Abstract
The maternal kidneys undergo numerous physiological changes during pregnancy to maintain a healthy pregnancy for mother and child. Over the past decade, interest in microRNAs (miRNAs) for regulating gene expression during pregnancy has expanded. However, the role of miRNAs in modulating kidney physiology during pregnancy has not been extensively investigated. In this study, miRNome profiling suggested differential expression of 163 miRNAs (of 887 miRNAs detected) in the kidneys from pregnant mice at 6.5 days gestation when compared to non-pregnant female mice, of which 35 and 128 miRNAs were potentially down- and up-regulated, respectively. We performed network and pathway analyses of the >1700 potential mRNA targets of the differentially expressed miRNAs using MiRNet, Gene Ontology, Reactome and KEGG analyses. The mRNA targets were over-represented in numerous cellular signalling pathways, including cellular protective responses. In addition, we explored 13 and 29 potential differentially expressed miRNAs to have putative binding sites in the Slc13a1 and Slc26a1 sulfate transporter mRNAs, respectively, and that decreased levels of mir-466k may potentially explain the increased expression of these sulfate transporters in early mouse gestation. Collectively, this study suggests altered expression levels of miRNAs during mouse gestation, which provides pilot data for future investigations into the molecular events that modulate kidney adaptsation to pregnancy.
Keywords: miRNA, miRNome, Renal, Pregnancy, Sulfate, Mouse, Adaptation to pregnancy
1. Introduction
During pregnancy, the maternal kidneys undergo numerous physiological changes, including increased glomerular filtration rate, renal plasma flow and kidney volume, as well as altered renal tubular handling of nutrients (glucose and amino acids) and wastes such as uric acid [1]. These changes in kidney haemodynamic, reabsorption and excretion functions are important for maintaining a healthy pregnancy for mother and child. Animal studies also show a protective effect of pregnancy on the kidney from existing kidney dysfunction and acute injury caused by ischemia [2]. Investigations into the mechanisms of kidney adaptation to pregnancy have mostly focused on the physiological responses of the kidneys to changes in circulating hormone levels, including relaxin, progesterone, estradiols, and the renin-angiotensin-aldosterone system [[3], [4], [5], [6]]. However, our knowledge of kidney adaptation at the gene expression level is less well advanced, as it is not feasible to sample kidney tissue from healthy pregnancies. As an approach to further understand these molecular processes, we investigated the kidneys of pregnant and non-pregnant female mice to explore potential differentially expressed microRNAs (miRNAs), which are small (≈22 nucleotides) non-coding regulatory RNAs involved in gene expression at both transcriptional and translational levels [7]. This approach provides pilot data for future investigations of the molecular pathways associated with kidney adaptation to pregnancy. In the present study, we initially focussed on miRNAs that potentially explain enhanced sulfate reabsorption during pregnancy.
Sulfate is an obligate nutrient for fetal growth and development, and is supplied from maternal circulation to the fetus via the placenta [8]. During gestation, maternal circulating sulfate levels increase by approximately 2-fold to meet the high fetal demands for sulfate [9,10]. The increased sulfate level in maternal circulation is due to enhanced renal reabsorption of sulfate in the pregnant mother, which is mediated by the SLC13A1 and SLC26A1 sulfate transporters [11,12]. SLC13A1 is a sodium-dependent sulfate co-transporter expressed on the apical membrane of renal epithelial cells in the proximal tubule where it mediates the first step of sulfate reabsorption, and the second step is mediated by the SLC26A1 sulfate-anion exchanger on the basolateral membrane [13]. Targeted disruption of either the Slc13a1 or Slc26a1 genes in mice leads to renal sulfate wasting and hyposulfataemia [14,15]. More recent studies showed that pregnant female Slc13a1 null mice exhibit hyposulfataemia and late gestational fetal demise, highlighting the importance of maintaining high maternal sulfate levels throughout pregnancy [10]. These findings in mice may be clinically relevant when considering the complete or partial loss-of-function variants reported in the human SLC13A1 and SLC26A1 genes that are associated with renal sulfate wasting and decreased circulating sulfate level [[16], [17], [18]]. However, the consequences of disrupted maternal kidney SLC13A1 and SLC26A1 on human fetal outcomes awaits further investigation.
Maternal circulating sulfate levels increase with a concomitant decrease in renal sulfate excretion in the first trimester of both human and mouse pregnancy [9,10]. In female mice, pregnancy-dependent increases in plasma sulfate level were found to be associated with 2-fold increased levels of Slc13a1 and Slc26a1 mRNA from 4 to 6 days of pregnancy [12]. The transcriptional regulation of Slc13a1 and Slc26a1 was initially investigated using in vitro cell culture assays to characterise putative transcription factor binding motifs in their 5′-flanking regions [19,20]. Those studies identified a thyroid hormone responsive element in the 5′-flanking sequences of Slc13a1 and Slc26a1, as well as a vitamin D responsive element in the Slc13a1 promoter, suggesting hormonal regulation of these genes and renal sulfate reabsorption [[21], [22], [23]]. Indeed, this was found to be the case with hypothyroidism or vitamin D deficiency in rats leading to renal sulfate wasting and decreased plasma sulfate level [24,25]. More recent studies investigated the potential contribution of ovarian hormones to enhanced expression of Slc13a1 and Slc26a1 in pregnancy using ovariectomized mice treated with estrogen (E2) and/or progesterone (P4) for 3 days [12]. However, those treatments neither increased renal Slc13a1 and Slc26a1 mRNA levels, nor recapitulated the pregnancy-induced increase in circulating sulfate level, suggesting that E2 and/or P4 do not regulate the expression of these sulfate transporters. Currently, the mechanism leading to increased expression of Slc13a1 and Slc26a1 in the maternal kidneys during pregnancy is unknown.
In recent years, interest in miRNAs as regulators of gene expression during pregnancy has expanded. Those studies have focussed on the role of miRNAs in endometrial receptivity, implantation, immune cells associated with maternal tolerance to the fetus, placentation and labour, as well as pregnancy-related pathologies including preeclampsia, fetal growth restriction and spontaneous fetal loss [26,27]. However, the role of miRNAs in the maternal kidneys during pregnancy for regulating renal physiology, including increased sulfate reabsorption, has not been reported.
In this pilot study, we explored the kidney profile of 940 miRNAs between pregnant and non-pregnant female mice. Our findings suggest differential expression of 163 miRNAs, including 13 down-regulated and 29 up-regulated miRNAs that have putative binding sites in Slc13a1 and Slc26a1 mRNAs.
2. Materials and methods
2.1. Animals and tissues
Male and virgin female CD1 mice were housed together overnight, and the presence of a copulatory plug the following morning indicated successful mating that was designated embryonic day 0.5 (E0.5). Pregnant female mice were singly housed in conventional conditions and fed a standard rodent chow and water ad libitum as previously described [12]. Non-pregnant diestrus (n = 3) and pregnant (n = 3) female mice at gestational day 6.5 (E6.5) were euthanized and then kidneys collected at 0900 h for RNA isolation [28]. We selected E6.5 as the gestational age because maternal kidney Slc13a1 and Slc26a1 mRNA levels increase at this time point in pregnancy [12], and the present study explored the potential involvement of miRNAs in regulating the mRNA levels of these 2 genes. All procedures were approved by the University of Queensland Animal Ethics Committee, and were performed in accordance with relevant guidelines and regulations.
2.2. Kidney miRNome profiling
Total RNA was isolated from whole kidneys using TRIzol® (Invitrogen) reagent and RNeasy mini spin columns (QIAGEN) according to the manufacturer's protocol. 500 ng of pooled RNA samples from the non-pregnant and pregnant mice (n = 3 each) were used for reverse transcription of total RNA (miScript II RT Kit, QIAGEN). First strand cDNA quality was assessed using a miScript miRNA QC PCR Array (QIAGEN). A miRNome miScript miRNA PCR Array (QIAGEN) containing primers for 940 miRNAs was used to amplify the kidney miRNome of non-pregnant and pregnant mice using a miScript SYBR green PCR Kit with a ViiA™ 7 Real-Time PCR System (Applied Biosystems). The thermal cycling protocol was: 15 min at 95 °C; and 40 cycles of 94 °C for 15 s, 55 °C for 30 s and 70 °C for 30 s. Sample quality control and data were analyzed using the manufacturer's online analysis software (http://www.qiagen.com/au/shop/genes-and-pathways/data-analysis-center-overview-page/). Raw data are available in 6 supplemental files upon request.
2.3. Identification of miRNA-target genes and gene networks
All miRNAs that showed potential differential expression ≥ or ≤ 2-fold in the kidneys of pregnant mice, together with their target coding genes known to be expressed in the mouse kidney [29], were functionally annotated via the following web-enabled tools: MiRNet (http://www.mirnet.ca/) for miRNA target gene network analysis [30]; Gene Ontology (http://www.geneontology.org/) for molecular function (MF) enrichment analysis; Reactome (http://www.reactome.org/) and KEGG (http://www.genome.jp/kegg/) for miRNA target gene pathway analysis [31]. To identify the mRNA targets of all miRNAs that had potential differential expression ≥ or ≤ 2-fold in the kidneys of pregnant mice, we used the miRNET validated gene-miRNA interaction information retrieval system. This search identified a total of 910- and 2054- genes targeted by 35 down-regulated and 128 up-regulated miRNAs, respectively. From this list of 2964 genes, we then manually filtered those genes known to be expressed in the mouse kidney [29], narrowing down to 357 and 899 target genes, respectively, for further miRNA target gene network analysis using miRNET. Genes that are incorporated in the network were further analyzed with Gene Ontology, Reactome and KEGG using a hypergeometric test after adjustment for false discovery rate (FDR) to provide a ranked representation of the most “enriched” molecular functions and biological pathways.
2.4. Identification of putative miRNA binding sites in Slc13a1 and Slc26a1 mRNA sequences
We determined whether any of the potential differentially expressed miRNAs have putative binding sites in the 5’-UTR, coding sequence and 3’-UTR of Slc13a1 and Slc26a1, using online miRNA tools: miRWalk2.0 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/index.html) [32]. To undertake a comparative analysis of putative miRNA binding sites in Slc13a1 and Slc26a1 mRNA sequences, three additional miRNA target prediction programs were used: miRanda (http://www.microrna.org/microrna/home.do) [33], Targetscan (http://www.targetscan.org/vert_71/) [34] and RNA22 (https://cm.jefferson.edu/rna22/Interactive/) [35]. Each of these tools vary in their algorithms and parameters used to predict miRNA targets [36] and we included all of the putative miRNA binding sites identified by at least 2 prediction tools to allow comprehensive coverage.
3. Results and discussion
3.1. Kidney miRNome profile of pregnant mice
Over the past decade, numerous studies have shown the involvement of miRNAs in regulating renal development and pathology [37]. Those findings demonstrate the importance of miRNAs in maintaining kidney homeostasis, suggesting that miRNAs may be involved in the numerous physiological changes that occur in the maternal kidneys during pregnancy, such as increased kidney length and volume, increased glomerular filtration rate (GFR), decreased fractional reabsorption of glucose and amino acids, and increased sulfate reabsorption [1,38]. However, the role of miRNAs in maternal kidney physiology during pregnancy has not been extensively researched. In this study, we explored 163 miRNAs (of 887 miRNAs detected) that showed potential differential expression (≥ ±2-fold change) in the kidneys of pregnant mice at 6.5 days gestation when compared to non-pregnant female mice (Fig. 1).
Fig. 1.

Comparison of miRNA levels in the kidneys of pregnant and non-pregnant mice. A total of 887 miRNAs were detected in the kidneys of pregnant and non-pregnant mice. Of the 887 mature miRNAs detected, 163 miRNAs were found to have potential differential expression (≥ ± 2-fold change, outside dotted line); 128 upregulated (red), 35 downregulated (blue), and 724 miRNAs remained unchanged in pregnancy (black). Plot was generated using commercially available data analysis software (QIAGEN, Data analysis centre). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Interestingly, the most up-regulated and down-regulated miRNAs identified in this pilot study have known responses to kidney injury, suggesting a potential protective role for altered miRNA expression as kidney physiology adapts to pregnancy. We found increased levels (by ≥2-fold) of 128 miRNAs, including 3 miRNAs (miR-342-5p, −494-5p and -216a-5p) that are potentially ≥7-times more abundant in the kidneys of pregnant mice (Table 1). Previous studies have shown increased kidney levels of these 3 miRNAs in response to acute tubular necrosis, ischemia-reperfusion injury and diabetic nephropathy, respectively [[39], [40], [41]]. We also explored 35 down-regulated miRNAs, including miR-384-5p which showed the greatest decrease by 13.5-fold (Table 1). In recent years, miR-384-5p has attracted attention in health and pathology, including its role in enhanced renal fibrosis after renal injury by decreasing BMP7 protein levels [42]. That study also demonstrated that suppression of miR-384-5p attenuates the severity of renal injury in mice, suggesting a protective response for the down-regulation of this miRNA (i.e. enabling BMP7 to preserve renal function).
Table 1.
miRNAs with potential differential expression in the kidneys of pregnant mice.
| aID | bChange | aID | bChange | aID | bChange | aID | bChange |
|---|---|---|---|---|---|---|---|
| Down-regulated | |||||||
| miR-384-5p | −13.5 | miR-411-3p | −3.2 | miR-672-3p | −2.7 | miR-291a-3p | −2.2 |
| miR-669e-3p | −5.2 | miR-302d-3p | −3.1 | *miR-452-5p | −2.7 | miR-101b-5p | −2.2 |
| miR-873a-3p | −4.8 | miR-3077-3p | −3.1 | miR-152-5p | −2.5 | miR-135b-5p | −2.1 |
| miR-154-3p | −4.5 | *miR-300-5p | −3.1 | miR-32-5p | −2.5 | miR-154-5p | −2.1 |
| miR-153-3p | −4.5 | miR-1190 | −3.1 | #miR-3106-3p | −2.5 | miR-300-3p | −2.1 |
| #miR-190b-3p | −4.4 | miR-421-5p | −3.0 | *miR-449c-5p | −2.5 | miR-376a-3p | −2.1 |
| *miR-3098-3p | −4.3 | *#miR-466k | −2.9 | miR-679-3p | −2.4 | miR-1b-5p | −2.1 |
| miR-3099-3p | −3.9 | *miR-539-5p | −2.7 | #miR-3086-3p | −2.3 | miR-3065-5p | −2.0 |
| #miR-409-5p | −3.9 | miR-302a-5p | −2.7 | miR-291b-3p | −2.3 | ||
| Up-regulated | |||||||
| #miR-342-5p | 7.5 | miR-1943-5p | 3.5 | *miR-3074-1-3p | 2.7 | miR-153-5p | 2.3 |
| miR-494-5p | 7.2 | *miR-1960 | 3.5 | miR-1947-3p | 2.7 | miR-379-5p | 2.3 |
| miR-216a-5p | 7.0 | miR-3071-5p | 3.3 | miR-654-5p | 2.6 | miR-760-5p | 2.2 |
| #miR-331-5p | 6.6 | miR-204-3p | 3.3 | miR-1968-5p | 2.6 | miR-1957a | 2.2 |
| miR-182-3p | 6.1 | miR-1930-3p | 3.3 | miR-3089-5p | 2.6 | miR-1948-3p | 2.2 |
| #miR-3082-3p | 5.5 | #miR-201-3p | 3.2 | miR-3066-3p | 2.6 | miR-344e-3p | 2.2 |
| miR-370-5p | 5.4 | miR-3067-5p | 3.2 | miR-431-3p | 2.6 | miR-1192 | 2.2 |
| miR-377-5p | 5.3 | miR-669 h-3p | 3.2 | *miR-1928 | 2.6 | #miR-34c-3p | 2.2 |
| miR-290a-3p | 5.2 | miR-96-3p | 3.1 | miR-3098-5p | 2.6 | miR-551b-3p | 2.2 |
| miR-3071-3p | 5.1 | miR-3472 | 3.1 | miR-148b-5p | 2.6 | #miR-3075-5p | 2.2 |
| miR-412-3p | 5.1 | miR-491-3p | 3.1 | miR-701-3p | 2.5 | miR-211-3p | 2.2 |
| miR-365-1-5p | 5.1 | #miR-92a-2-5p | 3.1 | #miR-693-5p | 2.5 | #miR-1b-3p | 2.1 |
| #miR-1953 | 4.9 | miR-434-5p | 3.1 | #miR-764-5p | 2.5 | miR-467 g | 2.1 |
| miR-1194 | 4.4 | miR-3063-5p | 3.0 | #miR-540-5p | 2.4 | miR-3102-5p | 2.1 |
| miR-668-3p | 4.3 | miR-878-5p | 3.0 | miR-1900 | 2.4 | miR-486b-5p | 2.1 |
| miR-294-3p | 4.3 | #miR-682 | 3.0 | miR-3095-3p | 2.4 | miR-713 | 2.1 |
| miR-344f-3p | 4.3 | miR-195a-3p | 3.0 | miR-1941-5p | 2.4 | miR-212-5p | 2.1 |
| miR-675-3p | 4.2 | #miR-1198-3p | 3.0 | #miR-1906 | 2.4 | miR-1951 | 2.1 |
| miR-1964-5p | 4.2 | #miR-679-5p | 3.0 | *miR-598-5p | 2.4 | let-7a-2-3p | 2.1 |
| #miR-3112-3p | 4.1 | miR-758-3p | 2.9 | miR-298-3p | 2.4 | miR-1198-5p | 2.1 |
| miR-468-3p | 4.1 | miR-410-5p | 2.9 | miR-466 h-5p | 2.3 | miR-543-5p | 2.1 |
| miR-3101-3p | 4.0 | miR-871-3p | 2.9 | *miR-369-3p | 2.3 | miR-511-5p | 2.1 |
| miR-3093-5p | 3.9 | #miR-758-5p | 2.9 | miR-695 | 2.3 | miR-7a-2-3p | 2.1 |
| miR-881-3p | 3.9 | miR-205-3p | 2.9 | *miR-1952 | 2.3 | miR-3087-3p | 2.1 |
| miR-193b-5p | 3.9 | miR-3070b-5p | 2.8 | miR-379-3p | 2.3 | miR-106a-3p | 2.1 |
| miR-128-1-5p | 3.9 | miR-3101-5p | 2.8 | miR-3110-3p | 2.3 | miR-486a-3p | 2.1 |
| miR-299b-5p | 3.7 | miR-3088-5p | 2.8 | miR-708-3p | 2.3 | #miR-3102-3p | 2.0 |
| miR-3090-3p | 3.7 | miR-1971 | 2.7 | #miR-3075-3p | 2.3 | miR-1247-3p | 2.0 |
| miR-3104-5p | 3.6 | miR-133b-3p | 2.7 | #miR-432 | 2.3 | #miR-667-5p | 2.0 |
| miR-208a-3p | 3.6 | miR-361-3p | 2.7 | miR-666-5p | 2.3 | miR-3094-3p | 2.0 |
| miR-486b-3p | 3.5 | miR-295-5p | 2.7 | miR-875-3p | 2.3 | miR-3066-5p | 2.0 |
| #miR-337-3p | 3.5 | *miR-466 l-3p | 2.7 | #miR-3474 | 2.3 | miR-1962 | 2.0 |
ID, miRNA names have the prefix mmu- to indicate Mus musculus sequence, with nomenclature based on the Rfam database of RNA families [43]
Fold-change compared to non-pregnant female mice. miRNA has a putative binding site in *Slc13a1 and #Slc26a1 sulfate transporter mRNAs.
3.2. miRNA targets, networks and pathways
As an approach towards understanding the molecular consequences of an altered kidney miRNA profile, we used miRNET to identify biological interactions between our pilot miRNA data and kidney expressed mRNA targets. MiRNA-gene network analysis demonstrated that 18 of 35 potentially down-regulated miRNAs to be involved in a network (Supplemental Table 1). This network (Fig. 2A) contained 366 nodes and 411 edges.
Fig. 2.
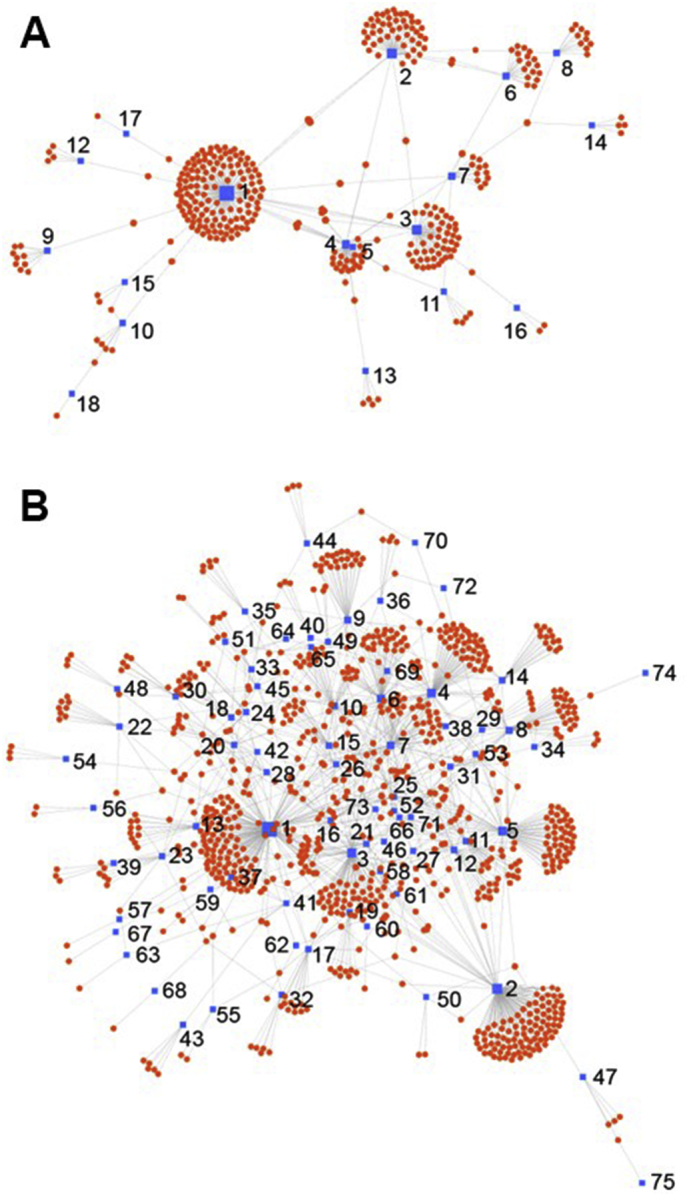
Node-link miRNET diagram of interactions between mRNA targets and potential differentially expressed miRNAs in the kidneys of pregnant mice. (A) Down-regulated and (B) up-regulated miRNAs (blue squares) and their predicted mRNA targets (red circles). Numbers in A and B refer to individual miRNAs in Supplemental Tables 1 and 2, respectively, with incremental numbering reflecting a decreasing number of mRNA targets. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Similarly, we identified 75 of 128 potentially up-regulated miRNAs to be involved in a network with 970 nodes and 1313 edges (Supplemental Table 2; Fig. 2B). Degree and betweenness are used in miRNet to evaluate the importance of each node (in this case, miRNAs as well as target mRNAs) in maintaining the overall connectivity within the network. The degree is a measure of the number of connections between nodes while betweenness within a network quantifies the number of shortest paths going through the node. Thus a node with higher degree and betweenness (which is also called a network hub) often indicates a more important role in the network. Analysing the mRNA targets within the network using Gene Ontology Enrichment Analysis identified a total of 350 Gene Ontology molecular functions with 4 being significantly enriched (p = 4.09e−02): zinc ion binding, protein domain specific binding, cation binding and transition metal ion binding (Supplemental Table 3). These enriched functions suggest a cellular protective response of the kidneys in pregnant mice. In particular, the functions of protein, cation and transition metal ion binding are cellular responses to protect against reactive oxygen species [44]. In addition, zinc binding protects cells against oxidative stress, stabilizes cellular membranes and induces metallothioneins that sequester reactive oxygen species and reduce hydroxyl radicals [45].
Interactions between the miRNAs and their mRNA targets are highly connected, with three miRNAs each having >100 targets (Fig. 2). In particular, mir-466 k targets 146 kidney expressed mRNAs (Fig. 2A) and has been linked to reduced angiogenesis and lymphangiogenesis, as well as osmoregulation and urine concentration in mice [46,47]. Of the potentially up-regulated miRNAs, mir-3089-5p and mir-758-3p have 172 and 115 mRNA targets, respectively (Fig. 2B). Whilst the role of mir-3089-5p is unknown, a recent study associated mir-758-3p with lupus nephritis in patients with systemic lupus erythematosus [48], suggesting that mir-758-3p levels increase in response to renal stress.
The most significantly enriched KEGG pathway was ErbB (p = 8.88e−05) signalling for the mRNA targets of potentially up-regulated miRNAs (Supplemental Table 4). The ErbB signalling pathway in the kidney plays a role in tubular regeneration following injury that can be caused by perfusion pressure or ischemia/reperfusion [49]. The enriched ErbB signalling pathway suggests a potential cellular protective response as the maternal kidney adapts to pregnancy. This finding complements a recent study in rats that showed pregnancy-associated upregulation of regeneration-associated pathways in the kidney, that protect the kidney from existing dysfunction and acute injury [2].
3.3. Putative miRNA binding sites in the Slc13a1 and Slc26a1 mRNA sequences
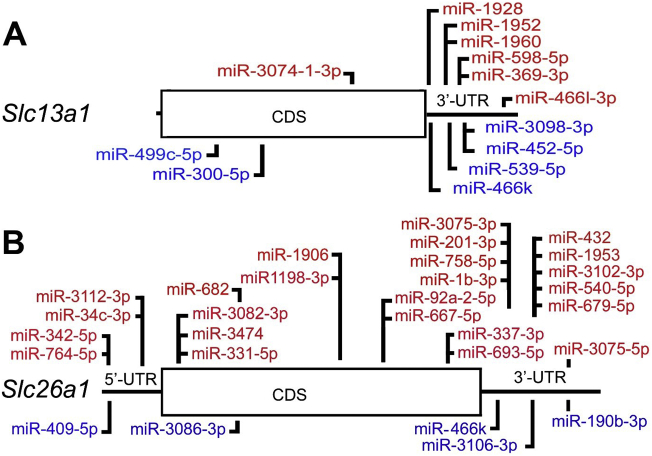
Increased sulfate reabsorption in the maternal kidneys during pregnancy is important for maintaining high circulating sulfate levels for healthy fetal growth and development. The increased plasma sulfate level is due to enhanced expression of the Slc13a1 and Slc26a1 sulfate transporters in the renal proximal tubule. In this study we considered the potential contribution of miRNAs to increasing Slc13a1 and Slc26a1 expression. Previous studies investigating the role of miRNAs in gene regulation have been focused on the 3’-UTR of mRNAs. However, we scanned the complete Slc13a1 and Slc26a1 mRNA sequences for potential miRNA binding sites as there is some evidence that both 5′- and 3′-untranslated mRNA sequences as well as coding sequences may also be important miRNA target regions for regulating gene expression [50,51]. In silico analysis using a combination of miRanda, Targetscan, RNA22 and miRWalk 2.0, identified a total of 186 and 294 putative miRNA binding sites in mouse Slc13a1 and Slc26a1 mRNAs, respectively (data not shown). By comparing these in silico findings to our list of potentially down- and up-regulated miRNAs (Table 1) we found a total of 13 and 29 differentially expressed miRNAs to have putative binding sites in the Slc13a1 and Slc26a1 mRNAs (Supplemental Table 5). These miRNA sites are located within the 5’-UTR of Slc26a1 (n = 5), as well as the coding sequences (n = 3 and 11) and 3’-UTRs (n = 10 and 11) of Slc13a1 and Slc26a1, respectively (Fig. 3).
Fig. 3.
Schematic showing the relative locations of putative miRNAs targets in the (A) Slc13a1 and (B) Slc26a1 mRNAs. miRNAs found to be potentially up-regulated (above red) and down-regulated (below blue) in the kidneys of pregnant mice when compared to non-pregnant mice. CDS, coding sequence. UTR, 5′- and 3′-untranslated regions. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Interestingly, miR-466k which has potential target sequences in both Slc13a1 and Slc26a1 3’-UTRs (Fig. 3) had the highest degree and betweenness scores in the down-regulated miRNAs list (Supplemental Table 1) suggesting that decreased miR-466k levels may be a potential contributor to the up-regulation of Slc13a1 and Slc26a1 mRNA levels in early mouse gestation. In addition, Reactome pathway analysis of other genes targeted by miRNAs which have putative target sites in Slc13a1 and Slc26a1 revealed that the most highly regulated pathways are “Transport of inorganic cations/anions and amino acids/oligopeptides” (p = 2.64e−02) suggesting that these miRNAs may have additional roles in regulating other solute-linked carrier (SLC) families in a similar manner.
4. Limitations
Limitations include the relatively small number of kidney samples, and the lack of validation of key genes of interest using independent samples. Accordingly, we consider this study as exploratory with the provided pilot data preparing the ground for more detailed investigations.
5. Summary
Collectively, this pilot study suggests differentially expressed miRNAs in the kidneys of pregnant mice, providing insights for future studies into a potential molecular mechanism of kidney adaptation to pregnancy. In particular, our preliminary findings suggest that the miRNA profile in the kidneys of pregnant mice, may be enriched for mRNA targets involved in cellular protective responses. Our pilot data also highlight mir-466k as a potential mechanism for enhanced Slc13a1 and Slc26a1 sulfate transporter expression during early gestation. Further studies are warranted to define the physiological roles of maternal kidney miRNAs in pregnancy, as well as their potential involvement in kidney pathology.
Author contributions
S.L. and P.A.D. conceived and designed the study, conducted data interpretation, and wrote the manuscript. S.L. and F.T.T. were involved in the acquisition and analysis of data. All authors drafted and revised the manuscript, and have read and approved the final manuscript for publication.
Competing interests
The authors declare that they have no financial and non-financial competing interests.
Acknowledgments
This work was supported by Mater Research and a Mater Foundation Principal Research Fellowship to P.A.D.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ymgmr.2019.100486.
Appendix A. Supplementary data
Supplemental Tables 1 to 5
References
- 1.Cheung K.L., Lafayette R.A. Renal physiology of pregnancy. Adv. Chronic Kidney Dis. 2013;20:209–214. doi: 10.1053/j.ackd.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Popkov V.A., Andrianova N.V., Manskikh V.N., Silachev D.N., Pevzner I.B., Zorova L.D., Sukhikh G.T., Plotnikov E.Y., Zorov D.B. Pregnancy protects the kidney from acute ischemic injury. Sci. Rep. 2018;8:14534. doi: 10.1038/s41598-018-32801-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman A.B., Abraham W.T., Zamudio S., Coffin C., Merouani A., Young D., Johnson A., Osorio F., Goldberg C., Moore L.G., Dahms T., Schrier R.W. Temporal relationships between hormonal and hemodynamic changes in early human pregnancy. Kidney Int. 1998;54:2056–2063. doi: 10.1046/j.1523-1755.1998.00217.x. [DOI] [PubMed] [Google Scholar]
- 4.Conrad K.P. Maternal vasodilation in pregnancy: the emerging role of relaxin. Am. J. Phys. Regul. Integr. Comp. Phys. 2011;301:R267–R275. doi: 10.1152/ajpregu.00156.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Irani R.A., Xia Y. Renin angiotensin signaling in normal pregnancy and preeclampsia. Semin. Nephrol. 2011;31:57–58. doi: 10.1016/j.semnephrol.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lafayette R.A., Hladunewich M.A., Derby G., Blouch K., Druzin M.L., Myers B.D. Serum relaxin levels and kidney function in late pregnancy with or without preeclampsia. Clin. Nephrol. 2011;75:226–232. doi: 10.5414/cnp75226. [DOI] [PubMed] [Google Scholar]
- 7.Cannell I.G., Kong Y.W., Bushell M. How do microRNAs regulate gene expression? Biochem. Soc. Trans. 2008;36:1224–1231. doi: 10.1042/BST0361224. [DOI] [PubMed] [Google Scholar]
- 8.Dawson P.A., Richard K., Perkins A., Zhang Z., Simmons D.G. Review: nutrient sulfate supply from mother to fetus: placental adaptive responses during human and animal gestation. Placenta. 2017;54:45–51. doi: 10.1016/j.placenta.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Dawson P.A., Petersen S., Rodwell R., Johnson P., Gibbons K., McWhinney A., Bowling F.G., McIntyre H.D. Reference intervals for plasma sulfate and urinary sulfate excretion in pregnancy. BMC Pregn. Childbirth. 2015;15:96. doi: 10.1186/s12884-015-0526-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawson P.A., Sim P., Simmons D.G., Markovich D. Fetal loss and hyposulfataemia in pregnant NaS1 transporter null mice. J. Reprod. Dev. 2011;57:444–449. doi: 10.1262/jrd.10-173k. [DOI] [PubMed] [Google Scholar]
- 11.Dawson P.A. Sulfate in fetal development. Semin. Cell Dev. Biol. 2011;22:653–659. doi: 10.1016/j.semcdb.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 12.Dawson P.A., Rakoczy J., Simmons D.G. Placental, renal, and Ileal Sulfate transporter gene expression in mouse gestation. Biol. Reprod. 2012;87:1–9. doi: 10.1095/biolreprod.111.098749. [DOI] [PubMed] [Google Scholar]
- 13.Dawson P.A. Role of sulphate in development. Reproduction. 2013;146:R81–R89. doi: 10.1530/REP-13-0056. [DOI] [PubMed] [Google Scholar]
- 14.Dawson P.A., Beck L., Markovich D. Hyposulfatemia, growth retardation, reduced fertility and seizures in mice lacking a functional NaSi-1 gene. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13704–13709. doi: 10.1073/pnas.2231298100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dawson P.A., Russell C.S., Lee S., McLeay S.C., van Dongen J.M., Cowley D.M., Clarke L.A., Markovich D. Urolithiasis and hepatotoxicity are linked to the anion transporter Sat1 in mice. J. Clin. Invest. 2010;120:702–712. doi: 10.1172/JCI31474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bowling F.G., Heussler H.S., McWhinney A., Dawson P.A. Plasma and urinary sulfate determination in a cohort with autism. Biochem. Genet. 2012;51:147–153. doi: 10.1007/s10528-012-9550-0. [DOI] [PubMed] [Google Scholar]
- 17.Dawson P.A., Sim P., Mudge D.W., Cowley D. Human SLC26A1 gene variants: a pilot study. Sci. World J. 2013:541710. doi: 10.1155/2013/541710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langford R., Hurrion E., Dawson P.A. Genetics and pathophysiology of mammalian sulfate biology. J. Genet. Genomics. 2017;44:7–20. doi: 10.1016/j.jgg.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Dawson P.A., Markovich D. Transcriptional regulation of the sodium-sulfate cotransporter NaS(i)-1 gene. Cell Biochem. Biophys. 2002;36:175–182. doi: 10.1385/CBB:36:2-3:175. [DOI] [PubMed] [Google Scholar]
- 20.Lee A., Dawson P.A., Markovich D. NaSi-1 and Sat-1: structure, function and transcriptional regulation of two genes encoding renal proximal tubular Sulfate transporters. Int. J. Biochem. Cell Biol. 2005;37:1350–1356. doi: 10.1016/j.biocel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Beck L., Markovich D. The mouse Na+-Sulfate Cotransporter gene Nas1: cloning, tissue distribution, gene structure, chromosomal assignment, and transcriptional regulation by vitamin D. J. Biol. Chem. 2000;275:11880–11890. doi: 10.1074/jbc.275.16.11880. [DOI] [PubMed] [Google Scholar]
- 22.Dawson P.A., Markovich D. Regulation of the mouse Nas1 promoter by vitamin D and thyroid hormone. Pflugers Arch. 2002;444:353–359. doi: 10.1007/s00424-002-0789-x. [DOI] [PubMed] [Google Scholar]
- 23.Lee A., Beck L., Markovich D. The mouse sulfate anion transporter gene Sat1 (Slc26a1): cloning, tissue distribution, gene structure, functional characterization, and transcriptional regulation thyroid hormone. DNA Cell Biol. 2003;22:19–31. doi: 10.1089/104454903321112460. [DOI] [PubMed] [Google Scholar]
- 24.Fernandes I., Hampson G., Cahours X., Morin P., Coureau C., Couette S., Prie D., Biber J., Murer H., Friedlander G., Silve C. Abnormal sulfate metabolism in vitamin D-deficient rats. J. Clin. Invest. 1997;100:2196–2203. doi: 10.1172/JCI119756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sagawa K., Murer H., Morris M.E. Effect of experimentally induced hypothyroidism on sulfate renal transport in rats. Am. J. Phys. 1999;276:F164–F171. doi: 10.1152/ajprenal.1999.276.1.F164. [DOI] [PubMed] [Google Scholar]
- 26.Cai M., Kolluru G.K., Ahmed A. Small molecule, big prospects: microRNA in pregnancy and its complications. J. Pregnancy. 2017;2017:6972732. doi: 10.1155/2017/6972732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morales Prieto D.M., Markert U.R. MicroRNAs in pregnancy. J. Reprod. Immunol. 2011;88:106–111. doi: 10.1016/j.jri.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 28.Dawson P.A., Gardiner B., Lee S., Grimmond S., Markovich D. Kidney transcriptome reveals altered steroid homeostasis in NaS1 sulfate transporter null mice. J. Steroid Biochem. Mol. Biol. 2008;112:55–62. doi: 10.1016/j.jsbmb.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 29.Cheval L., Pierrat F., Dossat C., Genete M., Imbert-Teboul M., Van Huyen J.P. Duong, Poulain J., Wincker P., Weissenbach J., Piquemal D., Doucet A. Atlas of gene expression in the mouse kidney: new features of glomerular parietal cells. Physiol. Genomics. 2011;43:161–173. doi: 10.1152/physiolgenomics.00093.2010. [DOI] [PubMed] [Google Scholar]
- 30.Fan Y., Siklenka K., Arora S.K., Ribeiro P., Kimmins S., Xia J. miRNet - dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 2016;44:W135–W141. doi: 10.1093/nar/gkw288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fabregat A., Sidiropoulos K., Garapati P., Gillespie M., Hausmann K., Haw R., Jassal B., Jupe S., Korninger F., McKay S., Matthews L., May B., Milacic M., Rothfels K., Shamovsky V., Webber M., Weiser J., Williams M., Wu G., Stein L., Hermjakob H., D'Eustachio P. The Reactome pathway knowledgebase. Nucleic Acids Res. 2016;44:D481–D487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dweep H., Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat. Methods. 2015;12:697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 33.John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 35.Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 36.Riffo-Campos Á.L., Riquelme I., Brebi-Mieville P. Tools for sequence-based miRNA target prediction: what to choose? Int. J. Mol. Sci. 2016;17 doi: 10.3390/ijms17121987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhatt K., Mi Q.S., Dong Z. MicroRNAs in kidneys: biogenesis, regulation, and pathophysiological roles. Am. J. Physiol. Ren. Physiol. 2011;300:F602–F610. doi: 10.1152/ajprenal.00727.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dawson P.A., Elliott A., Bowling F.G. Sulphate in pregnancy. Nutrients. 2015;7:1594–1606. doi: 10.3390/nu7031594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chung A.C., Lan H.Y. MicroRNAs in renal fibrosis. Front. Physiol. 2015;6:50. doi: 10.3389/fphys.2015.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan P.C., Chen C.C., Chen Y.C., Chang Y.S., Chu P.H. MicroRNAs in acute kidney injury. Hum. Genomics. 2016;10:29. doi: 10.1186/s40246-016-0085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pavkovic M., Vaidya V.S. MicroRNAs and drug-induced kidney injury. Pharmacol. Ther. 2016;163:48–57. doi: 10.1016/j.pharmthera.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun J., Yin A., Zhao F., Zhang W., Lv J., Lv J. Protection of tubular epithelial cells during renal injury via post-transcriptional control of BMP7. Mol. Cell. Biochem. 2017 doi: 10.1007/s11010-017-3063-4. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 43.Griffiths-Jones S., Bateman A., Marshall M., Khanna A., Eddy S.R. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monnier V.M., Neme T.I., Sell D.R., Weiss M.F. Transition metals and other forms of oxidative protein damage in renal disease. In: Miyata T., Eckardt K.U., Nangaku M., editors. Studies on Renal Disorders. Oxidative Stress in Applied Basic Research and Clinical Practice., Humana Press; 2011. [Google Scholar]
- 45.Marreiro D.D., Cruz K.J., Morais J.B., Beserra J.B., Severo J.S., de Oliveira A.R. Zinc and oxidative stress: current mechanisms. Antioxidants (Basel) 2017;6:E24. doi: 10.3390/antiox6020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo Y., Liu Y., Liu M., Wei J., Zhang Y., Hou J., Huang W., Wang T., Li X., He Y., Ding F., Yuan L., Cai J., Zheng F., Yang J.Y. Sfmbt2 10th intron-hosted miR-466(a/e)-3p are important epigenetic regulators of Nfat5 signaling, osmoregulation and urine concentration in mice. Biochim. Biophys. Acta. 2014;1839:97–106. doi: 10.1016/j.bbagrm.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 47.Seo M., Choi J.S., Rho C.R., Joo C.K., Lee S.K. MicroRNA miR-466 inhibits Lymphangiogenesis by targeting prospero-related homeobox 1 in the alkali burn corneal injury model. J. Biomed. Sci. 2015;22 doi: 10.1186/s12929-014-0104-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Navarro-Quiroz E., Pacheco-Lugo L., Lorenzi H., Díaz-Olmos Y., Almendrales L., Rico E., Navarro R., España-Puccini P., Iglesias A., Egea E., Aroca G. High-Throughput sequencing reveals circulating miRNAs as potential biomarkers of Kidney damage in patients with Systemic Lupus Erythematosus. PLoS ONE. 2016;11 doi: 10.1371/journal.pone.0166202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Campreciós G., Sánchez-Vizcaíno E., Soley M., Ramírez I. Chronic β-adrenergic stimulation increases ErbB receptors and cell proliferation in mouse kidney. Growth Factors. 2011;29:94–101. doi: 10.3109/08977194.2011.578576. [DOI] [PubMed] [Google Scholar]
- 50.Brümmer A., Hausser J. MicroRNA binding sites in the coding region of mRNAs: extending the repertoire of post-transcriptional gene regulation. Bioessays. 2014;36:617–626. doi: 10.1002/bies.201300104. [DOI] [PubMed] [Google Scholar]
- 51.Da Sacco L., Masotti A. Recent insights and novel bioinformatics tools to understand the role of microRNAs binding to 5′ untranslated region. Int. J. Mol. Sci. 2012;14:480–495. doi: 10.3390/ijms14010480. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Tables 1 to 5