Abstract
Lilies (Lilium) are among the most important floriculture crops, and to accelerate research regarding lily genetics, the development of reverse-genetics tools is necessary. However, Agrobacterium-mediated transformation in Lilium is time-consuming, since the plants require several years to progress from acclimation to flowering. Thus, virus-induced gene silencing (VIGS) is an attractive method for assaying gene function. In the present study, we modified a lily-derived strain of Cucumber mosaic virus (CMV-HL) as a VIGS vector and evaluated its effectiveness for inducing gene silencing in Lilium leichtlinii by introducing L. leichtlinii phytoene desaturase (LlPDS) gene fragments into an intercistronic region between the 3a and 3b genes of the CMV-HL RNA3 genome. At 30 days after inoculation (dpi) with LlPDS-containing CMV-HL, photo-bleaching was observed in the upper leaves of L. leichtlinii, and at 57 dpi, we observed that the natural orange color in flower tepals had faded. Reduced LlPDS expression and the detection of small interfering LlPDS RNA indicated that the color changes were the result of LlPDS gene silencing. In addition, the leaves also exhibited a mild photo-bleaching phenotype in the following year. Therefore, our results indicate that CMV-HL spreads systemically in the leaves and flowers of Lilium during the first year of infection, as well as in new shoots during the following year, and that the vector system can be successfully applied to induce short-term endogenous gene silencing in lilies.
Keywords: Cucumber mosaic virus (CMV)-HL, lily, phytoene desaturase (PDS), photo-bleaching, small interfering RNA (siRNA)
The genus Lilium (Liliaceae) comprises more than 90 species, which are classified into several sections. Since species belonging to the same section exhibit high interspecific crossing abilities, hybrid lilies, such as the Asiatic, Longiflorum, Trumpet, and Oriental hybrids, are principally developed by the interspecific hybridization of species within the same sections. However, more recently cultivars are also being developed by inter-section hybridization (Yamagishi 2013). In lily cultivars, variation in flower color is among the most important targets of cultivar breeding programs and is produced by variation in the accumulation of anthocyanin and carotenoid pigments (Yamagishi 2013). Several genes involved in anthocyanin and carotenoid biosyntheses have been studied in lilies (Jeknić et al. 2012; Lai et al. 2012; Nakatsuka et al. 2003, 2009; Suzuki et al. 2015; Yamagishi et al. 2010a, 2010b). Recently, the transcriptomes of several lily organs have been sequenced using next-generation sequencing (NGS), and a variety of potentially important novel genes have been identified (Huang et al. 2014; Suzuki et al. 2016; Villacorta-Martin et al. 2015; Wang et al. 2014), although their specific functions remain to be determined.
To accelerate research regarding lily genetics, it will be necessary to develop reverse-genetics tools that are capable of facilitating the functional screening of expressed sequences. In many plants, genetic transformation is an essential reverse-genetics tool that is used to validate the functions of endogenous genes, and Agrobacterium-mediated transformation is widely used to manipulate genes in both eudicots and monocots. However, in Lilium, although stable transformants have been successfully developed in a few laboratories (Azadi et al. 2010; Ogaki et al. 2008), the method is still challenging, and because lily plants usually require three or more years to progress from acclimation to flowering, the method is quite time-consuming, as well. Therefore, stable transformation is not an ideal solution for the functional screening of lily genes.
Alternatively, virus-induced gene silencing (VIGS) has been utilized for the knockdown of plant genes within a relatively short time frame. VIGS functions in a manner similar to that of post-transcriptional gene silencing (PTGS; Hamilton and Baulcombe 1999) and provides an attractive strategy to characterize the functions of endogenous genes (Senthil-Kumar and Mysore 2011a). In addition, the technology is well established in model plants, as well as in various floriculture crops. In eudicots, for example, VIGS has been applied to Aquilegia (Kramer et al. 2007), Eschscholzia californica (Wege et al. 2007), Thalictrum (Di Stilio et al. 2010), and Catharanthus roseus (Liscombe and O’Connor 2011), using Tobacco rattle virus (TRV) as the vector, and to Gentiana spp., using Apple latent spherical virus (Nakatsuka et al. 2015) and Broad bean wilt virus 2 (Tasaki et al. 2016). In monocots, VIGS has been applied to both Gladiolus and Phalaenopsis, using TRV (Zhong et al. 2014) and Cymbidium mosaic virus (Hsieh et al. 2013), respectively.
To use VIGS technology, the choice of viral vector is important, since VIGS is only possible when the viral vector can spread systemically in infected plants (Pflieger et al. 2013). Cucumber mosaic virus (CMV) is a member of the genus Cucumovirus, which is in the family Bromoviridae. A typical feature of CMV is its wide host range that includes the Liliaceae (Masuta et al. 2002), and the Lilium-derived CMV-HL strain (Hagita et al. 1989; Masuta et al. 2002) is expected to be able to induce gene silencing in lilies. CMV is well characterized as a VIGS vector in other plant species (Hong et al. 2012; Kanazawa et al. 2011; Liu et al. 2010; Matsuo et al. 2007; Nagamatsu et al. 2007; Otagaki et al. 2006). CMV possesses tripartite positive-sense RNA genomes (RNA1, 2, and 3) and the 2b protein in the RNA2 genome acts as a silencing suppressor (Nakahara and Masuta 2014). Conventional CMV vectors for VIGS are often constructed by inducing a premature stop codon into the 2b gene and fragments of target genes are ligated downstream of the truncated 2b gene (Nagamatsu et al. 2007). In a preliminary experiment, we modified the RNA2 genome of CMV-HL to induce a premature stop codon in the 2b gene and to insert a cloning site downstream of the 2b gene; however, the modified CMV-HL vector failed to spread systemically in infected lilies. Therefore, we used an alternative strategy for developing the CMV-HL vector by introducing target gene fragments into an intercistronic region between the 3a and 3b genes in the RNA3 genome. In the present study, we report that the new CMV-HL vector spread systemically in infected lilies, induced the silencing of the endogenous phytoene desaturase (PDS) gene, and was even capable of changing the carotenoid color of lily tepals. In addition, we discuss the utility of CMV-HL as a VIGS vector for reverse genetics in lilies.
Materials and methods
Plant materials
To avoid co-infection with another virus, we used in vitro-propagated virus-free bulbs of L. leichtlinii ‘Hakugin’. L. leichtlinii is one of the parental species used to develop Asiatic hybrid lilies and its bulbs are produced as edible lilies (Suzuki and Yamagishi 2016). The major pigments that accumulate in the tepals of L. leichtlinii are carotenoids. After cold treatment at 4°C for more than 2 months, the bulbs were potted and grown in a growth chamber kept at 24°C under a 16 : 8 h light : dark cycle (fluorescent light). To evaluate the transmission of gene silencing in the successive season, bulbs were harvested at the end of the first year, stored at 4°C, and then potted in the following year. Nicotiana benthamiana plants were sown and grown in a growth incubator at 24°C under a 16 : 8 h light : dark cycle (fluorescent light).
Vector construction
The CMV-HL strain, which was isolated from L. leichtlinii in Japan (Hagita et al. 1989), was modified and used as a viral vector. The cDNA of the CMV-HL tripartite genomes (RNA1, 2, and 3) were previously cloned into pPCR-Script Amp SK(+) or pGEM-T easy vectors (Masuta et al. 2002; Yamaguchi et al. 2005). To insert target gene fragments into the RNA3 genome, the StuI/BglII (SB) cloning site (aggcctGagatct) or EcoRV/BglII (EB) cloning site (gatatcGagatct) was introduced to the intercistronic region immediately upstream of the 3b (coat protein) gene sequence in the RNA3 genome (Supplementary Figure S1). The cloning sites were introduced using an overlap-extension PCR method, with the Y3-5-NheI and Y3-StuI-BglII-CP3 (for fragment I) or Y3-StuI-BglII-CP5 and Y3-3-EcoRI (for fragment II) primers for the SB cloning site, or the Y3-5-NheI and Y3-EcoRV-BglII-CP3 (for fragment I) or Y3-EcoRV-BglII-CP5 and Y-CP-3-100 (for fragment II) primers for the EB cloning site (Supplementary Table S1). Briefly, DNA fragments I and II were separately amplified from the plasmid pCHL3. Then, overlapping PCR was conducted using a mixture of fragments I and II as a template and primers Y3-5-NheI and Y3-3-EcoRI for the SB cloning site, and Y3-5-NheI and Y-CP-3-100 for the EB cloning site. To construct pCHL3-SB, the amplified fragments and pCHL3 were digested using NheI and EcoRI and then ligated. Similarly, to construct pCHL3-EB, the amplified fragments were digested using NheI and BglII and then ligated to predigested pCHL3-SB.
LlPDS cloning and modified CMV construction
Total RNA was isolated from L. leichtlinii leaves using the NucleoSpin RNAII Kit (Macherey-Nagel GmbH & Co. KG, Düren, Germany), and cDNA was synthesized using ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo Co., Ltd., Osaka, Japan). Next we PCR amplified a partial PDS sequence from the leaf cDNA (LlPDS), using the primers LlPDSdf and LlPDSer, which were designed from the PDS sequence of the Asiatic hybrid lily cultivar ‘Connecticut King’ (Yamagishi et al. 2010a), and directly sequenced the amplified products (DDBJ accession: LC155113). The LlPDS fragments used to insert into the CMV vector (Supplementary Figure S2) were amplified using the primers PDS33f and LhPDSbr or the primers LlPDShf and LlPDSdr to amplify a 33- or a 52-nucleotide (nt) fragment, respectively. After restriction digestion, each of the PDS fragments was ligated into the SB or EB cloning sites of the RNA3 genome (Supplementary Figure S1).
In vitro transcription and inoculation
Before in vitro transcription, the plasmids pCHL2 and pCHL3 were linearized using SphI and EcoRI, respectively, and a fragment that included the T7 promoter and RNA1 sequence was PCR amplified from the plasmid pCHL1 and the primers CY2T7 and HL-1-3. Then, CMV RNA1, RNA2, and RNA3 were in vitro-synthesized using T7 RNA polymerase (Takara Bio, Otsu, Japan) in the presence of the m7G(5′)ppp(5′)G cap analogue (Life Technologies Co., Tokyo, Japan), as described by Otagaki et al. (2006). Each of the three transcription products and sodium phosphate buffer (pH 7) were mixed at a ratio of 1 : 1 : 1 : 7. N. benthamiana leaves were dusted with carborundum (400 mesh; Nacalai Tesque, Inc., Kyoto, Japan) and inoculated with the virus by rubbing with the vector-containing mixture. At 6 to 10 day post inoculation (dpi), when mild mosaic symptoms were observed in N. benthamiana, viral infection was confirmed using reverse transcription-PCR (RT-PCR) with the primers HL3ff and HL3er. The N. benthamiana leaves harboring CMV-HL were harvested and ground with phosphate buffer to produce leaf sap. Then, the L. leichtlinii leaves were dusted with Celite 545 (Wako Pure Chemical Industries, Ltd., Osaka, Japan) and inoculated by rubbing with the leaf sap. The inoculation of L. leichtlinii was performed twice; the second inoculation was conducted one week after the first.
Analysis of gene expression and detection of the CMV-HL vector
Total RNA was isolated and purified from leaves and tepals and cDNA was synthesized as described above, and the expression levels of LlPDS were determined using quantitative RT-PCR (qRT-PCR). The 20-µl RT-PCR reactions contained 10 µl of SYBR Premix Ex Taq (Takara Bio), 0.2 µM each of the gene-specific primers (Supplementary Table S1), and 2 µl of first-strand cDNA (10 times diluted). The signals were monitored using the CFX Connect Real-Time System (Bio-Rad, Tokyo, Japan), and the expression levels were normalized using the corresponding level of 18SrRNA expression for each sample. The primer specificity was confirmed using melting curve analysis and agarose gel electrophoresis of the qRT-PCR products. Relative fold differences were calculated based on the 2−ΔCt method (Livak and Schmittgen 2001), where ΔCt=Ct (PDS)−Ct (18SrRNA). Relative fold values of three to seven replications were used to calculate mean values and standard error (SE). The systemic infection of the lilies by CMV-HL was confirmed using RT-PCR with the primers HL3ff and HL3er, which were designed at the flank of the cloning sites in order to detect the deletion of insert sequences (Supplementary Table S1), and total RNA isolated from the leaves above the inoculated leaves (hereafter, upper leaves).
Detection of siRNA by stem-loop pulsed RT and end-point PCR
Low molecular weight (LMW) RNA was isolated from leaves, as described by Goto et al. (2003), or using a High Pure miRNA Isolation Kit (Roche Diagnostics K. K., Tokyo, Japan), according to the manufacturer’s instructions. Short interfering RNAs (siRNAs) were transcribed into cDNA using the stem-loop pulsed RT protocol, and then, end-point PCR was conducted to amplify 60-nt fragments that included the siRNA sequences (Varkonyi-Gasic et al. 2007; Yamagishi et al. 2015) using primers shown in Supplementary Table S2. The primers were designed based on siRNA sequences that were predicted using the algorithm of Ui-Tei et al. (2004) and Reynolds et al. (2004); e.g., A or U at the 5′ terminal end of the guide strand, G or C at the 5′ terminal end of the passenger strand, and base preferences at positions 3 (A), 10 (U), and 13 (A, U, or C) of the passenger strand. To confirm the quality of LMW RNA, we amplified U6 small nuclear RNA by reverse transcribing LMW RNA with the U6br primer and then PCR amplifying with the primers U6af and U6br (Supplementary Table S1).
Results
VIGS in leaves and flowers of L. leichtlinii using CMV harboring a 33-nt LlPDS fragment
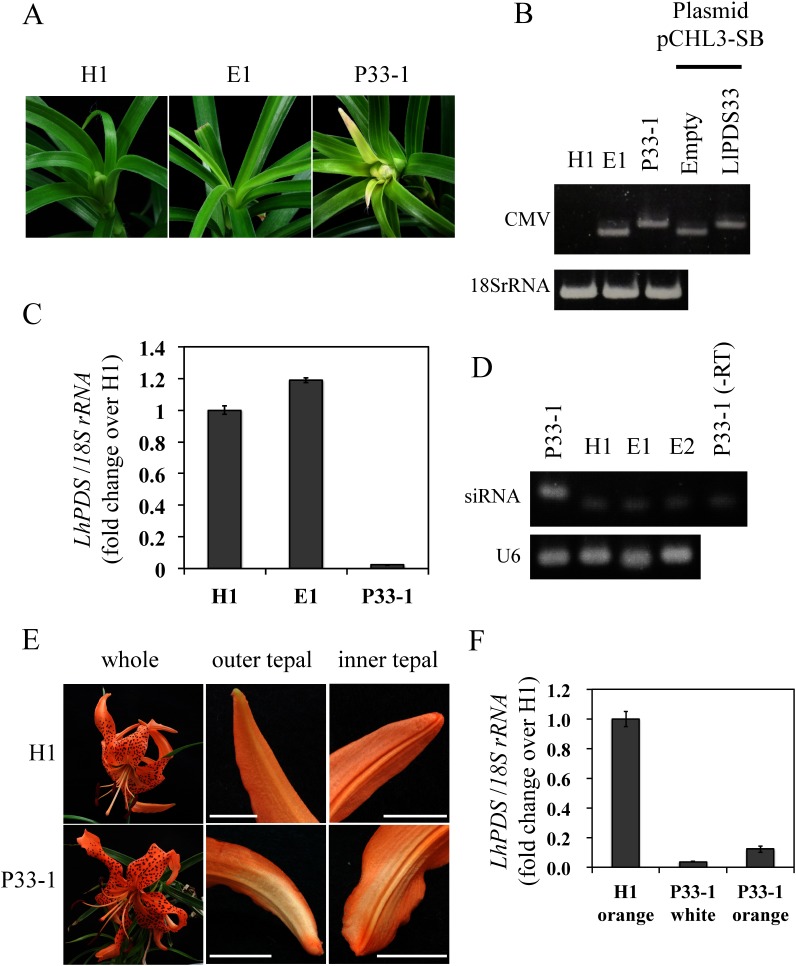
PDS silencing was induced by inoculating L. leichtlinii plants with CMV-HL that was modified to harbor LlPDS fragments. PDS encodes an enzyme that is essential for carotenoid biosynthesis and its suppression results in chlorophyll photo-bleaching in green tissues (Di Stilio et al. 2010). The photo-bleaching phenotype was observed in the leaves of one of the three plants that were infected with CMV-HL containing the 33-nt LlPDS fragment (named P33-1 plant) at 30 dpi (Figure 1A). RT-PCR confirmed the CMV-HL infection of upper leaves, and the length of the amplified fragments indicated that there was no deletion in the LlPDS fragment from the P33-1 plant (Figure 1B). The expression of LlPDS in the upper leaves of the P33-1 plant was reduced by 97.8% from that of an uninfected plant (hereafter, healthy plant, H1, Figure 1C). The production of siRNA is a hallmark of the occurrence of VIGS. The presence of siRNA from the 33-nt fragment of LlPDS was detected in the upper leaves of the P33-1 plant but was not detected in the upper leaves of plants inoculated with CMV-HL without the LlPDS inserts (hereafter, empty plants, E1 and E2) or a healthy plant (H1, Figure 1D). Sequencing of the 60-nt end-point PCR product confirmed that the amplicon contained the siRNA sequence.
Figure 1. Virus-induced gene silencing in Lilium leichtlinii (P33-1) inoculated with Cucumber mosaic virus (CMV) harboring a 33-nt LlPDS fragment. (A) Leaf phenotypes of healthy plant (H1), a plant inoculated with empty vector (E1), and a plant infected with CMV-HL harboring the 33-nt LlPDS fragment (P33-1) at 30 dpi. Leaf width was approx. 9 mm. (B) RT-PCR amplified the CMV RNA3 region including a cloning site to confirm insertion of the PDS fragment in upper leaves. Fragments amplified from the plasmid of pCHL3-SB without LlPDS fragment (Empty, 252 bp) and pCHL3-SB harboring 33-nt fragments of LlPDS (LlPDS33, 278 bp) are also shown. (C) Relative expression of LlPDS in upper leaves of H1, E1, and P33-1. Vertical bars indicate the standard error of three technical replicates. (D) End-point PCR amplification of siRNA (60-nt) in the upper leaves of the P33-1, H1 plants, and two plants inoculated with empty vector (E1 and E2). –RT: reverse transcriptase was not added at the pulsed RT reaction. U6: U6 small nuclear RNA (77 bp). (E) Phenotypes of flowers at 57 dpi. Orange color at the tip of both outer and inner tepals had faded in the P33-1 plant. Bar=1 cm. (F) Relative expression of LlPDS in the orange tepals of the H1 plant and color-faded (white) and orange tepal regions of the P33-1 plant. Vertical bars indicate the standard error of three technical replicates.
At 57 dpi, when the lilies were in full bloom, the orange tepal color had faded in P33-1 (Figure 1E); the color change was observed on the abaxial side of both the inner and outer tepals and detected near the tip of the tepals but not at other tepal regions. The expression levels of LlPDS in the white and orange tepal regions of the P33-1 plant were reduced to 3.8 and 12.4% of that in the orange tepals of the H1 plant, respectively (Figure 1F). These results indicated that the fading color in the tepals was caused by the suppression of carotenoid biosynthesis and that LlPDS was successfully silenced in the tepals of L. leichtlinii, as well.
Transmission of LlPDS silencing in the year following inoculation
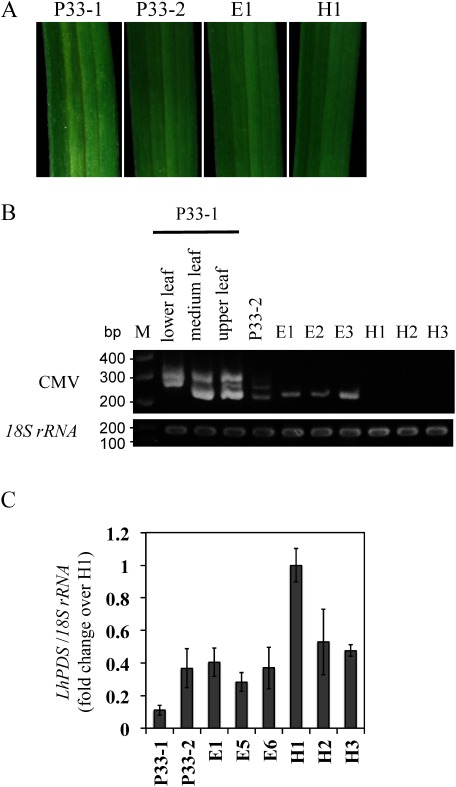
The P33-1 and P33-2 plants were grown in a succeeding year, in order to evaluate the transmission of PDS silencing. The P33-2 plant exhibited CMV-HL infection but no photo-bleaching in the first year, and three empty plants were newly generated in the successive season. Photo-bleaching was observed in the leaves of the P33-1 plant (Figure 2A); however, compared with the symptoms observed in the first year (Figure 1A), the photo-bleaching phenotype was mild, only observed around the veins, and was not observed in the upper leaves. In addition, no fading was observed in the orange color of the P33-1 flowers. No photo-bleaching was observed in leaves of the P33-2, empty (E1), and healthy (H1) plants. Viral fragments of the expected size (252 nt) were amplified from the empty plants, and no fragments were detected in the healthy plants. In the lower, middle, and upper leaves of the P33-1 plant, two or three bands appeared on a gel. According to their length, the smallest band should represent the viral fragment without the LlPDS insert, the intermediate-length band should represent the RNA3 fragment that includes the 33 nt LlPDS sequence, and the largest band should represent fragments generated by hybridization of the two smaller fragments. This result indicates that deletion of the LlPDS fragment occurs in the P33-1 plant. The P33-2 plant also yielded three bands, but the signal strength was weak when compared to that of the P33-1 plant, suggesting that the viral concentration was not high enough to induce PDS silencing (Figure 2B). The expression level of LlPDS in P33-1 leaves was low, whereas the expression of LlPDS in P33-2 leaves was similar to that in the empty and healthy plants (Figure 2C).
Figure 2. Transmission of LlPDS silencing in the year following inoculation. (A) Leaf phenotypes: Photo-bleaching appeared on the P33-1 leaf but not on the leaves of the P33-2, a plant inoculated with empty vector (E1), and a healthy plant (H1). Leaf width was approx. 9 mm. (B) RT-PCR amplified the CMV RNA3 region including a cloning site to confirm the possession of the LlPDS fragment in lower, middle, and upper leaves of the P33-1 plant, and the leaves of the P33-2, E1, E2, E3, H1, H2, and H3 plants. M: molecular weight marker. (C) Relative expression of LlPDS in leaves. Vertical bars indicate the standard error of three leaves.
VIGS in L. leichtlinii using CMV harboring a 52-nt LlPDS fragment
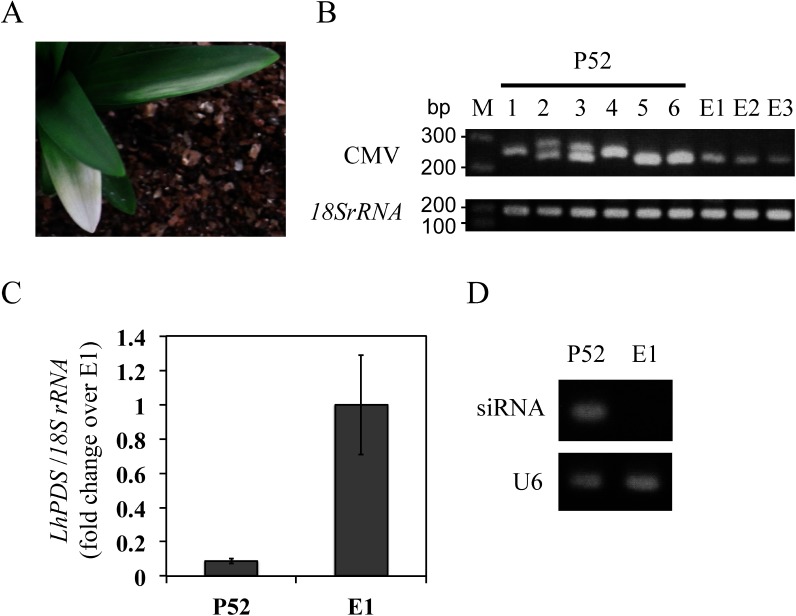
In the three L. leichtlinii plants infected with CMV-HL containing a 52-nt LlPDS fragment, no photo-bleaching phenotypes were observed. However, one of the plants (P52) exhibited the photo-bleaching phenotype in the successive season (Figure 3A). Systemic viral infection was confirmed using RT-PCR; the RNA3 sequence without the LlPDS fragment (252 nt) was amplified in empty plants E1, E2, and E3, whereas fragments with unexpected sizes were amplified from six randomly selected leaves (#1–6) of the P52 plant (Figure 3B). Sequence analysis indicated that at least three fragments were amplified from leaves #3 and #4. Fragment 1, which was detected in the both leaves, contained a 21-nt LlPDS sequence that was identical to the siRNA sequence. Meanwhile, fragments 2 and 3 were only detected in leaf #3, and fragment 2 only contained a 3-nt LlPDS fragment, whereas fragment 3 had lost the sequences of both LlPDS and the EcoRV restriction site (Supplementary Figure S3). These results indicate that intense rearrangements occurred around the cloning sites in the RNA3 viral genome during the process of dispersal. The expression level of LlPDS in the P52 leaves that exhibited photo-bleaching was reduced to 8.8% of that detected in the green leaves of the E1 plant (Figure 3C). Stem-loop pulsed RT and end-point PCR amplified a 60-nt fragment from the P52 plant (Figure 3D), and sequencing of the 60-nt fragment confirmed that the amplicon contained the siRNA sequence.
Figure 3. Virus-induced gene silencing in Lilium leichtlinii (P52) inoculated with Cucumber mosaic virus (CMV) harboring a 52-nt LlPDS fragment in the year after inoculation. (A) Photo-bleaching phenotype in the P52 plant. (B) RT-PCR amplified the CMV RNA3 region, including a cloning site, to confirm the insertion of the LlPDS fragment in six randomly selected leaves from the P52 plant (#1–6) and leaves of three plants (E1, E2, and E3) inoculated with empty vector. M: molecular weight marker. (C) Relative expression of LlPDS in six photo-bleached leaves from the P52 plant and four leaves of E1 plant. Vertical bars indicate the standard error of six (P52) and four (E1) leaves. (D) Detection of siRNA (stem-loop PCR, 60-nt) in leaves of the P52 and E1 plants. U6: U6 small nuclear RNA (77 bp).
Discussion
An abundance of Lilium gene sequences have been identified via NGS in an effort to characterize agriculturally important traits (Huang et al. 2014; Suzuki et al. 2016; Villacorta-Martin et al. 2015; Wang et al. 2014), and as a result, the establishment of reverse-genetics tools is an urgent issue for the functional characterization of the identified sequences. Because VIGS is able to alter plant phenotypes in relatively short periods of time and allows researchers to characterize phenotypes that might be lethal in stable transgenic plants, the technique should be valuable for the functional screening of expressed sequences. In the present study, we demonstrated that the newly developed CMV-HL vector was successful in silencing the expression of LlPDS in L. leichtlinii.
Systemic viral infection is essential for the successful application of VIGS. Although wild-type CMV-HL is able to infect lilies systemically, the modified CMV-HL vector with a truncated 2b gene failed to spread systemically in our preliminary experiment. Therefore, we introduced cloning sites just upstream of the 3b gene and demonstrated that the new vector successfully infected upper leaves and flowers (Figure 1). Moreover, the new CMV-HL vector was also preserved in dormant bulbs and spread into sprouting shoots in the following year (Figures 2 and 3). In CMV-derived viral vectors, the cloning sites of target genes are usually integrated at the downstream of 2b gene (Hong et al. 2012; Kanazawa et al. 2011; Otagaki et al. 2006) and in only one instance the cloning site has been introduced downstream of the 3b gene (Du et al. 2014). Therefore, the present study is the first to demonstrate that host plant gene fragments could be introduced just upstream of the 3b start codon without preventing systemic viral infection.
In the present study, we detected photo-bleaching leaf phenotypes and color-fading tepal phenotypes in L. leichtlinii plants infected with CMV-HL containing LlPDS fragments (Figures 1A, 1E, 2A, and 3A). A couple of pieces of evidence indicate that the silencing of the endogenous LlPDS gene contributed to the phenotypic changes. First, systemic virus infection was detected in the upper leaves. Second, the expression levels of LlPDS in the regions that exhibited phenotypic changes were notably decreased to 2.2–8.8% of the levels observed in the green leaves or orange tepals of healthy and empty plants, and finally, the siRNA molecules derived from the LlPDS fragments were detected in the upper leaves (Figures 1 and 3).
The observation of photo-bleaching at 30 dpi confirmed the ability of our VIGS system to influence host phenotypes within relatively short periods of time. Recently, VIGS was reported in an Oriental hybrid lily, in which the stem of cut lily flowers was immersed in a suspension of Agrobacterium tumefaciens containing TRV with a target lily gene fragment (Tong et al. 2013). However, although the authors observed reduced expression of the target gene, other indicators of successful VIGS, such as systemic viral infection and siRNA detection, were lacking. In particular, the confirmation of systemic viral infection should be indispensable, since lilies are not natural hosts for TRV (i.e., VIGS occurs only in regions infected by the virus; Pflieger et al. 2013; Senthil-Kumar and Mysore 2011b). Thus, it is not clear whether the suppression of endogenous gene expression in the Oriental hybrid lily was actually caused by gene silencing. In contrast, the CMV-induced PDS gene silencing observed in the present study was verified using several clear pieces of evidence.
LlPDS knockdown was induced in flowers of the P33-1 plant, which resulted in fading of the tepals’ orange color (Figure 1E). Because the colors, sizes, and shapes of flowers are among the most important characters in floriculture crops, the ability of CMV-HL to induce gene silencing in flower organs is undoubtedly valuable for functional studies of lily genes. In Figure 1E, a flower of P33-1 was crumpled. However, since the H1 flower was also mildly crumpled, this should be caused by the condition of a growth chamber, not caused by the vector inoculation.
In the faded regions of the P33-1 tepals, the expression level of LlPDS was 3.8% of that in the orange tepals of a healthy plant (H1). However, the faded regions were restricted to the tips of the tepals, and the expression level of LlPDS in the orange tepal regions of P33-1 was only reduced to 12.8% of that in the H1 tepals (Figure 1F). These results indicate that the expression levels of LlPDS were coincident with the fading of carotenoids and that threshold levels of LlPDS expression must be met for pigment biosynthesis, which is apparently somewhere between 3.8 and 12.4% of the steady-state expression level. Several pieces of evidence for the threshold mRNA levels of genes involved in flavonoid biosynthesis have been postulated. In CMV-induced silencing of the flavonoid 3′-hydroxylase (F3′H) gene in soybean pods, color changes occur when F3′H expression levels are lower than ∼3% of the steady-state expression level of the gene (Nagamatsu et al. 2009). Similarly, in the yellow seed coats of soybean (Senda et al. 2004) and white petal regions of the Red Star-type bicolor petunia (Koseki et al. 2005), the spontaneous PTGS of chalcone synthase (CHS) mRNA prevents anthocyanin biosynthesis, and even though low levels of CHS transcripts are detected at the yellow and the white regions, its levels are below the threshold, thus resulting in suppressed pigmentation.
In viral vector systems, constraints on the size of insert fragments have often been reported, where larger inserts are more susceptible to being lost from the vectors (Hsieh et al. 2013; Senthil-Kumar and Mysore 2011a; Yamagishi et al. 2015). In the present study, deletion of the 33-nt LlPDS fragment in the CMV-HL from the P33-1 plant was not detected in the first year, but was detected in the following year. On the other hand, intense rearrangements around the cloning site were observed in the CMV-HL from the P52 plant. In addition, in our preliminary experiment, we found that 67-, 69-, and 90-nt LlPDS fragments were all lost from the CMV-HL in N. benthamiana plants. These results indicate that insert size influences the stability of inserts in the CMV-HL vector and that ∼33-nt insertion sizes are suitable for the CMV-HL vector system.
Some reports have examined the transmittance of viral vectors to progenies. For example, Apple latent spherical virus is rarely transmitted via seeds to successive progenies in apples (Yamagishi et al. 2014), whereas the TRV vector can be transmitted via seeds to the progeny seedlings of N. benthamiana, which results in silencing phenotypes in ∼1% of the seedlings (Senthil-Kumar and Mysore 2011b). In the present study, we demonstrated that CMV-HL was transmitted via storage organs (bulbs) to shoots that developed in the year following inoculation and that the silencing phenotype was also observed in the second year (Figures 2 and 3). The transmittance of silencing phenotype will facilitate functional studies of the sequences expressed in bulbs and young shoots; e.g., sequences involved in bulb formation, bulb dormancy, and floral induction. However, in the present study, the photo-bleaching phenotypes observed during the second year were mild and the 33-nt LlPDS fragment was unstable (Figure 2B). Previous studies have reported that the frequency of gene silencing is usually reduced and the silencing phenotypes are usually weakened as the duration after inoculation increases, mainly owing to reduced concentrations of viral vectors or the loss of target gene sequences from the vector (Mei et al. 2016; Senthil-Kumar and Mysore 2011b). However, in the present study, we observed that a plant infected with CMV-HL harboring a 52-nt LlPDS fragment exhibited a clear photo-bleaching phenotype during the year following inoculation, even though the phenotype was not observed in the first year (Figure 3). Sequence analysis indicated that intense rearrangement had occurred around a cloning site and that the 21 nt LlPDS fragment, which had the same sequence as the siRNA, had been preserved in the CMV-HL vector. Because of size constraints, it is likely that 52-nt insert prevented viral propagation and movement, which prevented PDS silencing in the first year, and that, after rearrangement around the cloning site, the virus was able to spread and induce silencing.
The establishment of a VIGS system is important for the rapid analysis of gene function in plants that require a long period of growth. In the present study, we demonstrate that a VIGS system using the CMV-HL vector is able to rapidly induce photo-bleaching and color fading in the leaves and flowers of L. leichtlinii by silencing LlPDS. Future use of the CMV VIGS technique in lilies will facilitate the functional screening of genes involved in a variety floral characters.
Acknowledgments
The virus-free Lilium leichtlinii bulbs were kind gifts from the Hokuren Federation of Agricultural Cooperatives, Sapporo, Japan. This work was supported by a Grant-In-Aid for Scientific Research from the Japan Society for the Promotion of Science (nos. 24380014 and 15K14645).
Abbreviations
- CHS
chalcone synthase
- CMV
Cucumber mosaic virus
- dpi
days post inoculation
- LMW RNA
low molecular weight RNA
- NGS
next-generation sequencing
- PDS
phytoene desaturase
- PTGS
post-transcriptional gene silencing
- qRT-PCR
quantitative reverse transcription-PCR
- RT-PCR
reverse transcription-PCR
- siRNA
small interfering RNAs
- TRV
Tobacco rattle virus
- VIGS
virus-induced gene silencing
Supplementary Data
References
- Azadi P, Chin DP, Kuroda K, Khan RS, Mii M (2010) Macro elements in inoculation and co-cultivation medium strongly affect the efficiency of Agrobacterium-mediated transformation in Lilium. Plant Cell Tissue Organ Cult 101: 201–209 [Google Scholar]
- Di Stilio VS, Kumar RA, Oddone AM, Tolkin TR, Salles P, McCarty K (2010) Virus-induced gene silencing as a tool for comparative functional studies in Thalictrum. PLoS One 5: e12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Chen A, Chen W, Westwood JH, Baulcombe DC, Carr JP (2014) Using a viral vector to reveal the role of microRNA159 in disease symptom induction by a severe strain of Cucumber mosaic virus. Plant Physiol 164: 1378–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K, Kanazawa A, Kusaba M, Masuta C (2003) A simple and rapid method to detect plant siRNAs using nonradioactive probes. Plant Mol Biol Report 21: 51–58 [Google Scholar]
- Hagita T, Kodama F, Akai J (1989) The virus diseases of lily in Hokkaido. Ann Phytopathological Soc Jpn 55: 1–8 (in Japanese) [Google Scholar]
- Hamilton AJ, Baulcombe DC (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950–952 [DOI] [PubMed] [Google Scholar]
- Hong JS, Rhee SJ, Kim EJ, Kim TS, Ryu KH, Masuta C, Lee GP (2012) Application of a reassortant Cucumber mosaic virus vector for gene silencing in tomato and chili pepper plants. Plant Pathol J 28: 81–86 [Google Scholar]
- Hsieh MH, Lu HC, Pan ZJ, Yeh HH, Wang SS, Chen WH, Chen HH (2013) Optimizing virus-induced gene silencing efficiency with Cymbidium mosaic virus in Phalaenopsis flower. Plant Sci 201–202: 25–41 [DOI] [PubMed] [Google Scholar]
- Huang J, Liu X, Wang J, Lü Y (2014) Transcriptomic analysis of Asiatic lily in the process of vernalization via RNA-seq. Mol Biol Rep 41: 3839–3852 [DOI] [PubMed] [Google Scholar]
- Jeknić Z, Morré JT, Jeknić S, Jevremović S, Subotić A, Chen THH (2012) Cloning and functional characterization of a gene for capsanthin-capsorubin synthase from tiger lily (Lilium lancifolium Thunb. ‘Splendens’). Plant Cell Physiol 53: 1899–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa A, Inaba J, Shimura H, Otagaki S, Tsukahara S, Matsuzawa A, Kim BM, Goto K, Masuta C (2011) Virus-mediated efficient induction of epigenetic modifications of endogenous genes with phenotypic changes in plants. Plant J 65: 156–168 [DOI] [PubMed] [Google Scholar]
- Koseki M, Goto K, Masuta C, Kanazawa A (2005) The star-type color pattern in Petunia hybrida ‘red star’ flowers is induced by the sequence-specific degradation of the chalcone synthase RNA. Plant Cell Physiol 46: 1879–1883 [DOI] [PubMed] [Google Scholar]
- Kramer EM, Holappa L, Gould B, Jaramillo MA, Setnikov D, Santiago PM (2007) Elaboration of B gene function to include the identity of novel floral organs in the lower eudicot Aquilegia. Plant Cell 19: 750–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai YS, Shimoyamada Y, Nakayama M, Yamagishi M (2012) Pigment accumulation and transcription of LhMYB12 and anthocyanin biosynthesis genes during flower development in the Asiatic hybrid lily (Lilium spp.). Plant Sci 193–194: 136–147 [DOI] [PubMed] [Google Scholar]
- Liscombe DK, O’Connor SE (2011) A virus-induced gene silencing approach to understanding alkaloid metabolism in Catharanthus roseus. Phytochemistry 72: 1969–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Watanabe S, Uchiyama T, Kong F, Kanazawa A, Xia Z, Nagamatsu A, Arai M, Yamada T, Kitamura K, et al. (2010) The soybean stem growth habit gene Dt1 is an ortholog of Arabidopsis TERMINAL FLOWER1. Plant Physiol 153: 198–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Masuta C, Seshimo Y, Mukohara M, Jung HJ, Ueda S, Ryu KH, Choi JK (2002) Evolutionary characterization of two lily isolates of Cucumber mosaic virus isolated in Japan and Korea. J Gen Plant Pathol 68: 163–168 [Google Scholar]
- Matsuo K, Hong JS, Tabayashi N, Ito A, Masuta C, Matsumura T (2007) Development of Cucumber mosaic virus as a vector modifiable for different host species to produce therapeutic proteins. Planta 225: 277–286 [DOI] [PubMed] [Google Scholar]
- Mei Y, Zhang C, Kernodle BM, Hill JH, Whitham SA (2016) A foxtail mosaic virus vector for virus-induced gene silencing in maize. Plant Physiol 171: 760–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamatsu A, Masuta C, Matsuura H, Kitamura K, Abe J, Kanazawa A (2009) Down-regulation of flavonoid 3′-hydroxylase gene expression by virus-induced gene silencing in soybean reveals the presence of a threshold mRNA level associated with pigmentation in pubescence. J Plant Physiol 166: 32–39 [DOI] [PubMed] [Google Scholar]
- Nagamatsu A, Masuta C, Senda M, Matsuura H, Kasai A, Hong JS, Kitamura K, Abe J, Kanazawa A (2007) Functional analysis of soybean genes involved in flavonoid biosynthesis by virus-induced gene silencing. Plant Biotechnol J 5: 778–790 [DOI] [PubMed] [Google Scholar]
- Nakahara KS, Masuta C (2014) Interaction between viral RNA silencing suppressors and host factors in plant immunity. Curr Opin Plant Biol 20: 88–95 [DOI] [PubMed] [Google Scholar]
- Nakatsuka A, Izumi Y, Yamagishi M (2003) Spatial and temporal expression of chalcone synthase and dihydroflavonol 4-reductase genes in the Asiatic hybrid lily. Plant Sci 165: 759–767 [Google Scholar]
- Nakatsuka A, Yamagishi M, Nakano M, Tasaki K, Kobayashi N (2009) Light-induced expression of basic helix-loop-helix genes involved in anthocyanin biosynthesis in flowers and leaves of Asiatic hybrid lily. Sci Hortic (Amsterdam) 121: 84–91 [Google Scholar]
- Nakatsuka T, Saito M, Yamada E, Fujita K, Yamagishi N, Yoshikawa N, Nishihara M (2015) Isolation and characterization of the C-class MADS-box gene involved in the formation of double flowers in Japanese gentian. BMC Plant Biol 15: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogaki M, Furuichi Y, Kuroda K, Chin DP, Ogawa Y, Mii M (2008) Importance of co-cultivation medium pH for successful Agrobacterium-mediated transformation of Lilium×formolongi. Plant Cell Rep 27: 699–705 [DOI] [PubMed] [Google Scholar]
- Otagaki S, Arai M, Takahashi A, Goto K, Hong JS, Masuta C, Kazazawa A (2006) Rapid induction of transcriptional and post-transcriptional gene silencing using a novel Cucumber mosaic virus vector. Plant Biotechnol 23: 259–265 [Google Scholar]
- Pflieger S, Richard MMS, Blanchet S, Meziadi C, Geffroy V (2013) VIGS technology: An attractive tool for functional genomics studies in legumes. Funct Plant Biol 40: 1234–1248 [DOI] [PubMed] [Google Scholar]
- Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A (2004) Rational siRNA design for RNA interference. Nat Biotechnol 22: 326–330 [DOI] [PubMed] [Google Scholar]
- Senda M, Masuta C, Ohnishi S, Goto K, Kasai A, Sano T, Hong JS, MacFarlane S (2004) Patterning of virus-infected Glycine max seed coat is associated with suppression of endogenous silencing of chalcone synthase genes. Plant Cell 16: 807–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil-Kumar M, Mysore KS (2011a) New dimensions for VIGS in plant functional genomics. Trends Plant Sci 16: 656–665 [DOI] [PubMed] [Google Scholar]
- Senthil-Kumar M, Mysore KS (2011b) Virus-induced gene silencing can persist for more than 2 years and also be transmitted to progeny seedlings in Nicotiana benthamiana and tomato. Plant Biotechnol J 9: 797–806 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Suzuki T, Nakatsuka T, Dohra H, Yamagishi M, Matsuyama K, Matsuura H (2016) RNA-seq-based evaluation of bicolor tepal pigmentation in Asiatic hybrid lilies (Lilium spp.). BMC Genomics 17: 611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Tasaki K, Yamagishi M (2015) Two distinct spontaneous mutations involved in white flower development in Lilium speciosum. Mol Breed 35: 193 [Google Scholar]
- Suzuki T, Yamagishi M (2016) Aneuploids without bulbils segregated in F1 hybrids derived from triploid Lilium lancifolium and diploid L. leichtlinii crosses. Hort J 85: 224–231 [Google Scholar]
- Tasaki K, Atsumi G, Nishihara M, Sekine KT (2016) Development of a Broad bean wilt virus 2-based expression vector for gentian. Sci Hortic (Amsterdam) 201: 279–286 [Google Scholar]
- Tong Z, Li Q, Yang Y, Dai F, Gao J, Hong B (2013) Isolation and expression analysis of LoPIP2, a lily (Lilium Oriental Hybrids) aquaporin gene involved in desiccation-induced anther dehiscence. Sci Hortic (Amsterdam) 164: 316–322 [Google Scholar]
- Ui-Tei K, Naito Y, Takahashi F, Haraguchi T, Ohki-Hamazaki H, Juni A, Ueda R, Saigo K (2004) Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res 32: 936–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP (2007) Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villacorta-Martin C, Núñez de Cáceres González FF, de Haan J, Huijben K, Passarinho P, Lugassi-Ben Hamo M, Zaccai M (2015) Whole transcriptome profiling of the vernalization process in Lilium longiflorum (cultivar White Heaven) bulbs. BMC Genomics 16: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Yang Y, Liu X, Huang J, Wang Q, Gu J, Lu Y (2014) Transcriptome profiling of the cold response and signaling pathways in Lilium lancifolium. BMC Genomics 15: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wege S, Scholz A, Gleissberg S, Becker A (2007) Highly efficient virus-induced gene silencing (VIGS) in California poppy (Eschscholzia californica): An evaluation of VIGS as a strategy to obtain functional data from non-model plants. Ann Bot 100: 641–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi M (2013) How genes paint lily flowers: Regulation of colouration and pigmentation patterning. Sci Hortic (Amsterdam) 163: 27–36 [Google Scholar]
- Yamagishi M, Kishimoto S, Nakayama M (2010a) Carotenoid composition and changes in expression of carotenoid biosynthetic genes in tepals of Asiatic hybrid lily. Plant Breed 129: 100–107 [Google Scholar]
- Yamagishi M, Masuta C, Suzuki M, Netsu O (2015) Peanut stunt virus-induced gene silencing in white lupin (Lupinus albus). Plant Biotechnol 32: 181–191 [Google Scholar]
- Yamagishi M, Shimoyamada Y, Nakatsuka T, Masuda K (2010b) Two R2R3-MYB genes, homologs of petunia AN2, regulate anthocyanin biosyntheses in flower tepals, tepal spots and leaves of Asiatic hybrid lily. Plant Cell Physiol 51: 463–474 [DOI] [PubMed] [Google Scholar]
- Yamagishi N, Kishigami R, Yoshikawa N (2014) Reduced generation time of apple seedlings to within a year by means of a plant virus vector: A new plant-breeding technique with no transmission of genetic modification to the next generation. Plant Biotechnol J 12: 60–68 [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Seshimo Y, Yoshimoto E, Ahn HI, Ryu KH, Choi JK, Masuta C (2005) Genetic mapping of the compatibility between a lily isolate of Cucumber mosaic virus and a satellite RNA. J Gen Virol 86: 2359–2369 [DOI] [PubMed] [Google Scholar]
- Zhong X, Yuan X, Wu Z, Khan MA, Chen J, Li X, Gong B, Zhao Y, Wu J, Wu C, et al. (2014) Virus-induced gene silencing for comparative functional studies in Gladiolus hybridus. Plant Cell Rep 33: 301–312 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.