Abstract
Isoamylase (ISA) is a starch debranching enzyme that removes α-1,6-glucosidic linkages in α-polyglucans such as amylopectin. From previous studies, plant isoamylases have been shown to play a crucial role in amylopectin biosynthesis; however, little is known about their function in storage root tissues of plants such as cassava, yam and sweet potato. In this study, we isolated cDNA clones and characterized the cDNA nucleotide sequences of three genes (IbISA1, IbISA2, IbISA3) encoding isoamylase from sweet potato (Ipomoea batatas (L.) cv. White Star). Deduced amino acid sequences of the three isolated IbISAs have the specific regions that are highly conserved among the α-amylase family members. The product of IbISA2 is predicted to be enzymatically inactive, like other plant ISA2s, due to replacement of amino acid residues that are important for hydrolytic reaction. qRT-PCR analysis demonstrated that expression of IbISA2 was higher than that of the other two IbISAs (IbISA1 and IbISA3) in tuberous root at 109 days after planting, at which stage of tuberous root was at which stage tuberous roots were almost fully developed almost developed. This expression pattern observed in our experiments was different from that in other sink organs, such as seeds (endosperms), indicating that orchestration of ISA gene expression may depend on the differences in sink organ type between tuberous roots and seeds. The molecular characterization of three IbISA genes and their expression analysis in this study will contribute to further studies on starch biosynthesis in sweet potato, especially in storage root.
Keywords: genes encoding isoamylase-type starch debranching enzymes, Ipomoea batatas (L.), tuberous root
Plant starches, which are synthesized by photosynthesis, consist of two major types of glucose homopolymers: amylose and amylopectin (Smith et al. 1997). Amylose is essentially a linear molecule in which glucosyl monomers are joined via α-1,4 linkages, whereas amylopectin has a much more complex organization, in which linear α-1,4-glucan chains are regularly branched via α-1,6-glucosidic linkages (Dian et al. 2005; James et al. 2003). Many of the enzymes involved in starch metabolism have been investigated: at least four enzyme families have been found to participate in amylopectin biosynthesis, that is ADP-glucose pyrophosphorylase (AGPase, EC 2.7.7.27), soluble starch synthase (SS, EC 2.4.1.21), starch branching enzyme (SBE, EC 2.4.1.18), and starch debranching enzyme (DBE, EC 3.2.1.70), whereas amylose is synthesized by ADP-glucose pyrophosphorylase (AGPase) and granule-bound starch synthase (GBSS, EC 2.4.1.21) (Ball and Morell 2003; Hwang et al. 2005; Jeon et al. 2010; Myers et al. 2000; Nakamura 2002; Smith et al. 1997). Among the four enzyme families involved in amylopectin biosynthesis, DBEs are involved in the removal of α-1,6-glucosidic linkages. In plants, DBEs can be classified into two types: isoamylase (ISA, EC 3.2.1.68) and pullulanase (PUL, EC 3.2.1.41 or limit dextrinase, EC 3.2.1.42) (Nakamura et al. 1996). Both ISA and PUL hydrolyze α-1,6-glucosidic linkages, but they differ in their substrate specificity: ISA catalyzes amylopectin, glycogen and phytoglycogen but scarcely attacks pullulan, whereas PUL can catalyze pullulan and amylopectin, but not glycogen and phytoglycogen (Nakamura et al. 1996). Furthermore, the ISA type enzymes can be divided into three isoforms, designated ISA1, ISA2, and ISA3 (Hussain et al. 2003), whereas only one isoform of the PUL type is present (Dinges et al. 2003).
The importance of ISA for correct amylopectin biosynthesis has been suggested by several studies in maize (Beatty et al. 1999; James et al. 1995; Rahman et al. 1988), rice (Nakamura 1996; Nakamura et al. 1996; 1997), barley (Burton et al. 2002), Arabidopsis (Delatte et al. 2005; Wattebled et al. 2005), potato (Hussain et al. 2003) and Chlamydomonas (Ball et al. 1996; Mouille et al. 1996). Current evidence suggests that plant ISA is active as a complex containing different isoforms (Dauvillée et al. 2001; Fujita et al. 1999; Hussain et al. 2003). In the synthesis of potato storage starch, StISA1 and StISA2 are active as hetero-oligomers (Hussain et al. 2003). In Arabidopsis leaves, a source organ, AtISA1 and AtISA2 are active as hetero-oligomers, as in potato tubers (Delatte et al. 2005; Wattebled et al. 2005). In other sink organs, endosperms of rice and maize, both ISA1/ISA2 hetero-oligomer and ISA1 homo-oligomer were observed (Kubo et al. 2010; Utsumi and Nakamura 2006). When ISA1 and ISA2 form a hetero-oligomer, ISA1 has a direct catalytic role but the ISA2 subunit is likely to have a regulatory function because ISA2 is non-catalytic, due to substitutions of 6 out of 8 key amino acids within the active site (Hussain et al. 2003; Utsumi and Nakamura 2006). There are four regions (designated Region I, II, III and IV) that are highly conserved in the α-amylase family (GH13 family), and some amino acids in these conserved regions play an important role in activity of the enzyme (Lawson et al. 1994; Strokopytov et al. 1996). In particular, histidine (His (H)) in Region I, II, and IV (position His-137, His-269, and His-361 of Bacillus sp. strain TS-23 α-amylase) are important for correct catalytic activity (Chang et al. 2003). In contrast, ISA3 does not form a complex with ISA1 and ISA2, indicating that ISA3 functions as a monomer (Hishinuma et al. 2004; Takashima et al. 2007). However, in contrast to these findings, Arabidopsis AtISA2 was found to be co-expressed with AtISA3 in the absence of AtISA1 expression (Li et al. 2007).
In higher plants, there are specific sink organs, including seeds (endosperms), tuberous stems and tuberous roots. The expression of ISAs in relation to seed development has been investigated in several plant species, including rice, maize, barley, rye and amaranths (Kubo et al. 2010; Ohdan et al. 2005; Park et al. 2014; Sun et al. 1999; Zheng et al. 2013). However, in the case of tuberous root species, sweet potato (Convolvulaceae), cassava (Euphorbiaceae), and yam (Dioscoreaceae) (Scott et al. 2000), there are few reports of cloned and characterized ISAs (Beyene et al. 2010; Kim et al. 2005). Furthermore, the relationship between gene expression of ISAs and sink development is not well understood in tuberous root plants.
In the work reported here, we isolated the genes encoding IbISA1, IbISA2, and IbISA3 from one of the major tuberous root plant, sweet potato (Ipomoea batatas (L.)), and compared the amino acid sequences with those of other plant species with different types of sink (tubers and seed endosperms) and source (leaves). We concluded that IbISA2 does not have isoamylase enzyme activity, like ISA2 from other plant species. Furthermore, we determined the gene expression patterns of IbISAs by qRT-PCR, and examined their expression during tuberous root development. These three genes were spatiotemporally regulated in root (sink) and leaves (source) in sweet potato. Based on these results, we discuss the function of the three IbISAs in tuberous root hypertrophy.
Materials and methods
Plant Materials
Ipomoea batatas (L.), cultivar “White Star” plants were grown in the field. Mature leaves and storage roots were collected for cloning of genes for IbISAs. According to developmental stage, leaves and roots were also collected for qRT-PCR analysis.
Cloning and sequencing of genes encoding isoamylase-type starch debranching enzyme
Total RNAs were extracted from tuberous roots (ca. 2 g) using RNeasy Plant Mini Kit (Qiagen, Basel, Switzerland). From total RNAs, poly (A)+ RNAs were purified with Oligotex™-dT <Super> mRNA Purification Kit (TaKaRa Shuzo, Shiga, Japan) according to the manufacturer’s instructions. In order to isolate the full-length cDNA clones for genes encoding isoalmylase-type starch debranching enzymes 1, 2, and 3 (IbISA1, IbISA2, and IbISA3), the RACE method was used. Briefly, first-strand cDNAs for 5′-RACE and 3′-RACE analyses were synthesized with SMARTer™ RACE cDNA amplification kit (Clontech, Palo Alto, CA, USA) according to the manufacturer’s instructions.
The 5′-RACE and 3′-RACE fragments for IbISA1, IbISA2, and IbISA3 genes were amplified with first-strand cDNA as a template according to the manufacturer’s instruction for the SMARTer™ RACE cDNA amplification kit (Clontech). Specific primer sets of each gene were used, as shown in Table S1. The PCR products of the RACE reaction were cloned in pTA2 vector (TOYOBO, Osaka, Japan). Inserts in the plasmid vector were sequenced by the dideoxy chain-termination method by using a model 3130xl DNA sequencer (Applied Biosystems, Foster City, CA, USA) with BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems). Sequence data were analyzed by DNASIS for Windows (Hitachi Software Engineering, Yokohama, Japan).
On the basis of partial sequence cloned by the RACE method, additional primer sets were also designated to amplify the full-length cDNA of each IbISA gene. The nucleotide sequences of specific primer sets for each IbISA gene are shown in Table S1. The cloning and determination of the nucleotide sequence the PCR products were performed as described above.
Sequence analysis, sequence comparison and construction of phylogenetic trees
Sequence analysis was performed by using DNASIS for Windows (Hitachi Software Engineering) and BLAST program (http://ncbi.nlm.nih.gov/BLAST/; Altschul et al. 1990). For the phylogenetic analysis, amino acid sequences of ISAs from several plant species were obtained from the NCBI database (http://www.ncbi.nlm.nih.gov). Phylogenetic trees were constructed by the neighbor joining (NJ) method using GENETYX ver.11 (Software Development Co., Japan; Saitou and Nei 1987). Bootstrap values were calculated from 1,000 replications.
Quantitative real-time PCR
Total RNA and mRNA from leaves and tuberous roots were isolated as described above. Real-time PCR reactions were performed on a 7300 Real-time PCR system (Applied Biosystems, Foster City, CA, USA) with KOD SYBR® qPCR Mix (TOYOBO, Osaka, Japan), according to the manufacturer’s instructions. Primers sets used in qRT-PCR are also described in Table S1. IbCOX encoding cytochrome c oxidase subunit Vc and IbARF encoding ADP-ribosylation factor genes were used as reference genes.
Results
Isolation and characterization of IbISA1, IbISA2, and IbISA3 cDNA clones expressed in tuberous roots
cDNAs were reverse-transcribed from mRNA isolated from tuberous root of sweet potato, cultivar White Star. IbISA1, IbISA2, and IbISA3 sequences were then amplified from the cDNAs. To design primer sequences for amplification of IbISAs, the partial and/or full-length amino acid sequences of ISA1, ISA2, and ISA3 from several plant species (Arabidopsis thaliana: accession no. AEC09752, AEE27558, AEE82713, Zea mays: accession no. ACG43008, AAB97167, AAA91298, AAO17048, AAO17049, Oryza sativa: accession no. NP_001062271, BAA29041, ACY56088, ACY56099, BAC75533, Solanum tuberosum: accession no. AAN15317, AAN15318, AAN15319, Hordeum vulgare: accession no. AAM46866, Triticum aestivum: accession no. CAC82925, CAC41016, Pisum sativum: accession no. AAZ81835, AAZ81836, AAZ81837, Phaseolus vulgaris: accession no. BAF52941, BAF52943, BAF52942, Medicago truncatula: accession no. XP_003630623, XP_003602838, Aegilops tauschii: accession no. AAP44579, Secale cereale: accession no. ACM41701, and Ipomoea batatas: accession no. AAY84833) were obtained from public databases. These sequences were classified into three clusters (corresponding to ISA1, ISA2, and ISA3) by the neighbor joining method with GENETYX (data not shown).
To determine the conserved amino acid sequences of ISA1, ISA2, and ISA3, deduced amino acid sequences were aligned. In a BLAST search (Alschul et al. 1990) using each ISA conserved consensus sequence as a query in public databases, ISA1, ISA2, and ISA3 genes from I. batatas were identified as follows. In the case of IbISA1, a gene termed Ibisa1 (DQ074643) had previously been isolated from sweet potato, using a different cultivar, Kokei 14, from that used in the present study (Kim et al. 2005). Furthermore, in the transcriptome analysis data deposition (Tao et al. 2012), TSA (transcriptome shotgun assembly) for IbISA2 (accession number, JP111226) and IbISA3 (accession number, JP104934) were identified. A phylogenetic tree was constructed using these three I. batatas genes identified in databases and other ISA genes from several plant species. These three I. batatas genes were classified into the corresponding ISA1, ISA2, and ISA3 clusters, indicating that they are predicted to be functional genes. In the present study, from conserved sequences in three I. batatas ISA genes, degenerate primer sets (Table S1) were designed and used in PCR to obtain partial cDNA clones of IbISAs (IbISA1, IbISA2, and IbISA3) by 5′-RACE and 3′-RACE methods.
The nucleotide sequences of amplified fragments were determined and compared to ISA genes isolated from other plant species. This confirmed that the fragments amplified by 5′-RACE and 3′-RACE contained the partial sequence of IbISA1, IbISA2, and IbISA3 genes (data not shown). Based on the nucleotide sequence from the 5′-RACE and 3′-RACE experiments, full-length cDNA clones for IbISA1, IbISA2, and IbISA3 genes were amplified with primer sets as shown in Table S1. For each gene, the complete nucleotide sequences of eight independent clones were determined. Interestingly, for each gene, IbISA1, IbISA2, and IbISA3, the independent clone sequences were identical. The full-length cDNAs of IbISA1, IbISA2 and IbISA3 were 2,524 bp, 2,880 bp, and 2,919 bp long, respectively. The nucleotide sequence data for IbISA1, IbISA2 and IbISA3 genes have been deposited in the DDBJ, EMBL, and GenBank databases with accession numbers LC052789, LC052790, and LC052791, respectively.
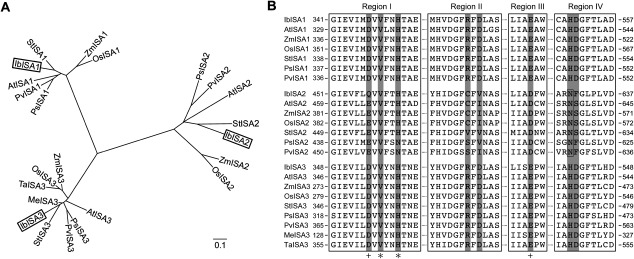
In order to discover whether these clones code for isoamylase enzymes, amino acid sequences deduced from the IbISA1, IbISA2 and IbISA3 cDNA clones were analyzed. The deduced amino acid sequences of the IbISAs cDNA clones suggested that they encode polypeptides of 799, 865 and 768 amino acid residues, respectively. These deduced amino acid sequences were then aligned to ISAs from other plant species. IbISA1, IbISA2 and IbISA3 isolated from sweet potato in this experiment formed clusters with ISA1, ISA2 and ISA3 from other species, respectively (Figure 1A). The deduced amino acid sequence of IbISA1 showed 95.1% similarity to Ibisa1 (accession no. AAY84833), 81.3% identity with StISA1 (accession no. AAN15317), 70.2% identity with AtISA1 (accession no. AEC09752) and 67.2% identity with ZmISA1 (accession no. EU970890) in the European Molecular Biology Open Software Suite (EMBOSS; Rice et al. 2000). The deduced amino acid sequence of IbISA2 showed 62.4% similarity with StISA2 (accession no. AAN15318), 52.1% identity with AtISA2 (accession no. AEE27558) and 43.6% identity with ZmISA2 (accession no. AAO17048). The deduced amino acid sequence of IbISA3 showed 74.7% similarity with StISA3 (accession no. AAN15319).
Figure 1. Phylogenetic tree of ISAs based on deduced amino acid sequences and sequence alignment of ISAs in four conserved regions. (A) Phylogenetic tree of ISAs based on deduced amino acid sequences. The tree was constructed using the neighbor-joining method (Saitou and Nei 1987). Scale bar represents evolutionary distance. Bootstrap values were calculated from 1,000 replications. The deduced amino acid sequences of sweet potato IbISAs protein (in box) were derived in this study (IbISA1, accession LC052789; IbISA2, accession LC052790; IbISA3, accession LC052791). Accession numbers for genes encoding ISAs of other plant spices are: Arabidopsis thaliana AtISA1, AEC09752; AtISA2, AEE27558; AtISA3, AEE82713; potato (Solanum tuberosum) StISA1, AAN15317; StISA2, AAN15318; StISA3, AAN15319; maize (Zea mays) ZmISA1, EU970890; ZmISA2, AAO17048; ZmISA3, AAO17049; rice (Oryza sativa) OsISA1, BAC75533; OsISA2, AAT93894; OsISA3, XP_450961; pea (Pisum sativum) PsISA1, AAZ81835; PsISA2, AAZ81836; PsISA3, AAZ81837; common bean (Phaseolus vulgaris) PvISA1, BAF52941; PvISA2, BAF52942; PvISA3, BAF52943; sweet potato (Ipomoea batatas) IbISA1, AAY84833; wheat (Triticum aestivum) TaISA3, AEV92948; cassava (Manihot esculenta) MaISA3, ADD10143. (B) Multiple sequence alignment of ISAs in four conserved regions. Amino acid sequences in Regions I to IV are highly conserved in the α-amylase family. In particular, eight amino acid residues (shaded dark gray) are highly conserved in all active members of the α-amylases, ISA1 and ISA3. Region IV of ISA2s shows replaced residues from H (His) to N (Asn).
Alignment of the amino acid sequences of the three IbISAs revealed that they have four conserved regions (Region I to IV), as present in other α-amylase family (GH13 family) starch hydrolytic enzymes (Beatty et al. 1999; James et al. 1995; Jespersen et al. 1993). In particular, within these four conserved regions, eight amino acid residues are highly conserved in all active members of the α-amylase family (Hussain and Martin 2009; Hussain et al. 2003). In comparison with other deduced amino acid sequences of ISAs, these eight amino acid residues were conserved in IbISA1 and IbISA3, as in other ISA1s and ISA3s, indicating that IbISA1 and IbISA3 are predicted to be active α-amylase enzymes. However, in the case of IbISA2, as in other plant ISA2, six of the eight conserved residues were replaced by different amino acid residues compared to the conserved residues in ISA1 and ISA3 (Figure 1B; dark gray shading). As shown in Figure 1B, in Region I, Val (V) and His (H), indicated by (*), were conserved in ISA2s, as in ISA1s and ISA3s. Other two sites in Region I and III indicated by (+) retained functionally similar amino acid residues, Asp (D) and Glu (E), in IbISA1 and IbISA3 but not in IbISA2, indicating that these four amino acids are sufficient for α-amylase enzyme activity. In contrast, at the other four positions, amino acid residues conserved in ISA1s and ISA3s were replaced in ISA2s by amino acids with different properties (Figure 1B). In particular, the replacement of His (H) by Asn (N) in Region IV is predicted to result in loss of function in hydrolytic enzymes, such as S-RNase (Royo et al. 1994).
Spatiotemporal expression of IbISA1, IbISA2, and IbISA3 genes in sweet potato
In order to determine whether IbISA1, IbISA2, and IbISA3 are regulated spatiotemporally in sweet potato, qRT-PCR was performed. To relate the spatiotemporal expression pattern of the three IbISA genes to plant development, we observed the morphology of roots. Until about 50 days after planting (DAP), no change in gross root morphology was observed (data not shown). At about 60 DAP, adventitious roots were present but the distinction between potential storage roots and normal roots was unclear. The average fresh weight of roots was 37 g at 60 DAP (Figure S1B). At 75 to 80 DAP, hypertrophy of roots was observed but there were individual differences within a plant of root size. The average fresh weight of roots was 133 g (Figure S1B). At 95 to 100 DAP, rapid growth of tuberous roots was observed and the average fresh weight of roots was 315 g (Figure S1B). At 110 DAP, larger tuberous roots were present and the average fresh weight of roots was 659 g (Figure S1B). Therefore, we collected leaves (source) and tuberous roots (sink) for qRT-PCR analysis at approximately 10 days intervals from 48 DAP (that is at 48, 57, 67, 78, 90, 99 and 109 DAP).
Next, the optimum set of reference genes for qRT-PCR was determined. Generally, genes encoding β-actin (ACT) and/or α-tublin (TUB) are used as reference genes (Osaka et al. 2013), but there are no ideal references genes able to fulfill all experimental requirements, and so the selection of optimum reference genes is essential (Bustin et al. 2009; Park et al. 2012). In the case of sweet potato, according to variety and/or environmental conditions, expression of reference genes differed (Park et al. 2012). Thus, for this study, to determine suitable reference genes in the variety White Star, under non-stress condition, seven genes encoding β-actin (ACT), α-tubulin (TUB), cytochrome c oxidase subunit Vc (COX), ubiquitin extension protein (UBI), ADP-ribosylation factor (ARF), phospholipase D1α (PLD), and histone (H2B) were surveyed for gene expression stability in qRT-PCR. From this survey, COX and ARF genes were found to be expressed at constant levels, and their expression levels were stable in different organs, growth stages, and sampling times (data not shown). Thus, COX and ARF were selected as reference genes in this experiment. In addition to primer sets for these two reference genes, three sets of specific primers, discriminating the three genes (IbISA1, IbISA2, and IbISA3) were designed, as shown in Table S1. Expression levels of each IbISAs, shown in Figures 2 and S1, were designated as relative values compared to expression levels of ARF and COX designated at 100.
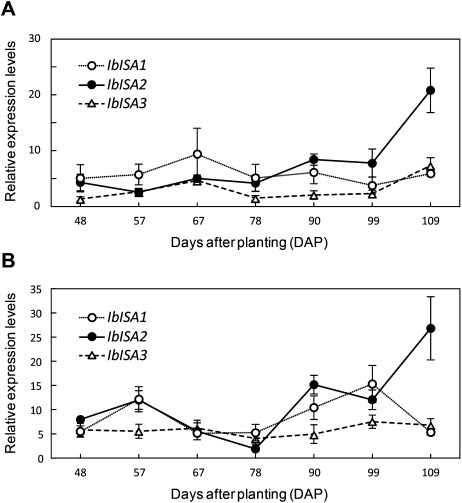
Figure 2. Spatiotemporal expression of ISAs in sweet potato root and leaf. (A) Relative transcript abundances in wild-type tuberous root. (B) Relative transcript abundances in wild-type leaves. (A and B) Results are mean±SE of four biological replicates. When absent, the error bars are smaller than the symbols.
From examination of gene expression patterns in tuberous roots (sink), expression levels of the three ISA genes were constant until 99 DAP (Figure 2A). However, at 109 DAP, the IbISA2 expression level was significantly up-regulated to more than 2-fold and approximately 2.5-fold compared to that of IbISA1 and IbISA3, respectively (Figure 2A). In the leaves (source), the IbISA3 expression pattern was relatively constant at all seven stages (Figure 2B). The expression pattern of IbISA1 and IbISA2 in leaves was similar until 99 DAP, followed by a higher level of IbISA2 expression at 109 DAP, the mature stage (Figure 2B). From a comparison between tuberous root development (indicated by DAP) and the expression of IbISA, the increase in tuberous root development correlated with the expression pattern of IbISA2, but not of IbISA1 and IbISA3 (Figure S1A). Thus, at least in the morphologically mature tuberous roots, the expression of the IbISA2 appears to be related to the accumulation of starch; that is, morphological maturation and gene function of IbISAs are linked during tuberous root development in I. batatas. In contrasts, the expression patterns of ISAs in seeds, another type of sink (Kubo et al. 2010; Ohdan et al. 2005; Park et al. 2014; Sun et al. 1999; Zheng et al. 2013), was different from our data in sweet potato, as discussed below.
Discussion
Molecular characterization of isoamylase genes in sweet potato
The genes encoding three isoforms of ISA in plants have been isolated and characterized in various plant species, including rice, maize and potato, which store starch in seeds and stems (Fujita et al. 1999; Hussain et al. 2003). However, few studies have focused on tuberous root plants, such as cassava, sweet potato, and yam, which store their starch in roots (Beyene et al. 2010; Kim et al. 2005). In this study, we isolated three genes encoding IbISA1, IbISA2, and IbISA3 from one of the tuberous root plants, sweet potato (Ipomoea batatas (L.)). Isolation of Ibisa1 (DQ074643) has been reported in a previous study, which used a different cultivar, Kokei 14 (Kim et al. 2005); however, the current study is the first to report cloning of genes encoding all three types of ISA from sweet potato. The sequence diversity (4.9%) between the two ISA1 genes from different cultivars (IbISA1 and Ibisa1) could be due to the self-incompatibility trait in sweet potato (Martin 1965). In addition to this characteristics, the three IbISA genes were more similar to corresponding ISA genes of other dicotyledonous plants (Solanum tuberosum, Arabidopsis thaliana, Phaseolus vulgaris, Pisum sativum and Manihot esculenta) than those of monocotyledonous plants (Zea mays and Oryza sativa) in the phylogenetic tree (Figure 1A), suggesting the ISA gene speciation occurred after diversification of dicotyledonous and monocotyledonous plants, as found in other functional genes (Nakayama et al. 2010). Furthermore, within dicotyledonous plants, the nucleotide sequences of ISAs of sweet potato have higher similarity to those of potato and cassava ISAs than those of other plant species. This high sequence similarity among these species may be related to the formation of starch storage organs.
From the sequence alignment of ISAs from sweet potato and other species (Figure 1B), deduced amino acid sequences of the three IbISA genes isolated in this study contain four highly conserved regions (Region I to IV), which are classified into the α-amylase enzyme family, as reported for other ISAs from several plants (Jespersen et al. 1993; James et al. 1995; Beatty et al. 1999). In the case of IbISA2, six of eight residues thought to be important for α-amylase activity were replaced by other amino acid residues (Figure 1B; gray shading), indicating that IbISA2 has no catalytic function, as proposed for ISA2s from other plant species (Hussain et al. 2003; Utsumi and Nakamura 2006). In particular, histidine residue (His) is important for the hydrolysis reaction catalyzed by several enzymes (Chang et al. 2003; Royo et al. 1994). When His was replaced by another amino acid residue, hydrolytic enzyme activity was lost in S-RNase (Royo et al. 1994) and α-amylase (Chang et al. 2003). In the enzymatically active ISA1 and ISA3, two His residues within Regions I and IV were conserved. In contrast, ISA2 had only one conserved histidine residue in Region I, but in Region IV, His was replaced by another residue (Figure 1B; Region IV box), suggesting that His in Region IV may be particularly important for hydrolysis of α-1,6-glucosidic linkages.
Spatiotemporal expression of IbISA genes during tuberous root development
In a previous report, the expression pattern of sweet potato ISA1 gene was observed up to 60 DAP (Kim et al. 2005). However, tuber development continues after 60 DAP and expression of all three ISAs in sweet potato are expected to play an important role in tuber maturation and starch storage after 60 DAP. The tuberous roots of sweet potato are harvested at around 90 DAP (Woolfe 1992), and the harvesting period varies from 90 to 120 DAP in different cultivars (Ravi et al. 2009). In the case of cultivar White Star used in this experiment, the dry weight of tuberous roots is maximum at about 105 to 119 DAP (La Bonte et al. 2000), and therefore at 109 DAP morphogenesis of roots would have reached the hypertrophy stage. In addition, the weight of the storage root was found to increase according to the accumulation of photosynthetic products such as starch (Wilson 1982), and the dry weight of sweet potato correlates with starch content of tuberous root at different plant development stages (Li and Liao 1983). As described by Noda et al. (1992), the percentage of amylose content against to dry weight does not increase according to growth of sweet potato tuberous root. In our experiment, the expression of IbISA2 gene was correlated to tuberous root development, indicating that the content of amylose and amylopectin should be precisely regulated by ISA2 and related genes in tuberous root development. From our gene expression data and the above information on root development, IbISA2 expression was highest in roots at the hypertrophy stage (around 109 DAP), and therefore IbISA2 is expected to function in amylopectin biosynthesis in the sink (roots) of sweet potato with tuberous roots.
The IbISA2 expression pattern in sweet potato was different from that of ISAs found in rice, rye, barley, maize, and amaranths (Kubo et al. 2010; Ohdan et al. 2005; Park et al. 2014; Sun et al. 1999; Zheng et al. 2013). In rice, both ISA2 and ISA3 were expressed at a very low level, and ISA1 had the highest expression around the middle stage of seed developmental, indicating that in the case of seeds as a sink, ISA1 is the main ISA active in amylopectin biosynthesis (Kubo et al. 2010). Thus the expression pattern of ISAs might be related to the pattern of the sink development. In the case of tuberous roots, sink size would be greater as roots develop. However, in the case of seeds (endosperms) of Poaceae, sink size is restricted by that of the lemma and palea. This difference in development patterns of sinks between seeds and tuberous roots may contribute to the different expression patterns of ISAs. In any case, orchestration of expression of the three ISAs genes, in both seeds and tuberous roots, is expected to be important for sink organ development.
As described above, IbISA2 is expected to be non-functional as an enzyme from the predicted amino acid sequence but showed a high level of expression at the mature stage of tuberous root development. This higher expression of non-enzymatically active ISA2 and low expression of enzymatically active ISA1 resembles the expression of cell-cell recognition genes, SLG and SRK, of the self-incompatibility system in Brassica species (Watanabe et al. 2012). The S-domain of SRK, the receptor for the pollen ligand SP11, is highly similar to SLG, and SRK interacts with SP11 but SLG does not. SLG expression is over 10 times higher than SRK. Interestingly, SLG is required for a full manifestation of the self-incompatibility response (Takasaki et al. 2000). Thus, like the SLG function in self-incompatibility in Brassica species, the actual function of ISA2 may be discovered by different approaches in future.
In conclusion, from our data and from other plant species, the three ISA genes appear to be regulated spatiotemporally in sinks (endosperms, tubers, or roots) and sources (leaves) for amylopectin biosynthesis in different plant species. From the present study, higher expression of IbISA2 may play an important role in amylopectin biosynthesis during root hypertrophy in sweet potato. Furthermore, orchestration of ISAs gene expression was different the different sink organs, that is seeds and tuberous roots, indicating that ISAs may contribute to the differences in quality and quantity of amylopectin in seeds and tuberous roots.
Supplementary Data
Acknowledgments
We thank all members of the DNA Analysis Technology Education Center in Ishikawa Prefectural University for helpful discussion on this work. The authors are also grateful to Kana Ito (Tohoku University) for technical assistance. This work was supported in part by JSPS KAKENHI Grant Numbers 23113006, 23113001, 25252001, 16H04854, 16K15085, 16H06470, 16H06464, 16J01836. M.N. is recipients of a Research Fellowship for Young Scientists from JSPS.
Abbreviations
- DAP
days after planting
- IbISAs
isoamylase-type starch debranching enzymes of Ipomoea batatas (L.) Lam.
- ISA
isoamylase
- qRT-PCR
quantitative RT-PCR
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Ball S, Guan HP, James M, Myers A, Keeling P, Mouille G, Buléon A, Colonna P, Preiss J (1996) From glycogen to amylopectin: A model for the biogenesis of the plant starch granule. Cell 86: 349–352 [DOI] [PubMed] [Google Scholar]
- Ball SG, Morell MK (2003) From bacterial glycogen to starch: Understanding the biogenesis of the plant starch granule. Annu Rev Plant Biol 54: 207–233 [DOI] [PubMed] [Google Scholar]
- Beatty MK, Rahman A, Cao H, Woodman W, Lee M, Myers AM, James MG (1999) Purification and molecular genetic characterization of ZPU1, a pullulanase-type starch-debranching enzyme from maize. Plant Physiol 119: 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyene D, Baguma Y, Mukasa SB, Sun C, Jansson C (2010) Characterisation and role of Isoamylase1 (MEISA1) gene in cassava. Afr Crop Sci J 18: 1–8 [Google Scholar]
- Burton RA, Jenner H, Carrangis L, Fahy B, Fincher GB, Hylton C, Laurie DA, Parker M, Waite D, Van Wegen S, et al. (2002) Starch granule initiation and growth are altered in barley mutants that lack isoamylase activity. Plant J 31: 97–112 [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, et al. (2009) The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55: 611–622 [DOI] [PubMed] [Google Scholar]
- Chang CT, Lo HF, Chi MC, Yao CY, Hsu WH, Lin LL (2003) Identification of essential histidine residues in a recombinant α-amylase of thermophilic and alkaliphilic Bacillus sp. strain TS-23. Extremophiles 7: 505–509 [DOI] [PubMed] [Google Scholar]
- Dauvillée D, Colleoni C, Mouille G, Buléon A, Gallant DJ, Bouchet B, Morell MK, D’Hulst C, Myers AM, Ball SG (2001) Two loci control phytoglycogen production in the monocellular green alga Chlamydomonas reinhardtii. Plant Physiol 125: 1710–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte T, Trevisan M, Parker ML, Zeeman SC (2005) Arabidopsis mutants Atisa1 and Atisa2 have identical phenotypes and lack the same multimeric isoamylase, which influences the branch point distribution of amylopectin during starch synthesis. Plant J 41: 815–830 [DOI] [PubMed] [Google Scholar]
- Dian W, Jiang H, Wu P (2005) Evolution and expression analysis of starch synthase III and IV in rice. J Exp Bot 56: 623–632 [DOI] [PubMed] [Google Scholar]
- Dinges JR, Colleoni C, James MG, Myers AM (2003) Mutational analysis of the pullulanase-type debranching enzyme of maize indicates multiple functions in starch metabolism. Plant Cell 15: 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Kubo A, Francisco PB Jr, Nakakita M, Harada K, Minaka N, Nakamura Y (1999) Purification, characterization, and cDNA structure of isoamylase from developing endosperm of rice. Planta 208: 283–293 [DOI] [PubMed] [Google Scholar]
- Hishinuma R, Komatsu A, Ichinose Y, Iwahashi Y, Kato T, Komae K (2004) Different expression of three HvISO genes and their subunits of barley isoamylase. Plant Cell Physiol 45: P601 [Google Scholar]
- Hussain H, Mant A, Seale R, Zeeman S, Hinchliffe E, Edwards A, Hylton C, Bornemann S, Smith AM, Martin C, et al. (2003) Three isoforms of isoamylase contribute different catalytic properties for the debranching of potato glucans. Plant Cell 15: 133–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain H, Martin C (2009) Comparative analysis of primary and secondary structure for pea isoamylase isoforms predicts different catalytic properties against glucan substrates. Starch-Starke 61: 570–577 [Google Scholar]
- Hwang JW, Kim SK, Lee JS, Kim IS (2005) Gene expression of the biosynthetic enzymes and biosynthesis of starch during rice grain development. J Plant Biol 48: 448–455 [Google Scholar]
- James MG, Robertson DS, Myers AM (1995) Characterization of the maize gene sugary1, a determinant of starch composition in kernels. Plant Cell 7: 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James MG, Denyer K, Myers AM (2003) Starch synthesis in the cereal endosperm. Curr Opin Plant Biol 6: 215–222 [DOI] [PubMed] [Google Scholar]
- Jeon JS, Ryoo N, Hahn TR, Walia H, Nakamura Y (2010) Starch biosynthesis in cereal endosperm. Plant Physiol Biochem 48: 383–392 [DOI] [PubMed] [Google Scholar]
- Jespersen HM, MacGregor EA, Henrissat B, Sierks MR, Svensson B (1993) Starch- and glycogen-debranching and branching enzymes: Prediction of structural features of the catalytic (β/α)8-barrel domain and evolutionary relationship to other amylolytic enzymes. J Protein Chem 12: 791–805 [DOI] [PubMed] [Google Scholar]
- Kim SH, Hamada T, Otani M, Shimada T (2005) Cloning and characterization of sweetpotato isoamylase gene (IbIsa1) isolated from tuberous root. Breeding Sci 55: 453–458 [Google Scholar]
- Kubo A, Colleoni C, Dinges JR, Lin Q, Lappe RR, Rivenbark JG, Meyer AJ, Ball SG, James MG, Hennen-Bierwagen TA, et al. (2010) Functions of heteromeric and homomeric isoamylase-type starch-debranching enzymes in developing maize endosperm. Plant Physiol 153: 956–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Bonte DR, Picha DH, Johnson HA (2000) Carbohydrate-related changes in sweetpotato storage roots during development. J Am Soc Hortic Sci 125: 200–204 [Google Scholar]
- Lawson CL, van Montfort R, Strokopytov B, Rozeboom HJ, Kalk KH, de Vries GE, Penninga D, Dijkhuizen L, Dijkstra BW (1994) Nucleotide sequence and X-ray structure of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 in a maltose-dependent crystal form. J Mol Biol 236: 590–600 [DOI] [PubMed] [Google Scholar]
- Li L, Ilarslan H, James MG, Myers AM, Wurtele ES (2007) Genome wide co-expression among the starch debranching enzyme genes AtISA1, AtISA2, and AtISA3 in Arabidopsis thaliana. J Exp Bot 58: 3323–3342 [DOI] [PubMed] [Google Scholar]
- Li L, Liao CH (1983) Studies on the variation in crude starch percentage in sweetpotato. J Agric Res 32: 325–335 [Google Scholar]
- Martin FW (1965) Incompatibility in the sweetpotato: A review. Econ Bot 19: 406–415 [Google Scholar]
- Mouille G, Maddelein ML, Libessart N, Talaga P, Decq A, Delrue B, Ball S (1996) Preamylopectin processing: A mandatory step for starch biosynthesis in plants. Plant Cell 8: 1353–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AM, Morell MK, James MG, Ball SG (2000) Recent progress toward understanding biosynthesis of the amylopectin crystal. Plant Physiol 122: 989–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y (1996) Some properties of starch debranching enzymes and their possible role in amylopectin biosynthesis. Plant Sci 121: 1–18 [Google Scholar]
- Nakamura Y (2002) Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: Rice endosperm as a model tissue. Plant Cell Physiol 43: 718–725 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Kubo A, Shimamune T, Matsuda T, Harada K, Satoh H (1997) Correlation between activities of starch debranching enzyme and α-polyglucan structure in endosperms of sugary-1 mutants of rice. Plant J 12: 143–153 [Google Scholar]
- Nakamura Y, Umemoto T, Takahata Y, Komae K, Amano E, Satoh H (1996) Changes in structure of starch and enzyme activities affected by sugary mutations in developing rice endosperm. Possible role of starch debranching enzyme (R-enzyme) in amylopectin biosynthesis. Physiol Plantarum 97: 491–498 [Google Scholar]
- Nakayama H, Yamaguchi T, Tsukaya H (2010) Expression patterns of AaDL, a CRABS CLAW ortholog in Asparagus asparagoides (Asparagaceae), demonstrate a stepwise evolution of CRC/DL subfamily of YABBY genes. Am J Bot 97: 591–600 [DOI] [PubMed] [Google Scholar]
- Noda T, Takahata Y, Nagata T (1992) Developmental changes in properties of sweet potato starches. Starch-Stärke 44: 405–409 [Google Scholar]
- Ohdan T, Francisco PB Jr, Sawada T, Hirose T, Terao T, Satoh H, Nakamura Y (2005) Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J Exp Bot 56: 3229–3244 [DOI] [PubMed] [Google Scholar]
- Osaka M, Matsuda T, Sakazono S, Masuko-Suzuki H, Maeda S, Sewaki M, Sone M, Takahashi H, Nakazono M, Iwano M, et al. (2013) Cell type-specific transcriptome of Brassicaceae stigmatic papilla cells from a combination of laser microdissection and RNA sequencing. Plant Cell Physiol 54: 1894–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SC, Kim YH, Ji CY, Park S, Jeong JC, Lee HS, Kwak SS (2012) Stable internal reference genes for the normalization of real-time PCR in different sweetpotato cultivars subjected to abiotic stress conditions. PLOS ONE 7: e51502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YJ, Nishikawa T, Tomooka N, Nemoto K (2014) Molecular characterization of an isoamylase 1-type starch debranching enzyme (DBEI) in grain amaranth (Amaranthus cruentus L.). Mol Biol Rep 41: 7857–7864 [DOI] [PubMed] [Google Scholar]
- Rahman MD, Richards GN (1988) Interactions of starch and other polysaccharides with condensed tannins in hot water extracts of ponderosa pine bark. J Wood Chem Technol 8: 111–120 [Google Scholar]
- Ravi V, Naskar SK, Makeshkumar T, Babu B, Krishnan BP (2009) Molecular physiology of storage root formation and development in sweet potato (Ipomoea batatas (L.) Lam.). J Root Crops 35: 1–27 [Google Scholar]
- Rice P, Longden I, Bleasby A (2000) EMBOSS: The European molecular biology open software suite. Trends Genet 16: 276–277 [DOI] [PubMed] [Google Scholar]
- Royo J, Kunz C, Kowyama Y, Anderson M, Clarke AE, Newbigin E (1994) Loss of a histidine residue at the active site of S-locus ribonuclease is associated with self-compatibility in Lycopersicon peruvianum. Proc Natl Acad Sci USA 91: 6511–6514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Scott GJ, Rosegrant MW, Ringler C (2000) Roots and Tubers for the 21st Century: Trends, Projections and Policy Options. Intl Food Policy Res Inst, Washington
- Smith AM, Denyer K, Martin C (1997) The synthesis of the starch granule. Annu Rev Plant Physiol Plant Mol Biol 48: 67–87 [DOI] [PubMed] [Google Scholar]
- Strokopytov B, Knegtel RM, Penninga D, Rozeboom HJ, Kalk KH, Dijkhuizen L, Dijkstra BW (1996) Structure of cyclodextrin glycosyltransferase complexed with a maltononaose inhibitor at 2.6 Å resolution. Implications for product specificity. Biochem 35: 4241–4249 [DOI] [PubMed] [Google Scholar]
- Sun C, Sathish P, Ahlandsberg S, Jansson C (1999) Analyses of isoamylase gene activity in wild-type barley indicate its involvement in starch synthesis. Plant Mol Biol 40: 431–443 [DOI] [PubMed] [Google Scholar]
- Takasaki T, Hatakeyama K, Suzuki G, Watanabe M, Isogai A, Hinata K (2000) The S receptor kinase determines self-incompatibility in Brassica stigma. Nature 403: 913–916 [DOI] [PubMed] [Google Scholar]
- Takashima Y, Senoura T, Yoshizaki T, Hamada S, Ito H, Matsui H (2007) Differential chain-length specificities of two isoamylase-type starch-debranching enzymes from developing seeds of kidney bean. Biosci Biotechnol Biochem 71: 2308–2312 [DOI] [PubMed] [Google Scholar]
- Tao X, Gu YH, Wang HY, Zheng W, Li X, Zhao CW, Zhang YZ (2012) Digital gene expression analysis based on integrated de novo transcriptome assembly of sweet potato [Ipomoea batatas (L.) Lam.]. PLOS ONE 7: e36234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsumi Y, Nakamura Y (2006) Structural and enzymatic characterization of the isoamylase1 homo-oligomer and the isoamylase1-isoamylase2 hetero-oligomer from rice endosperm. Planta 225: 75–87 [DOI] [PubMed] [Google Scholar]
- Watanabe M, Suwabe K, Suzuki G (2012) Molecular genetics, physiology and biology of self-incompatibility in Brassicaceae. Proc Jpn Acad Ser B Phys Biol Sci 88: 519–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattebled F, Dong Y, Dumez S, Delvallé D, Planchot V, Berbezy P, Vyas D, Colonna P, Chatterjee M, Ball S, et al. (2005) Mutants of Arabidopsis lacking a chloroplastic isoamylase accumulate phytoglycogen and an abnormal form of amylopectin. Plant Physiol 138: 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson LA (1982) Tuberization in sweet potato (Ipomoea batatas (L). Lam.). In: Villareal RL, Griggs TD (eds) Sweet Potato. Asian Vegetable Research and Development Center, Shanhua, Tainan, Taiwan, pp 79–94
- Woolfe JA (1992) Sweet potato: An untapped food resource. Cambridge University Press, Cambridge
- Zheng K, Xu J, Jiang Q, Laroche A, Wei Y, Zheng Y, Lu Z (2013) Isolation and characterization of an isoamylase gene from rye. Crop J 1: 127–133 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.