SPR experiments, was captured on a CAP sensor chip via a biotin at its 5′ extremity. The aptamer was captured on a biotin CAPture reagent. Integrins were injected at concentrations ranging from 8–130 nM. We tested human integrins α5β1 and αvβ3 to check the H02 aptamer’s specificity. The surface was then regenerated. CAP chips are designed to capture biotinylated molecules reversibly on the sensor surface, facilitating its regeneration.38 An SPR cycle thus consisted of injections of biotin CAPture reagent, biotinylated aptamer, integrin, and regeneration solution. Successive cycles were repeated, changing the integrin nature and concentration at each cycle (Figure S3A). Because of 2′-fluoro modifications, aptamer H02 was resistant to degradation over time, and aptamer injections were highly reproducible. The biotinylated aptamer reached the same level at each cycle with as low as 2% variation in responses over 20 cycles (Figure S3A). Therefore, the surface has the same properties during all experiments.
Figure 4.
Surface Plasmon Resonance Experiments for Target Validation
(A) Experimental scheme. The biotin CAPture reagent (green), composed of streptavidin conjugated with an oligonucleotide, is stably hybridized to a complementary sequence immobilized on the sensor chip (black). The biotinylated aptamer (orange) is captured to the biotin CAPture reagent. Integrins are used as analytes (black). The interaction between integrins and the captured aptamer is studied. The surface is then regenerated and rebuilt with new biotin CAPture reagent in the next cycle. (B and C). SPR sensorgrams obtained for the injections of integrins α5β1 (B) and αvβ3 (C) at 8.1 nM (yellow), 16.2 nM (light green), 32.5 nM (dark green), 65 nM (light blue), and 130 nM (dark blue).
Figures 4B and 4C show the sensorgrams obtained for integrins α5β1 and αvβ3, respectively, after double referencing (subtraction of reference channel and buffer injection). The sensorgrams show that, even when responses are low, aptamer H02 bound specifically to integrin α5β1 in a dose-dependent manner but failed to interact with integrin αvβ3. The equilibrium affinity parameter (KD) of the interaction between integrin α5β1 and aptamer H02 was 72 ± 11 nM. To ensure that integrin was active, positive controls were used. Figure S3B shows binding by SPR of integrin α5β1 to its natural ligand, fibronectin. We also demonstrated that only aptamer H02 was an α5β1 binder (but not, for example, aptamer B03; Figure S3C).
H02 Aptamer Stability, Specificity and Affinity for U87MG α5+ Cells
Because aptamer H02 is a non-modified RNA molecule, we tested its stability in the presence of cells at 4°C and 37°C in the buffer used for selection. The results (Figure S4A) confirmed that incubation on cells for 1 h, which corresponds to the maximum contact time of RNAs with cells during different assays, does not induce aptamer H02 degradation. However, this aptamer is extremely rapidly degraded when incubated on cells in a culture medium supplemented with 10% fetal bovine serum (FBS) (Figure S4A.). If this aptamer had to be used under more complex conditions than in a simple buffer, then it would have to be modified to increase its nuclease sensitivity. For example the 2′-fluoro-modified H02 aptamer is very stable in contact with cells at 4°C and at 37°C as it is not degraded in the selection buffer and in a complex medium for at least 1 h (Figure S4B).
The specificity of aptamer H02 for α5-overexpressing cells was confirmed by flow cytometry at 4°C by incubation of Cy5-coupled aptamers B03 and H02 at 500 nM for 1 h with detached CHO-B2 cells (Figure 5A) and by incubation of Cy5-coupled aptamers G11 and H02 at concentrations ranging from 0.15–5 μM with detached CHO-B2 α5+ cells (Figure S5). Although no shift in fluorescence was detected for CHO-B2 cells after incubation with the two Cy5-labeled H02 and B03 aptamers, a shift was detected for CHO-B2 α5+ cells with aptamer H02 but not with aptamer B03. Figure S5 also confirmed that aptamer H02 was our best hit from SELEX because it binds much better to α5-expressing cells than aptamer G11.
Figure 5.
Flow Cytometry Binding of Aptamers
(A) Comparison of the binding profiles of aptamers H02 (left) and B03 (center) on CHO-B2 cells (gray lines) and CHO-B2 α5+ cells (black lines) at 4°C. Profiles without aptamer labeling are shown on the right. (B) Titration of aptamer H02 resulted in determination of the equilibrium affinity parameter KD for the interaction between U87MG α5+ cells and aptamers H02 (277.8 ± 51.8 nM). Cy5-aptamer H02 at concentrations of 0.15, 0.3, 0.6, 1.25, 2.5, and 5 μM was incubated on ice with a constant amount of cells and analyzed by flow cytometry.
The equilibrium affinity parameter KD of the interaction between aptamer H02 and U87MG α5+ cells was determined using flow cytometry (Figure 5B). Binding events associated with the fluorescence signal of different concentrations of aptamer, ranging from 0.15–5 μM, to a constant number of cells were measured. A KD of 277.8 ± 51.8 nM was determined by plotting the mean fluorescence of U87MG α5+ cells against the concentration of the H02 aptamer.
H02 Aptamer Localization in U87MG α5+ Cells
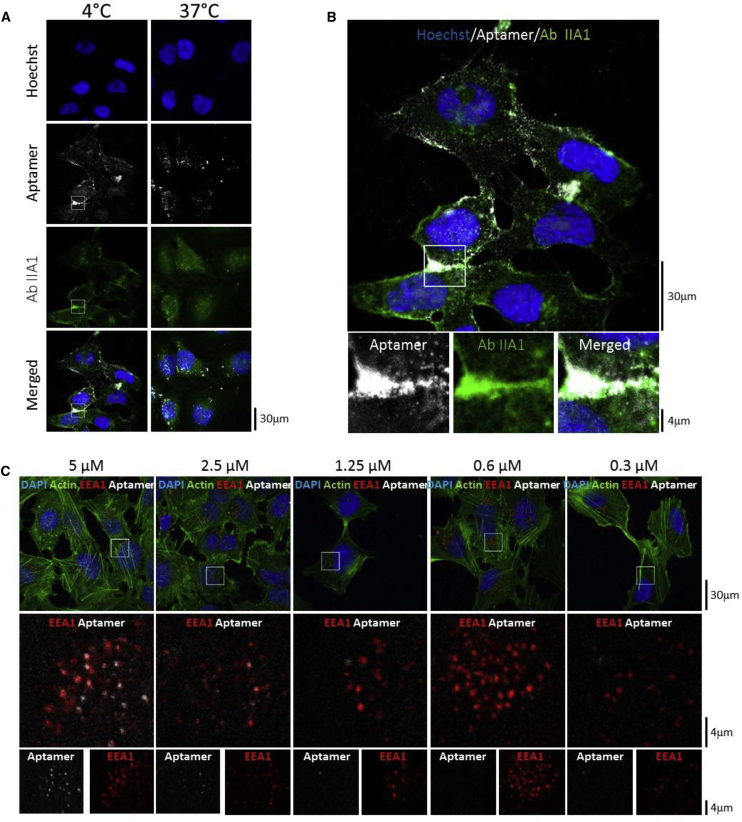
The localization of aptamer H02 on GBM U87MG α5+ cells was analyzed by confocal microscopy at 4°C and 37°C. Cells were incubated with the Cy5-labeled aptamer H02 and with the anti- α5 IIA1 antibody, followed by incubation of a secondary antibody labeled with Alexa 546. Spots of co-localization were detected between Alexa 546 and Cy5, which reflect spatial proximity between the α5 subunit and the aptamer H02, at 4°C and, to a lesser extent, at 37°C (Figures 6A and 6B). Aptamer H02 detected α5β1 mostly at the plasma membrane and at cell-cell junctions at 4°C, whereas punctuate labeling suggested internalized molecules at 37°C. The weaker co-localization observed with anti-β1 antibody and aptamer H02 (Figure 3B) is explained as the β1 subunit associates with different α subunits to form different integrins, whereas the α5 subunit associates only with the β1 subunit to form the fibronectin receptor.
Figure 6.
Confocal Microscopy Analysis of Aptamer H02 on U87MG α5+
(A) Confocal microscopy analysis with aptamer H02 at 5 μM and the anti-α5 antibody IIA1 on U87MG α5+ cells at 4°C and 37°C. The aptamer is labeled with Cy5 (white). Incubation of antibody IIA1 was followed by incubation with a secondary antibody labeled with Alexa 546 (green). Nuclei are stained with Hoechst (blue). (B) Top: enlargement of the merged image in (A) at 4°C. Bottom: magnified images acquired by zooming in on the indicated areas of the parental image. Scale bars are shown in the lower right corners of the images. (C) Co-localization of aptamer H02 and the endocytosis marker EEA1. Shown are confocal images of U87MG α5+ cells incubated at 37°C with the Cy5-labeled aptamer H02 at five different concentrations (5, 2.5, 1.25, 0.6, and 0.3 μM). After aptamer labeling (shown in white), cells were fixed, permeabilized, and then labeled to detect nuclei (DAPI, blue), actin (Phalloidin-Atto 488, green), and early endosome EEA1 (EEA1 immunolabeling, red). Shown in the first row are merged images. Shown in the second row are magnified images of selected areas (white squares) of the parental images. These images show co-labeling of EEA1 and aptamer H02. Shown in the third row are separate EEA1 and aptamer labeling. Images were captured at the same setting to allow direct comparison of staining patterns.
We next wanted to confirm the internalization of aptamer H02 in α5-expressing cells at 37°C. To this end, adherent U87MG α5+ cells were labeled for 30 min with the Cy5-coupled aptamer H02 at five different concentrations (5, 2.5, 1.25, 0.6, and 0.3 μM). After cell fixation, cells were immunolabeled with the anti-EEA1 antibody to detect early endosomes and then analyzed by confocal microscopy. Figure 6C shows clear co-localization of aptamer H02 at 5, 2.5, and 1.25 μM with the anti-EEA1 antibody in U87MG α5+ cytoplasm, suggesting aptamer H02 endocytosis. A 3D reconstruction of whole z stacks is shown in Video S1.
After aptamer labeling, cells were fixed, permeabilized and then labeled with the anti-EEA1 antibody labeling early endosomes. The 3.58 μm stack (12 images) was acquired by confocal microscopy. 3D animation was performed by Imaris 92.1 (bitplane). First all channels then only early endosomes and aptamers are displayed in maximum intensity projection. Finally these two channels are shown in normal shading rendering. DAPI staining in shown blue, actin labeling in green, EEA1 labeling in red and the Cy5-coupled aptamer H02 labeling in white.
Fluorescently labeled aptamers were not detected at lower concentrations (0.6 and 0.3 μM). The lower concentration limit of 1.25 μM corresponds to 4.5-fold the KD of the H02-cell interaction and, theoretically, to 82% receptor occupancy, governed by concentration and affinity. At 4°C, aptamers were not detected at concentrations lower than 5 μM (Figure S6), suggesting a different binding mechanism at 4°C and at 37°C. Figures 5A and S5 show that, in flow cytometry experiments, a difference could be detected at 4°C between H02 and aptamers B03 and G11 at concentrations lower than 1 μM. This difference may be due to the differences inherent to the two different techniques (flow cytometry versus confocal microscopy).39
Cyto- and Histofluorescence with Aptamer H02 on Different GBM Cell Lines and on Patient-Derived GBM Xenografts
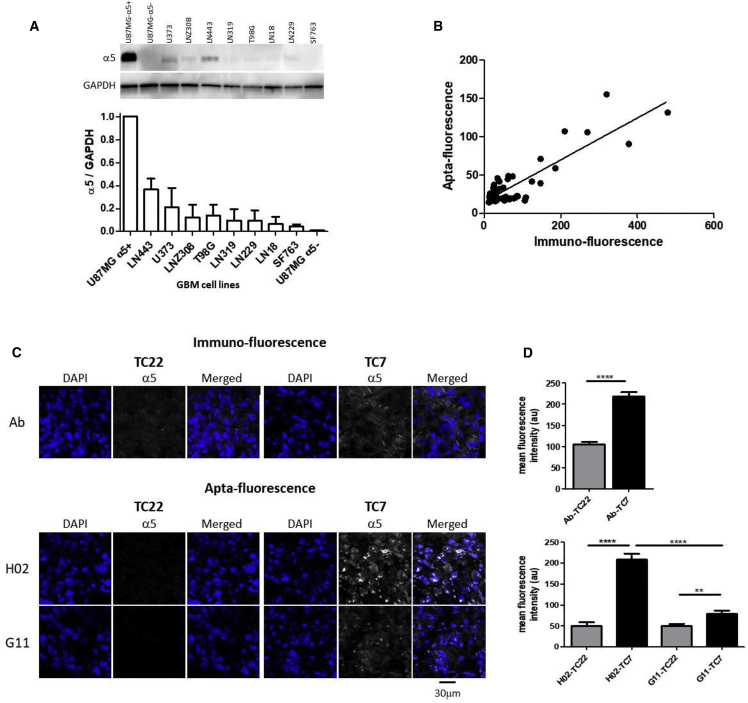
Aptamer molecules have the potential to revolutionize the field of diagnostics for the detection of cell-specific biomarkers.40 We evaluated the capacity of aptamer H02 to be a new tool for the characterization of GBM α5-expressing cells and tissues. We first characterized the ability of aptamer H02 to distinguish between 10 human GBM cell lines expressing different levels of the α5 subunit (Figure 7A). Among those are U87MG α5+ and U87MG α5−, used for cell-SELEX, and eight GBM cell lines, LN319, LN229, SF763, LN18, LNZ308, U373, T98G, and LN443. For aptacytochemical assays, confluent adherent cells were stained with the Cy5-labeled aptamer H02 at 4°C at 5 μM for 30 min. After cell fixation with paraformaldehyde, cells were immunolabeled with an anti-α5 primary antibody and a secondary antibody labeled with Alexa 546. Quantification of apta- and immunostaining of the different cell lines is shown in Figure 7B. A correlation coefficient of 0.78 has been determined between apta- and immunofluorescence. Integrin α5β1 expression in GBM cell lines can therefore be monitored in cytofluorescence with the Cy5-labeled aptamer H02.
Figure 7.
Aptafluorescence on GBM Cells and Tissues
(A–D) Aptafluorescence on GBM cells (A and B) and tissues (C and D). (A) Western blot analysis. One representative western blot of the GBM cell lines (U87MG α5+, U87MG α5−, LN319, LN229, SF763, LN18, LNZ308, U373, LN443, and T98G) used in this study is shown at the top. Histograms, at the bottom, show the quantification of α5 integrin expression normalized to GAPDH levels (mean ± SEM of 3 independent experiments). GAPDH was used as a loading control. (B) Immuno-quantification versus apta-quantification of confocal fluorescence experiments on ten GBM cell lines. Immuno-quantification was performed with an anti-α5 antibody, followed by a secondary antibody coupled to Alexa 546. Aptafluorescence was performed with the Cy5-labeled aptamer H02. Quantification of mean fluorescence intensity (a.u.) was performed on at least 5 randomly selected images per cell line. The correlation coefficient is 0.78, with p < 0.0001. (C) Immuno-fluorescence (top panel) and apta-fluorescence (bottom panel) of patient-derived tumor xenografts TC7 and TC22, showing high and low levels of α5 integrins, respectively. For immunofluorescence, detection of α5 (white) was performed with an antibody, followed by a secondary antibody coupled to Alexa 647. For aptafluorescence, detection of α5 (in white) was performed with the Cy5-labeled aptamers H02 and G11. DAPI staining is shown in blue. Images were captured at the same setting to allow direct comparison of staining patterns. One representative image per condition is shown. (D) Quantification of immuno-fluorescence (top panel) and apta-fluorescence (bottom panel) by confocal microscopy. Histograms show quantification of 10 to 26 different images per condition. Statistical analyses were done with Student’s t test (****p < 0.0001 and **p < 0.003).
Histochemical assays were performed on two patient-derived GBM xenografts, TC22 and TC7. These two tumors present a 7.6-fold difference in mRNA ITGA5 levels, with the TC7 level higher than that of TC22. These tissue sections, embedded in paraffin, were first deparaffinized and subjected to an antigen unmasking protocol. Histofluorescence analysis was performed on the two tissue sections to quantify the protein level of integrin α5 in both tumors and to compare immuno- and aptahistochemistry assays. For immunostaining, the indirect assay consisted of successive incubation of an anti-α5 antibody and a secondary fluorescent antibody. Immunohistochemical assays showed that the TC7 tumor presented a higher integrin α5 expression level than the TC22 tumor (Figures 7C and 7D, top panels), in accordance with the mRNA levels. Aptahistochemical assays were performed by incubation of tumor sections with the Cy5-labeled aptamers H02 and G11 at the lower concentration limit of 1 μM, as outlined in H02 Aptamer Localization in U87MG α5+ Cells (Figures 7C and 7D, bottom panels). With aptamer H02, the fluorescence intensity is 4-fold higher on TC7 than on TC22. Under the conditions used for these histochemical experiments, the TC7 versus TC22 discrimination capacity was better with aptamer H02 than with the anti-α5 antibody. Aptamer G11 was also able to discriminate between both tissue sections, but to a much lesser extent than aptamer H02 (difference of 1.6-fold).
Aptamer H02 is therefore an interesting and promising new tool to differentiate cells according to their α5 subunit expression levels in cyto- and histofluorescence experiments.
Discussion
Biomarkers are indicators used to establish a diagnosis and prognosis and predict susceptibility to targeted therapies. GBM is the most aggressive form of brain tumors in adults. Despite intensive treatments, the prognosis of GBM patients remains poor. There is an urgent need to incorporate known biomarkers into clinical trials and routine clinical practice, which may assist not only with patient selection but also with adjustment of the treatment schedule based on patient-specific biology.41 Because differential expression of cell surface proteins often occurs in tumor cells, and considering their accessibility to extracellular ligands, these proteins provide biomarkers of interest in oncology. The identification of molecular probes specific for cell surface protein biomarkers is of great importance.22 Because of their high affinity and specificity toward their targets, aptamers are attractive and promising tools, alternatives to antibodies for clinical applications. In this study, using a complex and highly stringent SELEX strategy, we showed that it was possible to select an RNA aptamer specific to a pre-identified heterodimeric cell surface protein embedded in its natural environment. Integrin α5β1 is a GBM cell surface biomarker.26 Aptamer H02 was able to differentiate between high and low expression of the α5 integrin on cells and tissues. This aptamer is suitable for tumor characterization.
Two main processes have been developed to select aptamers specific for pre-determined cell surface proteins: protein- and cell-based SELEX.22 In protein-based SELEX, a purified protein is used as target, usually full-length or truncated (generally recombinant ectodomains). The major issues with the protein-based SELEX process are that purification of membrane proteins is not easy and that purified proteins may not adopt the same conformational state as in their endogenous cellular environment. Some aptamers identified through protein-based SELEX failed to recognize their target when embedded in whole living cells.42, 43 In cell-based SELEX, targets are cell surface proteins in their cellular environment. This process is much more complex than protein-based SELEX. Modification of cell lines is often required to guide the selection toward the desired target, like over- and/or underexpression of the protein target for positive- and/or counterselection, respectively. Cell-SELEX generally employs cells genetically modified to overexpress a defined cell surface target for positive selection and mock cells for counterselection or mock cells for positive selection and isogenic cells underexpressing the cell surface target for counterselection.22 Cancer cell lines have been used for the cell-based SELEX process,9, 44 both for therapy and diagnostic purposes.
Only three integrins (β2, αvβ3, and α6β4) have so far been used as pre-identified targets for the SELEX processes. Blind et al.45 selected, by protein-SELEX, RNA aptamers targeting a 46-mer peptide corresponding to the complete cytoplasmic domain of the β2 subunit of integrin αLβ2. Cells infected with vaccinia viruses encoding β2-specific aptamers enabled high-level cytoplasmic expression of RNA aptamers. Intracellular integrin-binding aptamers reduced inducible cell adhesion to the intercellular adhesion molecule 1 (ICAM-1). To target integrin αvβ3, two different protein-based SELEX processes and a cell-based SELEX were used to identify 2′-fluoropyrimidine RNA aptamers. The 2′-fluoropyrimidine modification confers increased nuclease resistance to RNA molecules. Aptamer Apt-αvβ3, selected on αvβ3 purified by immunoaffinity chromatography, was able to bind αvβ3 integrin expressed on the surface of live cells and to impair endothelial cell growth and survival.46, 47 To select aptamers specific to homodimer αv and β3, Gong et al.48 developed a strategy called multivalent aptamer isolation (MAI)-SELEX. Two distinct selection stages were employed, the first being a classical affinity selection on the purified full-length αvβ3 integrin. The second, for specificity, leads selection to β3 because integrin αIIbβ3 served as a protein decoy. Two aptamers specific for αv and β3 were identified with affinities in the low nanomolar range. Takahashi et al.49 applied a process they called isogenic cell (Icell)-SELEX to identify RNA aptamers targeting αv integrins (integrin alpha subunit [ITGAV]), in which isogenic HEK293 cell lines were manipulated for counterselection by microRNA-mediated silencing and for positive selection by overexpression of target proteins. Integrin α6β4 has recently been the target of hybrid SELEX,50, 51 a combination of protein- and cell-based SELEX processes, for which five rounds of cell-SELEX on PC-3 cells were followed by 7 rounds of protein-based SELEX on a recombinant α6β4 protein.52 In this last study, despite introduction of counterselection on PC-3 β4 integrin (ITGB4) knockdown cells to deplete single-stranded DNA (ssDNA) aptamers specific for cell surface markers other than the β4 subunit, the cell-SELEX process alone was not sufficient to prevent enrichment of non-target-specific aptamers.
To allow discovery of highly selective but also conformation-dependent aptamers and to guide the selection toward α5β1 integrin, the complex SELEX strategy we adopted presents two originalities compared with other SELEX strategies toward cell surface biomarkers. In our study, a hybrid SELEX combining successive rounds of cell-SELEX, protein-SELEX, and then again cell-SELEX was performed. Usually, in hybrid SELEX, the first rounds of selection are performed by cell-SELEX, and then rounds of selection are performed on the same version of the target in its purified form by protein-based SELEX.50, 52 A reverse hybrid SELEX combines protein-SELEX followed by cell-SELEX.51, 53, 54 Consequently, our strategy combines hybrid SELEX and reverse hybrid SELEX (Figure 1). The second originality is that two different cell lines were used compared with one in previous studies. These two cell lines were the human GBM U87MG and the hamster CHO-B2 cell lines. Both cell lines were genetically transformed to express high levels of the human α5 subunit for positive selection rounds. These two cell lines do not express the same level of the α5 subunit (Figure 1). The first rounds of selection were performed on the U87MG α5+ cells to guide the selection toward cells highly expressing the α5 subunit. The last rounds of cell-SELEX were performed on CHO-B2 α5+ cells, a cell line expressing α5 levels similar to α5 expression of the wild-type GBM cell line U373, to guide the selection to a more natural expression and, probably, a more natural conformation and environment of the target. For counterselection rounds, the ideal cell line was CHO-B2 because it does not express the α5 subunit at all. U87MG α5− cells were stably transfected to repress the human α5 gene by transfecting a pSM2 plasmid coding for a short hairpin RNA (shRNA) targeting the α5 mRNA.26 Despite the fact that U87MG α5− cells were probably not fully depleted in α5, we showed that the differential expression level of α5 between U87MG α5+ and U87MG α5− cells (not detected in western blots) was high enough to permit a differential binding pattern of aptamer H02 (Figure 3B). The keys to this successful complex SELEX are the use of alternative rounds of cell- and protein-SELEX rounds, the use of at least two different cell lines to remove unspecific binding, and the high differential expression of the target expressed on cells used for positive selection compared with cells used for counterselection.
Aptamer H02 was selected after 18 rounds of a stringent SELEX process. This aptamer was the most represented sequence. It is not degraded in contact with cells under the conditions used for the experiments. As for aptamers G11 and B03, the predicted secondary structure of aptamer H02 is highly stable in imperfect hairpins. Using label-free SPR experiments, integrin α5β1 was validated as the target of aptamer H02 (Figure 4B). Aptamer H02 was also identified as a binder of α5-expressing cells on cells used for positive selection or on other GBM cells in aptacytochemistry assays (Figures 6, 7A, and 7B). U87MG α5+ cells incubated with aptamer H02 allowed internalization of aptamer H02 at 37°C by endocytosis of the α5β1 cell surface receptor. A KD value of 277.8 ± 51.8 nM was determined for the interaction between aptamer H02 and U87MG α5+ cells. This affinity value is of the same order of magnitude than the 100–400 nM that have been determined for aptamers characterized toward other integrins by cell- or hybrid SELEX.49, 52 By SPR, a KD of 72 nM was determined for the interaction between aptamer H02-2′F and integrin α5β1. The 3.8-fold difference in binding affinity between the aptamer-recombinant protein and aptamer-cell interaction can certainly be explained by the different techniques used (flow cytometry versus SPR) and by conformational differences between cell surface proteins and soluble recombinant proteins. However, it is not due to the use of the H02-2′F aptamer in SPR versus the non-modified aptamer in flow cytometry because the H02-2′F and non-modified H02 aptamers have the same affinity for U87MG α5+ cells (data not shown). Only a few studies compared aptamer binding with isolated proteins and with tumor cell surface protein biomarkers.22 The 5-fold difference, which is of the same order of magnitude than the difference observed in our study, has been described for the OX40-aptamer interaction and can be explained by conformational differences.55
In the field of precision oncology, histological detection of specific biomarkers is a crucial diagnostic tool. Immunohistochemistry is a cheap, easy method for detection of tumor biomarkers. Aptahistochemistry is a new option, still rarely described, for which aptamers, as a new class of probes, are used instead of antibodies. In our study, we used aptamer H02 directly end-labeled with a single cyanine 5 fluorescent dye. Aptamer H02 was able to specifically interact with α5-overexpressing tumor tissues from patient-derived xenografts (Figures 7C and 7D) because it efficiently differentiates TC7 (tissue with high α5 expression) from TC22 (tissue with low α5 expression). Aptamer H02 is an effective molecular probe for labeling histological tissue sections and detection of the α5β1 biomarker on tumor cells. Aptamer probes may become powerful tools for pathologists to characterize tumor tissues because the protocols are simple to implement, straightforward to automate, and can be applied to paraffin-embedded cancer tissue as well as to frozen tumor tissues.56, 57, 58 Aptahistochemistry could certainly be easily extrapolated to biomarker multiplexing detection59 to assess co-localization of different markers on the same tumor section. Aptamer H02 targeting integrin α5β1 as well as other aptamers targeting other GBM biomarkers might therefore help to better characterize GBM inter-tumoral as well as intra-tumoral heterogeneity, which would have implications for personalized targeted therapies. A recent review describes the technicalities of the current applications of aptahistochemistry.60 Aptafluorescence will probably reduce the cost, time, and cross-reactivity concerns compared with indirect immunofluorescence approaches generally based on primary and then secondary antibodies. Conjugation of dye on aptamers is easy and reduces the risk of disrupting the aptamer structure compared with antibodies generally labeled with multiple tags. Compared with an antibody, the aptamer’s smaller size (10-fold reduction in size) allows better penetration in tissues,61, 62 particularly in applications for which epitope accessibility is reduced, such as in fixed tissues. A further advantage of aptamers is that the target of interest is not limited to molecules that produce an immune response in the host animal, as for antibodies.40 Chemical synthesis of aptamers virtually eliminates the issue of batch-to-batch variation.
Conclusion
In conclusion, we characterized a new, original, and powerful aptamer tool to detect GBM tumoral cells and tissues expressing integrin α5β1. These detections might be extended for use in other cancers where α5β1 has proven to be a therapeutic target, such as colon cancer, ovarian cancer, breast cancer, lung tumors, and melanoma.25 For clinical translation, the structure of aptamer H02 will have to be confirmed, and aptamer H02 will be improved further by truncations to obtain the minimal active fraction and by increasing its resistance toward nucleases via modifications of its nucleic acid backbone and extremities.63 Internalized, an aptamer targeting integrin α5β1 might open roads for α5β1-specific therapeutic payload delivery. Endocytosis may be crucial for targeting and increasing the therapeutic efficacy of GBM drugs. Linked to a cytotoxic agent, an aptamer to integrin α5β1 could serve as a carrier for targeted therapeutic delivery. Such aptamers were very recently called “charomers.”64 Charomers internalized with integrin α5β1 would be very powerful carriers to deliver therapeutic agents into targeted cells.
Materials and Methods
Materials
The ssDNA library was synthetized and purified by Eurogentec (Seraing, Belgium). All RNA aptamers and chemicals were purchased from IBA and Sigma-Aldrich (Hamburg, Germany), respectively, unless otherwise stated. The sequences of all primers, the library, and aptamers from this study are described in Table S2.
Cell Culture and Transfection
Cell culture medium and reagents were from Lonza (Basel, Switzerland) or Gibco (Thermo Fisher Scientific, Waltham, MA, USA). U87MG cells were from the ATCC. U373MG and T98G cells were from ECACC (European Collection of Authenticated Cell Cultures, Sigma). LN319, LN229, LN443, LN18, and LNZ308 cells were kindly provided by Prof. Monika Hegi (Lausanne, Switzerland), SF763 by Frédéric André (Marseille, France), and CHO-B2 by Wolfram Ruf (La Jolla, CA, USA). All GBM cells were routinely cultured in Eagle’s minimum essential medium (EMEM), 10% heat-inactivated FBS, and 2 mM glutamine. For U373MG and T98G, 1% non-essential amino acids and 1 mM sodium pyruvate were added to the medium. For CHO-B2 cells, EMEM was substituted for DMEM (high glucose). U87MG cells were stably transfected to overexpress (U87MG α5+) and repress (U87MG α5−) the human α5 gene, as described previously.26 CHO-B2 cells lacked the α5 subunit. They were stably transfected by a pcDNA3.1 plasmid provided by Dr. Ruoshlati (La Jolla, CA, USA) to overexpress the human α5 integrin gene in fusion with the gene for GFP by using jetPRIME (Polyplus transfection) according to the manufacturer’s instructions and named CHO-B2 α5+ cells.
Expression and Purification of α5β1-Fc
The recombinant soluble human α5β1-Fc integrin (a gift from Martin Humphries, Manchester, UK) was produced from NSO culture supernatant and purified via the Fc domain on protein A-Sepharose as described previously.33 The purity of the protein was verified by Coomassie staining of SDS-polyacrylamide gels.
SELEX Strategy
The RNA library was obtained by transcription from a starting ssDNA library (Eurogentec) containing 30 random nucleotides (N30) and flanked by primer annealing sites: 5′-GTGTGACCGACCGTGGTGC-N30-GCAGTGAAGGCTGGTAACC-3′. Two primers, P3′ (5′-GTGTGACCGACCGTGGTGC-3′) and P5′ (5′-TAATACGACTCACTATAGGTTACCAGCCTTCACTGC-3′), containing the T7 transcription promoter (underlined) were used for PCR amplification as described previously.35 Synthesis of the RNA library and transcription followed by DNase I (Roche) treatment have been described previously.65 The RNA pool was gel purified by denaturing (7 M urea) gel electrophoresis on an 8% polyacrylamide gel. The band corresponding to the RNA was visualized by UV shadowing and cut out for overnight extraction (500 mM NH4OAc, 1 mM EDTA, and 20% phenol) at 4°C. The RNAs were then purified by phenol-chloroform extraction and ethanol precipitation. Prior to each round, the RNA pool was heated at 80°C for 2 min, immediately cooled down on ice for 5 min, and then kept at room temperature (RT) for 10 min to allow formation of the optimal conformation in selection buffer composed of 1 mM MgCl2 and 0.5 mM CaCl2 in PBS (pH 7.4). For cell-SELEX, adherent cells at confluency were washed 3 times in PBS and 3 times in selection buffer before incubation with the starting RNA library (1 nmol) at 37°C for 30 min under slow agitation (75 rpm). Cells were then washed once for 5 min and detached with a cell scraper. Binding RNA molecules were detached from cells by heating at 95°C for 2 min. Eluted RNA pools were extracted by phenol-chloroform and ethanol precipitated. They were then amplified by reverse transcription prior amplification.65 The dsDNA pool was then transcribed as described above. For the next rounds of selection, the number of washes was modified compared with the first round as described in Table S1. From the fourth round to the sixth round, the RNA pool was first incubated with cells used for counterselection, and unbound sequences were then incubated with cells used for positive selection (Figure 1). A protein-based SELEX process was applied during rounds 8−10. The recombinant α5β1-Fc integrin was coupled to protein A-Sepharose 4B conjugate (Invitrogen), ahead washed 3 times with selection buffer. We employed a negative selection step in which the RNA pools were pre-incubated with protein A-Sepharose beads alone prior to positive selection. Unbound sequences were incubated under agitation on α5β1-Fc-loaded beads for 20 mins. Eluted RNA was recovered, reverse transcribed, PCR amplified, and transcribed back into RNA for the subsequent round as described above. For rounds 9 and 10, counterselection was also performed on negative control immunoglobulin G (IgG; cetuximab, Merck Serono). Beginning with round 11, another cell-SELEX process was performed as described above. Cells for counterselection and positive selection and SELEX conditions are described in Figure 1 and Table S1. After the 14th round of selection, competitor yeast tRNA was added (Table S1). At the end of SELEX, the sequences of the 18th pools were cloned with the TOPO-TA cloning kit (Invitrogen Life Technologies) before sequencing. The sequences were compared by MultAlin.66 Prediction of secondary structure was obtained using the mfold software.67
Flow Cytometry
Flow cytometry was performed with individual aptamers directly coupled to Cy5 at their 3′ end. For comparison of the binding profile of different aptamers to cells, aptamers were used at a final concentration of 500 nM. For determination of equilibrium binding affinities, aptamer H02 was used at concentrations of 0.15, 0.3, 0.6, 1.25, 2.5, and 5 μM. After detachment with EDTA (0.2 M), 300,000 cells were incubated for 30 min at 4°C with Cy5-labeled aptamers. As a control, cells were incubated with a 1/100 dilution of an anti-α5 antibody (mouse anti-human CD49e, IIA1 antibody, BD Chemigen) for 30 min, followed by 30-min incubation with Alexa 647-conjugated affine pure goat anti-mouse IgG (Jackson ImmunoResearch) at 10 μg/mL. After washing, cells were analyzed using a FACSCalibur flow cytometer (Becton Dickinson), and the mean fluorescence intensity (counting 10 000 events) was measured using Flowing software 2.5.1. For KD determination, experiments were repeated three times, and data were evaluated using GraphPad Prism (version 5.04).
SPR Analyses of Aptamer H02-Integrin Interactions on a CAP Sensor Chip
All experiments were performed on a Biacore T200 instrument (GE Healthcare) at 25°C. The sensor surface and other Biacore consumables were purchased from GE Healthcare. Integrins α5β1 and αvβ3 were from R&D Systems. Running buffer was composed of PBS (10 mM), NaCl (150 mM), and MgCl2 (1 mM), filtered through a 0.22-μm membrane, and supplemented with surfactant P20 (0.005% v/v). The biotin CAPture reagent, composed of streptavidin conjugated with an oligonucleotide, was stably hybridized to the complementary sequence of a CAP sensor chip following the biotin CAPture kit instructions (GE Healthcare). The biotinylated aptamer was denatured at 95°C for 3 min, incubated on ice, and then injected onto the biotin CAP reagent at 100 nM for 5 min at a flow rate of 10 μL/min. Five different concentrations of integrin (ranging from 8–130 nM) were injected into the flow cells at 10 or 30 μL/min for 300 s. Dissociation followed for 300 s. After each measurement, the sensor chip was washed with an injection of 6 M guanidine hydrochloride in 0.25 M NaOH as recommended by the manufacturer. The reference surface was treated similarly except that aptamer injection was omitted. Binding curves were double-reference-subtracted from the buffer blank and reference flow cell (without the aptamer). The equilibrium response was recorded 5 s before the end of integrin injection. The KD was determined by fitting the equilibrium response versus the [integrin] curve to a simple 1:1 interaction model with the Biacore T200 evaluation software (GE Healthcare).
Western Blot
Cells were lysed (1% Triton X-100, NaF [100 mmol/L], NaPPi [10 mmol/L], and Na3VO4 [1 mmol/L] in PBS, supplemented with complete anti-protease cocktail; Roche), and 10 μg of protein was separated by SDS-PAGE (Bio-Rad) and transferred to polyvinylidene fluoride (PVDF) membranes (Amersham). Blots were probed with antibodies to α5 integrin (H104, Santa Cruz Biotechnology) and to glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Millipore). Horseradish peroxidase (HRP)-coupled secondary antibodies were from Promega. Proteins were visualized with enhanced chemiluminescence using the LAS4000 imager, and densitometry analysis was performed using the ImageJ software (GE Healthcare). GAPDH was used as housekeeping protein to serve as the loading control for cell lysate samples. Analyses were performed on three independent experiments.
Fluorescence-Based Assays on Cell Lines
The adherent CHO-B2 and GBM cell lines (U87MG α5+, U87MG α5−, LN319, LN229, LN443, SF763, LN18, LNZ308, U373, and T98G) were plated on sterile glass slides for one night at 37°C in culture medium, washed three times, and then saturated for 1 h at RT in selection buffer containing 2% BSA. Cy5-labeled aptamers were denatured at 95°C for 3 min and incubated on ice for 5 min and then on cells in selection buffer for 30 min on ice or at 37°C at concentrations dependent on the assay (5, 2.5, 1.25, 0.6, or 0.3 μM). Cells were then washed in selection buffer, fixed for 8 min in 4% paraformaldehyde (PFA), permeabilized for 2 min with 0.2% Triton, and washed again. Sequentially, when immunocytochemistry was performed, the primary antibodies used were the anti-α5 antibody (mouse anti-human CD49e, IIA1 antibody, BD Chemigen, 1/200), the anti-β1 antibody (mouse anti-human CD29 antibody, clone TS2/16, BioLegend, 1/500), or the anti-EEA1 (early endosome antigen 1) antibody (anti-mouse clone 14/EEA1, BD Transduction Laboratories, 1/1,000). Primary antibodies were added for 1 h at RT or overnight (O/N) at 4°C, followed by a secondary antibody coupled to Alexa 546 or 568 (Life Technology) at a 2 μg/mL final concentration. Hoechst or DAPI were added at 1/1,000 for 1 h at RT to visualize the nucleus. F-actin identification was performed using Phalloidin-ATTO 488 (Sigma) at 1/4,000. Washing steps were performed before mounting using fluorescent mounting medium (S3023, Dako).
Fluorescence-Based Histochemical Assays of Patient-Derived Xenografts
Two patient-derived heterotopic xenografts (PDXs) were selected for in vivo analysis.68 TC7 and TC22 GBM-PDXs presented high and low levels of α5 integrins, respectively. PDX mouse models were established using tissues surgically removed from patients as described previously.69, 70 Integrin α5 was apta- and immunostained using formalin-fixed paraffin-embedded xenografts mounted on glass slides. Sections were deparaffinized, rehydrated, and subjected to an antigen unmasking protocol. Briefly, sections were boiled at 100°C for 10 min in target retrieval solution (pH 9) (S2367, Dako), cooled down to RT for 20–40 min, and rinsed in H2O. For aptafluorescence, slides were rinsed for 5 min in selection buffer, dried, incubated in blocking buffer (2% BSA in selection buffer) for 1 h at RT, rinsed in H2O and then in selection buffer, and dried. RNA molecules were denatured at 95°C for 3 min and incubated on ice for 5 min before dilution in selection buffer to a 1 μM final concentration. Aptamers were incubated on tumor sections for 1 h at RT in a humid chamber, washed in selection buffer, dried, fixed in 4% PFA, and then washed three times in PBS. For immunofluorescence, slides were rinsed for 5 min in PBS-T (0.1% Tween 20 in PBS), dried, and then incubated in blocking buffer BB-I (5% goat serum in PBS-T) for 1 h at RT in a humid chamber. Overnight incubation with anti-integrin α5 mAb 1928 (6B8516, Millipore, 1/200) in BB-I was followed by a 5-min wash in PBS-T and by an incubation step with a 1/100 dilution of the goat anti-rabbit secondary antibody coupled to Alexa Fluor 647 (A21245, Life Technologies). Immuno- and apta staining was followed by DAPI (10 μg/mL) staining for 30 min at RT to visualize cell nuclei. The stained samples were then washed in PBS, and coverslips were mounted onto tissue sections using fluorescent mounting medium (S3023, Dako).
Confocal Imaging
Images were acquired using a confocal microscope (Leica TCS SPE II, 63× magnification, oil immersion).
Mean fluorescence intensity on cells and tissues was measured using ImageJ software. Statistical analysis of data was performed with Student’s t test. Data were analyzed with GraphPad Prism version 5.04 and are represented as mean ± SEM.
Author Contributions
Research was performed by P.F., A.-M.R. (aptamer identification), E.C.D.S. (apta- and immuno-fluorescence), and M.-C.M., F.N., C.S. (flow cytometry). D.G. and I.L.-R. provided useful reagents. P.F., E.C.D.S., M.D., and L.C. analyzed the data. P.F., M.L., R.V., N.E.-S., S.M., and M.D. provided expertise. L.C. designed, coordinated, and performed research and wrote the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We thank Martin Humphries (Manchester, UK) for the gift of NS0 cells expressing α5β1-Fc integrin and Julie Hémart and Antoine Schillinger for technical assistance. This work was supported by two contracts from the SATT Conectus Alsace (I14-057/01 and I18-020/01), a grant from the PEPS-IDEX program of the Université de Strasbourg (W15RPE24), the Ligue contre le cancer CCIR-Est (S19R417C), and the INTEGLIO program of the Cancéropôle Grand-Est.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.05.006.
Supplemental Information
References
- 1.Stupp R., Mason W.P., van den Bent M.J., Weller M., Fisher B., Taphoorn M.J.B., Belanger K., Brandes A.A., Marosi C., Bogdahn U., European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups. National Cancer Institute of Canada Clinical Trials Group Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Woodworth G.F., Dunn G.P., Nance E.A., Hanes J., Brem H. Emerging insights into barriers to effective brain tumor therapeutics. Front. Oncol. 2014;4:126. doi: 10.3389/fonc.2014.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K., Ohgaki H., Wiestler O.D., Kleihues P., Ellison D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 4.Hung A.L., Garzon-Muvdi T., Lim M. Biomarkers and Immunotherapeutic Targets in Glioblastoma. World Neurosurg. 2017;102:494–506. doi: 10.1016/j.wneu.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 5.Cloughesy T.F., Cavenee W.K., Mischel P.S. Glioblastoma: from molecular pathology to targeted treatment. Annu. Rev. Pathol. 2014;9:1–25. doi: 10.1146/annurev-pathol-011110-130324. [DOI] [PubMed] [Google Scholar]
- 6.Wadajkar A.S., Dancy J.G., Hersh D.S., Anastasiadis P., Tran N.L., Woodworth G.F., Winkles J.A., Kim A.J. Tumor-targeted nanotherapeutics: overcoming treatment barriers for glioblastoma. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2017 doi: 10.1002/wnan.1439. Published online November 4, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacobi N., Seeboeck R., Hofmann E., Eger A. ErbB Family Signalling: A Paradigm for Oncogene Addiction and Personalized Oncology. Cancers (Basel) 2017;9:33. doi: 10.3390/cancers9040033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciriello G., Miller M.L., Aksoy B.A., Senbabaoglu Y., Schultz N., Sander C. Emerging landscape of oncogenic signatures across human cancers. Nat. Genet. 2013;45:1127–1133. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen M., Yu Y., Jiang F., Zhou J., Li Y., Liang C., Dang L., Lu A., Zhang G. Development of Cell-SELEX Technology and Its Application in Cancer Diagnosis and Therapy. Int. J. Mol. Sci. 2016;17:E2079. doi: 10.3390/ijms17122079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoon S., Rossi J.J. Emerging cancer-specific therapeutic aptamers. Curr. Opin. Oncol. 2017;29:366–374. doi: 10.1097/CCO.0000000000000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pastor F. Aptamers: A New Technological Platform in Cancer Immunotherapy. Pharmaceuticals (Basel) 2016;9:E64. doi: 10.3390/ph9040064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu G., Chen X. Aptamer-based targeted therapy. Adv. Drug Deliv. Rev. 2018;134:65–78. doi: 10.1016/j.addr.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pastor F., Berraondo P., Etxeberria I., Frederick J., Sahin U., Gilboa E., Melero I. An RNA toolbox for cancer immunotherapy. Nat. Rev. Drug Discov. 2018;17:751–767. doi: 10.1038/nrd.2018.132. [DOI] [PubMed] [Google Scholar]
- 14.Zhou J., Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 2017;16:181–202. doi: 10.1038/nrd.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaur H., Bruno J.G., Kumar A., Sharma T.K. Aptamers in the Therapeutics and Diagnostics Pipelines. Theranostics. 2018;8:4016–4032. doi: 10.7150/thno.25958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruiz Ciancio D., Vargas M.R., Thiel W.H., Bruno M.A., Giangrande P.H., Mestre M.B. Aptamers as Diagnostic Tools in Cancer. Pharmaceuticals (Basel) 2018;11:86. doi: 10.3390/ph11030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bouvier-Müller A., Ducongé F. Application of aptamers for in vivo molecular imaging and theranostics. Adv. Drug Deliv. Rev. 2018;134:94–106. doi: 10.1016/j.addr.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Kim M., Kim D.M., Kim K.S., Jung W., Kim D.E. Applications of Cancer Cell-Specific Aptamers in Targeted Delivery of Anticancer Therapeutic Agents. Molecules. 2018;23:830. doi: 10.3390/molecules23040830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 20.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 21.Robertson D.L., Joyce G.F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature. 1990;344:467–468. doi: 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]
- 22.Mercier M.-C., Dontenwill M., Choulier L. Selection of Nucleic Acid Aptamers Targeting Tumor Cell-Surface Protein Biomarkers. Cancers (Basel) 2017;9:E69. doi: 10.3390/cancers9060069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hynes R.O. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 24.Desgrosellier J.S., Cheresh D.A. Integrins in cancer: biological implications and therapeutic opportunities. Nat. Rev. Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaffner F., Ray A.M., Dontenwill M. Integrin α5β1, the Fibronectin Receptor, as a Pertinent Therapeutic Target in Solid Tumors. Cancers (Basel) 2013;5:27–47. doi: 10.3390/cancers5010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janouskova H., Maglott A., Leger D.Y., Bossert C., Noulet F., Guerin E., Guenot D., Pinel S., Chastagner P., Plenat F. Integrin α5β1 plays a critical role in resistance to temozolomide by interfering with the p53 pathway in high-grade glioma. Cancer Res. 2012;72:3463–3470. doi: 10.1158/0008-5472.CAN-11-4199. [DOI] [PubMed] [Google Scholar]
- 27.Maglott A., Bartik P., Cosgun S., Klotz P., Rondé P., Fuhrmann G., Takeda K., Martin S., Dontenwill M. The small α5β1 integrin antagonist, SJ749, reduces proliferation and clonogenicity of human astrocytoma cells. Cancer Res. 2006;66:6002–6007. doi: 10.1158/0008-5472.CAN-05-4105. [DOI] [PubMed] [Google Scholar]
- 28.Martin S., Cosset E.C., Terrand J., Maglott A., Takeda K., Dontenwill M. Caveolin-1 regulates glioblastoma aggressiveness through the control of α(5)β(1) integrin expression and modulates glioblastoma responsiveness to SJ749, an α(5)β(1) integrin antagonist. Biochim. Biophys. Acta. 2009;1793:354–367. doi: 10.1016/j.bbamcr.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 29.Freije W.A., Castro-Vargas F.E., Fang Z., Horvath S., Cloughesy T., Liau L.M., Mischel P.S., Nelson S.F. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64:6503–6510. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- 30.Phillips H.S., Kharbanda S., Chen R., Forrest W.F., Soriano R.H., Wu T.D., Misra A., Nigro J.M., Colman H., Soroceanu L. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 31.Riemenschneider M.J., Mueller W., Betensky R.A., Mohapatra G., Louis D.N. In situ analysis of integrin and growth factor receptor signaling pathways in human glioblastomas suggests overlapping relationships with focal adhesion kinase activation. Am. J. Pathol. 2005;167:1379–1387. doi: 10.1016/S0002-9440(10)61225-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cosset E.C., Godet J., Entz-Werlé N., Guérin E., Guenot D., Froelich S., Bonnet D., Pinel S., Plenat F., Chastagner P. Involvement of the TGFβ pathway in the regulation of α5 β1 integrins by caveolin-1 in human glioblastoma. Int. J. Cancer. 2012;131:601–611. doi: 10.1002/ijc.26415. [DOI] [PubMed] [Google Scholar]
- 33.Coe A.P., Askari J.A., Kline A.D., Robinson M.K., Kirby H., Stephens P.E., Humphries M.J. Generation of a minimal alpha5beta1 integrin-Fc fragment. J. Biol. Chem. 2001;276:35854–35866. doi: 10.1074/jbc.M103639200. [DOI] [PubMed] [Google Scholar]
- 34.Schreiner C.L., Bauer J.S., Danilov Y.N., Hussein S., Sczekan M.M., Juliano R.L. Isolation and characterization of Chinese hamster ovary cell variants deficient in the expression of fibronectin receptor. J. Cell Biol. 1989;109:3157–3167. doi: 10.1083/jcb.109.6.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Da Rocha Gomes S., Miguel J., Azéma L., Eimer S., Ries C., Dausse E., Loiseau H., Allard M., Toulmé J.J. (99m)Tc-MAG3-aptamer for imaging human tumors associated with high level of matrix metalloprotease-9. Bioconjug. Chem. 2012;23:2192–2200. doi: 10.1021/bc300146c. [DOI] [PubMed] [Google Scholar]
- 36.Cibiel A., Quang N.N., Gombert K., Thézé B., Garofalakis A., Ducongé F. From ugly duckling to swan: unexpected identification from cell-SELEX of an anti-Annexin A2 aptamer targeting tumors. PLoS ONE. 2014;9:e87002. doi: 10.1371/journal.pone.0087002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu L., Zhang Z., Zhao Z., Liu Q., Tan W., Fang X. Cellular Internalization and Cytotoxicity of Aptamers Selected from Lung Cancer Cell. Am. J. Biomed. Sci. 2013;5:47–58. [Google Scholar]
- 38.Suenaga E., Mizuno H., Penmetcha K.K.R. Monitoring influenza hemagglutinin and glycan interactions using surface plasmon resonance. Biosens. Bioelectron. 2012;32:195–201. doi: 10.1016/j.bios.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 39.Basiji D.A., Ortyn W.E., Liang L., Venkatachalam V., Morrissey P. Cellular image analysis and imaging by flow cytometry. Clin. Lab. Med. 2007;27:653–670, viii. doi: 10.1016/j.cll.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bauer M., Macdonald J., Henri J., Duan W., Shigdar S. The Application of Aptamers for Immunohistochemistry. Nucleic Acid Ther. 2016;26:120–126. doi: 10.1089/nat.2015.0569. [DOI] [PubMed] [Google Scholar]
- 41.Szopa W., Burley T.A., Kramer-Marek G., Kaspera W. Diagnostic and Therapeutic Biomarkers in Glioblastoma: Current Status and Future Perspectives. BioMed Res. Int. 2017;2017:8013575. doi: 10.1155/2017/8013575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y., Kuan C.-T., Mi J., Zhang X., Clary B.M., Bigner D.D., Sullenger B.A. Aptamers selected against the unglycosylated EGFRvIII ectodomain and delivered intracellularly reduce membrane-bound EGFRvIII and induce apoptosis. Biol. Chem. 2009;390:137–144. doi: 10.1515/BC.2009.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chauveau F., Aissouni Y., Hamm J., Boutin H., Libri D., Ducongé F., Tavitian B. Binding of an aptamer to the N-terminal fragment of VCAM-1. Bioorg. Med. Chem. Lett. 2007;17:6119–6122. doi: 10.1016/j.bmcl.2007.09.046. [DOI] [PubMed] [Google Scholar]
- 44.Ohuchi S. Cell-SELEX Technology. BioRes. 2012;1:265–272. doi: 10.1089/biores.2012.0253. Open Access. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blind M., Kolanus W., Famulok M. Cytoplasmic RNA modulators of an inside-out signal-transduction cascade. Proc. Natl. Acad. Sci. USA. 1999;96:3606–3610. doi: 10.1073/pnas.96.7.3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mi J., Zhang X., Giangrande P.H., McNamara J.O., 2nd, Nimjee S.M., Sarraf-Yazdi S., Sullenger B.A., Clary B.M. Targeted inhibition of alphavbeta3 integrin with an RNA aptamer impairs endothelial cell growth and survival. Biochem. Biophys. Res. Commun. 2005;338:956–963. doi: 10.1016/j.bbrc.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 47.Ruckman, J., Gold, L., Stephens, A., and Janjic, N. (2001). Nucleic acid ligands to integrins. US patent US7094535 B2, filed December 18, 2001, and granted August 22, 2006.
- 48.Gong Q., Wang J., Ahmad K.M., Csordas A.T., Zhou J., Nie J., Stewart R., Thomson J.A., Rossi J.J., Soh H.T. Selection strategy to generate aptamer pairs that bind to distinct sites on protein targets. Anal. Chem. 2012;84:5365–5371. doi: 10.1021/ac300873p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takahashi M., Sakota E., Nakamura Y. The efficient cell-SELEX strategy, Icell-SELEX, using isogenic cell lines for selection and counter-selection to generate RNA aptamers to cell surface proteins. Biochimie. 2016;131:77–84. doi: 10.1016/j.biochi.2016.09.018. [DOI] [PubMed] [Google Scholar]
- 50.Pestourie C., Cerchia L., Gombert K., Aissouni Y., Boulay J., De Franciscis V., Libri D., Tavitian B., Ducongé F. Comparison of different strategies to select aptamers against a transmembrane protein target. Oligonucleotides. 2006;16:323–335. doi: 10.1089/oli.2006.16.323. [DOI] [PubMed] [Google Scholar]
- 51.Zhu G., Zhang H., Jacobson O., Wang Z., Chen H., Yang X., Niu G., Chen X. Combinatorial Screening of DNA Aptamers for Molecular Imaging of HER2 in Cancer. Bioconjug. Chem. 2017;28:1068–1075. doi: 10.1021/acs.bioconjchem.6b00746. [DOI] [PubMed] [Google Scholar]
- 52.Berg K., Lange T., Mittelberger F., Schumacher U., Hahn U. Selection and Characterization of an α6β4 Integrin blocking DNA Aptamer. Mol. Ther. Nucleic Acids. 2016;5:e294. doi: 10.1038/mtna.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilner S.E., Wengerter B., Maier K., de Lourdes Borba Magalhães M., Del Amo D.S., Pai S., Opazo F., Rizzoli S.O., Yan A., Levy M. An RNA alternative to human transferrin: a new tool for targeting human cells. Mol. Ther. Nucleic Acids. 2012;1:e21. doi: 10.1038/mtna.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boltz A., Piater B., Toleikis L., Guenther R., Kolmar H., Hock B. Bi-specific aptamers mediating tumor cell lysis. J. Biol. Chem. 2011;286:21896–21905. doi: 10.1074/jbc.M111.238261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pratico E.D., Sullenger B.A., Nair S.K. Identification and characterization of an agonistic aptamer against the T cell costimulatory receptor, OX40. Nucleic Acid Ther. 2013;23:35–43. doi: 10.1089/nat.2012.0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zamay G.S., Ivanchenko T.I., Zamay T.N., Grigorieva V.L., Glazyrin Y.E., Kolovskaya O.S., Garanzha I.V., Barinov A.A., Krat A.V., Mironov G.G. DNA Aptamers for the Characterization of Histological Structure of Lung Adenocarcinoma. Mol. Ther. Nucleic Acids. 2017;6:150–162. doi: 10.1016/j.omtn.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pu Y., Liu Z., Lu Y., Yuan P., Liu J., Yu B., Wang G., Yang C.J., Liu H., Tan W. Using DNA aptamer probe for immunostaining of cancer frozen tissues. Anal. Chem. 2015;87:1919–1924. doi: 10.1021/ac504175h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zeng Z., Zhang P., Zhao N., Sheehan A.M., Tung C.-H., Chang C.-C., Zu Y. Using oligonucleotide aptamer probes for immunostaining of formalin-fixed and paraffin-embedded tissues. Mod. Pathol. 2010;23:1553–1558. doi: 10.1038/modpathol.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blom S., Paavolainen L., Bychkov D., Turkki R., Mäki-Teeri P., Hemmes A., Välimäki K., Lundin J., Kallioniemi O., Pellinen T. Systems pathology by multiplexed immunohistochemistry and whole-slide digital image analysis. Sci. Rep. 2017;7:15580. doi: 10.1038/s41598-017-15798-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bukari B.A., Citartan M., Ch’ng E.S., Bilibana M.P., Rozhdestvensky T., Tang T.-H. Aptahistochemistry in diagnostic pathology: technical scrutiny and feasibility. Histochem. Cell Biol. 2017;147:545–553. doi: 10.1007/s00418-017-1561-9. [DOI] [PubMed] [Google Scholar]
- 61.Xiang D., Zheng C., Zhou S.-F., Qiao S., Tran P.H.-L., Pu C., Li Y., Kong L., Kouzani A.Z., Lin J. Superior Performance of Aptamer in Tumor Penetration over Antibody: Implication of Aptamer-Based Theranostics in Solid Tumors. Theranostics. 2015;5:1083–1097. doi: 10.7150/thno.11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun H., Tan W., Zu Y. Aptamers: versatile molecular recognition probes for cancer detection. Analyst (Lond.) 2016;141:403–415. doi: 10.1039/c5an01995h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang T., Chen C., Larcher L.M., Barrero R.A., Veedu R.N. Three decades of nucleic acid aptamer technologies: Lessons learned, progress and opportunities on aptamer development. Biotechnol. Adv. 2019;37:28–50. doi: 10.1016/j.biotechadv.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 64.Hahn U. Charomers-Interleukin-6 Receptor Specific Aptamers for Cellular Internalization and Targeted Drug Delivery. Int. J. Mol. Sci. 2017;18:E2641. doi: 10.3390/ijms18122641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dausse E., Cazenave C., Rayner B., Toulmé J.-J. In vitro selection procedures for identifying DNA and RNA aptamers targeted to nucleic acids and proteins. Methods Mol. Biol. 2005;288:391–410. doi: 10.1385/1-59259-823-4:391. [DOI] [PubMed] [Google Scholar]
- 66.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blandin A.-F., Noulet F., Renner G., Mercier M.-C., Choulier L., Vauchelles R., Ronde P., Carreiras F., Etienne-Selloum N., Vereb G. Glioma cell dispersion is driven by α5 integrin-mediated cell-matrix and cell-cell interactions. Cancer Lett. 2016;376:328–338. doi: 10.1016/j.canlet.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 69.Leuraud P., Taillandier L., Medioni J., Aguirre-Cruz L., Crinière E., Marie Y., Kujas M., Golmard J.L., Duprez A., Delattre J.Y. Distinct responses of xenografted gliomas to different alkylating agents are related to histology and genetic alterations. Cancer Res. 2004;64:4648–4653. doi: 10.1158/0008-5472.CAN-03-3429. [DOI] [PubMed] [Google Scholar]
- 70.Pinel S., Mriouah J., Vandamme M., Chateau A., Plénat F., Guérin E., Taillandier L., Bernier-Chastagner V., Merlin J.L., Chastagner P. Synergistic antitumor effect between gefitinib and fractionated irradiation in anaplastic oligodendrogliomas cannot be predicted by the Egfr signaling activity. PLoS ONE. 2013;8:e68333. doi: 10.1371/journal.pone.0068333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
After aptamer labeling, cells were fixed, permeabilized and then labeled with the anti-EEA1 antibody labeling early endosomes. The 3.58 μm stack (12 images) was acquired by confocal microscopy. 3D animation was performed by Imaris 92.1 (bitplane). First all channels then only early endosomes and aptamers are displayed in maximum intensity projection. Finally these two channels are shown in normal shading rendering. DAPI staining in shown blue, actin labeling in green, EEA1 labeling in red and the Cy5-coupled aptamer H02 labeling in white.