Abstract
The recent developments of transcription activator-like effector nucleases (TALEN) and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein-9 nuclease (Cas9) have expanded plant breeding technology. One technical issue related to the current genome editing process is residual transgenes for TALEN and CRISPR/Cas9 left in plant genomes after the editing process. Here, we aim to add transient kanamycin resistance into apple leaf cells by introducing neomycin phosphotransferase II (NPTII) into apple leaf cells using the fusion peptide system. At 75 mg/L of kanamycin for 2 days, apple JM1 leaf cells infiltrated with NPTII could be selected. Thus, we successfully demonstrated the first transient selection system of plant cells using a fusion peptide-mediated protein delivery system.
Keywords: apple, carrier peptide, neomycin phosphotransferase II (NPTII), protein delivery
New plant breeding technology is studied and used everywhere, largely because of the recent development of transcription activator-like effector nucleases (TALEN) and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein-9 nuclease (Cas9) (Li et al. 2012; Nekrasov et al. 2013). One technical issue related to genome editing is residual transgenes for TALEN or CRISPR/Cas9 left in plant genomes after the editing process. To avoid the problem of foreign genes remaining in plant genomes, Cas9 proteins with gRNA were introduced into plant protoplasts of Arabidopsis thaliana, tobacco, lettuce and rice to edit the nuclear genome without any insertion of foreign genes into the genomes (Woo et al. 2015). However, to select edited cells after the genome editing, we also need selection systems from the mixture of edited and non-edited (wild-type) cells. The presence of any selection markers in plant genome is not preferred for genome-edited crops and plants. To clear this technical issue, we aim to develop a transient selection system, which can select target cells without genome insertion of antibiotic resistance genes. Establishment of the transient selection system will enable complete DNA-free plant genome editing.
We recently developed and reported on a direct protein delivery system for intact plants (Ng et al. 2016). In this study, target proteins such as bovine serum albumin (66 kDa), alcohol dehydrogenase (150 kDa), and the yellow fluorescent protein (27 kDa) were complexed with fusion peptides and infiltrated into intact A. thaliana leaves. The fusion peptide used was designed to consist of a cationic sequence and a cell penetrating peptide, according to previous studies (Lakshmanan et al. 2013; Lakshmanan et al. 2015; Numata 2015; Numata et al. 2014). In the case of protein containing a signal peptide, such as a nuclear localization signal or a peroxisomal targeting signal, the fusion peptide did not disturb the signal peptide; therefore, the target protein could be localized based on the signal peptide (Ng et al. 2016). The fusion peptide system can deliver proteins with a wide range of molecular weights (27 to 150 kDa) into the cells of intact plants without preventing protein functions (Ng et al. 2016).
In this study, we aim to add transient kanamycin resistance into apple leaves by introducing neomycin phosphotransferase II (NPTII; EC 2.7.1.95) into JM1 apple (Malus prunifolia (Wild.) Borkh.264 ‘Seishi’×M. pumila Mill. var. paradisiaca Schneid. ‘M.9’) leaves using the fusion peptide system. JM1 was released in 1996 and registered as No. 7443 under the Plant Variety Protection and Seed Act of Japan in 1999. According to its orchard performance and resistance to disease and pests, JM1 is considered as rootstock for Fuji (Soejima et al. 2010). Trees of Fuji on JM1 rootstocks demonstrate better rooting ability and cumulative yield efficiency in comparison to existing rootstocks. The first genome editing in apple was reported (Nishitani et al. 2016), but the transient selection system is needed to solve the problem of foreign genes remaining in plant genome.
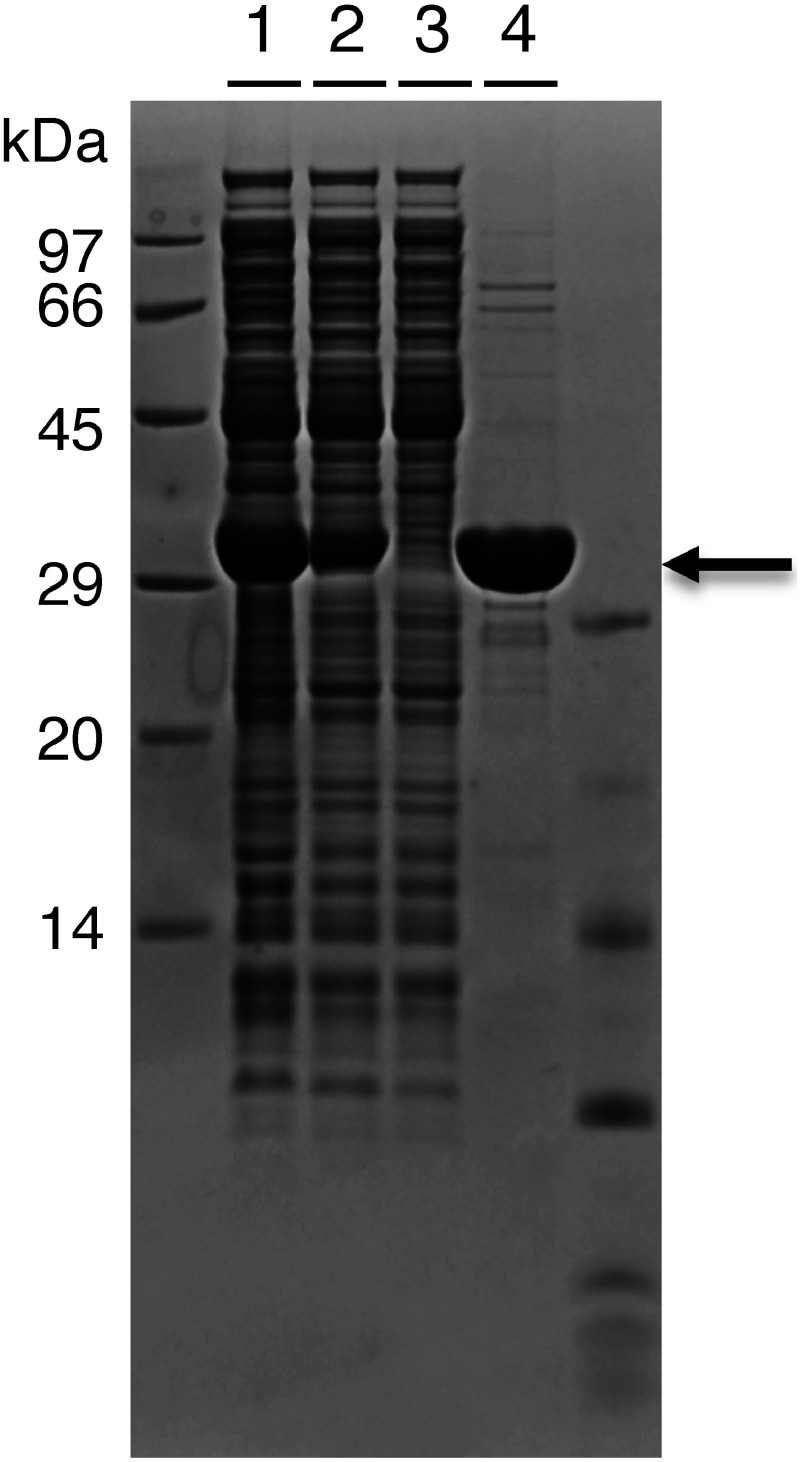
NPTII is one type of aminoglycoside phosphotransferase that inactivates aminoglycoside antibiotics, including kanamycin and neomycin (Yenofsky et al. 1990). NPTII inactivates kanamycin by catalyzing the transfer of the terminal phosphate of ATP to kanamycin and is used in plants, as well as in yeast and mammals (Bevan et al. 1983; Colbére-Garapin et al. 1981; Jimenez and Davies 1980). We synthesized NPTII using a cell-free system, as reported previously (Numata et al. 2015; Numata et al. 2012). NPTII gene for plant selection was amplified by PCR with pMpGWB401 binary vector (Ishizaki et al. 2015) as a template with the following primers: 5′-ATA TCC ATG GGG ATT GAA CAA GAT GGA TTG CAC GC-3′ and 5′-ATA TGG ATC CCG GAA GAA CTC GTC AAG AAG GCG AT-3′. The amplified DNA fragment was cloned into the NcoI and BamHI sites of pET28b(+), and the sequence was confirmed. The NPTII gene cloned was then amplified for cell-free synthesis by a two-step PCR, according to previous reports (Yabuki et al. 2007). Briefly, the first PCR was carried out in a 20 µl reaction mixture with 3 µl of a 50-fold diluted buffer, 50 nM each of forward (FW) and reverse (RV) unique primers for NPTII, 0.2 mM of each deoxyribonucleotide triphosphate (dNTP), 1×Expand Hi-Fi buffer (Roche, Basel, Switzerland), and 0.5 U Expand Hi-Fi enzyme (Roche) with a hot start. The FW and RV primer sequences are FW: 5′-ACT GAG AAC CTG TAC TTC CAG GGA ATG ATT GAA CAA GAT GGA TTG CAC G-3′ and RV: 5′-GGG CGG GGA TCA ATC AAT CAT TAT CAG AAG AAC TCG TCA AGA AGG CG-3′. The second PCR was carried out in a reaction mixture (20 µl) with 5 µl of 5-fold diluted first PCR product, 50 pM T7P fragment (5′-GCT CTT GTC ATT GTG CTT CGC ATG ATT ACG AAT TCA GAT CTC GAT CCC GCG AAA TTA ATA CGA CTC ACT ATA GGG AGA CCA CAA CGG TTT CCC TCT AGA AAT AAT TTT GTT TAA CTT TAA GAA GGA GAT ATA CAT ATG AAA GAT CAT CTC ATC CAC AAT CAT CAC AAA CAT GAG CAC GCT CAT GCC GAA CAT ACT GAG AAC CTG TAC TTC CAG GG-3′), 50 pM T7T fragment (5′-AAT GAT TGA TTG ATC CCC GCC CAG CTG AGT TGG CTG CTG CCA CCG CTG AGC AAT AAC TAG CAT AAC CCC TTG GGG CCT CTA AAC GGG TCT TGA GGG GTT TTT TGC TGA AAG GAG GAA CTA TAT CCG GAT AAC CTC GAG CTG CAG GCA TGC AAG CTT GGC GAA GCA CAA TGA CAA GAG C-3′), 1 µM U2 universal primer (5′-GCT CTT GTC ATT GTG CTT CG-3′), 0.2 mM of each dNTP, 1×Expand Hi-Fi buffer, and 0.5 U Expand Hi-Fi Enzyme with a hot start. The His-tag used in this study was a modified version of the natural poly-histidine tag (Yabuki et al. 2007). A dialysis-mode cell-free protein synthesis method, which was previously reported (Spirin et al. 1988), was used to synthesize NPTII protein. The experimental condition for the cell-free synthesis was based on our previous study (Numata, et al. 2015). The tagged NPTII protein was purified with nickel-nitrilotriacetic acid (Ni-NTA) agarose column by AKTA Express (GE Healthcare, Little Chalfont, UK) as previously reported (Numata et al. 2015). The yield of the purified protein was determined by the Bradford method using a Bio-Rad Protein Assay kit (Bio-Rad, Hercules, CA). Bovine serum albumin was used as the protein standard. The yield of the purified NPTII protein was 6.3 mg from 9 ml-scale cell-free synthesis. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using 15–20% precast Tris-HCl gels (DRC Co., Ltd., Kyoto, Japan). The gel was stained with Coomassie brilliant blue. Figure 1 shows the resultant SDS-PAGE and confirmed that NPTII was successfully prepared after Ni-NTA purification.
Figure 1. SDS-PAGE analysis of purified NTPII. (Lane 1) Total fraction of cell-free synthesis. (Lane 2) Supernatant fraction. (Lane 3) Flow through after Ni-NTA column. (Lane 4) Elution from Ni-NTA column. The molecular weight of NPTII with His-tag is 32.3 kDa, which is indicated by an arrow.
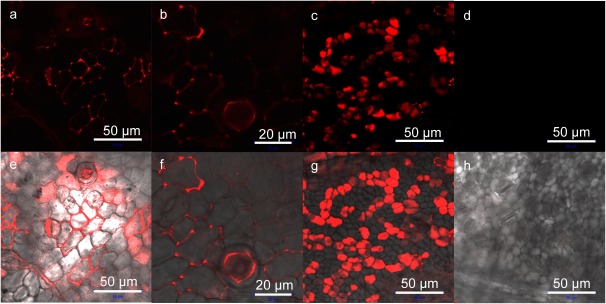
NPTII protein was labeled with Rhodamine B isothiocyanate (RhB) (Mw: 536.08 g/mol, Sigma-Aldrich. St. Louis, MO), according to a previous report (Ng et al. 2016). Fusion peptide 2BP100-K8 (Amino acid sequence: KKLFKKILKYL KKLFKKILKYLKKKKKKKK, Theoretical pI/Mw: 10.75/3851.13 Da) was custom synthesized at the Peptide Synthesis Facility, RIKEN Brain Science Institute, Wako, Japan by using standard 9-fluorenylmethoxycarbonyl solid-phase peptide synthesis (Fields and Noble 1990). The peptides were purified using high-performance liquid chromatography, and the molecular weights were confirmed by matrix-assisted laser desorption/ionization–time of flight mass spectrometry. We prepared an ionic complex of the fusion peptide 2BP100-K8 and NPTII by mixing 2 µg (approximately 0.062 nmol) of NPTII and the fusion peptide of approximately 2.4 µg (0.62 nmol). The complex solution of 15 µl containing 2 µg of NPTII was introduced into apple leaf cells. The apple leaves used were one to two weeks old and were collected from apple branches in hydroponic culture with tap water. After 6 h of infiltration, the localization of RhB labeled NPTII was observed by confocal laser scanning microscopy (CLSM, ZeissLSM 700, Carl Zeiss, Oberkochen, Germany). Figure 2 shows the localization of NPTII in the epidermal cells and sponge cells, indicating that NPTII was successfully introduced into the cells, but not all the cells were transfected. Based on the microscopic analysis, the cells were partially transfected with RhB labeled NPTII. According to our previous study (Ng et al. 2016), the peptide-mediated protein delivery was confirmed by several quantitative methods.
Figure 2. Localization of RhB-labeled NPTII after infiltration of the NPTII and fusion peptide complex in apple leaf cells based on CLSM observations. (a–c) Fluorescence images of RhB-labeled NPTII (red). (d) A fluorescence image of mock control. (e–g) Overlay images of the fluorescence and differential interference contrast images. (h) An overlay image of mock control. (a, e) Apple leaf epidermis cells after 6 h of infiltration. (b, f) Higher magnification image of apple leaf epidermis cells after 6 h of infiltration. (c, g) Spongy mesophyll cells of apple leaf after 6 h of infiltration. (d, h) apple leaf epidermis cells after 6 h of infiltration of the peptide without NPTII protein.
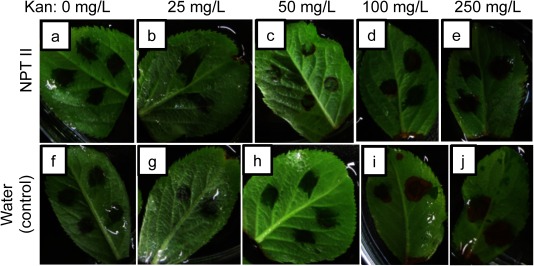
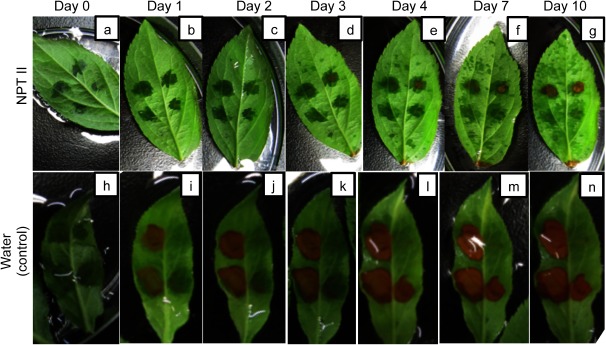
Using the non-labeled NPTII, we infiltrated the complex of NPTII and the fusion peptide into JM1 apple leaves and treated the leaves in kanamycin solutions for 10 days. The kanamycin concentrations were 0, 25, 50, 75, 100 and 250 mg/L. Five leaves were investigated in one experiments and the experiments were performed three times. Two to four spots were infiltrated in each leaf. Figure 3 shows 2-day resistance tests of NPTII-infiltrated JM1 leaves to kanamycin. The cell death was judged based on the color change of leaves from green to brown. This is because in our protein-based selection system we plan to select resistant cells by our eyes without any assays. After the incubation of kanamycin for 2 days, 50 mg/L of kanamycin was not enough to kill the infiltrated cells, whereas 100 mg/L of kanamycin seemed an excess amount to bring about kanamycin resistance for the selection process. We subsequently performed the 2-day resistance test of NPTII-infiltrated leaves exposed to 75 mg/L of kanamycin (Figure 4). As a result, the NPTII-infiltrated cells could survive with 75 mg/L of kanamycin after the 2-day incubation, but the water-infiltrated cells started to be killed by lethal toxicity of kanamycin. The time-course study on cell viability with 75 mg/L of kanamycin indicates that some of the NPTII-infiltrated cells were killed at day 3. The kanamycin condition showed significant cytotoxicity to the NPTII-infiltrated cells after 4 days (Figure 4e–g). As control experiments, without NPTII, the apple JM1 leaf was killed with 75 mg/L of kanamycin after just 1 day (Figure 4i). We also performed control experiments using either NPTII or the fusion peptide (Figure 5). The apple JM1 leaf cells infiltrated with NPTII alone did not show kanamycin resistance at 75 mg/L of kanamycin after 2 days incubation (Figure 5a). The result of NPTII alone (Figure 5a) clearly differed from that of the complex of NPTII and the fusion peptide (Figure 4c), indicating that NPTII alone did not function, while the fusion peptide introduces and protect NPTII in the cells (Figure 4c). This might be because NPTII was degraded or inactivated without the fusion peptide after the infiltration, according to our previous studies on protection assay of the fusion peptide’s complex (Ng et al. 2016; Numata and Kaplan 2010). Similar to NPTII alone, the apple JM1 leaf cells infiltrated with the fusion peptide alone did not show kanamycin resistance (Figure 5b). Based on the overall results, at 75 mg/L of kanamycin for 2 days, apple JM1 leaf cells infiltrated with NPTII could be selected. Thus, we successfully demonstrated the first transient selection system of plant cells using the fusion peptide-mediated protein delivery system.
Figure 3. Kanamycin resistance test of apple JM1 leaves infiltrated with the fusion peptide and NPTII complex (a–e) or distilled water (f–j) for 2 days. The amount of NPTII used is approximately 2 µg. The kanamycin concentrations are 0 (a, f), 25 (b, g), 50 (c, h), 100 (d, i) and 250 mg/L (e, j).
Figure 4. Time-course kanamycin resistance test of apple JM1 leaves infiltrated with the fusion peptide and NPTII complex (a–g) or distilled water (h–n). The amount of NPTII used is approximately 2 µg. The concentration of kanamycin is 75 mg/L.
Figure 5. Kanamycin resistance test of apple JM1 leaves infiltrated with either NPTII (a) or fusion peptide (b). The amount of NPTII used is approximately 2 µg. The concentration of kanamycin is 75 mg/L. The incubation time is 2 days.
In summary, we propose a transient selection system for modified plant cells using the peptide-mediated protein delivery system. This method potentially enables us to select the genome-edited cells transiently without any insertion of foreign genes, such as the selection marker gene. This technique will allow DNA-free plant genome editing in the near future. The current issues to realize the DNA-free plant genome editing are selectivity of the protein-based selection system and efficiency of regeneration from cells to plants. To improve this transient selection system, we expect more stable and active enzymes to enable antibiotic resistance.
Acknowledgments
This work was supported by Cabinet Office, Government of Japan, Cross-ministerial Strategic Innovation Promotion Program (SIP), “Technologies for creating next-generation agriculture, forestry and fisheries” (funding agency: Bio-oriented Technology Research Advancement Institution, NARO) (to K. N., C.N and Y. K.). The authors have no conflict of interest to declare.
Abbreviations
- PCR
polymerase chain reaction
- TALEN
transcription activator-like effector nucleases
- CRISPR
clustered regularly interspaced short palindromic repeats
- Cas9
CRISPR-associated protein-9 nuclease
- NPTII
neomycin phosphotransferase
- FW
forward
- RV
reverse
- dNTP
deoxyribonucleotide triphosphate
- Ni-NTA
nickel-nitrilotriacetic acid
- SDS-PAGE
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- RhB
Rhodamine B isothiocyanate
- CLSM
confocal laser scanning microscopy
References
- Bevan MW, Flavell RB, Chilton MD (1983) A chimaeric antibiotic resistance gene as a selectable marker for plant cell transformation. Nature 304: 184–187 [PubMed] [Google Scholar]
- Colbére-Garapin F, Horodniceanu F, Kourilsky P, Garapin AC (1981) A new dominant hybrid selective marker for higher eukaryotic cells. J Mol Biol 150: 1–14 [DOI] [PubMed] [Google Scholar]
- Fields GB, Noble RL (1990) Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int J Pept Protein Res 35: 161–214 [DOI] [PubMed] [Google Scholar]
- Ishizaki K, Nishihama R, Ueda M, Inoue K, Ishida S, Nishimura Y, Shikanai T, Kohchi T (2015) Development of gateway binary vector series with four different selection markers for the liverwort marchantia polymorpha. PLoS One 10: e0138876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez A, Davies J (1980) Expression of a transposable antibiotic resistance element in Saccharomyces. Nature 287: 869–871 [DOI] [PubMed] [Google Scholar]
- Lakshmanan M, Kodama Y, Yoshizumi T, Sudesh K, Numata K (2013) Rapid and efficient gene delivery into plant cells using designed peptide carriers. Biomacromolecules 14: 10–16 [DOI] [PubMed] [Google Scholar]
- Lakshmanan M, Yoshizumi T, Sudesh K, Kodama Y, Numata K (2015) Double-stranded DNA introduction into intact plants using peptide-DNA complexes. Plant Biotechnol 32: 39–45 [Google Scholar]
- Li T, Liu B, Spalding MH, Weeks DP, Yang B (2012) High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol 30: 390–392 [DOI] [PubMed] [Google Scholar]
- Nekrasov V, Staskawicz B, Weigel D, Jones JD, Kamoun S (2013) Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol 31: 691–693 [DOI] [PubMed] [Google Scholar]
- Ng KK, Motoda Y, Watanabe S, Sofiman Othman A, Kigawa T, Kodama Y, Numata K (2016) Intracellular delivery of proteins via fusion peptides in intact plants. PLoS One 11: e0154081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani C, Hirai N, Komori S, Wada M, Okada K, Osakabe K, Yamamoto T, Osakabe Y (2016) Efficient Genome Editing in Apple Using a CRISPR/Cas9 system. Sci Rep 6: 31481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numata K (2015) Poly(amino acid)s/polypeptides as potential functional and structural materials. Polym J 47: 537–545 [Google Scholar]
- Numata K, Kaplan DL (2010) Silk-based gene carriers with cell membrane destabilizing peptides. Biomacromolecules 11: 3189–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numata K, Motoda Y, Watanabe S, Osanai T, Kigawa T (2015) Co-expression of two polyhydroxyalkanoate synthase subunits from synechocystis sp PCC 6803 by Cell-Free synthesis and their specific activity for polymerization of 3-Hydroxybutyryl-Coenzyme A. Biochemistry 54: 1401–1407 [DOI] [PubMed] [Google Scholar]
- Numata K, Motoda Y, Watanabe S, Tochio N, Kigawa T, Doi Y (2012) Active intermediates of polyhydroxyalkanoate synthase from aeromonas caviae in polymerization reaction. Biomacromolecules 13: 3450–3455 [DOI] [PubMed] [Google Scholar]
- Numata K, Ohtani M, Yoshizumi T, Demura T, Kodama Y (2014) Local gene silencing in plants via synthetic dsRNA and carrier peptide. Plant Biotechnol J 12: 1027–1034 [DOI] [PubMed] [Google Scholar]
- Soejima J, Yoshida Y, Haniuda T, Bessho H, Tsuchiya S, Masuda T, Komori S, Sanda T, Ito Y, Sadamori S, et al. (2010) New dwarfing apple rootstocks JM1, JM7 and JM8. Bull Natl Inst Fruit Tree Sci 11: 1–16 [Google Scholar]
- Spirin AS, Baranov VI, Ryabova LA, Ovodov SY, Alakhov YB (1988) A continuous cell-free translation system capable of producing polypeptides in high yield. Science 242: 1162–1164 [DOI] [PubMed] [Google Scholar]
- Woo JW, Kim J, Kwon SI, Corvalan C, Cho SW, Kim H, Kim SG, Kim ST, Choe S, Kim JS (2015) DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol 33: 1162–1164 [DOI] [PubMed] [Google Scholar]
- Yabuki T, Motoda Y, Hanada K, Nunokawa E, Saito M, Seki E, Inoue M, Kigawa T, Yokoyama S (2007) A robust two-step PCR method of template DNA production for high-throughput cell-free protein synthesis. J Struct Funct Genomics 8: 173–191 [DOI] [PubMed] [Google Scholar]
- Yenofsky RL, Fine M, Pellow JW (1990) A mutant neomycin phosphotransferase II gene reduces the resistance of transformants to antibiotic selection pressure. Proc Natl Acad Sci USA 87: 3435–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]